Abstract
The 5S genes of the eight species of the D. melanogaster subgroup have been mapped. The spacers, in contrast with coding regions, differ markedly between most species. One 5S gene unit has been sequenced for both D. simulans and D. teissieri. The mature 5S RNA region in these two species is identical to the corresponding region of D. melanogaster. Only 5 nucleotide variations occur between the D. melanogaster and D. simulans 5S gene spacers. The spacer in D. teissieri is very different. Only two segments, located one at each side of the coding region, are clearly homologous to corresponding sequences of D. melanogaster and D. simulans.
Full text
PDF


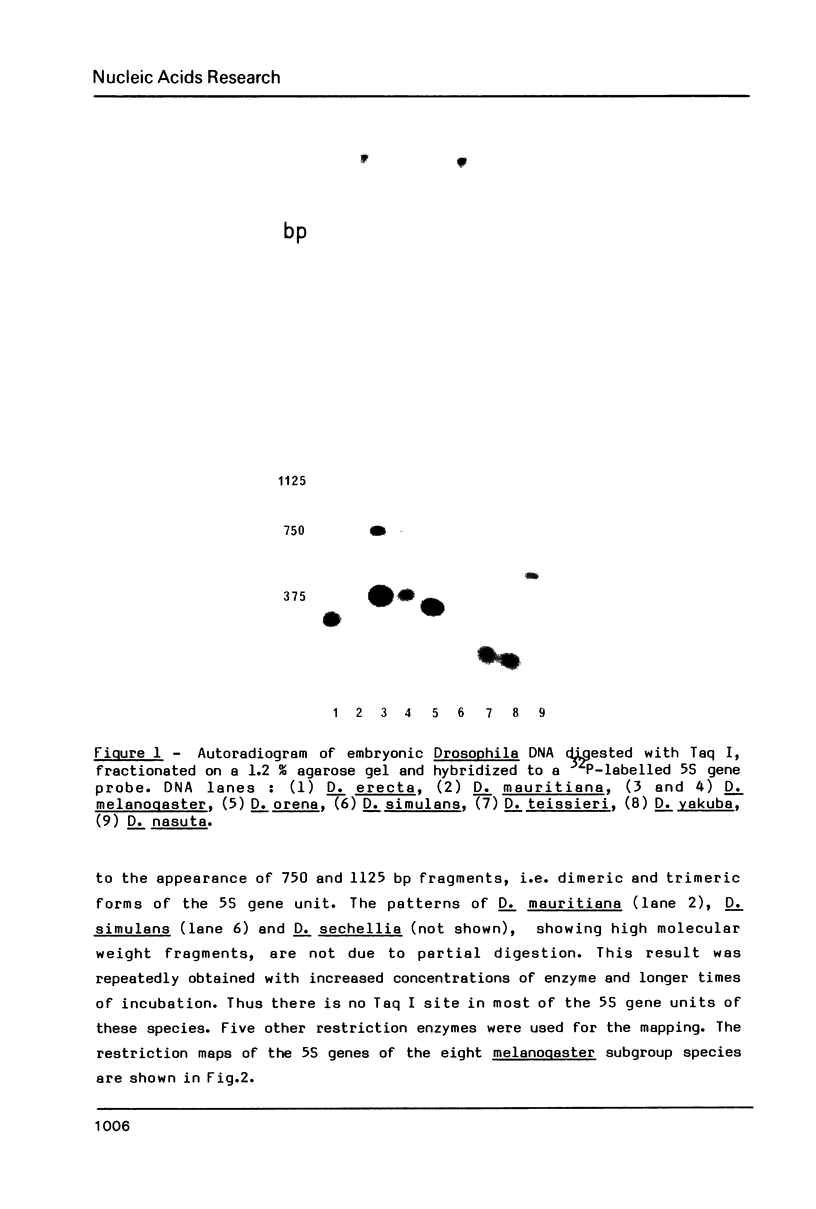








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Cartwright E. M., Brown D. D. Sequence studies of the 5 S DNA of Xenopus laevis. J Mol Biol. 1974 Nov 15;89(4):703–718. doi: 10.1016/0022-2836(74)90046-1. [DOI] [PubMed] [Google Scholar]
- Carroll D., Brown D. D. Adjacent repeating units of Xenopus laevis 5S DNA can be heterogeneous in length. Cell. 1976 Apr;7(4):477–486. doi: 10.1016/0092-8674(76)90199-9. [DOI] [PubMed] [Google Scholar]
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Egel R. Intergenic conversion and reiterated genes. Nature. 1981 Mar 19;290(5803):191–192. doi: 10.1038/290191a0. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r105–r133. [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indik Z. K., Tartof K. D. Glutamate tRNA genes are adjacent to 5S RNA genes in Drosophila and reveal a conserved upstream sequence (the ACT-TA box). Nucleic Acids Res. 1982 Jul 24;10(14):4159–4172. doi: 10.1093/nar/10.14.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Jacq B., Jourdan R., Jordan B. R. Structure and processing of precursor 5 S RNA in Drosophila melanogaster. J Mol Biol. 1977 Dec 15;117(3):785–795. doi: 10.1016/0022-2836(77)90069-9. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F., Ashburner M. A. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 1976 May 18;193(1112):275–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- Louis C., Schedl P., Samal B., Worcel A. Chromatin structure of the 5S RNA genes of D. melanogaster. Cell. 1980 Nov;22(2 Pt 2):387–392. doi: 10.1016/0092-8674(80)90349-9. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Strachan T., Coen E., Webb D., Dover G. Modes and rates of change of complex DNA families of Drosophila. J Mol Biol. 1982 Jun 15;158(1):37–54. doi: 10.1016/0022-2836(82)90449-1. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Evolution of transcribed and spacer sequences in the ribosomal RNA genes of Drosophila. Cell. 1979 Jul;17(3):607–614. doi: 10.1016/0092-8674(79)90268-x. [DOI] [PubMed] [Google Scholar]
- Tschudi C., Pirrotta V. Sequence and heterogeneity in the 5S RNA gene cluster of Drosophila melanogaster. Nucleic Acids Res. 1980 Feb 11;8(3):441–451. doi: 10.1093/nar/8.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer E. A., Martin S. L., Beverley S. M., Kan Y. W., Wilson A. C. Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2158–2162. doi: 10.1073/pnas.77.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]





