Abstract
Alternative processing of pre-mRNA plays an important role in protein diversity and biological function. Previous studies on alternative splicing (AS) often focused on the spatial patterns of protein isoforms across different tissues. Here we studied dynamic usage of AS across time, during murine retina development. Over 7000 exons showed dynamical changes in splicing, with differential splicing events occurring more frequently in early development. The overall splicing patterns for exclusive and inclusive exons show symmetric trends and genes with symmetric splicing patterns that tend to have similar biological functions. Furthermore, we observed that within the retina, retina-enriched genes that are preferentially expressed at the adult stage tend to have more dynamically spliced exons compared to other genes, suggesting that genes maintaining retina homeostasis also play an important role in development via a series of AS events. Interestingly, the transcriptomes of retina-enriched genes largely reflect the retinal developmental process. Finally, we identified a number of candidate cis-regulatory elements for retinal AS by analyzing the relative occurrence of sequence motifs in exons or flanking introns. The occurrence of predicted regulatory elements showed strong correlation with the expression level of known RNA binding proteins, suggesting the high quality of the identified cis-regulatory elements.
INTRODUCTION
The investigation of alternative splicing (AS) has attracted great attention in recent years. AS increases the diversity of mRNAs and proteins (1), and thereby plays an important role in increasing and fine-tuning the function of proteins (2). Disruption of AS has been linked to a number of human diseases, including retinal diseases (3–5). The leap from the pre-genomics studies of individual genes to high-throughput technologies, such as microarrays and deep sequencing, has enabled the investigation of transcription and splicing events on a more global, genomic scale (6–8). Application of these techniques virtually re-branded AS from being rare to being almost universal (9,10). Experimental evidence indicates that the majority (~95%) of human multiple-exon genes, which account for ~86% of the genome, are indeed alternatively spliced. Taken together, this collection of all possible alternatively spliced mRNAs, is referred to as the spliceome. Although substantial qualitative and quantitative information on a number of spliceomes has been gathered, most described splicing profiles are studied at the fully mature stage of the tested tissues. However, not only does AS generate a varying composition of isoforms in adult tissues, but splicing profiles also change during the development of these tissues.
Retinal tissue has one of the highest levels of AS (11,12), and dysregulation of AS and mutations in general splicing factors have been associated with retinal diseases (13–16). For example, among the many genes that can cause retinitis pigmentosa (RP), a form of retinal degeneration, three genes associated with dominant RP (PRPF3, PRPF8 and PRPF31) encode widely expressed proteins involved in function of the spliceosome, the molecular machine that carries out the splicing process (3–5). Moreover, mutation of PRPF31 has been implicated in dysregulation of the splicing of the rhodopsin gene and subsequent induction of apoptosis of retinal cells (17). Overall, it has been estimated that at least 15% of disease-causing mutations disrupt the cis-regulatory elements for splicing (18–20). Therefore, we expect AS to play a crucial role in retinal development and disease. However, despite its likely important role in the retina, global and detailed knowledge about AS in the retina is limited.
In this study of retinal RNA splicing, we used the unbiased methodology of exon microarrays to analyze the dynamics of AS events from embryonic day 15 (E15) through postnatal development. We focused on those AS exons whose inclusion rate underwent a significant change between the investigated developmental stages. Instead of enumerating all AS events at each given time point, we studied changes in AS by examining the relative inclusion level of exons during retinal development (Figure 1a). We observed that a number of AS exons had a significant dynamic change in their inclusion level at different stages. Interestingly, exons from retina-enriched genes show a higher frequency of dynamic AS during retina development than exons of genes whose expression is not enriched in the retina. The AS profiles of these retina-enriched genes showed strong association with special biological functions and exhibited a stage-specific signature. Through exhausted motif search, we found a number of cis-regulatory motifs over-represented for these AS events in intronic or exonic regions, providing the foundation for further mechanistic studies.
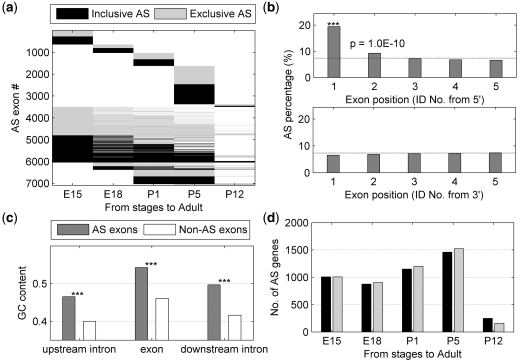
Figure 1.
Determination of AS exons. (a) Schematic representation of dynamic AS in our study. (b) SI of exons between any retina stage and adult, and their corresponding t-test P-value (minus log scale of base 10). The circle points represent AS exons identified in the study.
MATERIALS AND METHODS
Experimental design
Affymetrix Mouse Exon 1.0 arrays were used to investigate retinal gene expression and splicing at six developmental time points. Retinas from embryonic day 15 (E15), E18, postnatal day 1 (P1), P5, P12 and 2 month (adult) littermate embryos or pups were pooled so as to generate two independent biological replicates for each time point from E15 to P12 and triplicates for adult. Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and the samples were further purified using the RNeasy Mini Kit (Qiagen). On-column DNase I digestion was applied to all samples. For comparison, we analyzed brain gene expression at the exon level in tissue from E15 and adult mice, using two biological replicates per time point (pooled embryonic brain and individual adult). Normalization of exon and gene expression data was performed using GC pre-background adjustment, RMA background correction and quantile normalization (Partek Genomic Suite software). The overall expression level for a given gene is defined as the average expression level of all core probesets for the gene.
Quantitative study of AS
To focus on dynamic changes in relative exon usage, independent of overall gene expression variation, we used the normalized intensity (NI) of an exon relative to overall gene expression in a way similar to that defined by Clark et al. (21),
where Ie and Ig are the expression levels for a given exon and the gene that harbors the exon, respectively. The difference of NI for a given exon at two time points, or two samples, is called the splicing index (SI) of the two points, or samples (21),
where t1 and t2 are two time points or two samples. SI thus describes the temporal difference of exon inclusion levels or difference between the samples. Note that in this study we focus exclusively on AS on cassette exons, and ignore the splicing events that occur within exons.
Determination of dynamic AS exons
To obtain a reliable set of dynamically spliced exons, we first excluded 5944 genes that showed low levels of expression (<4.758 in log2 scale) at 5 or 6 developmental time points examined. The low expression cutoff was chosen as roughly the lowest one-third of all gene expression levels. We then determined the threshold at which to consider a significant difference between exon NIs at different time points by calculating the SI distribution between the replicates at the ‘same stage’. The hypothesis is that there are no significant dynamically spliced exons among replicates at the same time point. Since the SI distribution is close to a normal distribution (Supplementary Figure S1), we selected the conservative value of 3σ (where σ is the standard deviation of the SI distribution between replicates) as the minimal SI difference needed to define a significant AS event. Furthermore, we performed robust t-test on the NIs of all exons between each stage (duplicates) and adult (triplicates). The P-values were obtained for the significance of exon NI difference. The P-value cutoff was chosen as 0.01.
Differentially expressed genes
Differentially expressed genes were used in principal component analysis (PCA). To define the differentially expressed genes, we calculated P-value and fold change (FC) of gene expression between any stage and adult. We chose 0.01 as the cutoff of P-value, whereas the FC cutoff was chosen as three times the standard deviation of gene expression difference between replicates at the same time point. Genes that satisfied both the P-value and FC cutoffs for comparison between any stage and adult were defined as being differentially expressed during development.
Tissue-enriched genes
Similar to that described by Yu et al. (22), we obtained gene expression profiles across multiple tissues based on the NCBI UniGene database. We then calculated expression enrichment (EE) scores for each gene in 47 tissues. The corresponding P-value was calculated based on hypergeometric probability distribution and further modified by Bonferroni correction for multiple testing on the 47 tissues. If EE of one gene in a tissue was within the top 5% of all EE distributions and its P-value was <0.05, the gene was classified as a tissue-enriched gene.
Enriched Kyoto encyclopedia of genes and genomes pathway and gene ontology terms
A enriched Kyoto encyclopedia of genes and genomes (KEGG) pathway or gene ontology (GO) term was selected as an enriched KEGG pathway or GO term for a group of genes if its enrichment score (ratio of frequency in foreground to that in background) is >1.5 and its P-value was <0.05 after multiple-test Bonferroni correction. The P-value was calculated based on the hypergeometric distribution.
Dynamic AS regulatory motifs
Previous studies (6,23,24) indicated that splicing cis-regulatory elements fall in the vicinity of the splice site (SS), usually within 200 nt from the SS. Our motif search focused on four regions: (i) upstream intron: last 200 nt in the upstream intron from the 3′ splicing site (3′SS), or the sequence from the previous exon end to the 3′SS of the exon of interest, whichever is shorter; (ii) 5′ exon: first 200 nt sequence of the exon from the 5′-end or the whole exon, whichever is shorter; (iii) 3′ exon: 200 nt sequence from the 3′-end or the whole exon, whichever is shorter; (iv) downstream intron: first 200 nt in the downstream intron from the 5′SS or the sequence from the 5′SS to the next exon, whichever is shorter. Repeat elements were defined by RepeatMasker (http://repeatmasker.org) and excluded from the analysis.
All 5376 potential m-mer nucleotides (m = 4, 5 and 6) were studied as potential AS cis-regulatory elements because the lengths of most of known splicing regulatory motifs are around this range (25,26). We calculated the frequency of m-mer nucleotides in AS exons and non-AS exons. Non-AS exons are referred to exons which have high NI and have no significant change during development. The Δ value for each m-mer nucleotide, as defined below, represents a measure of the difference in frequency between the two groups (24):
 |
where fas and fnon are the frequencies of m-mer nucleotide in AS and non-AS exons, NAS and Nnon are the total numbers of single nucleotides in the two exon groups, and
A Δ-value greater than 0 means the m-mer nucleotide occurs more than expected in AS exons as compared to non-AS exons, whereas Δ < 0 means the m-mer element occurs less than expected.
To determine the statistical significance of m-mer occurrence, we calculated the enrichment score, defined as the ratio of its frequency in AS exons to that in non-AS exons. We focused on those sequences whose position-dependent frequencies in AS exons deviated significantly from those in non-AS exons. P-values were calculated using the Kolmogorov–Smirnov (KS) test. The m-mer nucleotide was selected as an enriched cis-element only if its Δ fell within the top 5%, its enrichment score was in the top quarter of all cis-elements, and its KS test P-value was <0.05 after Bonferroni correction.
To evaluate the predicted motifs, we calculated the false discovery rate (FDR) of the prediction. We shuffled AS and non-AS exons groups with the same group sizes as real data set. We then performed the same motif discovery algorithm on the randomized data set. The FDR was defined as M1/M0, where M1 is the average number of motifs from a number of randomized data sets, and M0 is total number of real motifs obtained.
Quantitative real-time PCR gene expression analysis of retinal cells and other tissues
Total RNAs were extracted from adult mouse retina, brain, heart, kidney, liver and testis using Trizol (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA from each tissue was used to synthesize cDNA using Superscript III RT (Invitrogen) with random hexamers. For more detailed analysis of regional gene expression patterns in the retina, laser capture microdissection (LCM) was used. Dissection of retina at earlier developmental time points, and the retinal layers [ganglion cell layer (GCL), inner nuclear layer (INL) and outer nuclear layer (ONL)], as well as cDNA preparation, were performed as described previously (27). PCR primers were designed specifically for each isoform of pleckstrin homology domain containing, family B (evectins) member 1 (Plekhb1). The primer sequences for amplification of the isoform containing exon 7 (chr7:107798442–107798546 from mm9 Assembly) are 5′-AAGGTCAGGTGTACAACCCTCTCA-3′ in exon7 and 5′-TGTGTCACACCAGGACCTGCATAA-3′, which spans the junction of exons 8 and 9. The primer sequences for the isoform not containing exon 7 are 5′- AAATTCCACCCCGGTACGCGTCTA-3′, which spans the junction sequence between exons 6 and 8 and 5′- ATAAGTGAACCAAGAGCAGCACCCGT-3′ in exon 9.
Quantitative real-time PCR (qPCR) was performed on an iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with iQ SYBR green supermix (Bio-Rad). The output data was analyzed with Bio-Rad iQ5 Standard Edition V 2.1 program. We used the ratio of relative transcript amount of Plekhb1 with the exon 7 to that without exon 7 to approximately represent the inclusion rate of exon 7.
RESULTS
Determining dynamic AS events
In this study we have studied the dynamic usage of exons during retinal development. Relative retinal exon expression levels were profiled using Affymetrix Mouse Exon arrays. Whole retina was harvested at six time points: embryonic day 15 (E15), E18, postnatal Day 1 (P1), P5, P12 and at adult age (2 months old). To determine the differential usage for each exon between two points, we used the SI to describe the relative expression level difference for the exon. In this work, the adult stage was chosen as the reference point (t2) for comparison. The SI value of the AS event can be either positive or negative. A positive SI represents an exon that is more included (inclusive exon) at one stage compared to adult. In contrast, a negative SI describes an exon that is less included (exclusive exon) at one particular stage compared to adult.
To obtain reliable AS events, we applied a series of filtering operations on the exon expression data (‘Materials and Methods’ section). Exons belonging to low expressed genes were excluded, since low expression of a gene could introduce high noise in SI. We calculated the P-value of SI using a t-test for each exon between retina developmental stages. Only exons with high absolute SI values and significant P-values were considered to be dynamically spliced exons (Figure 1b, magenta points). We denoted these exons as AS exons and the genes harboring these exons as AS genes.
Global view of AS events during retina development
In total we analyzed 96 787 exons from 10 686 expressed genes at the core level according to Affymetrix annotation. Among them, 7087 exons (8.2%) of 3566 genes (33.4%) underwent dynamical splicing at least at one stage compared to adult. We next examined the global properties of these dynamically spliced exons.
First, approximately half of the AS exons are stage-specific (top half in Figure 2a), suggesting that these genes may have a unique isoform at one particular developmental stage. The other AS exons show different exon usage compared to adult retina at multiple stages. The numbers of inclusive and exclusive AS exons at each stage are roughly equal (Figure 2a).
Figure 2.
Global view of AS events during development stages. (a) Stage-specific and non-specific AS events at each stage. (b) AS exon percentages at different locations, from 5′ and 3′, respectively. The dashed lines represent the ratio of AS exons to all exons at all locations. (c) GC contents for AS exons and constitutive exons (***P = 0). (d) Number of AS genes at each stage compared to adult.
Second, we checked the relative positions of the AS exons in the genes. We found that alternative promoter usage is a widespread mechanism for generating alternative transcripts during retinal development. In contrast, exons at poly(A) sites and other sites do not show enrichment for dynamical splicing (Figure 2b). Although individual examples have been shown for alternative promoter usage (28–31), here we have shown that this is the general trend for AS genes during development.
Third, we found that AS exons have higher GC content (46–54%) than do constitutive exons (40–46%), both in the exons themselves and in their flanking introns (Figure 2c). This finding is consistent with previous reports and is attributed to the fact that alternatively spliced sites tend to have a more stable structure (32,33). Another possible reason for higher GC-content in AS exons could be that the 5′ region in a gene tends to have higher GC-content and AS exons are enriched at the 5′-end of genes. We indeed found that the GC content of AS exons and their flanking introns are higher in 5′ regions than in other positions. In contrast, there is no obvious difference in GC content between constitutive exons at 5′-end and at other positions (Supplementary Table S1), so it seems a unique feature for AS exons that their higher GC-content is a function of their relative genomic position.
Fourth, pathway analysis of AS genes revealed that genes involved in phototransduction, ABC transporter, and extracellular matrix receptor interactions are over-represented as being alternatively spliced during retinal development. Further, GO analysis confirmed that membrane-associated and extracellular matrix proteins are enriched for AS genes (details in Supplementary Tables S2 and S3).
Finally, the number of AS genes ranges from 1500 to 2500 from E15 to P5 (Figure 2d). The number of AS genes decreases dramatically to less than 400 at P12, suggesting that the retinal spliceome at P12 closely resembles that in adult retina. Interestingly, although we used the spliceome at the adult stage as reference, the number of AS genes at P1 and P5 are greater than those at E15 and E18. This observation indicates that AS activity might be at its highest level at early postnatal days.
Splicing patterns and the associated biological functions
To examine whether the genes sharing similar splicing patterns during development share other attributes, we clustered genes based on their exon inclusion levels. We used k-means (k = 8) with metrics of Euclidian distance to cluster all AS exons based on their SI profiles across developmental stages (Figure 3a). The average profile for each cluster is shown in Figure 3b. Several major features emerge from this analysis.
Figure 3.
K-mean clustering of AS exon SI profiles during retina development. (a) SI profiles of AS exons clustered by k-means (k = 8). (b) Average SI of AS exons in each cluster. The number n in bracelet is number of unique harboring genes in the cluster. (c) Hierarchical clustering of AS exon clusters based on correlation of GO term enrichment scores between gene cluster pairs.
First, we observed symmetry between pairs of clusters with opposite SI profiles (exclusive and inclusive). The SI profiles of clusters C0 and C7, C1 and C6, C2 and C5 and C3 and C4 are highly symmetric. One possible explanation for such symmetric splicing patterns would be that two exons in one gene are mutually exclusive. However, only 149 pairs of mutually exclusive exons were identified in the symmetric clusters from >7000 AS exons. We also examined the gene expression profiles for these clusters and there is no obvious correlation between gene expression levels and exon SIs (Supplementary Figure S2). Therefore, we expect other intrinsic reasons for opposite splicing trends between genes and further investigation is needed to explore the underlying mechanism.
Second, the genes with opposite splicing patterns tend to share similar biological functions. We performed GO analysis on all clusters and calculated enrichment score of each GO term in each cluster. We then examined the similarity between these clusters in terms of biological functions by calculating the correlation coefficient of enrichment score profiles for all GO terms between the clusters. It is interesting to see that clusters with symmetric SI profiles always have high correlation coefficients of their GO enrichment score profiles (Figure 3c). This observation suggests that genes with certain biological functions do not have preference for direction (i.e. inclusion or exclusion) of AS. Rather, altered function for certain classes of genes is shared at particular developmental time points, but for some genes the altered function is brought about by inclusion of an exon, and in cases it is brought about by exclusion of an intron.
Third, these clusters have different temporal features during development. Clusters C2 and C5 show significant AS only in early stages (E15 and E18) and the SI level then rapidly approaches adult stage levels. In contrast, the genes in clusters C0, C1, C6 and C7 have significant AS changes from E15 to P5. The splicing patterns persist for a long period in the early time points so that even at P5, the SI level is still significantly different from that at the adult stage. The third type of clusters includes C3 and C4, which have significant SIs at P5 and modest SIs at P1. The different temporal splicing pattern genes exhibit different GO functions. For example, one cellular component, ‘photoreceptor outer segment’, and eight biological processes, such as ‘visual perception’ and ‘sensory perception of light stimulus’, are significantly over-represented in the C2 and C5 cluster pair. Genes in this group includes Arr3, Pcdh15 and Pde6c. The molecular function ‘structural constituent of eye lens’ is significantly enriched (P < 1.5 × 10−6) in the two cluster pairs (C0 and C7; C1 and C6). Example genes include Bfsp2, Cryaa and Cryab. The biological process ‘cell adhesion’ is enriched for genes in clusters of C3 and C4 (e.g. gene Robo2). The result suggests that genes with different splicing patterns tend to have distinct biological functions.
It is interesting to point out that those genes preferentially expressed in mature Muller and cone cells (34) tend to have different AS patterns. Genes expressed in cone photoreceptors are more significantly enriched in clusters with early AS changes (i.e. clusters of C2 and C5), whereas genes expressed in Muller cells are over-presented in clusters whose splicing patterns change at late stage (i.e. C0, C1, C6 and C7). This is consistent with the knowledge that cone cells are born before birth, while the birth of Muller cells peaks at early postnatal days in mouse (35).
Retina-enriched genes are enriched for AS during retina development
We then asked whether the genes that maintain retinal homeostasis in the adult retina also play a role in development. We define retina-enriched genes as genes that are preferentially expressed in ‘adult’ retina compared to other tissues. After applying our criteria (see ‘Materials and Methods’ section), 394 retina-enriched genes were identified in our study, <4% of all genes. We found that retina-enriched genes are indeed enriched for AS at all retinal developmental stages (Figure 4a). In particular, they are significantly enriched at stages E15 (P < 1.2 × 10−11), E18 (P < 1.5 × 10−11), and P1 (P < 2 × 10−4). In general, over 52% of retina-enriched genes underwent AS at least at one stage compared to ~31% of all the genes (P = 1.9 × 10−11, Figure 4b).
Figure 4.
Retina-enriched AS pattern as signature of retina development. (a) Number of retina-enriched AS genes at each stage, compared to expected number derived from all general AS genes, The double asterisks represent the P < 10−3. (b) Percentage of all AS genes and four tissue-enriched (retina, brain, spleen and muscle) AS genes. (c) Comparison between AS percentage of retina-enriched genes and all other genes at different intervals of gene expression level. (d) PCA on NI of all exons, AS exons and retina-enriched AS exons, respectively. (e) PCA on gene expression of all genes, differentially expressed genes, and retina-enriched differentially expressed genes, respectively. The circle sizes (from small to large) represent the retina developmental stages (from early to late).
As a control, we calculated the percentage of AS genes in other tissue-enriched gene sets, including brain, spleen and muscle. We found that spleen and muscle-enriched genes are not enriched for AS during retina development. In contrast, the percentage of AS genes in the brain-enriched gene set is much higher than average during retina development, indicating the strong association between these two neuronal tissues, retina and brain.
In order to examine whether the enrichment of AS exons in retina-enriched genes is due to the overall higher expression level of these genes, we compared the frequencies of dynamic AS exons for retina-enriched genes and other genes at the same expression level by grouping the genes with similar expression levels. We found that retina-enriched genes tend to have higher chance to harbor AS exons than other genes at almost all expression level intervals (Figure 4c). Therefore, the observation that retina-enriched genes tend to have more AS exons is not simply a function of their high expression level.
To test whether the observation is only applied to retina or it is a general rule for other tissues, we carried out a similar analysis by comparing AS profiles between E15 and adult brain (Supplementary Figure S3). In this case, genes that are preferentially expressed in adult brain show a higher percentage of AS genes, while retina-enriched genes have lower percentage of AS genes. This result suggests that the genes maintaining tissue homeostasis might play an important role in development by expressing different gene isoforms, resulting in the expression of different gene products.
Spliceome of retina-enriched genes can be a signature of developmental states
We have shown that retina-enriched genes tend to have a higher percentage of AS exons during development. Interestingly, their collective splicing pattern largely reflects their developmental stage. Specifically, we performed PCA on exon NI and gene expression, respectively, across retinal development to examine whether the developmental time points showed a pattern corresponding to linear time. When the PCA was based on all 96 787 exons, or the 7087 AS exons, respectively, the time points were not positioned according to developmental stage (Figure 4d). In contrast, when the PCA was based on the set of 598 retina-enriched AS exons, the relative position of the points closely followed the developmental stage of the samples from E15 to adult (Figure 4d). Similar trends were observed if we used expression of different gene sets to describe the developmental stages (Figure 4e). Interestingly, the AS analysis did not resolve the later time points so well and the gene expression analysis did not resolve the earlier time points so well. This finding suggests that the collective splicing of retina-enriched genes is a good indicator for developmental stage, and can complement gene expression patterns as a marker of retinal development.
AS sequence motifs for retina development
To understand the possible regulatory mechanisms controlling AS, we attempted to identify cis-regulatory elements connected to retinal AS by employing a splicing motif discovery algorithm (see ‘Materials and Methods’ section). We compared the occurrence of short sequence segments in AS exons and their flanking introns with that in non-AS counterparts. We searched over-represented sequence motifs in four regions: upstream intron, 5′ exon, 3′ exon, and downstream intron. Numerous significant cis-elements associated with AS were discovered. Similar to the finding that splicing enhancers at 5′ SS and 3′SS overlap extensively (24), most of them were observed to be enriched within multiple regions (Figure 5a). This finding suggests that many AS regulatory elements are not limited to a specific region. The FDR for the predicted motif is 1.24% based on random shuffling evaluation (see ‘Materials and Methods’ section for details). We then clustered all enriched sequence elements according to their sequence similarity. Consensus motifs (logos) were generated for all clusters (Figure 5b). We observed that many of the representative AS motifs in different regions (e.g. exons and introns) exhibit sequence similarity.
Figure 5.
Identification of cis-regulatory elements. (a) Venn-diagram of cis-regulatory elements discovered in upstream intron, AS exon and downstream intron. (b) Enriched representative motifs in three regions. Each motif's name consists of letter-number-letter. The first capital letter ‘E’ represents enriched. The number in the middle, ‘3′, ‘0′ and ‘5′ represents upstream intron, exon, and downstream intron, respectively. The lower-case letter is a random order number of the motif. The motif name in the bracelet represents the motif which is very similar (dissimilarity <1.7) to the motif with the logo shown. (c) Negative correlation between expression level of Ptbp2 (nPTB) and occurrence enrichment of CUCUCU in upstream intron of AS exons. (d) Positive correlation between expression of Mbnl1 and occurrence enrichment of GCGGG. (e) Distribution of correlation coefficients between gene expression of 209 RNA binding proteins and occurrence enrichment of all identified AS motifs. As comparison, the distribution for RNA binding proteins and all m-mer nucleotides are also shown.
Interestingly, we found that the occurrences of some motifs in AS exons or introns are correlated with the gene expression level of splicing regulatory factors during development. For example, the occurrence of CUCUCU negatively correlates with the expression of neural polypyrimidine tract binding protein (nPTB), which primarily functions as a splicing repressor in neuronal cells (Figure 5c). nPTB regulates AS events by binding to the sequence motif CUCUCU located in introns flanking the AS exons (36–38). The observed negative correlation between nPTB expression and CUCUCU occurrence is consistent with nPTBs inhibitory role. The study of correlation between AS motifs and genes provides us with the potential to find novel splicing regulators of retinal AS. For example, the expression of the gene muscleblind-like 1 (Mbnl1) is positively correlated with cis-regulatory elements GCGGG during retina development (Figure 5d). This finding suggests that Mbnl1 may play a role in AS events during retina development.
We then used the observed correlations to perform a global assessment of the quality of the predicted motifs. We calculated the correlation between the occurrence of identified cis-regulatory elements and the gene expression of 209 RNA-binding proteins with GO terms of ‘RNA binding’ and/or ‘RNA splicing’ and differentially expressed during retina development. The correlation between any pair of a predicted cis-regulatory element and an RNA binding protein were calculated and the overall distribution is shown in Figure 5e. The distribution is clearly enriched for highly correlated pairs compared to the correlation distribution for all 4, 5, 6-mer nucleotides and RNA binding proteins. As another negative control, we calculated the correlation between expression level of randomly selected genes and the occurrence of predicted motifs, and found that the correlation is significantly lower than the observed correlation between RNA binding proteins and motifs (P = 0). Taken together, our results suggest that the identified motifs are enriched for bona fide regulatory elements. These elements provide the foundation for further mechanistic studies of retinal AS.
Splicing patterns also show cell type specificity
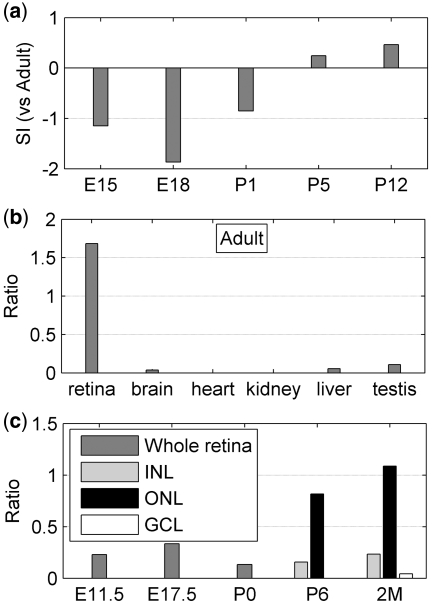
The above-described studies examined the dynamics of AS during development, which represents splicing changes in the temporal dimension. In a different dimension, AS patterns can also vary between different cell types even within the same tissue. For example Basigin 2 (Bsg2) has been reported to show an AS pattern that is specific to photoreceptor cells as compared to other retinal and non-retinal cells (39–41). Using a bioinformatics approach based on available expressed sequence tag (EST) data sets from retinal and non-retinal tissues, we independently identified Bsg2 as having an AS form that is preferentially expressed in the retina, and using QPCR analysis of retinal layer samples obtained by laser capture microdissection (LCM) also found that the retinal isoform is preferentially expressed in photoreceptors (T. Masuda et al., unpublished data). Using the same EST-based approach, coupled with our analysis described above, we searched for genes that might show both retinal cell type-specific AS as well as developmentally modulated AS. An example of such a gene is Plekhb1. The development-related microarray data indicated that exon 7 of Plekhb1 became more inclusive as retinal development proceeded (Figure 6a). Using a QPCR assay with specific primers that could distinguish the splice isoforms with and without exon 7, we found that the exon 7 containing isoform is strongly enriched in the retina compared to non-retinal tissues (Figure 6b).
Figure 6.
Differential splicing patterns (including or excluding exon 7) of gene Plekhb1 in different tissues, at different stage and in different retina cells. (a) SI of Plekhb1 exon 7 for retina development stages based on microarray data. (b) Ratio of qPCR data with exon 7 to that without exon 7 for different tissues. (c) Ratio at different developmental time points and in different LCM-derived retina layers. The ONL is composed exclusively of photoreceptor cells.
We then used the QPCR assay with LCM-derived RNA from retinal samples from E 11.5, E17.5, P0, P6 and 2 month old mice to dissect splicing patterns along both the temporal and spatial dimensions. At the earlier developmental time points we could not dissect specific retinal layers because at these stages the retina is consists of a morphologically homogeneous neuroblastic layer. At P6 we separated distinct INL and outer nuclear (photoreceptor) layers. At the 2 month stage we dissected distinct INL, ONL and retinal ganglion cell (RGC) layers. The LCM/QPCR results showed a general pattern of increasing exon 7 inclusion over developmental time (Figure 6c) that was similar to the microarray data (Figure 6a). More significantly, the LCM results showed that the exon 7 containing splice isoform of Plekhb1 is highly enriched in photoreceptors. These results, taken together, demonstrate the potentially complex landscape of spatiotemporal RNA splicing regulation.
DISCUSSION
In this work, we used the mouse retina as a model system to study the dynamics of AS during development. Our study provides insight into the molecular basis of the retinal development, identifying a large number of exons (>7000) that undergo statistically significant splicing changes during development. A global clustering of exons based on their splicing patterns revealed a symmetric relationship between exon clusters. The genes in opposite splicing trends (more inclusive versus more exclusive compared to adult) tend to share similar biological functions. Interestingly, retina-enriched genes are more significantly enriched in AS genes than other genes during retina development. In fact, 52% of retina-enriched genes compared to 31% general genes underwent such AS events, suggesting that the genes regulating homeostasis in the adult retina also play an important role during development, and part of this activity may be regulated by modulation of the expression of alternative isoforms. We also identified many cis-regulatory elements that are likely to regulate splicing events during retinal development, providing the foundation for further studies of AS in the retina.
We made several interesting observations and these findings could be generalized to other systems. For example, we found that retina-enriched genes are enriched for AS changes during retinal development. We performed a similar analysis on mouse brain and found that genes preferentially expressed in adult brain are also enriched for AS during brain development. This suggests that our observation might be a general rule that genes maintaining tissue/cell-type homeostasis also play an important role during development via numerous AS events. Another possible general rule is that genes with opposite symmetric splicing patterns tend to share similar biological functions. This finding indicates that even though functions of individual genes are of course dependent on the inclusion or exclusion of a particular exon, the biological functions are not related to the direction of AS, either inclusive or exclusive, on a global scale.
We also found that genes specific for different cell types tend to have different temporal AS profiles. For example, Muller-specific genes tend to have different isoform patterns from adult until postnatal stage P5, while cone-enriched genes tend to have significant AS only at early developmental stages. Another interesting example is Plekhb1, which shows differential splicing both during development and across cell types. This suggests that different cell types within the retina may have distinct AS patterns during development and have different critical time points during their differentiation. In order to deconvolute average signals obtained using a complex system such as the retina we plan to analyze AS in retinal development at the cellular level. In other words, we will perform a similar analysis on different retinal cell types, rather than in whole retina, as we did in this study. We believe that such analysis will provide more biological insights into the role of AS in retinal development.
In an effort to define the motifs responsible for retinal AS, we identified short RNA sequences that are enriched in AS exons or their flanking introns as compared to their occurrences in non-AS exons. The discovery of these putative motifs represents the first step towards a mechanistic study of AS regulation in the retina. The next step will be experimental validation of these motifs and identification of the potential splicing factors that recognize these motifs. Protein microarrays provide an ideal approach for this problem. We will synthesize the putative RNA motifs and probe them individually on protein microarrays that contain thousands of proteins. Such an approach has proven to be successful in identification of novel protein–DNA interactions (42). We expect that this approach will provide insights into protein–RNA interaction specificity and lead to the discovering of novel splicing factors and regulatory molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work, performed in the Guerrieri Center for Genetic Engineering and Molecular Ophthalmology, was supported by grants from the NIH (EY017589, EY01765, EY009769 and 5P30EY001765), Research to Prevent Blindness and by funds generously provided by the Guerrieri Family Foundation and Mr and Mrs Robert and Clarice Smith. Funding for open access charge: EY017589.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This manuscript is dedicated, with thanks, in memory of Mrs Patti Guerrieri.
REFERENCES
- 1.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG, Inglehearn CF, Keen TJ, Willis C, Moore AT, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 4.Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 5.McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum. Mol. Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 6.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 9.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 11.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beit-Ya’acov A, Mizrahi-Meissonnier L, Obolensky A, Landau C, Blumenfeld A, Rosenmann A, Banin E, Sharon D. Homozygosity for a novel ABCA4 founder splicing mutation is associated with progressive and severe Stargardt-like disease. Invest. Ophthalmol. Vis. Sci. 2007;48:4308–4314. doi: 10.1167/iovs.07-0244. [DOI] [PubMed] [Google Scholar]
- 14.Haywood-Watson RJ, 2nd, Ahmed ZM, Kjellstrom S, Bush RA, Takada Y, Hampton LL, Battey JF, Sieving PA, Friedman TB. Ames Waltzer deaf mice have reduced electroretinogram amplitudes and complex alternative splicing of Pcdh15 transcripts. Invest. Ophthalmol. Vis. Sci. 2006;47:3074–3084. doi: 10.1167/iovs.06-0108. [DOI] [PubMed] [Google Scholar]
- 15.Comitato A, Spampanato C, Chakarova C, Sanges D, Bhattacharya SS, Marigo V. Mutations in splicing factor PRPF3, causing retinal degeneration, form detrimental aggregates in photoreceptor cells. Hum. Mol. Genet. 2007;16:1699–1707. doi: 10.1093/hmg/ddm118. [DOI] [PubMed] [Google Scholar]
- 16.Kanadia RN, Clark VE, Punzo C, Trimarchi JM, Cepko CL. Temporal requirement of the alternative-splicing factor Sfrs1 for the survival of retinal neurons. Development. 2008;135:3923–3933. doi: 10.1242/dev.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan L, Kawada M, Havlioglu N, Tang H, Wu JY. Mutations in PRPF31 inhibit pre-mRNA splicing of rhodopsin gene and cause apoptosis of retinal cells. J. Neurosci. 2005;25:748–757. doi: 10.1523/JNEUROSCI.2399-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 19.Nlend Nlend R, Meyer K, Schumperli D. Repair of pre-mRNA splicing: prospects for a therapy for spinal muscular atrophy. RNA Biol. 7:430–440. doi: 10.4161/rna.7.4.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 21.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34:4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: the splicing regulatory machinery. Cell. 2008;133:192 e191. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Wahlin KJ, Moreira EF, Huang H, Yu N, Adler R. Molecular dynamics of photoreceptor synapse formation in the developing chick retina. J. Comp. Neurol. 2008;506:822–837. doi: 10.1002/cne.21582. [DOI] [PubMed] [Google Scholar]
- 28.Andersen M, Lenhard B, Whatling C, Eriksson P, Odeberg J. Alternative promoter usage of the membrane glycoprotein CD36. BMC Mol. Biol. 2006;7:8. doi: 10.1186/1471-2199-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SX, Searcy TR, Wu Y, Gozal D, Wang Y. Alternative promoter usage and alternative splicing contribute to mRNA heterogeneity of mouse monocarboxylate transporter 2. Physiol. Genomics. 2007;32:95–104. doi: 10.1152/physiolgenomics.00192.2007. [DOI] [PubMed] [Google Scholar]
- 30.Morgan MA, Magnusdottir E, Kuo TC, Tunyaplin C, Harper J, Arnold SJ, Calame K, Robertson EJ, Bikoff EK. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol. Cell Biol. 2009;29:5813–5827. doi: 10.1128/MCB.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, et al. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Kuo CJ, Chen L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC Genomics. 12:90. doi: 10.1186/1471-2164-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mollet IG, Ben-Dov C, Felicio-Silva D, Grosso AR, Eleuterio P, Alves R, Staller R, Silva TS, Carmo-Fonseca M. Unconstrained mining of transcript data reveals increased alternative splicing complexity in the human transcriptome. Nucleic Acids Res. 38:4740–4754. doi: 10.1093/nar/gkq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc. Natl Acad. Sci. USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan RC, Black DL. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 38.Gooding C, Roberts GC, Smith CW. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 39.Ochrietor JD, Moroz TP, van Ekeris L, Clamp MF, Jefferson SC, deCarvalho AC, Fadool JM, Wistow G, Muramatsu T, Linser PJ. Retina-specific expression of 5A11/Basigin-2, a member of the immunoglobulin gene superfamily. Invest. Ophthalmol. Vis. Sci. 2003;44:4086–4096. doi: 10.1167/iovs.02-0995. [DOI] [PubMed] [Google Scholar]
- 40.Ochrietor JD, Linser PJ. 5A11/Basigin gene products are necessary for proper maturation and function of the retina. Dev. Neurosci. 2004;26:380–387. doi: 10.1159/000082280. [DOI] [PubMed] [Google Scholar]
- 41.Clamp MF, Ochrietor JD, Moroz TP, Linser PJ. Developmental analyses of 5A11/Basigin, 5A11/Basigin-2 and their putative binding partner MCT1 in the mouse eye. Exp. Eye Res. 2004;78:777–789. doi: 10.1016/j.exer.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.