Abstract
Infectious bronchitis virus (IBV) causes highly contagious respiratory or urogenital tract diseases in chickens. The Maryland 27(Md27) strain was first isolated in 1976 from diseased chicken flocks in the Delmarva Peninsula region. To understand the genetic diversity and phylogenetic relationship of existing strains with Md27, the complete nucleotide sequence of the 3′end coding region (∼7.2 kb) of Md27 was determined and compared with other IBV strains and coronaviruses. It has the same S-3-M-5-N-3′ gene order, as is the case of other IBV strains. The spike gene of Md27 exhibits 97% identity with the SE17 strain. There are deletions at the spike gene, non-coding region between M and 5 genes, and at the 3′ untranslated region (UTR), which is different from Ark-like strains. Phylogenetic analysis and sequence alignments demonstrate that Md27 is a chimera containing different gene segments that are most closely related to the SE17, Conn and JMK strains. This current study provides evidence for genomic mutations and intergenic recombination that have taken place in the evolution of IBV strain Md27.
Keywords: infectious bronchitis virus, structural genes, sequencing, phylogenetic analysis, recombination
1. Introduction
Infectious bronchitis virus (IBV), a member of the Coronaviridae, order Nidovirales, is a pathogen of domestic chickens that causes acute, highly contagious respiratory disease [1,2]. The IBV genome contains a single, positive-strand RNA molecule, which is about 27.6 kb long and has a cap at the 5′end and poly (A) tail at the 3′end [3]. It comprises ten open reading frames (ORFs) and the first 20 kb genome is made up of ORF1, which is a replicase gene. The replicase has two ORFs, 1a and 1b [4]; among which 1b is produced as 1ab polyprotein by a -1 ribosomal frame-shifting mechanism [4]. The ORF1 encodes non-structural proteins associated with RNA replication and transcription. The IBV genome codes for four major structural proteins; the spike (S) glycoprotein, the small envelope (E) protein, the membrane (M) glycoprotein, and the nucleocapsid (N) protein [5]. In addition to this, IBV has other genes that encode for non-structural proteins interspersed among structural genes, namely 3a, 3b, 5a and 5b [6].
Numerous IBV serotypes, such as Arkansas, Massachusetts, Connecticut, Florida, Georgia and others, referred to as variants, exist in the United States of America (USA). These variants have practical significance for controlling the disease because immunity following infection or vaccination with one serotype often is not protective against subsequent infections with heterologous serotypes [7,8]. Many serotypes have been described for IBV, probably due to the frequent point mutations that occur in RNA viruses and also due to recombination events [9–11]. It is essential to characterize field isolates for the selection of appropriate vaccine strains.
The late Dr. Warren Marquardt (Department of Veterinary Medicine, University of Maryland) received samples from poultry diagnostic laboratories on the Delmarva Peninsula from 1971 through 1974 [12]. Out of 106 isolates made, three were not neutralized by any kind of serum. Two of these appear to be identical on the basis of a serum neutralization test and had been designated as Maryland 27 (Md27). The Md27 antiserum has an unusually broad spectrum of minor cross-reactions with the other viruses [12]. The Md27 strain has never been characterized by sequencing and it is essential to sequence these typical strains to understand the evolution of IBV geographically and also to implement an effective vaccination program to control new variant IBV strains. Here, we describe the complete sequence analysis of the 7.27 kb of 3′end of the genome of Md27 IBV strain and its comparison and phylogenetic relationship with many heterologous IBV strains, and other coronaviruses from different parts of the world.
2. Results and Discussion
The structural organization of Md27 and nucleotide identity with other IBV strains is shown in Tables 1 and 2, respectively. The spike protein gene of Md27 is 3507 nucleotides long (1168aa) and codes for a protein of approximately 128.6 kDa. The transcription regulating sequence (TRS) CTTAACAAA is located 49 nucleotides from the start codon of the spike gene. The spike protein is most likely glycosylated and has 22 potential N-glycosylation sites (Asn-Xaa-Ser/Thr) which are predicted by NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/). Hydrophobicity analysis (http://www.cbs.dtu.dk/services/TMHMM) predicted the occurrence of one transmembrane (TM) domain at (1100–1122aa) in the C-terminus of the spike protein and one endodomain (1122–1168aa). The Spike protein is cleaved into S1 and S2, of which S1 produces neutralizing and serotype specific antibodies [13,14]. The spike protein cleavage site for Md27 (His-Arg-Ser-Arg-Arg/Ser) is located between residues 544 and 545. The S1 protein is closely related to the SE17 strain (96% identity) and exhibits <86% identity with other IBV strains. The S2 gene is more conserved than S1 and has 98% identity with Ark DPI, Gray, Jilin, JMK and SE17 strains. The S2 subunit may also induce serotype specific neutralizing antibodies and S2 subunits are conserved within a serotype but not between serotypes [15]. There is a notable deletion of 3 nucleotides in the S1 gene between nucleotides 69 and 70, which is different from other Ark-like strains.
Table 1.
Structural genome organization of Md27*.
| Gene | Frame | Start | Stop | Size (nucleotides) | Size (amino acids) |
|---|---|---|---|---|---|
| S | +1 | 1 | 3507 | 3507 | 1168 |
| 3a | +3 | 3507 | 3680 | 174 | 57 |
| 3b | +2 | 3680 | 3874 | 195 | 64 |
| 3c(E) | +3 | 3855 | 4178 | 324 | 107 |
| M | +1 | 4156 | 4827 | 672 | 223 |
| 5a | +3 | 5178 | 5375 | 198 | 65 |
| 5b | +2 | 5372 | 5620 | 249 | 82 |
| N | +1 | 5563 | 6792 | 1230 | 409 |
| 3′ UTR | 6793 | 7276 | 483 |
The first nucleotide of start codon of spike gene is counted as nucleotide 1 of the structural genome.
Table 2.
Percent (%) identity of Md27 structural genes with other IBV strains.
| IBV Strains | S1 | S2 | 3a | 3b | 3c(E) | M | 5a | 5b | N | 3′UTR | 7kb* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ArkDPI | 86(84) | 98(98) | 91 | 98 | 98(96) | 99(98) | 99 | 98 | 96(96) | 98 | 95 |
| Ark99 | 85(84) | 96(95) | 97 | 98 | 95(91) | 97(96) | NA | NA | 96(95) | 96 | NA |
| Beaudette | 83(80) | 94(93) | 86 | 84 | 88(82) | 91(93) | 93 | 96 | 93(94) | 97 | 91 |
| BJ | 77(76) | 85(89) | 86 | 76 | 87(81) | 90(93) | 86 | 93 | 89(91) | 88 | 85 |
| Cal99 | 82(79) | 94(93) | 97 | 98 | 93(88) | 96(95) | 99 | 98 | 94(95) | 90 | 91 |
| CK/CH/LSD/05I | 78(75) | 87(90) | 85 | 84 | 92(87) | 96(96) | 99 | 99 | 96(96) | 91 | 89 |
| Connecticut | 83(80) | 95(96) | 97 | 98 | 98(97) | 99(98) | 99 | 98 | 96(95) | NA | NA |
| CU-T2 | 85(83) | 94(93) | 94 | 97 | 91(87) | 88(86) | 98 | 98 | 94(93) | 97 | NA |
| DE072 | 62(50) | 75(76) | NA | NA | NA | NA | 98 | 98 | NA | 98 | NA |
| Gray | 86(82) | 98(97) | NA | NA | 96(92) | 97(97) | 98 | 98 | 95(95) | 97 | NA |
| GAV-92 | 83(79) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| H120 | 83(80) | NA | NA | NA | NA | 91(94) | NA | NA | 92(94) | 82 | NA |
| H52 | 83(80) | NA | NA | NA | NA | NA | NA | NA | 95(95) | NA | NA |
| HK | 83(80) | 95(95) | 91 | 98 | 98(95) | 99(98) | 99 | 98 | 96(95) | 44 | NA |
| Holte | 83(78) | 94(94) | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Ind/TN/92/03 | NA | NA | NA | NA | NA | NA | NA | NA | 94(94) | NA | NA |
| IS/1366 | 77(74) | NA | NA | NA | NA | NA | NA | NA | 92(94) | NA | NA |
| Jilin | 86(84) | 98(98) | 91 | 98 | 98(96) | 99(98) | 99 | 98 | 96(96) | 98 | 95 |
| JMK | 86(84) | 98(98) | NA | NA | NA | 97(96) | NA | NA | NA | NA | NA |
| KB8523 | 83(80) | 90(91) | NA | NA | NA | 93(94) | NA | NA | 93(95) | NA | NA |
| LX4 | 76(75) | 85(88) | 81 | 76 | 89(81) | 90(91) | 83 | 91 | 88(91) | NA | 26 |
| M41 | 83(79) | 94(94) | 84 | 85 | 88(82) | 91(94) | 90 | 96 | 95(94) | 97 | 91 |
| Qu16 | 84(81) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SAIBK | 81(79) | 87(90) | 82 | 82 | 85(81) | 89(92) | 85 | 96 | 87(92) | 90 | 86 |
| SAIBb2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 85 | NA |
| SE 17 | 96(93) | 98(98) | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| TW2296/95 | 80(78) | 86(89) | 81 | 85 | 88(83) | 90(91) | 86 | 95 | 88(89) | NA | NA |
| UK/2/91 | 77(75) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Vic | 82(79) | 89(91) | 84 | 88 | 88(87) | 89(94) | 88 | 94 | 90(93) | NA | NA |
Sequences with >95% identity are indicated in bold letters; Amino acid sequences identity are within the parenthesis; NA-Not available for Analysis.
Entire structural genome, starting from start codon of spike gene to last nucleotide of IBV genome.
Gene 3 codes for two non-structural proteins (3a and 3b) and one structural protein (E), which is a small membrane protein of coronavirus. Gene 3a and 3b start at the last nucleotide of the S gene and 3a genes, respectively. Gene 3b overlaps 3c by 20 nucleotides and 3a has a signal sequence at the very 5′ end (nucleotides 1–22). The small membrane protein E, encoded by a 107 nucleotides long ORF, is approximately 12 kDa in size. A TRS (CTGAACAAT) is located 20 nucleotides upstream of the ATG codon. Gene 3a of Md27 virus shows 97% identity with Conn, Cal99 and Ark99 strains. The 3b gene has 98% identity with Ark DPI, Ark99, Cal99, Conn, HK, and Jilin strains, whereas gene E shows 98% identity with Ark DPI, Conn, HK and Jilin. The comparative sequence alignment indicates that the E protein gene is more conserved than the genes coding for 3a and 3b.
Gene 4 codes for the matrix (M) protein of 223aa, which is the most abundant envelope glycoprotein of coronaviruses, and it overlaps the E gene by 23 nucleotides. Its molecular mass depends on the glycosylation and varies from 25 to 33 kDa [16]. The M protein of Md27 contains a single putative N-linked glycosylation site at amino acid position 4. The M protein of coronaviruses differs in glycosylation types. In case of IBV it is N-linked, whereas in MHV, it is O-linked [13,17]. Gene M of Md27 shows 99% identity with Ark DPI, Conn, HK and Jilin. RNA 5 of Md27 is dicistronic and it encodes for two nonstructural proteins, 5a and 5b, respectively.
There is a 350 nucleotides long non-coding region between gene M and 5 with a deletion of 9 nucleotides, which is similar to Mass-type IBV strains. Gene 5 is more conserved than any of the other genes of Md27, and it is nearly identical (98–99%) to other strains.
RNA 6, the smallest but the most abundant RNA in IBV-infected cells, encodes for the N protein. The N protein is the only phosphorylated structural protein in coronaviruses [18]. The N protein has many basic residues, and serine residues accounting for 8 to 10% of the total amino acids in the N protein. The abundance of serine residues accounts for the specific phosphorylation of serine residues [19,20]. The N protein of Md27 contains 33 serine residues. It has 96% identity with Ark DPI, Ark99, CK/CH/LSD/05I, Conn, HK and Jilin strains.
The Md27 has a 483 nucleotides long 3′UTR. Interestingly, there is a deletion of 22 nucleotides in the 5′ terminus of 3′UTR (Figure 1), which shows some similarity to the Chinese strains A2, BJ and SAIBb2. In the case of IBV, C- and N-terminal regions of the N protein (but not the central region) interact with the 3′end of the non-coding region of IBV genomic RNA [21]. This deletion suggests possible involvement of 3′UTR in replication, which may influence pathogenesis of the virus. The 3′UTR of Md27 is closely related (97–98% identity) to Ark DPI, DE072, Beaudette, Gray, M41, Jilin and CU-T2. The 3′UTR (∼161nucleotides) immediately downstream of the N gene is U-rich and it is a highly variable region of the coronavirus genome. It has been observed that the 3′ UTR is involved in genome replication of coronaviruses, despite its apparent ability to possess quite variable sequence and sequence lengths [22].
Figure 1.
Comparison of 5′ terminus of 3′ untranslated region (UTR) of Md27 with some selected strains of IBV*.
Nucleotide deletion of Md27 at 3′UTR is clearly shown. Sequences were compared using Gendoc software.
*Nucleotides of Md27 shown are starting from 86th nucleotide after the stop codon of N gene.
The phylogenetic relationship of specific genes of the Md27 with other IBV strains is illustrated in Figures 2 and 3. Comparisons are made for the structural genes (S1, S2, E, M and N) because nucleotide differences between strains mainly occur in these regions [23–26] The S1 and S2 gene of Md27 clusters with the SE-17 strain and in the E and M gene phylogeny it clusters with Ark DPI, Jilin, Conn, and HK. In the N gene phylogeny Md27 clusters with HK, Conn, Ind/TN/92/03 and H52 (Figure 3). Comparison of the structural genes of Md27 with other coronaviruses demonstrates high sequence identity with turkey coronavirus (TCoV) and IBVs from peafowl and partridge (Table 3). Among the four structural genes, M is highly conserved between coronaviruses, whereas the E protein is less conserved.
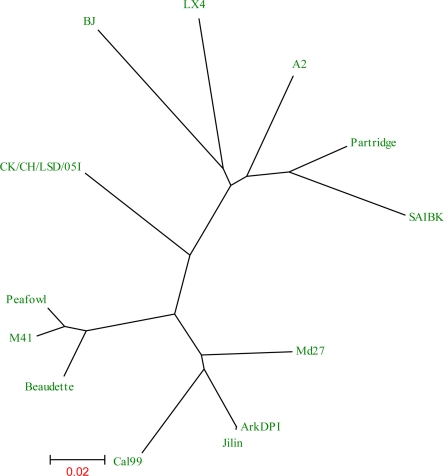
Figure 2.
Comparison of 7.27 kb of the 3′end of the Md27 genome sequence with other IBV strains by an unrooted phylogenetic tree. Sequences are aligned by neighbor-joining method using MEGA4 program. Md27 clustered with ArkDPI, Jilin and Cal99 IBV strains. The scale at the bottom indicates the number of substitution events.
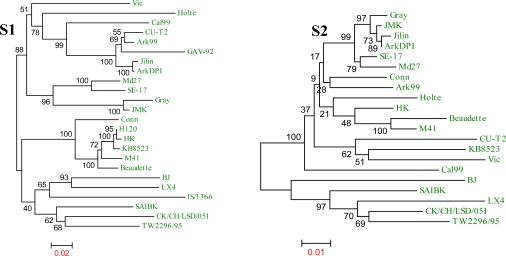
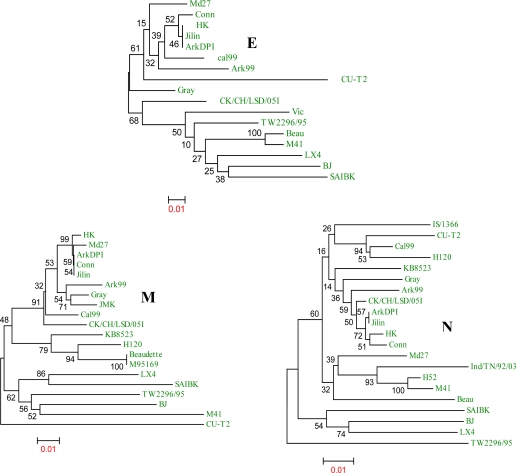
Figure 3.
Phylogenetic analyses of the structural genes of Md27 virus with those of other IBV strains. The Md27 strain had clustered with ArkDPI, HK, Conn, and Cal99 in most of the trees. The tree is constructed from aligned nucleotide sequences of structural genes (S1, S2, spike gene; E, envelope gene; M, matrix gene; N, nucleocapsid gene) by neighbor-joining method with 1000 bootstrap replicates using the MEGA4 program. For each tree, the bootstrap value for each branch is indicated and the scale at the bottom indicates the number of substitution events.
Table 3.
Percent (%) amino acid identity of Md27 structural proteins to other coronavirusesa.
| Coronavirusesb | S | E | M | N |
|---|---|---|---|---|
| BatCoV | 21 | 11 | 29 | 23 |
| BCoV | 21 | 13 | 30 | 24 |
| ECoV | 21 | 13 | 30 | 24 |
| FCoV | 26 | 16 | 23 | 22 |
| HCoV 229E | 26 | 10 | 25 | 22 |
| TGEV | 26 | 20 | 25 | 25 |
| MHV A59 | 22 | 14 | 31 | 25 |
| Peafowl, Avian | 86 | 82 | 94 | 95 |
| Partridge | 84 | 81 | 92 | 92 |
| SARS CoV | 20 | 16 | 29 | 23 |
| SW1 | 25 | 28 | 35 | 34 |
| TCoV | 33 | 91 | 97 | 94 |
Sequences with >50% identity are in bold letters;
BatCoV, Bat coronavirus; FCoV, feline coronavirus; HCoV, human coronavirus; BCoV, bovine coronavirus; MHV, mouse hepatitis virus; SARS-CoV, severe acute respiratory syndrome coronavirus; Partridge, Avian infectious bronchitis virus partridge/GD/S14/2003, Peafowl, Avian infectious bronchitis virus isolate Peafowl/GD/KQ6/2003, SW1, beluga whale coronavirus; TCoV, turkey coronavirus.
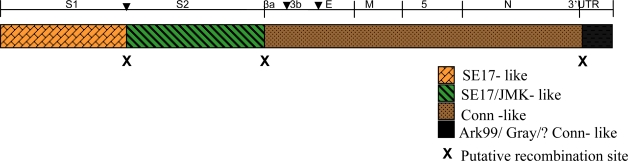
The complete structural genome analysis of Md27 suggests that it is a chimera of virus genomes represented by the SE17, Conn, JMK and Ark 99 IBV strains (see Figure 4), which were circulating during that time period [12,27]. The S1 and S2 genes of Md27 exhibit 96% and 98% identity, respectively, with SE17. Most of the nucleotide differences between Md27 and SE17 are in the S1 gene, especially in the hypervariable regions (HVR). The remainder of the structural sequence is not available for SE17, so it is difficult to conclude the genetic relationship of other genes of Md27 with SE17. Based on the limited data available (as many other strains are not yet sequenced), it is conceivable that the spike gene is derived from SE17 by recombination.
Figure 4.
Schematic representation of the structural genome of Md27. Entire structural genome of Md27 was analyzed for its similarity with other IBV strains. Structural genes and their ORFs are marked by ▾. Putative recombination sites in Md27 genome are denoted by x. Picture is not drawn to scale.
The Md27 shares close homology (96–99%) with the Conn strain, starting from gene 3a to the end of the N gene, which suggests that most of the Md27 structural genome was derived from a Conn strain. Table 2 shows the comparison of the entire 7 kb genome of Md27, which reveals that Ark DPI and Jilin share high levels of sequence identity with Md27. Earlier we reported that Jilin is actually an Ark DPI strain [28]. Ark DPI was isolated during the 1980s [27], and it was demonstrated that Ark DPI is a direct derivative of the Conn strain [28]. This study suggests that Md27 and Ark DPI are derived from the same ancestor, but diverged independently by recombination with different strains. Several factors determine IBV evolution, mainly high mutation rate due to the absence of proof-reading mechanisms, and recombination between strains because of widespread use of live vaccines, immunological pressure and frequent mixed infections [29–31]. Our data has provided the evidence that both genomic mutations and intergenic recombination have taken place in the evolution of Md27 IBV strain and its descendents.
3. Materials and Methods
3.1. Virus
Virus isolation was done in Dr. Marquardt’s laboratory by passaging tissue (trachea or kidney) homogenates into 9–10-day-old chicken embryos by the allantoic cavity route of inoculation and at least three passages were done before declaring samples as negative. Two of the isolates were named as Md27, and these were propagated by inoculating into 9–day-old embryonated specific-pathogen-free (SPF) chicken eggs and allantoic fluid was collected 72 h post-inoculation. The fluid was clarified by low speed centrifugation and clear supernatant was stored at −80 °C until further use.
3.2. RNA extraction and amplification
Genomic RNA was extracted from virus-infected allantoic fluid with Qiagen (Valencia, CA, USA) RNAeasy kit, following the manufacturer’s instructions and stored at −80 °C until further use. Oligonucleotides were designed based on the Ark DPI11 sequence (GenBank accession No. EU418976). Overlapping primers were designed in a manner such that each pair of primer covered approximately 2 kb of the genome. All gene fragments were amplified using RT-PCR kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions and the RT-PCR products were cloned into the pCR2.1 TOPO TA® vector (Invitrogen, CA). To determine 3′-terminus of Md27 genomic RNA, reverse transcription was carried out with a reverse poly (T) primer (5′-GCGGCCGCTTTTTTTTTTTTTTTT TT-3′) and an internal gene specific forward primer.
3.3. DNA sequence analysis
DNA from various clones was sequenced by dideoxy chain termination method using an automated DNA sequencer (Applied Biosystems Inc., Foster City, CA). At least three independent clones were sequenced for each amplicon to exclude errors that can occur from RT and PCR reactions. The assembly of contiguous sequences was performed with the GeneDoc software [32]. Comparative sequence analyses of MD27 with other IBVs and coronaviruses were conducted using the BLAST search, NCBI, and Vector NTI Advance 10. Phylogenetic analyses were carried out using the MEGA4 program [33]. The phylogenetic tree was constructed from aligned nucleotide and amino acid sequences using the neighbor-joining method with 1000 bootstraps.
3.4. GenBank accession numbers
The GenBank accession number for the MD27 sequence is FJ008695. The accession numbers for other IBV gene sequences used in this study are as follows: (a) complete structural genomes: TCoVMG10, NC_010800; Beaudette, NC_001451; CK/CH/LSD/05I, EU637854; A2, EU526388; Peafowl/GD/KQ6/2003, AY641576; Cal99, AY514485; M41, DQ834384; partridge/GD/S14/2003, AY646283; BJ, AY319651; LX4, AY338732; SAIBK, DQ288927; HK, AY761141; Vic, DQ490221; KB8523, M21515; TW2296/95, DQ646404 (b) S1: Jilin, AY839144; Gray, L18989; Holte, L18988; UK/2/91, Z83976; Qu16, AF349620; JMK, L14070; H52, AF352315; H120, M21970; GAV-92, AF094817; DE072, AF274435; IS/1366, EU350550; (c) S2: JMK, AF239982; Jilin, AY839146; Holte, AF334685; DE072, AY024337; Conn, AF094818; Gray, AF394180; H120, AF239982; (d) S: Ark99, L10384; CU-T2, U04739; (e) Gene 3: Jilin, AY846833; Conn, AY942752; CU-T2, U46036; Ark99, AY942751; Gray, AF318282 (f) M: Jilin, AY846833; JMK, AF363608; H120, AY028295; Gray, AF363607; (g) Gene 5: Jilin, AY839142; Gray, AF469011; DE072, AF203000; (h) N: Jilin, AY839145; Ind/TN/92/03, EF185916; H52, AF352310; H120, AY028296; Gray, M85245; (i) 3′UTR: H120, AJ278336.
4. Conclusions
In this study we analyzed ∼7.2 kb of the 3′ genomic end of the IBV strain Md27 and compared it with other IBV strains and coronaviruses from different parts of the world. This analysis suggests that Md27 evolved by recombination of different strains which were circulating during the same time period. This study demonstrates that recombination is the major evolutionary mechanism for infectious bronchitis virus to create new strains and variants.
Acknowledgments
The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service; grant number 2004-35204-14814.
References
- 1.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 2.Cavanagh D, Naqi SA. Infectious bronchitis. In: Saif YM, Barnes HJ, Glisson JR, Fadley AM, McDougald LR, Swane DE, editors. Diseases of poultry. 11th ed. Iowa State University Press; Ames, IA, USA: 2003. pp. 101–119. [Google Scholar]
- 3.Boursnell MEG, Brown TDK, Foulds IJ, Green PF, Tomely FM, Binns MM. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 4.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutou S, Sato S, Okabe T, Nakai M, Sasaki N. Cloning and sequencing of genes encoding structural proteins of avian infectious bronchitis virus. Virology. 1988;165:589–595. doi: 10.1016/0042-6822(88)90603-4. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D. Coronaviruses in poultry and other birds. Avian. Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 7.Gelb J, Jr, Wolff JB, Morna CA. Variant serotypes of infectious bronchitis virus isolated from commercial layer and broiler chickens. Avian Dis. 1991;35:81–87. [PubMed] [Google Scholar]
- 8.Gelb J, Jr, Jackwood MW. Infectious bronchitis. In: Swayne DE, editor. A laboratory manual for the isolation and identification of avian pathogens. 4th ed. American Association of Avian Pathologists; Kennett Square, PA, USA: 1998. pp. 169–173. [Google Scholar]
- 9.Cavanagh D, Davis PJ, Cook JKA. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21:401–408. doi: 10.1080/03079459208418858. [DOI] [PubMed] [Google Scholar]
- 10.Gelb J, Jr, Weisman Y, Ladman BS, Meir R. S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) Avian Pathol. 2005;34:194–203. doi: 10.1080/03079450500096539. [DOI] [PubMed] [Google Scholar]
- 11.Kottier SA, Cavanagh D, Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213:569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RB, Marquardt WW. Strains of infectious bronchitis virus on the Delmarva Peninsula and in Arkansas. Avian Dis. 1976;20:382–386. [PubMed] [Google Scholar]
- 13.Cavanagh D, Davis PJ, Cook JKA, Li D, Kant A, Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- 14.Parr RL, Collisson EW. Epitopes on the spike protein of a nephropathogenic strain of infectious bronchitis virus. Arch Virol. 1993;133:369–383. doi: 10.1007/BF01313776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callison SA, Jackwood MW, Hilt DA. Infectious bronchitis virus S2 gene sequence variability may affect S1 subunit specific antibody binding. Virus Genes. 1999;19:143–151. doi: 10.1023/A:1008179208217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu DX, Cavanagh D, Green P, Inglis SC. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locker JKG, Griffiths MC, Horzinek Rottier PJ. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N- and O-linked glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilbur SM, Nelson GW, Lai MM, McMillan M, Stohlman SA. Phosphorylation of the mouse hepatitis virus nucleocapsid protein. Biochem. Biophys. Res. Commun. 1986;141:7–12. doi: 10.1016/S0006-291X(86)80326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakhartchouk AN, Viswanathan S, Mahony JB, Gauldie J, Babiuk LA. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J Gen Virol. 2005;86:211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]
- 20.Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 21.Collisson EW, Williams AK, Chung SI, Zhou M. Interactions between the IBV nucleocapsid protein and RNA sequences specific for the 3′ end of the genome. Adv Exp Med Biol. 1995;380:523–528. doi: 10.1007/978-1-4615-1899-0_83. [DOI] [PubMed] [Google Scholar]
- 22.Williams AK, Wang L, Sneed LW, Collisson EW. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28:19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavanagh D, Mawditt K, Adzhar A, Gough RE, Picault JP, Naylor CJ, Haydon D, Shaw K, Britton P. Does IBV change slowly despite the capacity of the spike protein to vary greatly. Adv Exp Med Biol. 1998;440:729–734. doi: 10.1007/978-1-4615-5331-1_94. [DOI] [PubMed] [Google Scholar]
- 24.Koch G, Hartog L, Kant A, van Roozelaar DJ. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol. 1990;71:1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- 25.Jia W, Karaca K, Parrish CR, Naqi SA. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams AK, Wang L, Sneed LW, Collisson EW. Comparative analyses of the nucleocapsid genes of several strains of infectious bronchitis virus and other coronaviruses. Virus Res. 1992;25:213–222. doi: 10.1016/0168-1702(92)90135-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelb J, Jr, Leary JH, Rosenberger JK. Prevalence of Arkansas-type infectious bronchitis virus in Delmarva Peninsula chickens. Avian Dis. 1983;27:667–678. [PubMed] [Google Scholar]
- 28.Ammayappan A, Upadhyay C, Gelb J, Jr, Vakharia VN. Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination. Virol J. 2008;5:157. doi: 10.1186/1743-422X-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavanagh D, Picault JP, Gough R, Hess M, Mawditt K, Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- 30.Lai MMC, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Yuan X, Collisson EW. Experimental confirmation of recombination upstream of the S1 hypervariable region of infectious bronchitis virus. Virus Res. 1997;49:139–145. doi: 10.1016/S0168-1702(97)01466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas KB, Nicholas HBJ, Deerfield DW. GeneDoc: Analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- 33.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]