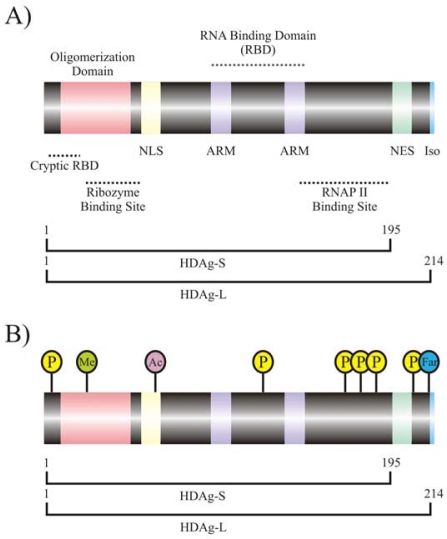
Figure 2.
Schematic representation of HDAg. A) Representation of HDAg showing significant functional regions. HDAg encompasses amino acids 1 to 195; the C-terminal expansion of HDAg-L (amino acids 196 to 214) is also indicated. NLS: nuclear localization signal (amino acids 66–75); NES: nuclear export signal (amino acids 198–210); ARM: arginine-rich motifs (amino acids 97–107 and 136–146); Iso: isoprenylation signal for the addition of farnesyl (amino acids 211–214). The cryptic RNA-binding domain (RBD) spans amino acids 2–27 while the main RBD spans both ARMs. The coiled-coil oligomerization domain spans amino acids 12–60. Ribozyme binding occurs at amino acids 24–50 and the RNAP II binding is localized to the C terminus at amino acids 150–195. B) Modification of HDAg. All the known naturally-occuring modifications are shown concurrently, though not every modification is present simultaneously in vivo. Lettering for the modifications is P: phosphorylation, Me: methylation, Ac: acetylation, Far: farnesylation. Sumoylation of multiple lysine residues of HDAg-S has also been reported.