MicroRNAs comprise a large family of small (∼22 nt) regulatory molecules found in all multicellular eukaryotes. Emerging data indicate that miRNA production can be regulated at many steps including post-transcription addition of nucleotides to the 3′ end of partially processed molecules. This paper shows that nontemplated addition of nucleotides to the 3′ end is a widespread phenomenon that is itself subject to regulation.
Keywords: microRNA, miRNA, deep-sequencing
Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that post-transcriptionally regulate gene expression. An emerging mechanism to control miRNA production is the addition of an oligo-uridine tail to the 3′ end of the precursor miRNA. This has been demonstrated for the Let-7 family of miRNAs in embryonic cells. Additionally, nontemplated nucleotides have been found on mature miRNA species, though in most cases it is not known if nucleotide addition occurs at the precursor step or at the mature miRNA. To examine the diversity of nucleotide addition we have developed a high-throughput sequencing method specific for miRNA precursors. Here we report that nontemplated addition is a widespread phenomenon occurring in many miRNA families. As previously reported, Let-7 family members are oligo-uridylated in embryonic cells in a Lin28-dependent manner. However, we find that the fraction of uridylated precursors increases with differentiation, independent of Lin28, and is highest in adult mouse tissues, exceeding 30% of all sequence reads for some Let-7 family members. A similar fraction of sequence reads are modified for many other miRNA families. Mono-uridylation is most common, with cytidine and adenosine modification less frequent but occurring above the expected error rate for Illumina sequencing. Nucleotide addition in cell lines is associated with 3′ end degradation, in contrast to adult tissues, where modification occurs predominantly on full-length precursors. This work provides an unprecedented view of the complexity of 3′ modification and trimming of miRNA precursors.
INTRODUCTION
The microRNA (miRNA) pathway has emerged as an important component of gene regulation (Du and Zamore 2005). This pathway was first described in Caenorhabditis elegans with the discovery of the developmental regulator lin-4 (Lee et al. 1993; Wightman et al. 1993). This gene was shown to encode a small, noncoding RNA that could base-pair with target mRNAs, leading to reduced target gene expression. More recently, thousands of distinct miRNAs have been discovered in many organisms, leading to the model that most, if not all, biological pathways are impacted by miRNA regulation (Griffiths-Jones et al. 2008). In addition, a large number of human diseases have been linked to misregulation of miRNA expression and/or function (Chang and Mendell 2007).
Recent efforts have focused on the regulation of miRNA production (Krol et al. 2010; Newman and Hammond 2010). miRNA expression begins with transcription of the primary transcript, or pri-miRNA (Kim et al. 2009). This long RNA is processed by the nuclear endonuclease Drosha, releasing the precursor RNA (pre-miRNA). The precursor has a characteristic stem–loop structure, with one or both strands of the stem representing the biologically active, mature species. Processing by the cytoplasmic nuclease Dicer removes the loop, allowing the mature strand(s) to be loaded into the RNA-induced silencing complex (RISC) effector complex (Kim et al. 2009). In principle, miRNA expression can be regulated at transcription, at either nuclease processing step, or at turnover of the mature miRNA, and in fact all of these mechanisms have been reported (Krol et al. 2010). Notably, the conserved miRNA family Let-7 is regulated in a complex manner at all of these steps. The RNA binding protein Lin28 blocks both processing steps during Let-7 biogenesis, while also directing the terminal uridylyl transferase Tut4 to add a uridine tail to the Let-7 precursor (Heo et al. 2008; Newman et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008; Hagan et al. 2009; Heo et al. 2009; Lehrbach et al. 2009). The tail promotes degradation of the precursor, further limiting production of mature Let-7.
To further our understanding of miRNA regulation, we sought to analyze precursor tailing and degradation for other miRNA families. Such efforts have been done indirectly by analyzing nontemplated nucleotide presence on mature miRNAs (Burroughs et al. 2010; Chiang et al. 2010; Fernandez-Valverde et al. 2010; Berezikov et al. 2011). It has demonstrated that many miRNA families have nontemplated nucleotide addition to the 3′ end. The increased occurrence on 3′ strand (3P) mature miRNAs suggests the addition occurs at the precursor; however, this has not been directly demonstrated. In addition, tailing will likely affect Dicer processing; thus, evidence of tailing on the mature miRNA may substantially underrepresent the extent of tailing at the precursor. To address these shortcomings, we have developed a high-throughput sequencing strategy specific for miRNA precursors.
RESULTS
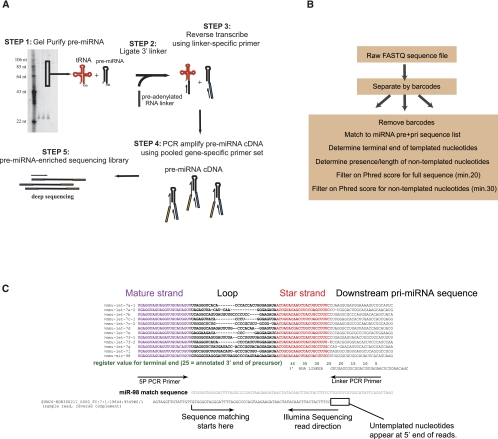
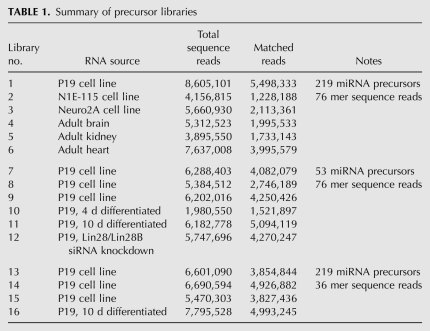
Protocols for sequencing mature miRNAs are well established (Hafner et al. 2008). Essentially, RNA linkers are ligated to the 5′ and 3′ end of small RNA fractions, and RT-PCR is performed to amplify the pool of miRNAs. This approach presents problems for precursor miRNAs for two reasons. First, precursors are present at 103–106-fold lower abundance than mature miRNAs. Secondly, the size of precursors (70 nt) is similar to the highly abundant tRNA pool. Thus, any sequence-independent method of library construction will lead to an unacceptably high fraction of tRNA contamination. To overcome these problems, we developed an approach that uses gene-specific primers at the 5′ end of the precursor population (Fig. 1A). Specifically, RNA was size-fractionated to remove mature miRNA species. A universal RNA linker was ligated to the 3′ end of the RNA population. Reverse transcription (RT) was carried out using a primer directed at the universal RNA linker sequence. The RT population was then amplified with a universal 3′ linker-primer and a pool of gene-specific 5′ primers. This step also adds the Illumina cluster generation sequence. This was followed by 19 additional PCR cycles using nested primers that are directed against the Illumina cluster sequences. For the first version of our protocol, the primer pool was designed to amplify 53 commonly studied mouse miRNA precursors. Analyses of the libraries generated from this smaller primer pool indicated that the protocol was robust enough to allow a greater number of precursor directed primers. We thus prepared additional libraries with a primer pool directed at 219 miRNA precursors, expanding the number of miRNA families in the analysis. For all libraries, SYBR green incorporation was measured to confirm linear amplification during PCR (data not shown). Libraries were sequenced to 36-nt-length using an Illumina Genome Analyzer II. Some libraries were additionally sequenced to 76-nt-length for characterization of longer polynucleotide tails. Table 1 summarizes library RNA source, read length, and primer pool size.
FIGURE 1.
Precursor-seq as a method of analyzing miRNA precursors. (A) Overview of experimental protocol for precursor-seq. See text for details. (B) Overview of bioinformatic analysis for precursor-seq. See text for details. (C) Sequence match algorithm overview. An alignment of the Let-7 family is shown, with mature strand in violet and star strand in red. The 3′ RNA linker is shown and both PCR primer sites. The match sequence for miR-98 is shown aligned with a sample read. Sequence matching starts at the indicated arrow and proceeds to the 3P arm. The terminal end of the precursor is determined by the last nucleotide adjacent to the RNA linker sequence. The terminal end is defined by “register,” shown in green, which corresponds to nucleotide position based on the genomic sequence. A value of “25” is defined based on the predicted terminal end in miRbase, if annotated. If not annotated, the end is defined on the expected overhang from the Drosha reaction. All registers were adjusted if necessary for the most abundant nonmodified terminal end.
TABLE 1.
Summary of precursor libraries
The data analysis pipeline for precursor libraries is summarized in Figure 1B. Since the sequence read should match one of the 219 precursors that the PCR primer pool was directed against, we developed a custom matching algorithm that searches a list of expected precursor matches. The matching strategy is illustrated in Figure 1C, using the Let-7 family as an example. The libraries were designed to sequence from the 3′ end of the precursor to provide maximum information about 3′ addition and trimming. Therefore, the matching algorithm scans sequence reads against the candidate list, beginning in the loop region of the precursors. Using the loop region for matching avoids miscalling reads due to PCR amplication artifacts and/or mispriming (the primers are directed at the 5P stem) (see Fig. 1C). This strategy has the added benefit of using the poorly conserved precursor regions for mapping, allowing discrimination within miRNA gene families. This is illustrated in Figure 1C, whereby the sample read is correctly matched to miR-98 and not other Let-7 family members, due to poor conservation in the loop region. The 3′ terminal end of the precursor is adjacent to the linker RNA sequence which allows for precise 3′ terminal end identification and nontemplated nucleotide determination. Notably, the library design does not provide information about the 5′ end of miRNA precursors. After matching, sequence reads were filtered based on the standard “Phred-20” cutoff for expected error rate. Data was further filtered to a higher Phred quality score (30) within nontemplated nucleotide tails. This was designed to prevent a miscalled base from being scored as a nontemplated addition. Tailing was only called if it was nontemplated and only if it was a homopolymeric tail extending to the linker junction in the sequence read. Mixed nucleotide tails are much less frequent and present an additional level of complexity for scoring algorithms. Thus, the high modification fractions we report are conservative but contain a very high degree of confidence.
Libraries were prepared from RNA from several mouse cell lines and tissues (Table 1). In most cases, over 50% of reads were successfully mapped to a precursor. The unmapped reads may be due to a miscalled base (only exact matches were allowed by the algorithm, minimum 23-nt-length) or may be due to mispriming during the PCR step. The lower mapping rate for libraries prepared with the large primer pool is likely due to increased mispriming. As stated above, the matching algorithm searches against loop sequences which are beyond the PCR primer site; therefore, misprimed products will not score a match and will not affect data quality. The correlation of precursor counts among replicate libraries was over 0.95 (Supplemental Fig. S1; cf. “undiff.219” to another “undiff.219”). The correlation between libraries of the same RNA source but different primer pool size was somewhat reduced (cf. “undiff.291” to “undiff.53”). The correlation between libraries from different cell types was low, as expected (“undiff.219” versus “10day.219”). Since libraries prepared from different size primer pools were less correlative, all comparative analyses in this study were done from libraries prepared with the same primer pool and sequenced to the same read length.
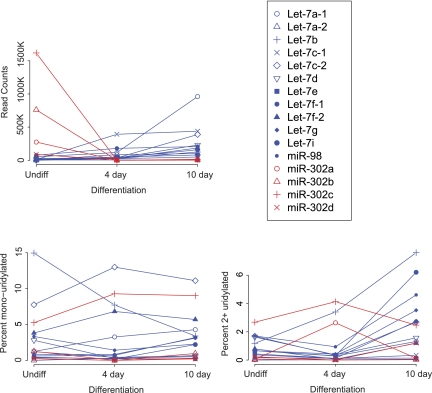
We first analyzed precursor expression during differentiation of P19 teratocarcinoma cells. This cell line resembles embryonic stem cells and can be differentiated with retinoic acid (Rudnicki et al. 1990). In undifferentiated cells, Let-7 biogenesis is blocked by the RNA binding protein Lin28 (Newman et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008). In differentiated cells, Lin28 is not expressed, allowing mature Let-7 production. In contrast, the embryonic miRNA family miR-302 is highly expressed in undifferentiated P19 cells and rapidly decreases during differentiation. Precursor levels followed this paradigm (Fig. 2). Precursor counts of Let-7 family members were variable but exhibited an overall increase during differentiation. A further mechanism of Let-7 regulation is uridylation of the precursor by the terminal uridylyl transferase Tut4 (Heo et al. 2008; Hagan et al. 2009; Heo et al. 2009; Lehrbach et al. 2009). This widely expressed enzyme is recruited to Let-7 precursors in embryonic cells by Lin28. Surprisingly, nontemplated addition of uridine to Let-7 precursors is higher in differentiated cells (which lack Lin28 expression) than in undifferentiated cells (Fig. 2; Supplemental Fig. S2). This includes the total fraction of uridylated reads (1.7% versus 5.9% for undifferentiated versus differentiated cells, respectively) and the fraction with two or more uridines added (0.55% versus 2.0%) (Fig. 2; Supplemental Fig. S3). The fraction of precursors uridylated varied for Let-7 family members, with Let-7c-2 the highest fraction of mono-uridine (over 10% of reads) and Let-7b the highest fraction with two or more uridines (increasing to 7% during differentiation). Replicate libraries displayed similar uridylation increases during differentiation (1.5% total, increasing to 5.8%, and 0.45% 2+U, increasing to 2.0%; data not shown).
FIGURE 2.
Let-7 precursor expression during P19 cell differentiation. Expression of Let-7 and miR-302 family member precursors during differentiation are shown. Raw sequence reads are plotted. Libraries 7, 10, and 11 were used. Percent reads with a mono-uridine addition, and uridine tails of two or more nucleotides, are shown.
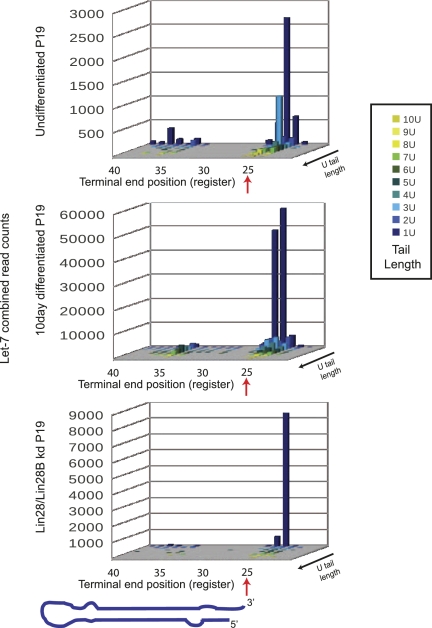
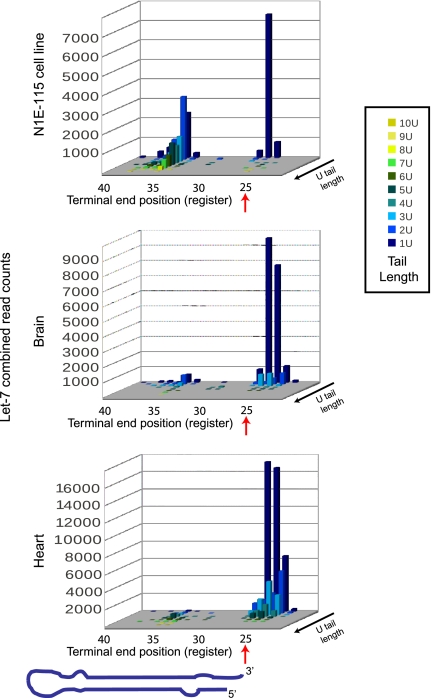
In addition to nontemplated nucleotide addition, our sequencing strategy also provided details about the 3′ terminal end of precursors. Uridine tail length versus terminal end register (the last templated nucleotide match) is plotted in three dimensions in Figure 3. Let-7 family member sequence reads are plotted as a composite for undifferentiated P19 cells. Mono-uridine addition is the most common, at the predicted full-length position of the precursor (defined arbitrarily as register position 25) (see Fig. 1C for further explanation of registers). For Let-7 family members, a peak of longer uridine tails was visible at the full-length position, representing precursors tailed after Drosha processing. In addition, a peak of tailed precursors ∼11 nt recessed into the stem was also visible. These presumably represent nuclease products that have been re-uridylated in the recessed position. As described above, uridine tailing is present in differentiated P19 cells in the absence of the cofactor Lin28. When viewed as a three-dimensional plot, differentiated cells also display tailing from a recessed position (Fig. 3).
FIGURE 3.
Let-7 precursors are uridylated in a Lin28 dependent and independent mechanism. The number of reads for all Let-7 family members were combined, counted by uridine tail length, and by nucleotide position (register) of the terminal end preceding the tail. Nucleotide position is arbitrarily set at 25 for full-length precursors, at Drosha cleavage site (see Fig. 1C for further description of registers). A cartoon precursor is shown for orientation. Precursor-seq data is shown for undifferentiated P19 cells (top), P19 cells differentiated for 10 d (middle), and undifferentiated P19 cells after 3× transfection with Lin28+Lin28B siRNAs (bottom). Total counts for Let-7 family members varied for each library, reflecting the different scales of the plots. Libraries 9, 11, and 12 are shown. Lin28 knockdown does not affect pluripotency markers. Thomson et al. 2006).
To further define the role of Lin28 in oligo-uridine modification, we depleted this protein and its related family member Lin28B from P19 cells using siRNAs. Precursor-seq demonstrated that oligo-uridylation was strongly reduced for the composite Let-7 family (Fig. 3). In addition, uridine addition from the recessed position was reduced. It should be noted that highly expressed Let-7 family members contribute more to the 3D composite figures; however, all individual Let-7 members have decreased oligo-uridylation upon Lin28 knockdown. Mono-uridylation changes are more variable for family members. (Let-7f-2 mono-uridylation increases from 3.9% to 12.7% in Lin28 knockdown cells; miR-98 decreases from 3.3% to 1.8%). Thus, while Lin28 promotes oligo-uridylation in P19 cells, it is not necessary for mono-uridylation. Furthermore, these data demonstrate a novel Lin28-independent pathway that is present in differentiated cells.
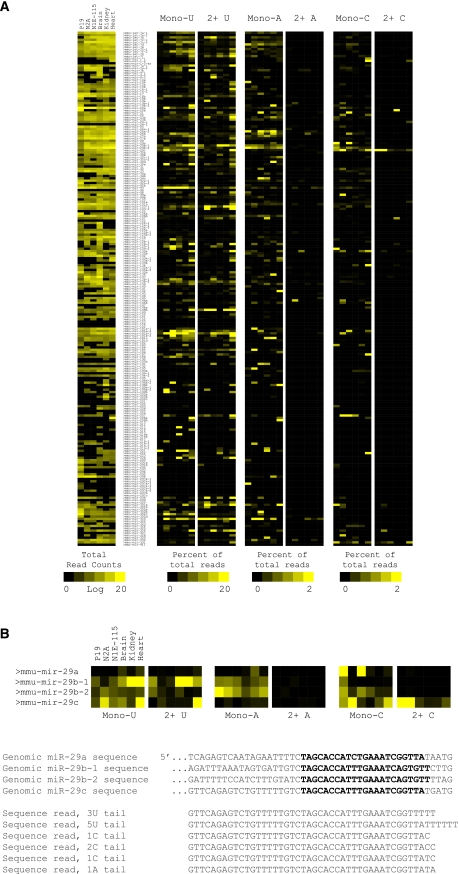
We next analyzed the full precursor data set. Figure 4A shows the expression map of precursor reads across cell lines and tissues analyzed. A majority of the precursors were widely expressed, with the exception of tissue-specific miRNAs (e.g., miR-124, miR-1, miR-133). The percent reads with uridine, adenosine, and cytidine addition are shown in the expression map (Fig. 4A). Guanosine addition did not occur after the Phred score filtering. Uridine was the most common addition, followed by adenosine and cytidine (note different scaling for A and C). Mono-uridine addition was more common; however, tails of two uridines and longer were frequent in many miRNA families. For example, 64.5% of all reads for miR-103-1 in heart had a uridine tail 2 nt or longer (3180 reads out of 4923 total). Addition of two or more cytidines or adenosines was rare. Of the tissues tested, heart had the greatest fraction of nucleotide addition, with 16.6% of all reads for all miRNA families combined. Interestingly, P19 cells had the lowest combined fraction, at 2.3% of total reads (this data point may be skewed by the low Let-7 counts in this cell line).
FIGURE 4.
Precursor expression and modification in cells and tissues. (A) Total precursor counts were normalized to 5M total, log-transformed, and plotted as a heat map. Libraries 1–6 were used. Percent modification for each untemplated nucleotide, mono, or two and longer, for each precursor, was calculated. The data is plotted as a heat map, linear scale, as percent of total reads for that precursor. All ordering of heat maps is the same. Note the scaling for U is different than A and C. (B) Section of the heat map for miR-29 family is expanded. Shown is the genomic sequence for all four family members. The mature strand (in this case, on the 3P arm) is in bold. Six representative sequence reads from the heart library are shown. The 5′ end of the read is not fully shown. Sequencing was performed from the 3′ end of the precursor; thus, the 3′ end represents the junction with the cloning linker.
MiRNA families and family members showed surprising preference for nucleotide addition. An example is shown in Figure 4B, which illustrates miR-29 family members containing adenosine, uridine, and cytidine addition. miR-29c has high fractions of uridine and cytidine addition but very little adenosine addition. Other members of this family, in contrast, have more abundant mono-adenosine addition. The genomic sequence alignment shows that miR-29c could score a nontemplated adenosine addition (if the nucleotides directly after the cleavage site were templated as adenosine, a nontemplated addition could not be scored). And in fact, a sequence read from heart is shown that has a mono-adenosine addition. These, however, were unexpectedly rare.
Mature miRNA duplexes are loaded into the RISC according to thermodynamic rules, with one strand loaded more frequently (in most cases) (Hutvagner 2005). Of the miRNAs in our data set, 57% preferentially load the 5P strand. We found no significant preference for nucleotide addition versus strand loaded (P-values 0.852, 0.3, and 0.451 for U, A, and C addition, two tailed unpaired t-test). Examination of the nucleotide directly preceding the nontemplated addition did reveal preference for uridine (Supplemental Fig. S4; cf. fractions to nontailed reads).
An analysis of terminal end location and modification in mouse tissues shows extensive mono-, di-, and tri-uridylation of Let-7 family members, predominantly located at the full-length precursor (Fig. 5). Interestingly, the neuroblastoma cell line N1E-115 has a very large peak of recessed, tailed precursors (Fig. 5).
FIGURE 5.
Tailing and trimming of Let-7 precursors in tissues and cell lines. The number of reads for all Let-7 family members were combined, counted by uridine tail length, and by nucleotide position (register) of the terminal end preceeding the tail. Nucleotide position is arbitrarily set at 25 for full length precursors, at Drosha cleavage site (see Fig. 1C for further description of registers). A cartoon precursor is shown for orientation. Total counts for Let-7 family members varied for each library, reflecting the different scales of the plots. Libraries 2, 4, and 6 shown.
DISCUSSION
Nontemplated nucleotide addition represents a further layer of complexity for the regulation of miRNA production and activity. Nucleotide addition to mature miRNAs has been demonstrated (Ameres et al. 2010). However, in most cases, addition is presumed to occur on the precursor species, and such additions have been inferred from mature miRNA sequence data. Furthermore, the increase in uridine addition to 3P loaded mature miRNAs suggested that addition could occur at the precursor species (Burroughs et al. 2010; Chiang et al. 2010; Fernandez-Valverde et al. 2010; Berezikov et al. 2011). This has never been directly analyzed, however. Data we present here bridge that gap, providing direct information about precursor modifications.
The direct mechanism of nucleotide addition is largely unknown. A family of terminal-uridylyl transferase enzymes has been described, and one member, Tut4, is responsible for tailing Let-7 family member precursors (Hagan et al. 2009; Heo et al. 2009; Lehrbach et al. 2009). This is promoted by the RNA binding protein Lin28. Our data supports the role of Lin28 in promotion of oligo-uridine addition, but not mono-uridine addition, to Let-7 precursors in embryonic cells. Unexpectedly, our data uncovers the existence of Lin28-independent tailing in differentiated cell lines and tissues. It should be noted that the uridine tail composition of endogenous Let-7 precursors has not been previously measured in embryonic cells but has been measured in HEK293 and Huh-7 cells and inferred from cell free activities (Heo et al. 2008, 2009; Hagan et al. 2009). Thus, the mechanisms responsible for Let-7 tailing are more complex than previously realized. There are likely to be other cofactors that can recruit TUT enzymes in nonembryonic cells and in tissues. Addition of adenosine to mature miRNAs has been described and is catalyzed by the cytoplasmic poly(A) polymerase GLD2 (Katoh et al. 2009; Lu et al. 2009; Burroughs et al. 2010). It is not known whether TUT family member proteins can add adenosine or cytidine.
Oligo-uridine tailing is known to promote degradation of Let-7 precursors and reduces Dicer processing efficiency (Heo et al. 2009). It is not known if this is a general feature of uridine tails, or tails of other nucleotide composition. It is certainly apparent from this work that widespread tailing occurs in miRNA families that are known to be highly expressed. Perhaps in some contexts, the tail does not promote degradation. In the case of Let-7, in P19 and neuroblastoma cells, the uridine-tailed precursors are recessed into the stem. This may represent a degradation intermediate. The mechanism of degradation is unknown, but it is interesting to note that Histone mRNAs are tailed with oligo-uridine prior to Lsm7/Exosome degradation (Mullen and Marzluff 2008). Alternatively, a mechanism has been described whereby Ago2 can cleave the 3P stem, leading to a truncated precursor (Diederichs and Haber 2007). Such truncated precursors could be tailed and further degraded, leading to reduced mature Let-7 production. In embryonic cells and cell lines, such a model is plausible, since mature Let-7 expression is often reduced. In adult tissues, however, mature Let-7 is very abundant and, based on our work, is uridylated at a very high frequency. In these contexts, uridylation is presumably not a signal for degradation, a point supported by the low frequency of trimmed species. Whether a tail can be removed, allowing later Dicing, is an interesting point of speculation.
MATERIALS AND METHODS
Cell culture and transfection
P19 embryonic carcinoma (EC), N1E-115, and Neuro2A cells were cultured as prescribed by ATCC.org. P19s were differentiated with all-trans retinoic acid (RA) as previously described (Thomson et al. 2006). Lin28 and Lin28B were depleted by siRNA trasfection, 3 times in 3 consecutive days. siRNA sequences are found in Newman et al. (2008). Mouse tissue RNA was made from newly dissected organs. RT-PCR was performed as described, using primers as described in Newman et al. (2008).
Precursor-seq cDNA library preparation
RNA from all cells and tissues was isolated using Trizol (Invitrogen). High molecular weight (MW) RNA from 50 ug of total RNA was partially depleted by precipitation with 5% PEG8000 / 0.5M NaCl; the lower MW fraction was separated by electrophoresis on an 8% denaturing acrylamide/urea gel. RNA of 50–100 nt was isolated; gel slices were crushed and eluted by incubation overnight in 0.3M sodium acetate, followed by isopropanol precipitation.
Cloning linker RNAs were purchased from IDT. The RNAs contained 5′ phosphates, were blocked on the 3′ end by di-deoxy-cytidine, and had the following sequences (barcodes are italicized and underlined; it should be noted that an oligo starting with the 5′ sequence “CCG…” was also purchased but failed to be pre-adenylated):
Barcode AAC:
5′-pGUUGAUCGUCGGACUGUAGAACUCUGAACddC-3′
Barcode TTC:
5′-pGAAGAUCGUCGGACUGUAGAACUCUGAACddC-3′
Barcode CCC:
5′-pGGGGAUCGUCGGACUGUAGAACUCUGAACddC-3′
Linkers were pre-adenylated as previously described with some modifications (Vigneault et al. 2008). Eight hundred pmols of RNA linker was incubated for 5 h at 37°C in a reaction containing 200 units of T4 RNA ligase I [New England Bio labs (NEB)], 1× RNA ligase I buffer [50mM Tris-HCl pH 7.8 / 10mM MgCl2 / 1mM ATP / 10mM DTT], 4 units of Ribolock RNase A inhibitor (Fermentas), and 25% DMSO. The RNA was then extracted with phenol-chloroform and electrophoresed on an 8% denaturing gel. Adenylated RNA oligo (which migrates ∼1–2 nucleotides higher than unadendylated oligo) was gel-purified away from unadenylated oligo and eluted as described above.
Five hundred ng of gel-purified RNA was ligated to 10 pmol pre-adenylated linker for 2 h at 25°C in a reaction containing 200 units RNA ligase II, truncated (NEB), 1× PNK buffer [70mM Tris-HCl pH 7.6 / 1mM MgCl2 / 0.5 mM DTT], and 2 units of Ribolock. RNA was then extracted with phenol-chloroform and precipitated with ethanol, 0.3M sodium acetate, and 2 uL glycogen. Ligated RNA was reverse-transcribed using the primer, 5′-GTTCAGAGTTCTACAGTCCGA-3′ and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions.
The resulting cDNA was then amplified in two PCR reactions:
miRNA-specific, 10 cycles: 4 uL of undiluted cDNA was amplified with the Illumina small RNA forward primer, 5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA-3′ (2.5uM), a mixture of 42 or 190 reverse primers (1.25uM each or 0.5uM each, respectively, for libraries corresponding to 53 or 219 pre-miRNAs, respectively). These primers contained the Illumina small RNA reverse primer appended to the sense sequence of the 5P arm of the precursor RNA (see Supplemental Data for full list of primer sequences), and Phusion DNA polymerase (Finnzymes) according to the manufacturer's instructions.
Illumina primers only, 19 cycles: 1% of the first PCR reaction was amplified with the forward primer above and the Illumina small RNA reverse primer, 5′-CAAGCAGAAGACGGCATACGA-3′, and Phusion DNA polymerase.
Thermo-cycling profile was as follows: 98°C for 30 sec followed by cycles of 98°C for 10 sec, 59°C for 30 sec, and 72°C for 15 sec.
PCR reactions were phenol-extracted and ethanol-precipitated.
High-throughput DNA sequencing and data analysis
Seventy-six or 36 nucleotide sequence reads were generated using an Illumina Genome Analyzer II and saved in the FASTQ format. Individual libraries were separated according to the barcode using a Perl script. Reads were matched to the precursor match file using a C program. This program scans each read against each candidate precursor match, starting from the loop into the 3P arm. It should be noted that sequencing occurs from the 3′ end of the miRNA precursor. Thus, the matching algorithm begins far into the read, scanning backward toward the first nucleotide, matching to a reverse complement list of precursor sequences. A minimum of 23 nt perfect match is required to indicate a match. Tail length, composition, and terminal nucleotide are counted. Raw data is deposited into a match file, with Phred scores, sequence name, and read sequence. A series of Perl scripts collated the raw data into text files for figure generation. Figures were generated in OpenOffice and R. Scripts, executables, and C source code are available upon request. Further discussion of bioinformatic analysis is provided in the Supplemental Material.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Hammond, Randell, and Hayes lab for advice and support. Work was funded by the NIH (NIGMS and ARRA) and by support from the Department of Cell and Developmental Biology, UNC. Sequence data uploaded to the NCBI-SRA, accession number SRP005265.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2713611.
REFERENCES
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. 2011. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Mendell JT 2007. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215–239 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD 2005. microPrimer: The biogenesis and function of microRNA. Development 132: 4645–4652 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS 2010. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hutvagner G 2005. Small RNA asymmetry in RNAi: Function in RISC assembly and gene regulation. FEBS Lett 579: 5850–5857 [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function, and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL 2009. Adenylation of plant miRNAs. Nucleic Acids Res 37: 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligo-uridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM 2010. Emerging paradigms of regulated microRNA processing. Genes Dev 24: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Sawtell NM, Reuhl KR, Berg R, Craig JC, Jardine K, Lessard JL, McBurney MW 1990. Smooth muscle actin expression during P19 embryonal carcinoma differentiation in cell culture. J Cell Physiol 142: 89–98 [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault F, Sismour AM, Church GM 2008. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat Methods 5: 777–779 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]