Abstract
Star–PAP is a non-canonical, nuclear poly(A) polymerase (PAP) that is regulated by the lipid signaling molecule phosphatidylinositol 4,5 bisphosphate (PI4,5P2), and is required for the expression of a select set of mRNAs. It was previously reported that a PI4,5P2 sensitive CKI isoform, CKIα associates with and phosphorylates Star–PAP in its catalytic domain. Here, we show that the oxidative stress-induced by tBHQ treatment stimulates the CKI mediated phosphorylation of Star–PAP, which is critical for both its polyadenylation activity and stimulation by PI4,5P2. CKI activity was required for the expression and efficient 3′-end processing of its target mRNAs in vivo as well as the polyadenylation activity of Star–PAP in vitro. Specific CKI activity inhibitors (IC261 and CKI7) block in vivo Star–PAP activity, but the knockdown of CKIα did not equivalently inhibit the expression of Star–PAP targets. We show that in addition to CKIα, Star–PAP associates with another CKI isoform, CKIε in the Star–PAP complex that phosphorylates Star–PAP and complements the loss of CKIα. Knockdown of both CKI isoforms (α and ε) resulted in the loss of expression and the 3′-end processing of Star–PAP targets similar to the CKI activity inhibitors. Our results demonstrate that CKI isoforms α and ε modulate Star–PAP activity and regulates Star–PAP target messages.
INTRODUCTION
In eukaryotes, the generation of messenger RNA (mRNA) is a highly orchestrated process comprised of a number of events, including transcription, splicing, capping and 3′-end processing followed by export to the cytoplasm for translation (1–4). The 3′-end processing of mRNA precursors (pre-mRNA) is an essential step in eukaryotic gene expression which involves two tightly coupled processes—endonucleolytic cleavage followed by the subsequent addition of a poly(A) tail (200–300 adenosine nucleotides) to the cleaved RNA (5–11). The polyadenylation of mRNAs is required for their export, stability, and efficient translation (5,10,12).
Polyadenylation of pre-mRNA is carried out by a class of enzymes called poly(A) polymerases (PAPs), which add a poly(A) tail to the 3′-end of cleaved RNA (13,14). PAPs function as a part of 3′-end processing complex (5,10) comprising of cleavage and polyadenylation specificity factors (CPSF-160, -100, -73 and -30 subunits) (15), cleavage stimulatory factors (CstF-77, -64 and -50 subunits) (16), cleavage factors I and II (CF I and CF II) (17,18), the scaffolding protein symplekin (19,20) and poly(A) binding protein (PABP) (21). There are multiple genes encoding PAPs in mammalian cells, including Trf4, GLD2 and PAPα, which are involved in degradation, mRNA stability, and the polyadenylation of newly transcribed mRNAs respectively (10,22,23).
Star–PAP is a newly identified, nuclear PAP that is regulated by the lipid messenger phosphatidylinositol-4,5-bisphosphate (PI4,5P2). Star–PAP is required for the expression of select target messages, including heme oxygenase 1 (HO-1) (24). Star–PAP couples to the transcriptional machinery to polyadenylate messages. Like PAPα, Star–PAP has a PAP domain and an RNA recognition motif; but the PAP domain of Star–PAP is split by a proline-rich region. Additionally, Star–PAP contains a zinc finger motif at the N-terminus (24). Star–PAP binds its target pre-mRNA that requires both ZF and RRM domains, recruits CPSF subunits to the poly(A) site and assembles into a distinct 3′-end processing complex (25). Star–PAP 3′-end processing complex contains unique proteins, such as phosphatidylinositol-4-phosphate 5-kinase I alpha (PIPKIα) and the PI4,5P2-senstive protein kinase casein kinase Iα (CKIα) both of which are also required for the expression of some of its target mRNAs (24,26).
CKIα is a PI4,5P2-sensitive protein kinase, a member of the CKI family of Serine/Threonine (S/T) specific protein kinases (27–29). CKIs are ubiquitous monomeric protein kinases in the size range from 35 to 55 KDa present in the membrane, nucleus, cytoplasm, cytoskeleton and spindles of mammalian cells (30,31). CKI phosphorylates a large number of proteins and its substrate selection in vivo is regulated by subcellular localization and/or docking sites on the specific substrate (27,28). CKI activity is regulated through a distinct recognition motif (S/T)(P)XX(S/T), where (S/T)(P) represents a phosphoserine or phosphothreonine residue, and the underlined S/T represents the CKI target site. This indicates that modification of serine or threonine residues by CKI requires the preceding phosphorylation of amino acid residues N-terminal of the target sequence (32–34). CKI also phosphorylates a related unprimed site, which optimally contains a cluster of acidic amino acids N-terminal to the target S/T, including an acidic residue at N − 3 and a hydrophobic region C-terminal to the target S/T (35–38). However, for several important CKI targets such as NF-AT4 and beta-catenin, CKI does not require N − 3 priming, but instead phosphorylates the first serine in the non-canonical sequence S-L-S, which is followed by a cluster of acidic residues, albeit less efficiently than the optimal sites (39–41). In addition, a CKI docking sequence (SDASSCE) has also been defined on the transcription factor NF-AT4 (A-2 domain), which is required for its phosphorylation (41,42).
There are at least four splice variants of CKIα (CK1α, CK1αS, CK1αL and CK1αLS), characterized by the presence or absence of two additional coding sequences (L or S insertions) in the catalytic domain and/or in the regulatory domain (43–47). Exon ‘L’ encodes a 28-amino acid stretch that is inserted after lysine 152, in the center of the catalytic domain. The ‘S’ insert encodes 12 amino acid residues and is located close to the carboxyl terminus of the protein. The various splice isoforms of CKIα differ in their substrate preference, kinase activity, biochemical properties, subcellular localization and biological functions in the cell (28,43,44,46,47). CKIα has been reported to associate with cytosolic vesicular structures, including small synaptic vesicles, the centrosome during interphase, and with the mitotic spindle during mitosis (30,48). In addition, CKIα localizes in the nucleus to nuclear speckles (26,49) which harbor various proteins required for RNA processing and transcription (50). Inhibitors of CKI activity, such as IC261 and CKI7, inhibit CKIα and other isoforms of CKI, including CKIδ and CKIε (51). While CKI7 equally inhibits all isoforms of CKI, IC261 has higher preference to inhibit CKIδ and CKIε than CKIα (52–54). These CKI isoforms have been implicated in circadian rhythm, DNA damage and various other physiological functions in the cell (51,55–57).
Previously, we have reported that Star–PAP associates with multiple kinases, including CKIα (24,26). Here, we show that tBHQ treatment stimulates CKI-mediated phosphorylation of Star–PAP, and that, in addition to CKIα, Star–PAP associates with another CKI isoform, CKIε in the complex, which can complement for the loss of CKIα in regulating Star–PAP activity. We demonstrate that CKI activity is necessary for in vitro Star–PAP polyadenylation activity and the 3′-end processing of its target messages in vivo. Our data indicate that the kinase activities of both the isoforms of CKI—α and ε modulate Star–PAP polyadenylation activity and target mRNAs.
MATERIALS AND METHODS
Cell culture, transfections and cell stimulation
HEK-293 and HeLa cells were obtained from ATCC and maintained in DMEM with 10% FBS at 37°C in 5% CO2. HeLa cells were transfected using Oligofectamine for siRNA knockdown (Invitrogen). In HEK-293 cells, transfections were accomplished by the calcium phosphate method with 5–10 µg of plasmid DNA. The knockdown of CKIα or Star–PAP was carried out as described previously (24,26). To induce a transcriptional antioxidant response, cells were treated for 4 h with 100 μM tert-butylhydroquinone (tBHQ) in DMSO. Control cells were treated with solvent control DMSO to a final concentration of 0.1%. The CKI activity inhibitors CKI7 (53) and IC261 (54) were dissolved in DMSO and used at a final concentration as indicated.
Expression and affinity purification of Star–PAP
Affinity purified human Flag–Star–PAP was obtained from HEK-293 cells with stably expressed Flag–Star–PAP as described previously (24). For antioxidant response, cells were treated with 100 μM tBHQ in DMSO for 4 h and DMSO for the control cells.
Protein kinase assays
Protein kinase assays were performed in a 20-μl reaction volume with 1× kinase buffer (25 mM Tris–HCl, pH 7.5; 5 mM β-glycerol phosphate; 10 mM MgCl2; 0.1 mM sodium vanadate and 0.5 mM DTT) as described previously (26). The reaction was initiated by adding 10 μCi of γ32P-ATP. For the inhibition studies, all reaction components except ATP were incubated with the inhibitors for 45 min on ice prior to starting the kinase assay. CKI7 or IC261 were resuspended in DMSO and used at a final concentration of 5–100 μM as indicated. Synthetic PI4,5P2 was resuspended in 20 mM Tris–HCl, subjected to bath sonication to form micelle, and used at a concentration of 25–100 μM as indicated.
Immunoprecipitation and immunoblot analysis
Total cell lysates were prepared in buffer A [50 µM Tris (pH 7.4), 100 mM KCl, 5 mM EDTA, 5% NP-40, 5 mM NaF, 100 µg/ml RNase A, 1 mM NaVO4, 50 mM β-glycerol phosphate, and protease inhibitors] and gently sonicated. Rabbit polyclonal anti-Star–PAP and anti-PIPKIα (24); mouse monoclonal anti-β-tubulin (Upstate Biotechnology); rabbit polyclonal anti-actin (Bethyl Laboratories); rabbit polyclonal anti-RNA polymerase II (N-20); rabbit polyclonal anti-CPSF-73 (Bethyl Laboratories); mouse monoclonal anti-Flag M5 (Sigma); rabbit polyclonal anti-CKIα (30); rabbit polyclonal anti-CKIε (Bethyl Laboratories); mouse monoclonal anti-CKIδ (Abcam); and rabbit polyclonal anti-CKII (Bethyl Laboratories) were used for western blot and RIP analysis. Antibodies were used between 1:1000–1:2000 dilutions for immunoblot experiments.
3′-RACE assay
Total RNA was extracted as described previously (24) and 3′-RACE was carried out as described previously (25). The cDNAs for the 3′-RACE assays were synthesized using the Smart RACE Kit (Clontech) according to manufacturer’s instructions with 1 µg of total RNA isolated from HEK-293 cells. The gene specific forward primers for HO-1 or GAPDH are 5′-GACCTGCCCAGCTCTGGCGAG-3′ and 5′-GTATGACAACGAATTTGGCTACAGCAAC-3′. The RACE products were confirmed by sequencing.
Quantitative real-time PCR (RT–qPCR)
Total RNA was extracted and reverse transcribed as described previously (24). Target mRNA was quantified with the MyiQ single-color real-time PCR detection system and iQ SYBR Green Supermix (Bio-Rad). Single-product amplification was confirmed by melting-curve analysis, and primer efficiency was near 100% in all experiments. Quantification is expressed in arbitrary units, and target mRNA abundance was normalized to the expression of GAPDH with the Pfaffl method. All real-time RT–qPCR results were representative of at least three independent experiments. Primers used for the PCR were CCACCAAGTTCAAGCAGCTCTA (forward) and GCTCCTGCAACTCCTCAAAGAG (reverse) for HO-1; GAAGGTCGGAGTC AACGGATTT (forward) and GAATTTGCCATGGGTGGAAT (reverse) for GAPDH; AAGTT CTTGAAACTCTGCAAGAGAAGG (forward) and (GCCTCAACTGTATTGAACTCGGAC) for GCLC respectively.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) experiments were performed as described previously (25). The primers specific to HO-1 used to study the association of CKIα and CKIε were 5′-TTCTGTTGTTTTTATAGCAG-3′ as forward and 5′-TCAAACAGACCAGCTCCTG-3′ as reverse.
In vitro polyadenylation assay
In vitro polyadenylation assay with Star–PAP purified from the HEK-293 cells were performed using the 45-mer RNA oligonucleotide (UAGGGA)5A15 as an RNA substrate as described previously (24). For inhibition studies, the Star–PAP complex was isolated after the treatment of the cells with CKI inhibitors for 3 h. For the polyadenylation assay with phosphatase treatment, the samples were incubated with lamda phosphatase in RT for 30 min before the initiation of the polyadenylation reaction. Control samples were equally incubated but without phosphatase.
RESULTS
The oxidative stress agonist tBHQ stimulates Star–PAP phosphorylation
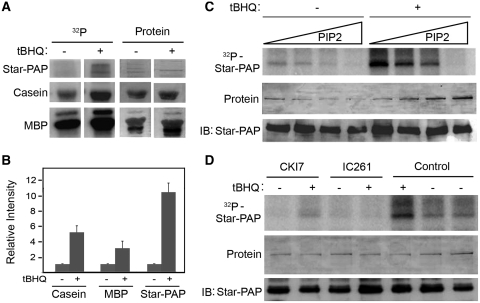
Previously, it had been shown that the Flag–Star–PAP complex purified from cells retained associated kinase activity toward Star–PAP and the generic protein kinase substrates casein and myelin basic protein (MBP) (26). One of the associated kinases that phosphorylated Star–PAP was identified as the PI4,5P2-sensitive isoform of CKI, CKIα (26), but it was not clear how these kinases integrate into signaling pathways that regulate Star–PAP. Since Star–PAP purified from cells treated with the oxidative stress agonist, tert-Butylhydroquinone (tBHQ) stimulated its polyadenylation activity (24), the effect of tBHQ treatment on the phosphorylation of Star–PAP was investigated. To assess kinase activity associated with the Star–PAP complex, the Flag–Star–PAP complex was isolated from HEK-293 cells stably expressing Flag–Star–PAP after treatment with tBHQ or the DMSO solvent control. The purified Star–PAP complex was then subjected to an in vitro kinase assay using purified Star–PAP and added casein or MBP as substrates. Treatment of the cell with tBHQ stimulated a dramatic increase in the phosphorylation of Star–PAP and exogenous casein or MBP by the kinase activity associated with the cell purified Star–PAP complex (Figure 1A). The phosphorylation of Star–PAP increased ~10-fold, whereas casein and MBP increased 3–6-fold upon tBHQ stimulation compared to the untreated samples (Figure 1A and B). Since the previously reported Star–PAP associated kinase, CKIα is a PI4,5P2 sensitive kinase (29), we analyzed the kinase assay in the presence of increasing concentration of PI4,5P2 (Figure 1C). As previously observed, the addition of PI4,5P2 to the kinase assay with Star–PAP purified from the unstimulated cells inhibited the phosphorylation of Star–PAP. Significant inhibition of phosphorylation was observed at 25–50 μM of PI4,5P2 and a complete inhibition was obtained at 100 μM of PI4,5P2 (Figure 1C and Supplementary Figure S1A). The stimulated phosphorylation of Star–PAP after the pre-treatment of the cell with tBHQ was equally inhibited by PI4,5P2. The significant inhibition of kinase activity required ~50 μM of PI4,5P2 with a complete inhibition obtainable at ~100 μM (Figure 1C and Supplementary Figure S1A).
Figure 1.
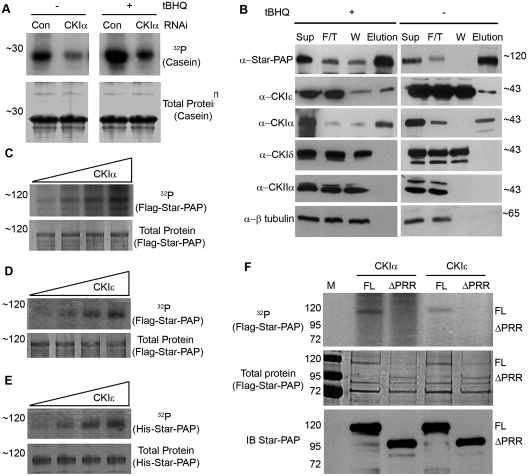
CKI-mediated Star–PAP phosphorylation is induced by tBHQ. (A) In vitro kinase assay of the Flag–Star–PAP complex purified from HEK-293 cells stably expressing Flag–Star–PAP after treatment with tBHQ (100 μM for 4 h) or the solvent control, DMSO using Star–PAP, Casein and Myelin basic protein (MBP) as substrates. The 32P incorporated and the total protein (coomasie-stained) are indicated. (B) Quantification of the kinase activity toward Star–PAP, Casein and MBP from the autoradiogram in A. The intensity in arbitrary unit is expressed relative to the tBHQ untreated samples. (C) In vitro kinase assay as in A of the Flag Star–PAP complex purified from HEK-293 cells stably expressing Flag Star–PAP after treatment with tBHQ (100 μM for 4 h) or the solvent control, DMSO with no exogenous substrate in presence of increasing amounts of PI4,5P2 (0, 25, 50 and 100 μM). The 32P incorporated, total protein and the immunoblot for Star–PAP are indicated. (D) In vitro kinase assay as in C but after the treatment with 100 μM of CKI7 or 100 µM IC261 prior to initiation of the kinase reaction. Control represents the untreated sample.
To confirm the role of CKI (or CKIα) in tBHQ-induced Star–PAP phosphorylation, in vitro kinase assays were carried out using the Star–PAP complex in the presence or absence of the CKI activity inhibitors CKI7 and IC261. Phosphorylation of Star–PAP was inhibited by 100 μM of either CKI7 or IC261, equally in the presence or absence of tBHQ treatment (Figure 1D). At higher concentration, the CKI inhibitors CKI7 and IC261 have been reported to inhibit other kinases (53,54,58). To confirm that the inhibition of Star–PAP phosphorylation by CKI7 and IC261 was specific, in vitro kinase assays were further carried out with low concentrations (5–15 μM) of CKI7 and IC261. We consistently observed inhibition of Star–PAP phosphorylation equally at 10 or 15 μM of CKI7 or IC261 in presence or absence of tBHQ treatment (Supplementary Figure S2A and B). At a concentration ≤5 μM of the inhibitors, there was only partial inhibition of Star–PAP phosphorylation (Supplementary Figure S2A and B). However, the association of CKIα with Star–PAP was not enhanced upon tBHQ treatment (Supplementary Figure S1B), suggesting an increase in the kinase specific activity or association of another kinase. Taken together, these data indicate that the phosphorylation of Star–PAP mediated by CKI activity is stimulated by tBHQ treatment of the cell.
Phosphorylation of Star–PAP is critical for its polyadenylation activity
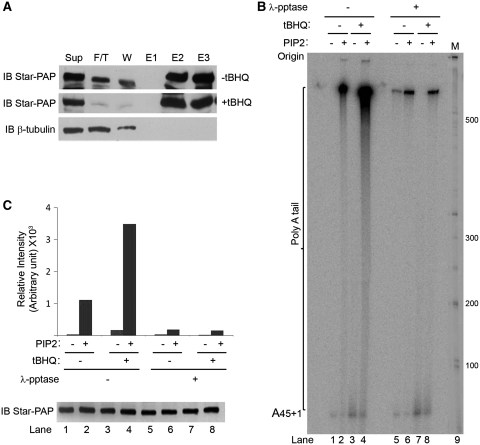
Previously, we reported that tBHQ treatment stimulated Star–PAP polyadenylation activity, and here we showed that tBHQ-induced CKI-mediated Star–PAP phosphorylation. Therefore, phosphorylation may play an important role in the regulation of Star–PAP activity. To test this, the Flag–Star–PAP complex was purified from the HEK-293 cells stably expressing Flag–Star–PAP after tBHQ treatment or solvent DMSO (Figure 2A). The purified Star–PAP complex was then treated with λ-phosphatase for 30 min at RT and then assayed for its polyadenylation activity using an A15 RNA substrate by an in vitro polyadenylation assay (24) in the presence or absence of PI4,5P2. Treatment with tBHQ resulted in a dramatic stimulation of the Star–PAP polyadenylation activity upon PI4,5P2 addition when compared to untreated cells (Figure 2B, lane 4), which is consistent with the previous finding that tBHQ priming is required for the stimulation of Star–PAP polyadenylation activity by PI4,5P2. When phosphatase treated, the purified Star–PAP complex showed minimal activity regardless of PI4,5P2 addition or tBHQ treatment (Figure 2B, lanes 5–8). There was a ~6-fold reduction in the Star–PAP polyadenylation activity after phosphatase treatment in presence of PI4,5P2 from tBHQ unprimed and ~20-fold reduction from the tBHQ primed samples compared to that of untreated Star–PAP (Figure 2C). Western blot analysis showed equal amounts of Star–PAP in each reaction lane (Figure 2C). The loss of Star–PAP activity after phosphatase treatment indicates that phosphorylation is critical for both Star–PAP polyadenylation activity and PI4,5P2 stimulation.
Figure 2.
Phosphorylation of Star–PAP is critical for its polyadenylation activity. (A) Immunoblot of Flag–Star–PAP purification. Sup = supernatant, F/T = Flow through, W = wash and E1-E3 = elution fractions of Star–PAP the complex. (B) In vitro polyadenylation assay of the Flag–Star–PAP complex purified from HEK-293 cells stably expressing Flag–Star–PAP after treatment with tBHQ (100 μM for 4 h) or the solvent control, DMSO with or without the addition of PI4,5P2 and λ-phosphatase treatment of the protein. The cell purified Star–PAP was treated with λ-phosphatase at RT for 30 min and subjected to polyadenylation assay using (UAGGGA)5A15 oligo as described (24). The poly(A) tail and the origin of the gel are indicated. (C) Quantification of the poly(A) tail addition in B (upper panel) and western blot analysis of Star–PAP in each reaction lane as of the PAP assay in B (lower panel). The intensity of radioactive 32P incorporation is expressed in arbitrary units.
CKI activity mediated phosphorylation of Star–PAP modulates Star–PAP polyadenylation activity in vitro
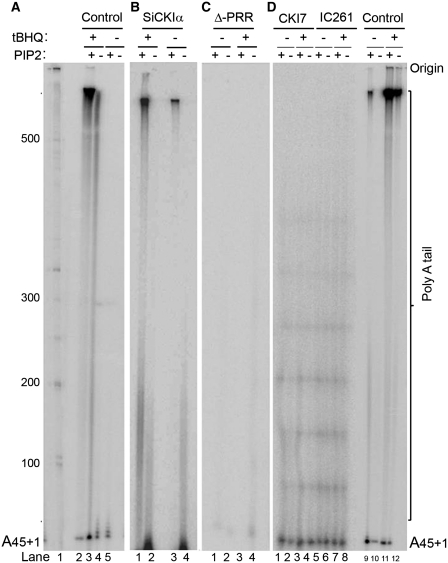
Star–PAP phosphorylation is inhibited by CKI inhibitors in vitro. The expression of Star–PAP target mRNAs, such as HO-1 was also blocked after the cells were treated with CKI inhibitors (26). To determine if CKI (or CKIα) is the critical kinase regulating Star–PAP activity, the Flag–Star–PAP complex was purified from the HEK-293 cells stably expressing Flag–Star–PAP following the knockdown of CKIα with siRNA (Supplementary Figure S3A for western) or pre-treatment with CKI inhibitors, CKI7 and IC261, and the subsequent treatment with tBHQ or DMSO (Supplementary Figure S3B). The purified Star–PAP complex (Supplementary Figure S3B) was then subjected to in vitro polyadenylation assays (24). Star–PAP purified from the CKIα inhibitor treated cells (CKI7 and IC261) diminished the Star–PAP polyadenylation activity compared to the control cells (Figure 3A and D). There was little polyadenylation activity, which failed to be stimulated by tBHQ treatment or the addition of PI4,5P2 (Figure 3D, lanes 1–8). The addition of the CKI inhibitors (CKI7 and IC261) to the polyadenylation reaction, however, did not affect the Star–PAP polyadenylation activity (data not shown). The loss of CKIα had a modest effect on the Star–PAP polyadenylation activity and was less efficient in the inhibition of Star–PAP polyadenylation activity compared to the CKI inhibitor treatments (Figure 3B and D) consistent with the previous observation (26). The addition of PI4,5P2 and/or tBHQ pre-treatment did show significant enhancement of the polyadenylation (Figure 3B). Additionally, the Flag–Star–PAP complex purified from cells transiently expressing a Star–PAP mutant with a deletion within the proline rich region (residues 223–274) (26), exhibited no polyadenylation activity (Figure 3C). Taken together, these data suggest that phosphorylation of Star–PAP by CKI modulates the Star–PAP polyadenylation activity downstream of stimulation by oxidant stress and phosphorylation primes Star–PAP, so that it can be stimulated by PI4,5P2.
Figure 3.
CKI-activity modulates Star–PAP polyadenylation activity. In vitro polyadenylation assays of cell-purified Flag–Star–PAP (A) after treatment with tBHQ or DMSO in presence and absence of PI4,5P2 addition; (B) purified after knockdown of CKIα with siRNA followed by treatment with tBHQ (100 μM for 4 h) or solvent control DMSO; (C) purified from HEK-293 cells transfected with a Star–PAP mutant construct that contains a deletion of proline rich region; (D) purified after the treatment of the cells with CKIα inhibitors CKI7 and IC261. Control refers to the polyadenylation assays carried out with untreated Star–PAP.
CKI activity is required for the efficient 3′-end processing and expression of Star–PAP target mRNAs in vivo
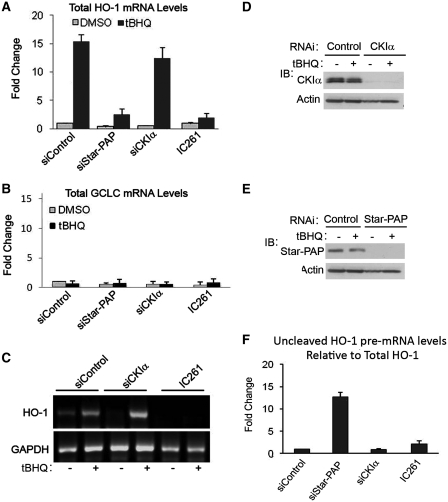
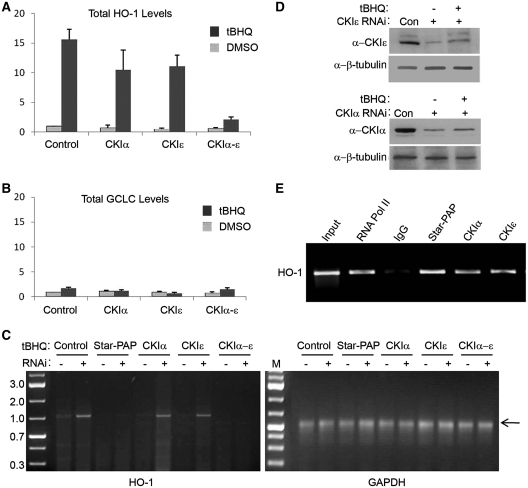
We have shown that CKIα phosphorylates Star–PAP and modulates Star–PAP’s polyadenylation activity. To further examine the role of CKI activity in the expression and 3′-end processing of Star–PAP target messages, total RNA was isolated from HEK-293 cells after transfection with siRNA (control siRNA or siRNA specific for Star–PAP or the associated kinase CKIα) (Figure 4D and E for western) or treatment with the CKI inhibitor IC261 in the presence and absence of tBHQ stimulation of the cells. We then analyzed the total mRNA expression and the measure of 3′-end processed (polyadenylated, mature) mRNA of Star–PAP direct target heme oxygenase-1 (HO-1) by RT–qPCR and 3′-RACE assay, respectively. As expected, RT–qPCR demonstrated an induction of ~15-fold of HO-1 expression after tBHQ treatment and knockdown of endogenous Star–PAP or treatment with CKI inhibitor IC261 ablated both basal and stress-induced HO-1 mRNA levels (Figure 4A) (24,26). Knockdown of CKIα did not inhibit tBHQ-induced HO-1 message unlike the CKI inhibitor, IC261, which efficiently blocked the tBHQ-induced HO-1 expression (Figure 4A) (26). The expression of the Star–PAP non-target glutamate-cysteine ligase, catalytic subunit (GCLC) was not affected by tBHQ treatment, CKIα knockdown, or CKI inhibitor treatments (Figure 4B).
Figure 4.
CKI-activity is required for the expression and efficient 3′-end processing of Star–PAP target message. HEK-293 cells were transfected with siRNA specific for endogenous Star–PAP or CKIα or treated with IC261 (100 µM for 2.5 h), then stimulated with the antioxidant tBHQ (100 µM for 4 h). Total RNA was extracted and analyzed by quantitative real-time RT–PCR for (A) HO-1 mRNA levels (B) and GCLC mRNA levels. Normalized to GAPDH in each data set, ± SEM, N ≥ 6. (C) 3′-RACE assay of HO-1 and GAPDH after HEK-293 cells were transfected with siRNA specific for endogenous CKIα or inhibitors of CKI-activity and then stimulated with the antioxidant tBHQ (100 µM for 4 h). Data is representative of N ≥ 3. (D and E) Western blot analysis of the siRNA knockdown of CKIα and Star–PAP. (F) Levels of uncleaved HO-1 pre-mRNA normalized to total mRNA from HEK 293 cells transfected with control, Star–PAP or CKIα specific siRNA, or after treatment with CKI inhibitor IC261. Data is representative of N ≥ 3.
To further examine the role of CKI activity in the 3′-end processing of HO-1 pre-mRNA, the knockdown and inhibitor approach described above was employed, followed by 3′-RACE analysis from the total RNA to measure the mature polyadenylated mRNA in the cell. Distinct RACE products for HO-1 (~700 bp in length) were observed in control cells with a large induction upon tBHQ stimulation. HO-1 production was lost after the inhibition of cellular CKI activity with IC261, demonstrating that CKI activity is necessary for the processing of this Star–PAP target mRNA (Figure 4C). Consistent with the expression profile, CKIα knockdown was not effective in blocking the 3′-end processed (polyadenylated) mRNA after tBHQ stimulation (Figure 4C). The non-target glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was not affected by tBHQ treatment, CKIα knockdown, or CKI inhibitor treatment (Figure 4C). These results suggest that CKI activity is required for the expression of Star–PAP target and that Star–PAP complex perhaps contains other isoforms of CKI in addition to CKIα.
Star–PAP is an essential factor for the cleavage of its target mRNA and Star–PAP recruits the cleavage factors to the pre-mRNA (25). Therefore, we measured the HO-1 uncleaved mRNA levels to study the possible role of CKI activity in the cleavage of Star–PAP target pre-mRNAs. Consistent with the previous report (24), Star–PAP knockdown resulted in ~12-fold increase in the quantity of uncleaved HO-1 mRNA relative to total HO-1 mRNA (Figure 4F). Loss of CKIα or treatment with IC261 did not exhibit any significant change in the cleavage of HO-1 pre-mRNA (Figure 4F); suggesting that the regulation Star–PAP target messages by CKI activity regulates polyadenylation. Similarly, the association and assembly of Star–PAP polyadenylaton complex did not appear to be affected by CKI activity as demonstrated by western analysis of key Star–PAP associated factors after the inhibition of CKI in the cell (Supplementary Figure S4A).
Another CKI isoform, CKIε associates with and phosphorylates Star–PAP
CKIα knockdown only partially blocked the basal expression of HO-1 (26) and poorly blocked tBHQ stimulated Star–PAP polyadenylation activity and the expression of Star–PAP targets. Therefore it is possible that Star–PAP complex associates with additional isoforms of CKI that compensate for the loss of CKIα. To test this hypothesis, we knocked down CKIα from the cell and isolated the Flag–Star–PAP complex and subjected it to kinase assay with casein as a substrate. We observed that even after the knock down of CKIα, significant amount of kinase activity was still associated with the Star–PAP complex toward casein, which was equally-induced by tBHQ treatment similar to the wild type cells (Figure 5A). This suggests that Star–PAP complex contains additional casein kinase(s) other than CKIα. Since, the kinase inhibitors IC261 and CKI7 employed for our study also inhibits CKIε and δ (51) in addition to CKIα (26), we tested the association of these isoforms in the Flag–purified Star–PAP complex from the cell in the presence or absence of tBHQ treatment. We observed the association of CKIε but not δ in the Star–PAP complex purified from cells both with tBHQ treatment and without tBHQ treatment (Figure 5B). Other casein kinases such as CKII did not associate with the Star–PAP complex (Figure 5B). Control protein, β-tubulin was also not detected in the Star–PAP complex purified from the cell (Figure 5B), suggesting the specificity of the proteins associated with the Star–PAP complex.
Figure 5.
CKI isoform, CKIε associates with and phosphorylates Star–PAP. (A) In vitro kinase assay of the Flag–Star–PAP complex purified from HEK-293 cells stably expressing Flag–Star–PAP after knockdown of CKIα with specific RNAi or control RNAi followed by treatment with tBHQ (100 μM for 4 h) or DMSO using generic kinase substrate Casein. The 32P incorporated and the total protein (coomasie-stained) are indicated. (B) Western analysis to detect the association of various CKI isoforms as indicated in the Flag-purified Star–PAP complex from the cell in presence and absence of tBHQ treatment of the cell. Sup = Supernatant, F/T = Flow through, W = Wash, E = Elution fraction of the purification. (C) In vitro kinase assay of increasing amounts of CKIα with heat inactivated Flag purified Star–PAP as substrate. (D) In vitro kinase assay of increasing amounts of CKIε with heat inactivated cell-purified Flag–Star–PAP as substrate. (E) In vitro kinase assay of increasing amounts of CKIε with heat inactivated recombinant His-tagged Star–PAP as substrate. The 32P incorporated and the total protein (coomasie-stained) are indicated. (F) In vitro kinase assay of CKIα and CKIε with Flag–Star–PAP full length (FL) and deleted for proline rich region (ΔPRR) as substrate. The 32P incorporated, the total protein (coomasie-stained) and the immunoblot are indicated.
We further investigated if CKIε phosphorylates Star–PAP by an in vitro kinase assay. GST-tagged recombinant full length CKIε was tested for its ability to phosphorylate the heat inactivated Flag–tagged Star–PAP isolated from the cell or His-tagged recombinant Star–PAP. We also tested the activity of GST-CKIε with its generic kinase substrate casein (Supplementary Figure S4B). We observed the phosphorylation of both cell-purified (Flag-tagged) (Figure 5D) and recombinant (His-tagged) (Figure 5E) Star–PAP by CKIε. There was a dose dependent increase in phosphorylation with increased CKIε concentration for both the substrates (Figure 5D and E). The control Flag-tagged CKIα also phosphorylated the heat inactivated cell-purified Flag–Star–PAP (Figure 5C) as previously described (26). These results indicate that the associated CKIε can directly phosphorylate Star–PAP.
Since CKIα phosphorylates Star–PAP in the proline rich region, which splits the catalytic domain, we tested if CKIε also shares the similar region of phosphorylation on Star–PAP. Kinase assays were performed to test the activity of GST-tagged CKIε toward Flag-purified Star–PAP with deletion of the proline rich region, with CKIα as control. As expected, both CKIα and CKIε exhibited kinase activity toward full length (FL) Flag–Star–PAP (Figure 5F). Remarkably, the GST–CKIε failed to phosphorylate the Star–PAP with deletion of proline rich region (ΔPRR) like CKIα, while there was intact phosphorylation of full length Star–PAP (Figure 5F). This suggests that both isoforms (α and ε) share similar region of phosphorylation on Star–PAP.
CKI isoforms α and ε are required for the stressed induced expression of Star–PAP target mRNA
Since CKIε phosphorylates Star–PAP and shares the similar region of phosphorylation with CKIα, we investigated if CKIε has any role in the regulation of Star–PAP target messages. RT–qPCR analysis was performed with total RNAs isolated from cells after the single knockdowns of CKIα and CKIε or together in the presence or absence of tBHQ treatment (Figure 6D for western). Consistent with above observations, there was no loss of oxidative stress-induced expression after CKIα knockdown (Figure 6A). Similarly, CKIε knockdown did not significantly impact tBHQ-induced expression of HO-1 mRNA (Figure 6A). However, strikingly, when both the isoforms (CKIα and CKIε) were knocked down together, this blocked the expression of HO-1 (Figure 6A) demonstrating that both kinases regulate expression of the Star–PAP target HO-1, or that the two isoforms have redundant function to regulate Star–PAP. The control mRNA GCLC was not affected by either single or double knockdowns of CKIα and CKIε (Figure 6B).
Figure 6.
CKI isoforms α and ε are required for the stress-induced expression and efficient 3′-end processing of Star–PAP target message. Quantitative real-time RT–PCR analysis for (A) HO-1 mRNA levels (B) and GCLC mRNA levels with the total RNAs from HEK-293 cells transfected with siRNA specific for endogenous CKIα and CKIε separately or together followed by treatment with the antioxidant tBHQ (100 µM for 4 h) or solvent DMSO. (C) 3′-RACE assay of HO-1 and GAPDH after HEK-293 cells were transfected with siRNA specific for endogenous CKIα and CKIε separately or together then stimulated with the antioxidant tBHQ (100 µM for 4 h) or DMSO. Data is representative of N ≥ 3. (D) Western blot analysis of the siRNA knockdown of CKIα and CKIε. (E) RIP analysis of HO-1 UTR by IP with antibodies specific to RNA Pol II (RNAPII), Star–PAP, CKIα, and CKIε followed by reverse transcription-PCR with primers specific to HO-1 UTR region.
Similarly, by the 3′-RACE assay which measures the mature polyadenylated message from the cell, we observed distinct signals from control resting cells, stimulated by tBHQ treatment that were lost upon the knockdown of Star–PAP (Figure 6C) as previously described (25). Consistent with the total mRNA levels, there was no significant reduction in the RACE products upon CKIα or CKIε knockdowns in presence of tBHQ treatment compared to the control samples (Figure 6C). Yet, when both isoforms (α and ε) were knocked down, there was complete loss of the RACE product corroborating the above observation that CKIα and CKIε are required for the expression of Star–PAP target mRNA. The control non-target GAPDH was not affected by either single or double knockdowns of CKIα and CKIε (Figure 6C).
Star–PAP is a PAP that binds to pre-mRNA along with other cleavage factors, assembling a distinct processing complex on the 3′-end of its target RNAs (25). In addition, CKIα associates with HO-1 pre-mRNA (26) perhaps by virtue of its interaction with Star–PAP 3′-end processing complex. Therefore, we investigated if CKIε exhibits association with HO-1 pre-mRNA by an RIP experiment. The cells were cross-linked with formaldehyde and IP’ed using the respective antibodies followed by detection of the associated RNA by RT–PCR using primers specific to HO-1 3′-UTR. As expected, we saw the association of Star–PAP and CKIα similar to the control RNA Pol II (Figure 6E). Interestingly, we also observed the association of CKIε with HO-1 3′-UTR RNA (Figure 6E) supporting the above observations that CKIε also acts as a part of Star–PAP 3′-end processing complex.
DISCUSSION
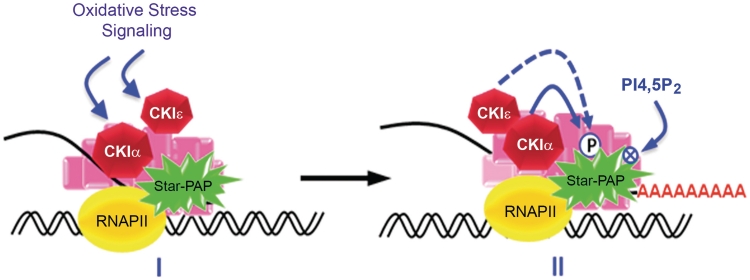
Star–PAP activity is regulated by its phosphorylation likely downstream of phosphoinositide and oxidative stress signals. CKI protein kinase activity regulates Star–PAP. A PI4,5P2 sensitive isoform, CKIα was demonstrated to associate with and phosphorylate Star–PAP within its catalytic region (26). Here, we show that CKI activity mediated the phosphorylation of Star–PAP that was stimulated by oxidative stress and this phosphorylation was critical for both Star–PAP’s polyadenylation activity and priming required for PI4,5P2 stimulation of polymerase activity. Moreover, Star–PAP associated with an additional CKI isoform, CKIε that compensated for the loss of CKIα. CKIε, like CKIα, also phosphorylated Star–PAP in the proline rich region (PRR). This indicated that CKI kinase activity is critical for Star–PAP activity and that CKIε acts as a functionally redundant isoform of CKI that can complement for the loss of CKIα in the cell to regulate Star–PAP. A single isoform of CKI may regulate Star–PAP phosphorylation and HO-1 expression. CKIα may be the major CKI isoform that modulates Star–PAP downstream of oxidative stress. CKIε could act as a substitute kinase to regulate Star–PAP’s control of HO-1 processing and could act more specifically in other pathways that regulate Star–PAP. A model depicting CKIα/CKIε-mediated regulation of Star–PAP and target messages is shown in Figure 7.
Figure 7.
A model depicting CKIα−ε mediated regulation of Star–PAP activity to modulate the 3′-end processing of Star–PAP target mRNA.
Gonzales et al. (26) showed that the CKIα phosphorylation site within the PRR of Star–PAP interrupts the catalytic domain and this implied that phosphorylation of Star–PAP could affect its enzymatic activity. Here, we show that in addition to CKIα, CKIε also phosphorylates Star–PAP in a similar region in the catalytic domain. This phosphorylation modulates Star–PAP polyadenylation activity and the expression of the Star–PAP target HO-1. Similar regulation of PAP activity by phosphorylation was also reported for canonical PAPα (59,60), although the mechanism of regulation is different. These differences can be attributed to the distinct kinases and stimuli responsible for the phosphorylation of the two polymerases (59). PAPα is phosphorylated at the C-terminal end by ERK kinase, whereas Star–PAP is phosphorylated within the catalytic region by both CKIα and CKIε (24,59,60). Inhibition of PAPα phosphorylation resulted in a loss of polyadenylation activity. Star–PAP phosphorylation is required for both polyadenylation activity and its stimulation by PI4,5P2. Since Star–PAP activity is specifically regulated by PI4,5P2 binding, an inhibition in activity may indicate that the PI4,5P2 binding site overlaps with the CKI phosphorylation sites or be regulated indirectly (24). These data indicate that phosphorylation modulates substrate recognition, PI4,5P2 binding or both to regulate Star–PAP activity. The proline rich region in the catalytic domain where both CKIα and ε phosphorylate Star–PAP contains a cluster of about 15 Ser/Thr residues which are putative sites for CKI phosphorylation. Therefore, it is likely that there is a complex phosphorylation pattern involving multiple sites that are common or overlapping for both the CKI isoforms.
Both the phosphorylation of Star–PAP by CKI isoforms and Star–PAP activity are regulated by oxidative stress. Yet, the association of CKIα or CKIε with Star–PAP did not increase upon stimulation with oxidative stress, suggesting that enhanced CKI kinase activity was responsible for the increase in Star–PAP activity. The mechanism of oxidant-induced CKI-regulated Star–PAP activity remains elusive. CKI isoforms act as a downstream target of oxidative stress signal; however, the current literature indicates that members of the CKI family are constitutively active kinases (49). Alternatively, an oxidative stress signal may induce yet-to-be-identified nuclear factors that cooperate with CKIα or CKIε to induce the phosphorylation of Star–PAP. One similar example is the phosphorylation of mammalian IFNARI by CKIα, which is greatly induced by ER stress (61).
CKIα or ε are regulated by hierarchical priming in that they can require prior phosphorylation by other protein kinases to generate their substrate site for phosphorylation (27). The CKI isoforms also spatially integrate into protein complexes and this modulates their activity toward proteins in those complexes (27,28,49). Both of these properties may influence CKI control of Star–PAP function. There are additional protein kinases that associate with the Star–PAP complex that are stimulated by oxidative stress and these may define the hierarchical priming of CKI isoforms for Star–PAP phosphorylation and activity. The kinase that phosphorylates MBP has not yet been identified, but may be a protein kinase that defines hierarchical priming of the associated CKI protein kinases.
Star–PAP regulation is integrated into multiple signaling pathways (62). Different Star–PAP target genes are specifically modulated by distinct signaling pathways individually or synergistically (62). In addition, Star–PAP-dependent mRNAs are specific to a select set of messages (24–26). However, it is still unclear how the Star–PAP dependent 3′-end processing complex identifies the target mRNAs (25,62). One possibility is the direct interaction of Star–PAP with certain sequence elements on its target mRNAs as evidenced by its footprint on the target HO-1 mRNA (25). Phosphorylation of Star–PAP at distinct sites by various protein kinases could also determine the target gene specificity or the signaling to regulate Star–PAP. Moreover, the Star–PAP 3′-end processing complex associates with PIPKIα (24) and protein kinases including CKIα and CKIε. It has been reported that PIPKIα or CKIα are required for the expression of some but not all Star–PAP target mRNAs (24,26). Therefore, such proteins could also be involved in modulating Star–PAP target specificity in addition to regulating its polyadenylation activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) (Grant number R01 GM051968 to R.A.A.); (Award number F32 GM082005 to C.A.B.); American Heart Association award (0920072G to R.S.L.). Funding for open access charge: NIH RO1 GM051968.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Andrew Hedman, Suyong Choi and Weimin Li for carefully reading the manuscript.
REFERENCES
- 1.Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O’Malley BW. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 3.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 4.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 6.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 7.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proudfoot N, O’Sullivan J. Polyadenylation: a tail of two complexes. Curr. Biol. 2002;12:R855–R857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- 9.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Ryner LC, Takagaki Y, Manley JL. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol. Cell. Biol. 1989;9:4229–4238. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 15.Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 17.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 18.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 20.Manley JL. A complex protein assembly catalyzes polyadenylation of mRNA precursors. Curr. Opin. Genet. Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 21.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 22.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 25.Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 2010;29:4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J. Biol. Chem. 2008;283:12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- 28.Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Bazenet CE, Brockman JL, Lewis D, Chan C, Anderson RA. Erythroid membrane-bound protein kinase binds to a membrane component and is regulated by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 1990;265:7369–7376. [PubMed] [Google Scholar]
- 30.Brockman JL, Gross SD, Sussman MR, Anderson RA. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc. Natl Acad. Sci. USA. 1992;89:9454–9458. doi: 10.1073/pnas.89.20.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuazon PT, Traugh JA. Casein kinase I and II–multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 1991;23:123–164. [PubMed] [Google Scholar]
- 32.Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 33.Flotow H, Roach PJ. Synergistic phosphorylation of rabbit muscle glycogen synthase by cyclic AMP-dependent protein kinase and casein kinase I. Implications for hormonal regulation of glycogen synthase. J. Biol. Chem. 1989;264:9126–9128. [PubMed] [Google Scholar]
- 34.Meggio F, Perich JW, Marin O, Pinna LA. The comparative efficiencies of the Ser(P)-, Thr(P)- and Tyr(P)-residues as specificity determinants for casein kinase-1. Biochem. Biophys. Res. Commun. 1992;182:1460–1465. doi: 10.1016/0006-291x(92)91898-z. [DOI] [PubMed] [Google Scholar]
- 35.Flotow H, Roach PJ. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 1991;266:3724–3727. [PubMed] [Google Scholar]
- 36.Pulgar V, Marin O, Meggio F, Allende CC, Allende JE, Pinna LA. Optimal sequences for non-phosphate-directed phosphorylation by protein kinase CK1 (casein kinase-1)–a re-evaluation. Eur. J. Biochem. 1999;260:520–526. doi: 10.1046/j.1432-1327.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- 37.Marin O, Meggio F, Sarno S, Andretta M, Pinna LA. Phosphorylation of synthetic fragments of inhibitor-2 of protein phosphatase-1 by casein kinase-1 and -2. Evidence that phosphorylated residues are not strictly required for efficient targeting by casein kinase-1. Eur. J. Biochem. 1994;223:647–653. doi: 10.1111/j.1432-1033.1994.tb19037.x. [DOI] [PubMed] [Google Scholar]
- 38.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc. Natl Acad. Sci. USA. 2003;100:10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 42.Holland PM, Cooper JA. Protein modification: docking sites for kinases. Curr. Biol. 1999;9:R329–R331. doi: 10.1016/s0960-9822(99)80205-x. [DOI] [PubMed] [Google Scholar]
- 43.Burzio V, Antonelli M, Allende CC, Allende JE. Biochemical and cellular characteristics of the four splice variants of protein kinase CK1alpha from zebrafish (Danio rerio) J. Cell Biochem. 2002;86:805–814. doi: 10.1002/jcb.10263. [DOI] [PubMed] [Google Scholar]
- 44.Green CL, Bennett GS. Identification of four alternatively spliced isoforms of chicken casein kinase I alpha that are all expressed in diverse cell types. Gene. 1998;216:189–195. doi: 10.1016/s0378-1119(98)00291-1. [DOI] [PubMed] [Google Scholar]
- 45.Tapia C, Featherstone T, Gomez C, Taillon-Miller P, Allende CC, Allende JE. Cloning and chromosomal localization of the gene coding for human protein kinase CK1. FEBS Lett. 1994;349:307–312. doi: 10.1016/0014-5793(94)00679-2. [DOI] [PubMed] [Google Scholar]
- 46.Yong TJ, Gan YY, Toh BH, Sentry JW. Human CKIalpha(L) and CKIalpha(S) are encoded by both 2.4- and 4. 2-kb transcripts, the longer containing multiple RNA-destablising elements. Biochim. Biophys. Acta. 2000;1492:425–433. doi: 10.1016/s0167-4781(00)00146-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Gross SD, Schroeder MD, Anderson RA. Casein kinase I alpha and alpha L: alternative splicing-generated kinases exhibit different catalytic properties. Biochemistry. 1996;35:16319–16327. doi: 10.1021/bi9614444. [DOI] [PubMed] [Google Scholar]
- 48.Gross SD, Hoffman DP, Fisette PL, Baas P, Anderson RA. A phosphatidylinositol 4,5-bisphosphate-sensitive casein kinase I alpha associates with synaptic vesicles and phosphorylates a subset of vesicle proteins. J. Cell Biol. 1995;130:711–724. doi: 10.1083/jcb.130.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gross SD, Loijens JC, Anderson RA. The casein kinase Ialpha isoform is both physically positioned and functionally competent to regulate multiple events of mRNA metabolism. J. Cell. Sci. 1999;112(Pt 16):2647–2656. doi: 10.1242/jcs.112.16.2647. [DOI] [PubMed] [Google Scholar]
- 50.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 51.Grozav AG, Chikamori K, Kozuki T, Grabowski DR, Bukowski RM, Willard B, Kinter M, Andersen AH, Ganapathi R, Ganapathi MK. Casein kinase I delta/epsilon phosphorylates topoisomerase IIalpha at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res. 2009;37:382–392. doi: 10.1093/nar/gkn934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behrend L, Milne DM, Stoter M, Deppert W, Campbell LE, Meek DW, Knippschild U. IC261, a specific inhibitor of the protein kinases casein kinase 1-delta and -epsilon, triggers the mitotic checkpoint and induces p53-dependent postmitotic effects. Oncogene. 2000;19:5303–5313. doi: 10.1038/sj.onc.1203939. [DOI] [PubMed] [Google Scholar]
- 53.Chijiwa T, Hagiwara M, Hidaka H. A newly synthesized selective casein kinase I inhibitor, N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase I from bovine testis. J. Biol. Chem. 1989;264:4924–4927. [PubMed] [Google Scholar]
- 54.Mashhoon N, DeMaggio AJ, Tereshko V, Bergmeier SC, Egli M, Hoekstra MF, Kuret J. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J. Biol. Chem. 2000;275:20052–20060. doi: 10.1074/jbc.M001713200. [DOI] [PubMed] [Google Scholar]
- 55.Eide EJ, Virshup DM. Casein kinase I: another cog in the circadian clockworks. Chronobiol. Int. 2001;18:389–398. doi: 10.1081/cbi-100103963. [DOI] [PubMed] [Google Scholar]
- 56.Ning K, Li L, Liao M, Liu B, Mielke JG, Chen Y, Duan Y, El-Hayek YH, Wan Q. Circadian regulation of GABAA receptor function by CKI epsilon-CKI delta in the rat suprachiasmatic nuclei. Nat. Neurosci. 2004;7:489–490. doi: 10.1038/nn1236. [DOI] [PubMed] [Google Scholar]
- 57.Sprouse J, Reynolds L, Kleiman R, Tate B, Swanson TA, Pickard GE. Chronic treatment with a selective inhibitor of casein kinase I delta/epsilon yields cumulative phase delays in circadian rhythms. Psychopharmacology. 2010;210:569–576. doi: 10.1007/s00213-010-1860-5. [DOI] [PubMed] [Google Scholar]
- 58.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 60.Lee SH, Choi HS, Kim H, Lee Y. ERK is a novel regulatory kinase for poly(A) polymerase. Nucleic Acids Res. 2008;36:803–813. doi: 10.1093/nar/gkm1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Carvalho LP, Bhattacharya S, Carbone CJ, Kumar KG, Leu NA, Yau PM, Donald RG, Weiss MJ, Baker DP, et al. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol. Cell. Biol. 2009;29:6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell. Biol. 2010;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.