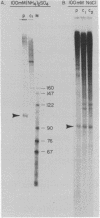
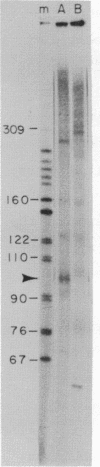
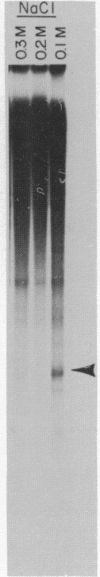
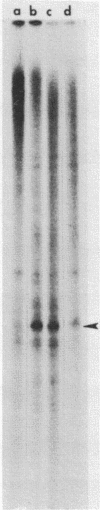
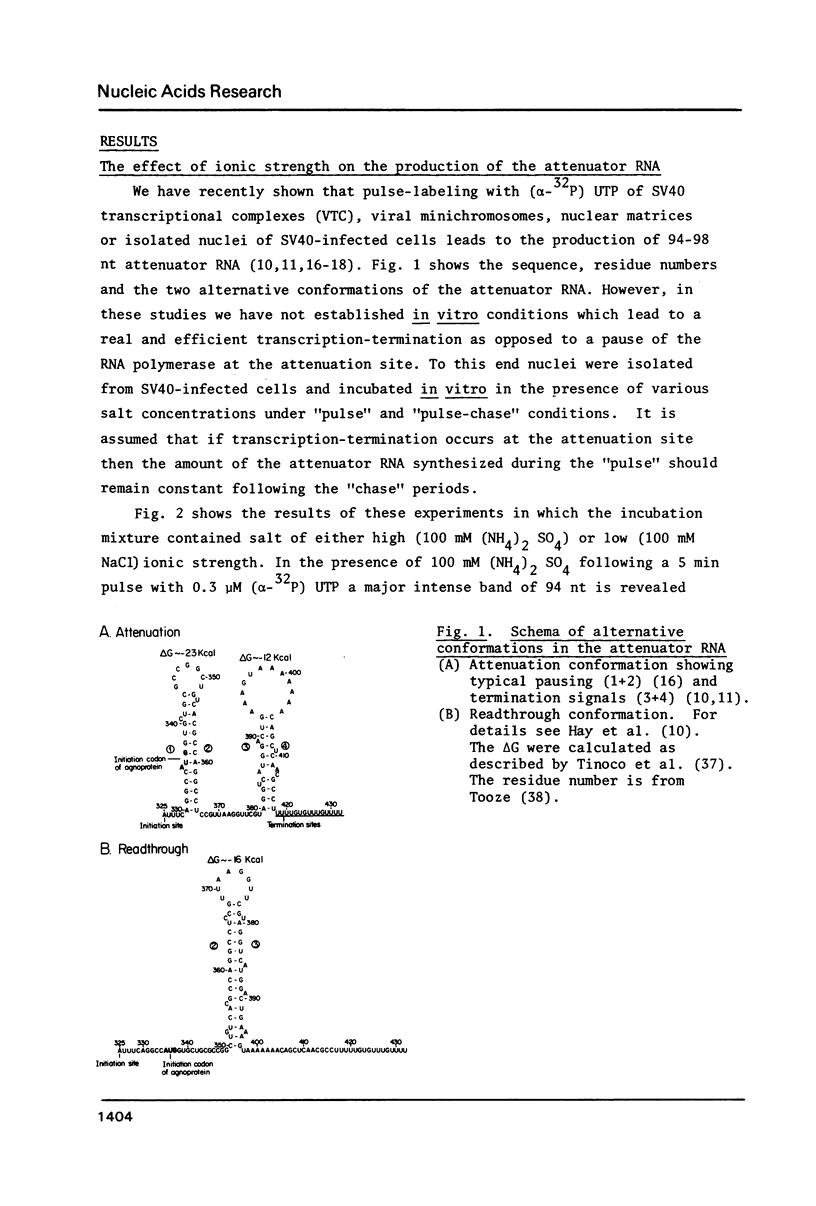
Abstract
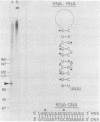
Nuclei which were isolated from SV40 infected cells with a hypotonic detergent-free buffer were used to establish in vitro conditions which lead to transcription-termination at the attenuation site of SV40. This system allowed us to identify regulatory elements involved in transcription-termination by RNA polymerase B transcribing SV40. Transcription-termination at the attenuation site was found to be ionic strength dependent. Efficient termination occurred at low (100 mM NaCl) but not at high (100 mM (NH4)2 SO4 or 300 mM NaCl) ionic strength. When nuclei were prewashed with 300 mM NaCl, the efficiency of transcription-termination was low even when transcription was carried out at low ionic strength (100 mM NaCl). Efficient transcription-termination in the high salt prewashed nuclei was reconstituted by complementation with a high salt (300 mM NaCl) soluble factor extracted from nuclei of uninfected cells. In addition, the efficiency of transcription-termination was significantly reduced when ITP replaced GTP in the transcription reaction mixture. Our data indicate that a nuclear factor and RNA secondary structure are essential regulatory elements involved in transcription-termination by RNA polymerase B.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Adhya S., Sarkar P., Valenzuela D., Maitra U. Termination of transcription by Escherichia coli RNA polymerase: influence of secondary structure of RNA transcripts on rho-independent and rho-dependent termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1613–1617. doi: 10.1073/pnas.76.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y., Hay N. Attenuation and modulation of mRNA secondary structure in a feedback control system regulating SV40 gene expression. Mol Biol Rep. 1983 May;9(1-2):91–100. doi: 10.1007/BF00777479. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Khoury G. Simian virus 40-associated small RNA: mapping on the simian virus 40 genome and characterization of its synthesis. J Virol. 1980 Dec;36(3):701–708. doi: 10.1128/jvi.36.3.701-708.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig M. M., Hentschel C. C. Transcription of sea urchin histone genes in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2337–2346. doi: 10.1093/nar/11.8.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Montoya J., Timko K. D., Attardi G. Sequence analysis and precise mapping of the 3' ends of HeLa cell mitochondrial ribosomal RNAs. J Mol Biol. 1982 May 5;157(1):1–19. doi: 10.1016/0022-2836(82)90510-1. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Greenblatt J., Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982 Jul;29(3):945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Hatfield G. W., Sharp J. A., Rosenberg M. Pausing and termination of human RNA polymerase II transcription at a procaryotic terminator. Mol Cell Biol. 1983 Oct;3(10):1687–1693. doi: 10.1128/mcb.3.10.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Jay G., Nomura S., Anderson C. W., Khoury G. Identification of the SV40 agnogene product: a DNA binding protein. Nature. 1981 May 28;291(5813):346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- Laub O., Aloni Y. Transcription of simian virus 40. VI. SV 40 DNA-RNA polymerase complex isolated from productively infected cells transcribed in vitro. Virology. 1976 Dec;75(2):346–354. doi: 10.1016/0042-6822(76)90033-7. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer J. C., Tiryaki D., Westergaard O. Termination of transcription in nucleoli isolated from Tetrahymena. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5563–5566. doi: 10.1073/pnas.76.11.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. T., Burgess R. R., Dahlberg J. E., Lund E. Transcription of a gene for human U1 small nuclear RNA. Cell. 1982 May;29(1):265–274. doi: 10.1016/0092-8674(82)90111-8. [DOI] [PubMed] [Google Scholar]
- Neff N. F., Chamberlin M. J. Termination of transcription by Escherichia coli RNA polymerase in vitro is affected by ribonucleoside triphosphate base analogs. J Biol Chem. 1978 Apr 10;253(7):2455–2460. [PubMed] [Google Scholar]
- Penman S. RNA metabolism in the HeLa cell nucleus. J Mol Biol. 1966 May;17(1):117–130. doi: 10.1016/s0022-2836(66)80098-0. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Hay N., Pruzan R., Jakobovits E. B., Aloni Y. In vitro premature termination in SV40 late transcription. EMBO J. 1983;2(2):185–191. doi: 10.1002/j.1460-2075.1983.tb01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Darnell J. E., Jr A precise termination site in the mouse beta major-globin transcription unit. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4694–4698. doi: 10.1073/pnas.80.15.4694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Skolnik-David H., Aloni Y. Pausing of RNA polymerase molecules during in vivo transcription of the SV40 leader region. EMBO J. 1983;2(2):179–184. doi: 10.1002/j.1460-2075.1983.tb01402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik-David H., Hay N., Aloni Y. Site of premature termination of late transcription of simian virus 40 DNA: enhancement by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole. Proc Natl Acad Sci U S A. 1982 May;79(9):2743–2747. doi: 10.1073/pnas.79.9.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg H. G., Birnstiel M. L. Bioassay for components regulating eukaryotic gene expression: a chromosomal factor involved in the generation of histone mRNA 3' termini. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6201–6204. doi: 10.1073/pnas.79.20.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wickens M. P., Gurdon J. B. Post-transcriptional processing of simian virus 40 late transcripts in injected frog oocytes. J Mol Biol. 1983 Jan 5;163(1):1–26. doi: 10.1016/0022-2836(83)90027-x. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]