Abstract
The transcriptional response to epidermal growth factor (EGF) was examined in a cultured cell model of adhesion. Gene expression was monitored in human embryonic kidney cells (HEK293) after attachment of cells to the extracellular matrix (ECM) proteins, laminin, and fibronectin, by using complementary DNA micorarrays printed with 1,718 individual human genes. Cluster analysis revealed that the influence of EGF on gene expression, either positive or negative, was largely independent of ECM composition. However, clusters of EGF-regulated genes were identified that were diagnostic of the type of ECM proteins to which cells were attached. In these clusters, attachment of cells to a laminin or fibronectin substrata specifically modified the direction of gene expression changes in response to EGF stimulation. For example, in HEK293 cells attached to fibronectin, EGF stimulated an increase in the expression of some genes; however, genes in the same group were nonresponsive or even suppressed in cells attached to laminin. Many of the genes regulated by EGF and ECM proteins in this manner are involved in ECM and cytoskeletal architecture, protein synthesis, and cell cycle control, indicating that cell responses to EGF stimulation can be dramatically affected by ECM composition.
Cell–extracellular matrix (ECM) interactions transduce intracellular signals that regulate cell cycle progression, movement, survival, gene expression, and physical support (1, 2). The control of these processes is mediated by cell surface integrin receptors binding to ECM proteins. Integrins are heterodimeric proteins formed from α and β subunits. There are 17 α and 8 β subunits, and each αβ combination confers a specific binding property to cells (1, 2). For example, of the αβ combinations that contain β1 integrin, four bind to fibronectin (α4β1, α5β1, α8β1, and αvβ1), and five (α1β1, α2β1, α3β1, α6β1, and α7β1) bind to laminins (2). Many signaling molecules are activated when integrins bind to the ECM, including adapter proteins, cytoplasmic kinases, small GTPases, and growth factor receptor tyrosine kinases (RTKs) (3).
In addition to activating their own complement of signaling molecules, integrins have another major role: functional modulation of RTKs. One of the most widely studied examples is the epidermal growth factor (EGF) receptor (EGFR), which continues to represent a paradigm for RTK signal transduction in general. A high degree of functional interdependence exists between ECM- and EGFR-activated signaling pathways. For example, EGFR autophosphorylation is enhanced in a number of cell types, including fibroblasts, smooth muscle, and kidney epithelial cells, when they interact with ECM proteins (4–6). This kind of overlapping signal transduction is thought to support or enhance a number ECM- and RTK-controlled cell functions, including proliferation and survival (7, 8). In addition to modulating EGFR signaling, ECM interaction has been found to be an obligatory requirement for many EGF-mediated biological responses. For example, EGF regulates integrin-mediated cell migration, an actin-based process that depends entirely on copresentation with ECM components (9, 10).
Many of the changes in cell physiology stimulated by ECM interaction and EGFR activation will undoubtedly arise from changes in gene expression (11). EGF is known to regulate the level of expression of a variety of genes, including those involved in ECM maintenance, e.g., syndecan-1, collagens, and matrix metalloproteinases (11–13). EGFR-controlled changes in gene expression are thought to reflect either a necessary step in the progression through the cell cycle or a response related to complex cellular processes, such as wound repair and embryogenesis. A major challenge is to completely classify genes that are regulated by EGF and determine the contribution of ECM in controlling their expression. This classification will give a clearer insight into the functional significance of ECM–EGFR crosscommunication. One means to achieve this classification is to survey the changes in expression of large numbers of genes simultaneously. In the present study, we have used cDNA microarrays to monitor the effects of cell adhesion and EGF stimulation on the expression of 1,718 human genes. Our findings demonstrate that many gene responses were common to both ECM and EGF stimulation, suggesting the involvement of shared intracellular signaling pathways. Intriguingly, we also found that the responses of certain genes were specifically programmed by the composition of the ECM. These data suggest that genetic responses of cells to growth factors depend on and are modulated by the ECM.
Materials and Methods
Materials.
Newborn calf serum and FCS were from GIBCO/BRL. Anti-rabbit IgG horseradish peroxidase was from Sigma. Anti-mouse IgG horseradish peroxidase antibody, CY3 and CY5 fluorescent dyes, and enhanced chemiluminescence reagents were from Amersham Pharmacia. Complete protease inhibitor tablets were from Boehringer Mannheim. Antiphospho-protein kinase B (PKB), antiphospho-ERK, anti-PKB, and anti-ERK protein antibodies were from New England Biolabs. Antibodies to EGFR and phosphotyrosine (PY99) were from Santa Cruz Biotechnology.
Cell Culture.
Human embryonic kidney (HEK)293 cells were grown at 37°C in an atmosphere of 5% CO2, 95% air in complete growth medium containing DMEM supplemented with 0.1% penicillin/streptomycin (10,000 units/ml) and 10% FCS. Confluent cells were serum-starved for 16 h and then detached by gentle washing with versene, 0.2 g/liter (wt/vol) EDTA in PBS. Cells were then resuspended in DMEM containing 0.1% BSA and kept in suspension with gentle shaking for 3 h. Cells were then either kept in suspension or plated onto prewarmed cell culture dishes coated with either polyL-lysine (10 μg/ml), fibronectin (10 μg/ml), or laminin (10 μg/ml). In all cases, cells were incubated at 37°C for an additional 3 h before stimulation with either 10 ng/ml epidermal growth factor (EGF) (GIBCO/BRL) or vehicle (0.1% wt/vol BSA in dH2O).
Cell Lysis, Immunoprecipitation, and Immunoblotting.
Detergent extracts of whole cells were prepared by solubilization in 55 mM Tris/HCl, pH 7.4/132 mM NaCl/22 mM NaF/1 mM Na3VO4/11 mM sodium pyrophosphate/0.1% Triton X-100 containing complete protease inhibitor mixture (Boehringer Mannheim). Cell lysates were centrifuged at 13,000 × g for 5 min and supernatants assayed for protein content by using the Bradford dye-binding method (Bio-Rad). Aliquots of 10 μg of cell extract were separated by SDS/PAGE followed by transfer to nitrocellulose, reaction with phospho-specific PKB and ERK or phosphotyrosine antibodies, and detection with horseradish peroxidase-conjugated secondary antibodies and the enhanced chemiluminescence detection system from Amersham Pharmacia. Where appropriate, membranes were stripped according to the manufacturer's recommendations and reprobed with primary and secondary antisera. For immunoprecipitation experiments, 500 μg of cell lysates was precleared with 60 μl of preequilibrated protein A beads followed by immunoprecipitation for 16 h at 4°C with anti-EGFR antibodies and 60 μl of protein A beads. Immuunoprecipitates were collected by centrifugation, washed three times with lysis buffer, and subjected to SDS/PAGE followed by immunoblotting with phosphotyrosine- or EGFR-specific antibodies.
RNA Expression Analysis.
Individual gene expression changes were detected by using spotted DNA microarrays (14, 15). We used DNA microarrays printed with 1,718 distinct human transcripts obtained from the Ontario Cancer Institute Microarray Centre (http://www.uhnres.utoronto.ca/services/microarray/products.html). Complete protocols for the generation of fluorescence-labeled sample from whole cell RNA, hybridization to DNA microarrays, and data after processing can by found at the array web site. Briefly, total RNA was extracted from HEK293 cells according to the manufacturer's instructions by using the Qiagen (Chatsworth, CA) Rneasy kit and was used to create CY3- or CY5- (Amersham Pharmacia) labeled cDNA by using Superscript II (GIBCO/BRL) reverse transcriptase primed with a polyT mRNA primer. Samples were prepared for hybridization by combining fragmented transcripts with sonicated salmon sperm DNA (0.5 μg/μl; Sigma) and yeast tRNA (0.5 μg/μl; GIBCO/BRL) in Easy Hyb solution (Roche Diagnostics). Samples were hybridized for 16 h at 37°C to cDNA micorarrays containing 1,718 known human expressed sequence tags. Arrays were then washed three times at room temperature with 0.1% (vol/vol) standard saline citrate (SSC) containing 0.1% (vol/vol) SDS followed by one wash with 0.1% SSC alone. Fluorescence intensities were captured by using a ScanArray 4000 (GSI Lumonics) laser confocal scanner. Expression data were analyzed as described (16) by clustering genes into groups on the basis of similarities in their expression profiles.
Results and Discussion
HEK293 cells (17) are a permanent cell line of primary human kidney cells that express at least five β1 integrin containing subunits on their cell surface β1αv, β1α2, β1α3, β1α5, and β1α6 (18). This diversity in integrin expression allows HEK293 cells to adhere to a wide range of ECM proteins, including vitronectin, fibronectin, laminin, and collagen (18), and therefore these cells represent a highly tractable model for ECM-interaction studies. Intracellular signaling from RTKs is known to be modified by cell adhesion to ECM in a wide range of primary and immortalized cell types (4–6). We therefore examined the genetic response cell adhesion confers on EGFR signaling in HEK293 cells. Cells that were held in suspension for 3 hours became refractory to EGF stimulation. Cells regained responsiveness to EGF if they were allowed to readhere to tissue culture plastic coated with polyL-lysine. Stimulation of polyL-lysine-attached cells with EGF promoted tyrosine phosphorylation of the 170-kDa EGFR (Fig. 1). In agreement with Moro et al. (8), attachment of cells to polyL-lysine alone did not stimulate phosphorylation of the EGFR (Fig. 1). Interaction of adhesive cells with Arg-Gly-Asp (RGD)-containing ECM proteins has also been shown to modulate RTK activity (4, 5). Attachment of HEK293 cells to the RGD-protein fibronectin or laminin markedly enhanced the ability of EGF to stimulate EGFR tyrosine phosphorylation, when compared with cells attached to polyL-lysine (Fig. 1). This enhancement is consistent with an enhancement of EGFR tyrosine phosphorylation by fibronectin, vitronectin, collagens types I and IV, and specific integrin engagement seen in other cell systems (4, 6, 8, 19). These results demonstrate that cell adhesion is an obligatory requirement for EGF to elicit intracellular responses in HEK293 cells; however, the quality of the response can be modified by interaction of adherent cells with ECM components.
Figure 1.
ECM and EGF stimulate tyrosine phosphorylation of the EGFR in HEK293 cells. HEK293 cell lysates were immunoprecipitated with EGFR polyclonal antibody, separated on 10% polyacrylamide gels and immunoblotted with mAb PY99 (P-tyr) or EGFR polyclonal Ab. Immunoblots were quantified densitometrically and expressed graphically (Lower). Data were normalized to the amount of EGFR immunoreactivity in precipitates from polyL-lysine-attached cells and expressed as means ± SEM (n = 3).
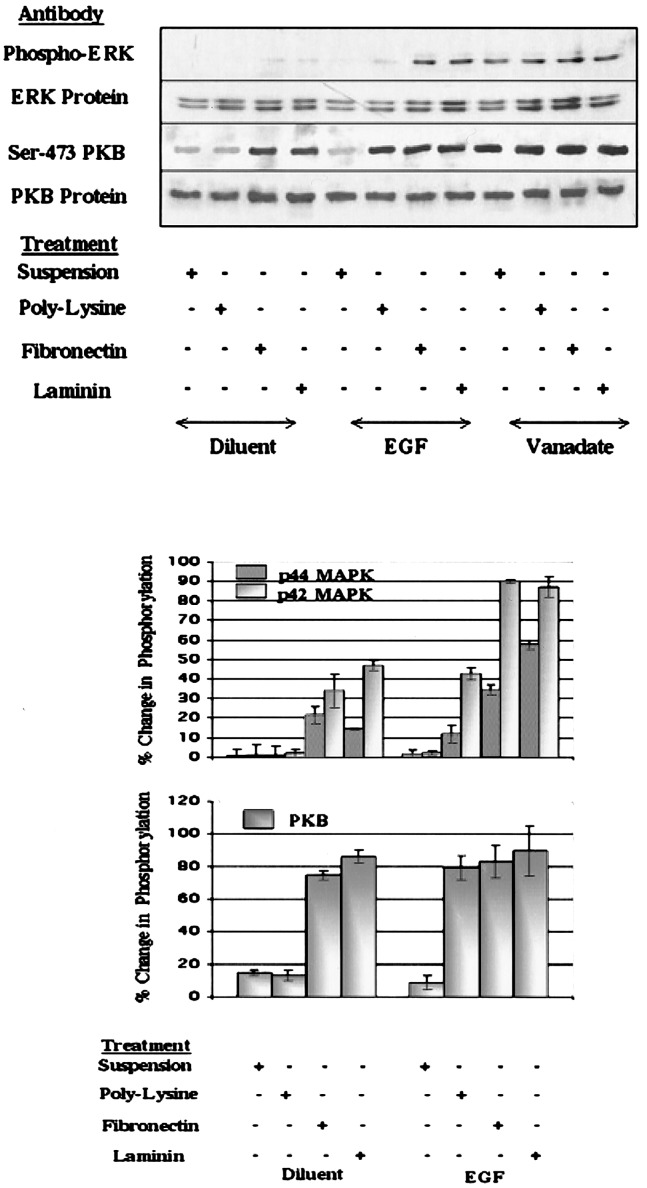
We next investigated whether cell adhesion is also sufficient for the coupling of tyrosine-phosphorylated EGFR to downstream signaling events. We used antibodies that specifically recognize the phosphorylated forms of protein kinase (PK)B/Akt and the erk-gene products p42 and p44 mitogen-activated PKs (MAPKs) to examine the effects of cell adhesion to polyL-lysine. Activation-associated phosphorylation of MAPKs on Thr-183 and Tyr-185 and PKB at Ser-473 are widely recognized as downstream markers of EGFR activation (20–22). We noted that in suspended HEK293 cells, EGF did not stimulate a significant increase in the phosphorylation of MAPK or PKB proteins (Fig. 2). In contrast, EGF stimulation of cells that were attached to polyL-lysine did provoke an increase (≈10 and 40%, respectively) in the phosphorylation of the 42- and 44-kDa forms of ERK (Fig. 2). Similarly, cell adhesion to polyL-lysine facilitated EGF-stimulated phosphorylation of PKB (Fig. 2). In this case, attachment of cells to polyL-lysine was sufficient for EGF to stimulate a near-maximal increase in PKB phosphorylation, provoking levels of PKB phosphorylation that were similar to those stimulated by the tyrosine phosphatase inhibitor sodium orthovanadate (Fig. 2). Interaction of cells with fibronectin or laminin enhanced the ability of EGF to stimulate phosphorylation of MAPK proteins by a factor of ≈2.5 (Fig. 2). In contrast, ECM attachment was not found to significantly increase the ability of EGF to stimulate PKB phosphorylation (Fig. 2). Indeed, simple attachment of cells to fibronectin or laminin was alone sufficient to induce near-maximal PKB phosphorylation.
Figure 2.
ECM and EGF stimulate phosphorylation of MAPK and PKB proteins in HEK293 cells. Serum-starved HEK293 cells were either held in suspension or replated for 3 h on tissue culture plastic coated with polyL-lysine (10 μg/ml), fibronectin (10 μg/ml), or laminin (10 μg/ml). Cells were then stimulated with EGF (10 ng/ml) or pervanadate (1 μM) for 1 h, as indicated. Aliquots of cell lysate (30 μg) were then separated on 10% polyacrylamide gels and immunoblotted with the indicated antibodies. Densitometric values from immunoblots are shown (Lower) and are expressed as a percentage of the values from pervanadate-treated cells (mean ± SEM for three observations).
These results demonstrate that the substrate that cells are attached to can alter the intracellular signaling responses to EGFR stimulation in HEK293 cells. Implicit in these observations is the suggestion that the EGFR can transduce at least two types of intracellular response in adherent cells. The first type is characterized by phosphorylation of PKB (Fig. 2); interaction of cells with ECM provokes similar responses to those observed in EGF-stimulated cells. The second type of response is substrata-specific, for example, adhesion-dependent/ECM-independent responses (maximal PKB phosphorylation and low-level phosphorylation of EGFR and MAPK) and adhesion-dependent/ECM-dependent (maximal MAPK phosphorylation and EGFR autophosphorylation). Moreover, because of its heterogeneous nature, ECM also has the potential to tailor unique responses to RTK activation that are composition-dependent. For example, attachment of cells to laminin was found to stimulate consistently higher levels of EGFR phosphorylation than attachment to fibronectin.
EGFR Activation and ECM Attachment Provoke Similar Transcriptional Responsiveness.
To further explore the signaling differences, we used DNA microarrays to study the effects of cell adhesion and ECM interaction on the transcriptional responsiveness of HEK293 cells. From the results presented in Figs. 1 and 2, we predicted that genes would respond to EGF stimulation in at least two different ways: (i) EGF responses that are similar in cells adherent to both polyL-lysine and ECM proteins, and (ii) responses that are unique to ECM-adherent cells.
Quiescent HEK293 cells were held in suspension before being allowed to readhere to polyL-lysine, fibronectin, or laminin-coated tissue culture plastic for an additional 3 h. This incubation period was chosen because it corresponded with a resumption of cell responsiveness to EGF as determined by EGFR, PKB, and MAPK phosphorylation (Figs. 1 and 2). Adherent cells were then stimulated for 1 h with 10 ng/ml EGF. A 1-h stimulation period was chosen because this corresponds with a resumption of transcriptional activity in quiescent human fibroblasts stimulated with serum (23). After EGF stimulation, mRNA was extracted and used to reverse transcribe cDNAs labeled with fluorochrome CY5 (treatment). These were mixed with an equal amount of CY3-labeled cDNA synthesized from RNA extracted from cells attached to polyL-lysine (control). PolyL-lysine-adherent cells were judged to be a good source of reference sample because EGFR-activated signaling pathways are inactive in these cells (Figs. 1 and 2). To control for possible incorporation bias because of differences in fluorescent dyes, samples were also prepared in which treatment and control dyes were reversed. In this case, cDNAs prepared from cells attached to polyL-lysine (control) were labeled with the CY5 dye, and cDNAs from cells stimulated with EGF and attached to fibronectin or laminin were labeled with CY3. This process is termed “fluor reversal.” For each treatment, at least three independent hybridizations to different microarrays were carried out, including at least one fluor reversal. Color representations of the results were produced by nominating the control fluorescent images as green and fluorescent images from treatment samples as red and combining the two color images. The data analysis package, cluster and tree view (14, 15), was then used to group genes according to similar patterns of responsiveness.
Alignment of results from experimental replicates revealed diverse patterns of responsiveness among the 1,718 genes tested. The full dataset from these experiments can be found in Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org. In some cases, there was variability in the magnitude of responsiveness of some genes between experiments likely because of experimental variation. However, for a large number of genes, the clustering and display methods applied revealed clusters that displayed unidirectional changes that were consistent through multiple of hybridizations as well as the fluor-reversal process. Two such groupings are shown in Fig. 3. Cluster A represents those genes that were induced by all treatment, whereas subset B contains genes that were generally suppressed. Group A genes (Fig. 3) include those involved in cytoskeletal architecture (e.g., plastin and coronin), protein translation (60S ribosomal proteins L6 and L35AG), and cell–matrix and cell–cell interaction (syndecan-1 and connexin). Intriguingly, EGF stimulation has been shown to increase the cellular levels of gap-junction connexin protein in kidney epithelial cells in parallel with an enhancement of intercellular communication (24), and EGF increases syndecan-1 mRNA in keritinocytes (25). These observations suggest that concomitant regulation of genes involved in extracellular adhesion may represent wide spread cellular adaptation to EGFR activation.
Figure 3.

EGF and ECM treatment provokes similar transcriptional responses. Clustered display of data from HEK293 cells. Cells were attached for 3 h to polyL-lysine (10 μg/ml), fibronectin (10 μg/ml), or laminin (10 μg/ml), and then some cells were stimulated with EGF (10 ng/ml) for an additional hour. CY5- and CY3-labeled cDNAs were prepared and hybridized to cDNA microarrays, as described in Materials and Methods. Microarrays were then subjected to confocal scanning and colored images generated as described (16). Images are labeled with the corresponding treatment identifiers (1, polyL-lysine plus EGF; 2, fibronectin; 3, fibronectin plus EGF; 4, laminin; and 5, laminin plus EGF). Two separate clusters of genes are indicated (subsets A and B), which show similar responsiveness to all of the treatments applied.
We also identified a smaller group of genes, subset B, that were down-regulated by the treatments applied (Fig. 3). Predominantly, this group contains genes known to be involved in the regulation of the cell cycle, including the tumor suppresser genes p53, retinoblastoma-like proteins, p18-INK (26), and MCC (27). EGF treatment has also been associated with decreased expression of the p53 gene in A431 cells, an event associated with control of cell proliferation (28). It appears, therefore, that the intracellular signals elicited by EGFR stimulation and attachment of cells to ECM can provoke similar nuclear responses. Among these is the unidirectional control of genes involved in cell cycle, cytoskeletal structure, and cellular adhesion. Moreover, EGF was found to stimulate qualitatively similar transcriptional responses in cell adherent to polyL-lysine as in cells attached to ECM fibronectin and laminin. These responses demonstrate that the expression of at least some genes can be controlled by EGFR stimulation in the absence of ECM protein interaction.
ECM Interaction Determines the Response of Genes to EGF Stimulation.
From our studies in HEK293 cells, it would appear that ECM interaction can alter the quality of intracellular signals initiated at the EGFR (Figs. 1 and 2). These findings are substantiated by the work of others using a variety of cell systems (4, 29, 30). The effects of ECM may be through the licensing of RTK activity or through mechanisms that have yet to be defined (4, 29, 30). What is not known is whether ECM-induced changes in the sensitivity of signaling pathways to RTK stimulation are conveyed to the nucleus in terms of altered gene activity. We found one group of 24 genes that responded to EGF in an ECM-specific manner (Fig. 4). This group contains two subsets, C and D. Subset C contains genes for an inward rectifying potassium channel, ribosomal S6 kinase, and the vitronectin receptor α subunit (integrin α-V). EGF promoted an increase in the expression of these genes in cells that were attached to polyL-lysine or fibronectin. In contrast, the same group of genes was down-regulated in cells on laminin and was unresponsive to further EGF stimulation (Fig. 4). The second subset in Fig. 4, subset D, also represents a diverse group of genes, including sodium/potassium-dependent ATPase, receptor tyrosine kinase RET, and stimulatory G-protein (Gs) α subunit. Subset D genes differ from those in subset C in that an increase in gene expression in both laminin- and fibronectin-attached cells. In contrast, copresentation of cells with laminin and EGF resulted in a generalized down-regulation of subset D genes. The effects of EGF on gene expression therefore seem to depend very much on the composition of the ECM.
Figure 4.
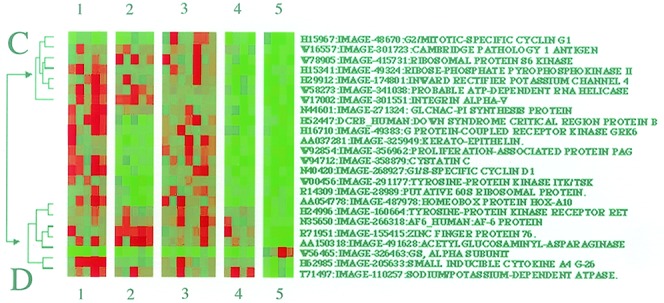
Gene responses specific to cells adhered to laminin. HEK293 cells were treated with ECM components and EGF, as described in Fig. 3 legend. Images are labeled with the corresponding sequence-verified gene names, image, and GenBank accession nos. and treatment identifiers (1, polyL-lysine plus EGF; 2, fibronectin; 3, fibronectin plus EGF; 4, laminin; and 5, laminin plus EGF). Two subsets of genes are indicated (C and D), which show similar responses to the treatments applied.
The results presented in Fig. 4 are examples of gene regulation that are specific to interaction of cells with laminin. Fig. 5 shows that fibronectin can also shape transcriptional responses to EGF stimulation. The first way in which responses are manifest (Fig. 5, subset E) is typified by the responses of the transcripts for DNA topoisomerase II and ATP-dependent RNA helicase. These genes were down-regulated in cells attached to all of the substrata tested. However, we observed that EGF stimulated an increase in gene expression only in cells that were specifically attached to fibronectin. In addition to topoisomerase II, which is involved in chromosome segregation at mitosis, fibronectin attachment was also found to increase the expression of cyclin G and D1 genes, which are involved in the transition from G1 to S and G2 to mitotic phases of the cell cycle, respectively (Fig. 4). Cyclin D1 expression is jointly regulated by growth factors and cell adhesion to the ECM in many cell types (31, 32), and cyclin G expression is markedly reduced on aggregate formation in embryonic carcinoma (33). These observations suggest that changes in the ability of a cell to interact with ECM may contribute to the control of cell growth by regulating expression of genes involved in cell cycle progression. Moreover, that RTK signaling can be programmed by specific interaction with ECM proteins may go some way to explaining differential effects of ECM on cell morphology, migration, and proliferation reported in a number of cell types (34, 35).
Figure 5.
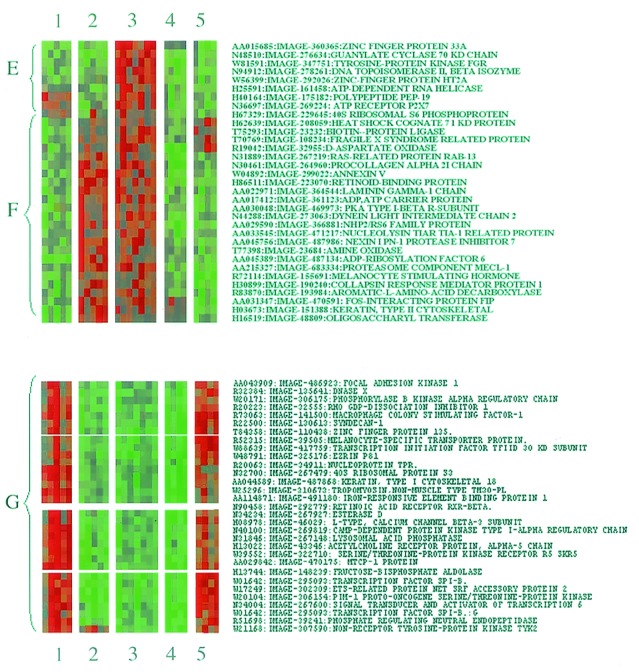
Fibronectin-specific changes in gene expression in EGF-stimulated HEK293 cells. Color cluster display of gene responses in HEK293 cells. Three subsets of gene responsiveness are indicated (E–G). The display contains annotations for treatment identifiers (1–5; see Fig. 3 legend).
Adherence of cells to fibronectin was found to up-regulate another group of genes independently of EGF stimulation (Fig. 5, subset F), in contrast to the other substrata tested, which promoted a generalized down-regulation of the same group of genes. Genes specifically up-regulated by fibronectin are implicated in ECM remodeling (collagen α-2, laminin γ-1, and the metalloprotease inhibitor nexin-1) and cytoskeletal architecture (type II cytokeratin and dynein light chain). These observations support a previous study by Lafrenie et al. (36). Here the effects of ECM on gene expression were examined in epithelial cells by using PCR-based subtractive hybridization (36). The majority of gene transcripts were up-regulated by adhesion of cells to both fibronectin and collagen; however, a small number of genes were up-regulated only in cells attached to collagen, and the expression of one unidentified gene was up-regulated only in cells plated on fibronectin (36). In the present study, we identify 14 genes that are also specifically up-regulated by interaction of cells with a fibronectin.
In addition to stimulating expression of some genes, we also found that cell attachment to fibronectin, like laminin (Fig. 4), can neutralize incoming signals from the EGFR. We identified a cluster of genes (Fig. 5, subset G) in which EGF stimulated an increase in the expression of genes in cells plated on laminin or polyL-lysine. These include genes involved in cytoskeletal structure (e.g., cytokeratin, ezrin, and nonmuscle tropomyosin) and signal transduction (e.g., FAK, PKA R-subunit, and STAT6). However, in cells attached to fibronectin, EGF was unable to provoke an increase in the expression of the same group of genes. Fibronectin, therefore, appears to neutralize or dampen the response of certain genes to EGF stimulation. These results further demonstrate that interaction of cells with different extracellular substrates can alter how genes respond to RTK stimulation.
A significant contribution to this specificity may arise from physical interaction between RTKs and integrins. EGFR is known to form a physical complex with active β1 integrins after attachment to fibronectin (8). Insulin, platelet-derived growth factor, and V-EGF receptors do not interact with β1 integrins but instead form complexes with the vitronectin receptor, integrin αVβ3 (37). This specificity of interaction is thought to underlie vitronectin-enhanced mitogenicity in some cell types (37). Recent observations by Tomatis et al. (38) show that the activation of the α7β1 laminin receptor negatively regulates the activity of the α5β1 fibronectin receptor (38). Because RTKs are thought to interact only with active integrins, the composition of ECM has the potential to determine the types of RTK/integrin liaisons that occur in the plasma membrane. This selectivity could lead to changes in the intracellular location or conformation of the EGFR, thereby changing the accessibility of the receptor intracellular domain to downstream signaling molecules. One downstream protein might be focal adhesion kinase, which is targeted to sites of integrin/RTK complex formation and is essential for the transmission of motility signals from the EGFR (39).
Overall, the observations presented here contribute to the emerging awareness that cell–ECM interaction is intimately involved in the development and propagation of cell responsiveness to the external environment. Moreover, a full appreciation of the transcriptional responses to growth factor receptor activation will not be gained until expression analysis is carried out under different conditions of cell attachment. Accordingly, recent work demonstrating that activation of PDGF and FGF receptors induces very similar changes in gene expression in fibroblasts should be reevaluated, because only one form of cell adherence, attachment to tissue culture plastic, was examined (40).
Mutation of ECM proteins and receptors results in a wide range of tissue defects in humans, including muscular dystrophy and osteogenesis imperfecta (41), and the molecules that mediate cell-ECM adhesion undergo dramatic alteration during conversion of cells to a malignant phenotype (41). Often cellular transformation is accompanied by down-regulation of ECM proteins, their cognate receptors, and cytoskeletal components. Further expression analysis will lead to the identification of gene expression patterns that may serve as “predictive signatures” of the transformed phenotype or other tissue abnormalities. For example, part of the cellular response to transforming mutations may be alterations in gene expression, similar to those induced by ECM association. These alterations may contribute to metastasis by enabling cancer cells to grow and survive in the absence of ECM. Indeed, the gene expression patterns conferred by ECM attachment in the present study (Figs. 4 and 5) may also be part of the genetic adaptations of metastatic tumors. By examining the relevant in vivo interactions and corresponding gene expression changes in tumor samples, we may gain a betterunderstanding of the contribution of ECM to the development of human diseases such as cancer.
Supplementary Material
Acknowledgments
We thank the staff of the Ontario Cancer Institute microarray facility for support and advice. S.J.Y. is a Human Frontier Science Program Research Fellow, and this work was supported by grants from the Medical Research Council and National Cancer Institute of Canada to J.R.W.
Abbreviations
- RTK
receptor tyrosine kinase
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ECM
extracellular matrix
- HEK
human embryonic kidney
- PKB
protein kinase B
References
- 1.Hynes R O. Trends Cell Biol. 1999;9:M33–M37. [PubMed] [Google Scholar]
- 2.Humphries M J. Trends Pharmacol Sci. 2000;21:29–32. doi: 10.1016/s0165-6147(99)01410-8. [DOI] [PubMed] [Google Scholar]
- 3.Giancotti F G, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 4.Cybulsky A V, McTavish A J, Cyr M D. J Clin Invest. 1994;94:68–78. doi: 10.1172/JCI117350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto S, Teramoto H, Gutkind J S, Yamada K M. J Cell Biol. 1996;135:1633–1642. doi: 10.1083/jcb.135.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones P L, Crack J, Rabinovitch M. J Cell Biol. 1997;139:279–293. doi: 10.1083/jcb.139.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodeck U, Jost M, Kari C, Shih D T, Lavker R M, Ewert D L, Jensen P J. J Cell Sci. 1997;110:113–121. doi: 10.1242/jcs.110.2.113. [DOI] [PubMed] [Google Scholar]
- 8.Moro L, Venturino M, Bozzo C, Silengo L, Altruda F, Beguinot L, Tarone G, Defilippi P. EMBO J. 1998;17:6622–6632. doi: 10.1093/emboj/17.22.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J D, Kim J P, Zhang K, Sarret Y, Wynn K C, Kramer R H, Woodley D T. Exp Cell Res. 1993;209:216–223. doi: 10.1006/excr.1993.1304. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Lin M L, Wiepz G J, Guadarrama A G, Bertics P J. J Biol Chem. 1999;274:11209–11219. doi: 10.1074/jbc.274.16.11209. [DOI] [PubMed] [Google Scholar]
- 11.Maatta A, Jaakkola P, Jalkanen M. J Biol Chem. 1999;274:9891–9898. doi: 10.1074/jbc.274.14.9891. [DOI] [PubMed] [Google Scholar]
- 12.Grande J P, Melder D C, Zinsmeister A R. J Lab Clin Med. 1997;130:476–486. doi: 10.1016/s0022-2143(97)90124-4. [DOI] [PubMed] [Google Scholar]
- 13.Leco K J, Khokha R, Pavloff N, Hawkes S P, Edwards D R. J Biol Chem. 1994;269:9352–9360. [PubMed] [Google Scholar]
- 14.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 16.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 18.Bodary S C, McLean J W. J Biol Chem. 1990;265:5938–5941. [PubMed] [Google Scholar]
- 19.Yu X, Miyamoto S, Mekada E. J Cell Sci. 2000;113:2139–2147. doi: 10.1242/jcs.113.12.2139. [DOI] [PubMed] [Google Scholar]
- 20.Peraldi P, Scimeca J C, Filloux C, Van Obberghen E. Endocrinology. 1993;132:2578–2585. doi: 10.1210/endo.132.6.8389283. [DOI] [PubMed] [Google Scholar]
- 21.Burgering B M, de Vries-Smits A M, Medema R H, van Weeren P C, Tertoolen L G, Bos J L. Mol Cell Biol. 1993;13:7248–7256. doi: 10.1128/mcb.13.12.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Weering D H, de Rooij J, Marte B, Downward J, Bos J L, Burgering B M. Mol Cell Biol. 1998;18:1802–1811. doi: 10.1128/mcb.18.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 24.Rivedal E, Mollerup S, Haugen A, Vikhamar G. Carcinogenesis. 1996;17:2321–2328. doi: 10.1093/carcin/17.11.2321. [DOI] [PubMed] [Google Scholar]
- 25.Jaakkola P, Jalkanen M. Prog Nucleic Acid Res Mol Biol. 1999;63:109–138. doi: 10.1016/s0079-6603(08)60721-7. [DOI] [PubMed] [Google Scholar]
- 26.Guan K L, Jenkins C W, Li Y, Nichols M A, Wu X, O'Keefe C L, Matera A G, Xiong Y. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 27.Matsumine A, Senda T, Baeg G H, Roy B C, Nakamura Y, Noda M, Toyoshima K, Akiyama T. J Biol Chem. 1996;271:10341–10346. doi: 10.1074/jbc.271.17.10341. [DOI] [PubMed] [Google Scholar]
- 28.Gulli L F, Palmer K C, Chen Y Q, Reddy K B. Cell Growth Differ. 1996;7:173–178. [PubMed] [Google Scholar]
- 29.Porter J C, Hogg N. Trends Cell Biol. 1998;8:390–396. doi: 10.1016/s0962-8924(98)01344-0. [DOI] [PubMed] [Google Scholar]
- 30.Aplin A E, Howe A K, Juliano R L. Curr Opin Cell Biol. 1999;11:737–744. doi: 10.1016/s0955-0674(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Ohtsubo M, Bohmer R M, Roberts J M, Assoian R K. J Cell Biol. 1996;133:391–403. doi: 10.1083/jcb.133.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roovers K, Davey G, Zhu X, Bottazzi M E, Assoian R K. Mol Biol Cell. 1999;10:3197–3204. doi: 10.1091/mbc.10.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto K, Prives C. Oncogene. 1999;18:4606–4615. doi: 10.1038/sj.onc.1202821. [DOI] [PubMed] [Google Scholar]
- 34.Ichinose M, Yahagi N, Matsubara Y, Tsukada S, Oka M, Shimizu Y, Yonezawa S, Kageyama T, Miki K, Fukamachi H. Biochem Biophys Res Commun. 1997;230:537–541. doi: 10.1006/bbrc.1996.5977. [DOI] [PubMed] [Google Scholar]
- 35.Hirst S J, Twort C H, Lee T H. Am J Respir Cell Mol Biol. 2000;23:335–344. doi: 10.1165/ajrcmb.23.3.3990. [DOI] [PubMed] [Google Scholar]
- 36.Lafrenie R M, Bernier S M, Yamada K M. J Cell Physiol. 1998;175:163–173. doi: 10.1002/(SICI)1097-4652(199805)175:2<163::AID-JCP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Soldi R, Mitola S, Strasly M, Defilippi P, Tarone G, Bussolino F. EMBO J. 1999;18:882–892. doi: 10.1093/emboj/18.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomatis D, Echtermayer F, Schober S, Balzac F, Retta S F, Silengo L, Tarone G. Exp Cell Res. 1999;246:421–432. doi: 10.1006/excr.1998.4315. [DOI] [PubMed] [Google Scholar]
- 39.Sieg D J, Hauck C R, Ilic D, Klingbeil C K, Schaefer E, Damsky C H, Schlaepfer D D. Nat Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 40.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 41.Lukashev M E, Werb Z. Trends Cell Biol. 1998;8:437–441. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.