Mitochondrial pre-mRNA editing in kinetoplastid protozoans requires the use of so-called guide RNAs. However, the details of guide RNA biogenesis are largely unknown. This paper identifies and characterizes a novel endoribonuclease, mRPN1. mRPN1 is shown to be in some way involved in guide RNA maturation. Intriguingly, mRNP1 shows similaritiesto bacterial RNase III, which cleaves dsRNAs.
Keywords: guide RNA processing, RNase III endonuclease, mitochondria; RNA editing, Trypanosoma brucei
Abstract
The mitochondrial genome of kinetoplastids, including species of Trypanosoma and Leishmania, is an unprecedented DNA structure of catenated maxicircles and minicircles. Maxicircles represent the typical mitochondrial genome encoding components of the respiratory complexes and ribosomes. However, most mRNA sequences are cryptic, and their maturation requires a unique U insertion/deletion RNA editing. Minicircles encode hundreds of small guide RNAs (gRNAs) that partially anneal with unedited mRNAs and direct the extensive editing. Trypanosoma brucei gRNAs and mRNAs are transcribed as polycistronic precursors, which undergo processing preceding editing; however, the relevant nucleases are unknown. We report the identification and functional characterization of a close homolog of editing endonucleases, mRPN1 (mitochondrial RNA precursor-processing endonuclease 1), which is involved in gRNA biogenesis. Recombinant mRPN1 is a dimeric dsRNA-dependent endonuclease that requires Mg2+, a critical catalytic carboxylate, and generates 2-nucleotide 3′ overhangs. The cleavage specificity of mRPN1 is reminiscent of bacterial RNase III and thus is fundamentally distinct from editing endonucleases, which target a single scissile bond just 5′ of short duplexes. An inducible knockdown of mRPN1 in T. brucei results in loss of gRNA and accumulation of precursor transcripts (pre-gRNAs), consistent with a role of mRPN1 in processing. mRPN1 stably associates with three proteins previously identified in relatively large complexes that do not contain mRPN1, and have been linked with multiple aspects of mitochondrial RNA metabolism. One protein, TbRGG2, directly binds mRPN1 and is thought to modulate gRNA utilization by editing complexes. The proposed participation of mRPN1 in processing of polycistronic RNA and its specific protein interactions in gRNA expression are discussed.
Introduction
Kinetoplastid protozoa include early-branched parasites that cause Leishmaniasis and Trypanosomiasis, such as Chagas' disease and sleeping sickness. Each cell has one mitochondrion and a single mitochondrial genome (kDNA) in an unusual catenated structure containing a dozen copies of a maxicircle and hundreds of different minicircles. Trypanosoma brucei maxicircle genes are tightly packed in both strands, including two rRNAs, 18 (edited and never-edited) mRNAs, and two guide RNAs (gRNAs). rRNAs and mRNAs are similar to those in other mitochondria, but the former are the smallest known examples in eukaryotes, and 12 mRNAs require editing. U insertion alone may account for more than half of the final sequence in some cases (Supplemental Fig. S1A). Since several maxicircle genes overlap, the formation of mature ends must be controlled (Clement et al. 2004). Each T. brucei minicircle encodes three to five gRNAs on only one strand, but low-level transcription of an unknown function in the complementary (antisense) strand has been detected (Supplemental Fig. S1B; Aphasizheva and Aphasizhev 2010). There are about 80–100 different minicircle sequence classes in this species (Simpson et al. 2000; Hong and Simpson 2003; Ochsenreiter et al. 2007), with no correlation between editing and expression (Koslowsky et al. 1992). Each gRNA gene may be able to initiate transcription, producing a polycistronic precursor (pre-gRNA) extending past downstream gRNAs. Maturation of the 5′-most gRNA is consistent with its detection via capping of the 5′ triphosphate in the primary transcript, implying post-transcriptional 3′ cleavage of precursor sequence. This 3′ processing would be followed by the addition and trimming of a short U tail. (Blum and Simpson 1990; Grams et al. 2000; Clement et al. 2004; Aphasizheva and Aphasizhev 2010; Zimmer et al. 2011). However, the precise structure of pre-gRNAs, including their transcription initiation and termination sites, and processing mechanisms are unknown.
U-insertion/deletion editing is catalyzed by the extensively studied multiprotein RNA editing core complex (RECC; the 20 S editosome or L-complex). Several mRNAs are edited at over a hundred sites, and at each site, editing starts by a nuclease cleavage directed by a complementary gRNA (Rusche et al. 1997; Stuart et al. 2005; Li et al. 2009; Simpson et al. 2010). There are three accepted specialized editing endonucleases, REN1, REN2, and REN3 (Carnes et al. 2005; Trotter et al. 2005; Kang et al. 2006; Simpson et al. 2010), although only REN1 has been produced in an active recombinant form (Kang et al. 2006). RENs have a conserved RNase III domain and two domains of unknown function (Worthey et al. 2003). After cleavage, U-addition or U-deletion occurs, followed by re-ligation. Since one gRNA directs the editing of only a few sites, multiple gRNAs are needed to edit an entire mRNA.
Several ancillary mitochondrial factors, often in multiprotein complexes, copurify with editing complexes via unidentified RNA linkers. Some factors affect editing indirectly, for example, by impacting the level of pre-mRNA precursors or the turnover of gRNA or mRNA in vivo, or possibly through associated activities identified in vitro: RNA annealing, unwinding, or 3′ remodeling (Aphasizhev et al. 2002; Pelletier and Read 2003; Mingler et al. 2006; Etheridge et al. 2008; Fisk et al. 2008; Hashimi et al. 2008; Panigrahi et al. 2008; Weng et al. 2008; Koslowsky 2009; Aphasizheva and Aphasizhev 2010; Hernandez et al. 2010). Finally, a mitochondrial activity was found to catalyze 3′ processing of polycistronic pre-gRNAs, but the responsible nucleases are not known (Grams et al. 2000).
Here, we report an endonuclease mRPN1 (mitochondrial RNA precursor-processing endonuclease 1) that is involved in gRNA metabolism. mRPN1 is a close homolog of the REN endonucleases but is not present in RECC or in the known multiprotein ancillary factors. Cleavage by recombinant mRPN1 requires Mg2+, a critical catalytic carboxylate, and its specificity is reminiscent of bacterial RNase III toward duplexed RNA. In contrast, RECC specifically cleaves at single/double-stranded mRNA junctions with gRNA. Inducible knockdown of mRPN1 results in the loss of gRNAs and accumulation of precursor sequences (pre-gRNAs), consistent with a role in processing. Finally, cell purified mRPN1 has nuclease-resistant interactions with three other proteins that were previously identified in relatively large MRB (mitochondrial RNA binding)-related complexes, which do not have mRPN1, and appear to have multiple roles in mitochondrial RNA metabolism (Hashimi et al. 2008; Panigrahi et al. 2008; Weng et al. 2008; Hernandez et al. 2010; Ammerman et al. 2011). One of these factors (TbRGG2) directly binds mRPN1 and was previously linked with gRNA utilization during editing, which may include entry into the editing pathway. The proposed role of mRPN1 and its protein associations in gRNA metabolism is discussed.
Results
mRPN1 is a homolog of the REN endonucleases, but mRPN1 and editing complexes exhibit fundamentally different specificity
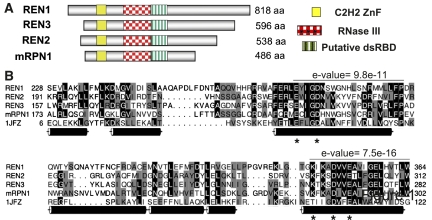
A search of the T. brucei genome revealed mRPN1 (Tb11.01.0150) as a homolog of REN editing endonucleases (Fig. 1). The overall average amino acid sequence homology of mRPN1 with these protein homologs is ∼16%–18% (Supplemental Fig. S2A). mRPN1 and REN proteins are (class I) RNase III family members, which also include Drosha (class II) and Dicer (class III) involved in the maturation of miRNAs (both) and the creation of siRNAs (the latter) in a number of organisms. The RNase III domain of mRPN1 is between 27% and 35% identical to that of RENs (Supplemental Fig. S2A). This domain includes four invariant catalytic residues and has been associated in most family members with double-stranded RNA (dsRNA) cleavage activity (Blaszczyk et al. 2001; Pertzev and Nicholson 2006). RENs and mRPN1 share two other domains of unknown function and a predicted N-terminal mitochondrial leader (Fig. 1; Supplemental Fig. S2B; data not shown). These domains represent a significantly divergent U1-like zinc finger, with the signature C2H2 residues, and a possible dsRBD (Worthey et al. 2003). We found an mRPN1 ortholog in Trypanosoma cruzi, but not in the related Leishmania genus.
FIGURE 1.
Similar domain organization of mRPN1 and REN proteins. (A) Aligned RNase III endonuclease domain and two other conserved sequences representing a U1-like C2H2 zinc finger and a putative dsRBD, which is more divergent in mRPN1 and REN1. Other regions have no significant homology. (B) Multiple sequence alignment of the RNase III domain, including the prototype domain of A. aeolicus, shows the most conserved motifs, their e-values, four invariant catalytic carboxylates, and a lysine residue (*) that corresponds to a catalytic residue in the mouse Dicer (Zhang et al. 2004). Cylinders denote the α-helices in the crystal structure (1JFZ) of the A. aeolicus domain (Blaszczyk et al. 2001).
Importantly, mRPN1 was not detected in reported purifications of either native or tandem-affinity purified (TAP) editing complexes, whether RENs or other core subunits were tagged, indicating a distinction of function between mRPN1 and REN proteins (Rusche et al. 1997; Panigrahi et al. 2006, 2007; Hernandez et al. 2008).
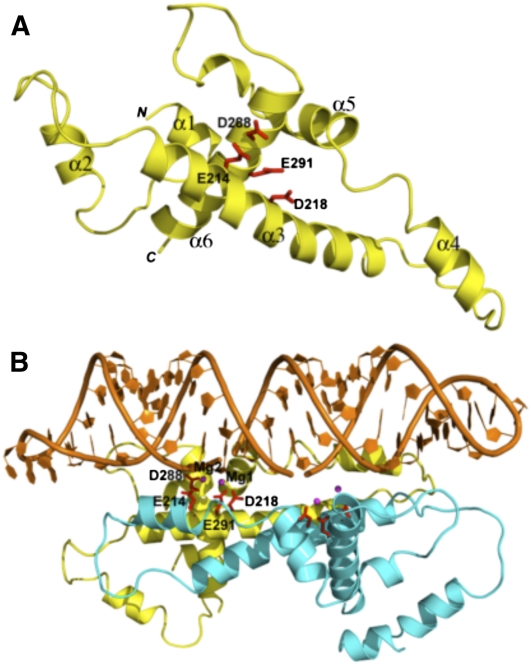
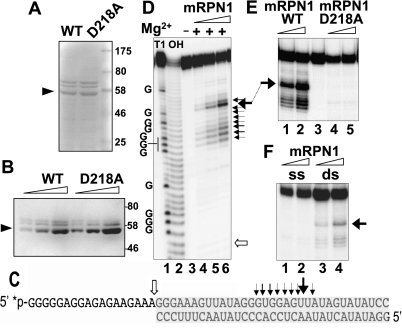
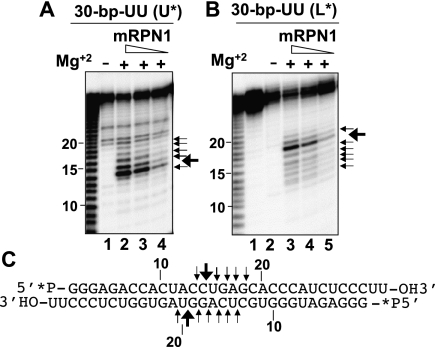
We investigated the possibility that mRPN1 may be processive. We first performed structure homology-modeling of the putative nuclease domain, using the Aquifex aeolicus (Aa) RNase III and RNase III–dsRNA structures as templates (Blaszczyk et al. 2001; Gan et al. 2008). The alignment showed that the RNase III fold indeed brings the catalytically important mRPN1 side-chains in a cluster, mimicking the catalytic Aa–RNase III structure, including the potential to dimerize (Fig. 2). Based on this information, we expressed in bacteria tagged mRPN1 wild-type (WT) or with a substitution of the invariant Aspartate 218 for Alanine (D218A) and tested these proteins for activity in vitro. A bacterially expressed recombinant protein of the expected size reacted with anti-mRPN1 antibodies, and size exclusion indeed indicates that mRPN1 forms a dimer (Fig. 3A,B; Supplemental Fig. S3). WT mRPN1 catalyzed a major cleavage and a few nearby cuts within the duplex in a model dsRNA with a 5′ overhang (Fig. 3C). This activity required Mg2+, both substrate strands, and D218 (Fig. 3D–F). In contrast to the mRPN1 activity, purified editing complexes cleave specifically at the dsRNA–ssRNA junction of this minimized substrate (Hernandez et al. 2008). Also, mRPN1 cleaved both strands of a short 30-bp dsRNA, which has been tested with RNase III and Dicer in vitro (Zhang et al. 2004). Interestingly, the mRPN1 specificity with the latter substrate was closer to RNase III than Dicer, and this activity also required Mg2+ (Fig. 4). Like these enzymes, mRPN1 induced a major cleavage that leaves 2-nucelotide (nt) 3′ overhangs, as well as minor nearby cuts in the duplex. Thus, mRPN1 is a dsRNA-dependent endonuclease that exhibits an RNase III–like specificity.
FIGURE 2.
Homology model of mRPN1. The A. aeolicus (Aa)-RNase III (PDB entry 1JFZ) and the Aa-RNase III-dsRNA (2NUF) structures as starting models (Blaszczyk et al. 2001; Gan et al. 2008). (A) Ribbon diagram of the mRPN1 RNase III domain with helices (α1–α6). The four conserved catalytic side-chains (E214, D218, D288, E291, clustered on α3 and α6) are in red. (B) Model of the mRPN1–dsRNA complex. One subunit is in yellow, the other in cyan, and the dsRNA in orange. The acidic catalytic side-chains and magnesium ions (labeled in one cleavage site only) are in red and magenta, respectively. Size exclusion chromatography of recombinant mRPN1 indicates a dimer, as expected for RNase III family members (see ahead Supplemental Fig. S3).
FIGURE 3.
mRPN1 cleavage within duplexed RNA. Coomassie blue (A) and Western (B) of 57.3 kDa His-tag purified mRPN1 wild-type (WT) and D218A mutant from bacteria. Two higher molecular weight bands present in this preparation but not after further purification by gel filtration appear to be contaminants (see Supplemental Fig. S3). (C) Model 33-bp duplex with a 18-nt 5′ overhang. Cleaved bonds and relative efficiency (black arrows). Editing complexes cleave the ssRNA–dsRNA junction in this minimized substrate (white arrow) (Hernandez et al. 2010). (D) Cleavage assay with ∼10 fmol of gel-purified dsRNA (32P = *), ∼1.2, 2.4, or 3.6 pmol of recombinant WT mRPN1 and Mg2+. A −Mg2+ lane has 3.6 pmol of WT mRPN1. Alkaline hydrolysis or T1 RNase cuts 3′ at Gs. (E) Cleavage with increasing mRPN1 WT or D218A. Lane 3 is RNA input. (F) Cleavage assay with WT mRPN1 without or with the bottom strand, i.e., using single-stranded (ss) or double-stranded (ds) RNA, respectively. The same major cut reproduced in all assays is indicated by an arrow.
FIGURE 4.
RNase III–like specificity of mRPN1. (A,B) Processing of a 30-bp dsRNA with a 5′ 32P on the upper (30 bp U*) or lower (30 bp L*) strand. Cleavage sites by increasing mRPN1 and relative efficiency (arrows). (C) dsRNA substrate and cleaved bonds. Reactions ±Mg2+. Lane 1 in B is RNA input.
mRPN1 down-regulation results in a loss of gRNA and accumulation of pre-gRNA transcripts
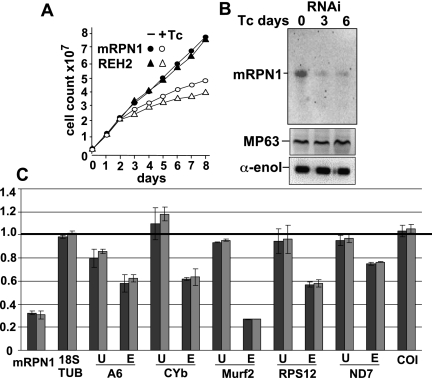
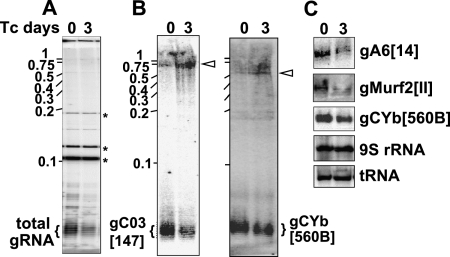
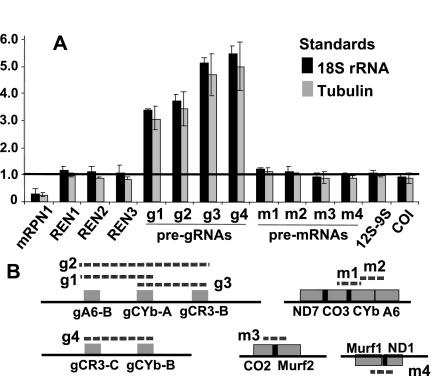
We examined the role of mRPN1 in trypanosomes via RNAi-mediated repression through the tetracycline induction of a dsRNA construct targeting mRPN1. This RNAi construct induced a growth defect that became apparent by day 3 and continued over 10 d without recovery. Similar results were obtained with independent clonal lines, a hairpin RNAi construct for mRPN1, and an RNAi construct for another factor, REH2, which is also important in gRNA metabolism (Fig. 5A; Hashimi et al. 2009; Hernandez et al. 2010; data not shown). mRPN1 was diminished, but control mitochondrial and cytosolic proteins were unaffected after several days (Fig. 5B). Since cells appeared normal at day 3, implying limited or no secondary effects, we performed all experiments at this post-induction time. Real-time RT-PCR (qPCR) of several editing substrates showed a decreased abundance of edited mRNAs upon mRPN1 suppression (Fig. 5C). Changes in unedited mRNAs were not statistically significant, as in other studies showing sublethal editing effects, including with our REH2 construct above, where unedited RNAs are not always evidently affected (Supplemental Fig. S4; Hashimi et al. 2008; Acestor et al. 2009). The never-edited mRNA COI was not affected, while the A6 mRNA may only be slightly decreased. We asked whether gRNA biogenesis was affected. Pre-gRNA transcripts are rarely detected in Northern blots or radioactive capping of a 5′ triphosphate, due to their low steady-state level and apparent short half-life (Riley et al. 1994; Clement et al. 2004; Aphasizheva and Aphasizhev 2010). Examination of the total gRNA pool, which is abundant and readily detected in the capping assay, showed an over 40% decrease upon mRPN1 RNAi (Fig. 6A). Induced cells accumulated a few large RNA species, which were only detected at long exposure times, suggesting heterogeneous precursor sizes (data not shown). Although potential pre-gRNAs are generally difficult to detect, examples of the blots exhibiting accumulation of long species after induction are shown in Figure 6B. Specific minicircle gRNAs and a maxicircle gRNA examined were evidently decreased, whereas mitochondrial 9S rRNA and an imported tRNA were not affected (Fig. 6B,C). Previous studies have used qPCR to detect low-level unprocessed pre-mRNA fragments (Acestor et al. 2009). We established similar assays for pre-gRNAs (Fig. 7). Importantly, polycistronic pre-gRNA fragments spanning two or three gRNA genes in different minicircles examined increased three- to fivefold after mRPN1 RNAi. In contrast, unprocessed mRNA precursors appeared largely normal. These pre-gRNAs were not affected by down-regulation of REH2 (Supplemental Fig. S4), indicating that mRPN1 and REH2 have different roles in gRNA expression. Importantly, despite a ∼70% repression of mRPN1, all three RENs appear unaffected (Fig. 7A), implying specific RNAi rather than off-targets on related proteins and suggesting that catalysis of editing is normal in these cells.
FIGURE 5.
mRPN1 knockdown induced with a dsRNA construct inhibits RNA editing. (A) Growth curve of trypanosomes (±) Tc-induction. A knockdown of REH2 was included as control. (B) Immunoblots of whole extract. MP63 (KREPA2; mitochondrial) and α-enolase (cytosolic) markers. (C) qPCR in duplicates at day 3 of induction: edited (E), unedited (U), and never-edited COI and mRPN1 mRNAs, normalized to tubulin (black) or nuclear 18S rRNA (gray). Uninduced cells (1.0). Bars above 1 (increase) or below 1 (decrease) after induction.
FIGURE 6.
mRPN1 down-regulation induced with a dsRNA construct results in loss of gRNA transcripts. Total RNA at day 0 and day 3 of induction. (A) Capping of total gRNA with guanylyl transferase and γ-32P ATP. The characteristic gRNA size heterogeneity is due to a variable 3′ U tail. A typical artifact in the assay is used to normalize the samples (*). (B) Northern blot of minicircle mature gRNAs gCO3[147] and gCYb[560B] and extended RNA species (arrowhead). (C) Blots of mature gRNAs (minicircle) gA6[14] and gCYb[560B], and (maxicircle) mMurf2[II], mitochondrial 9S rRNA, and imported tRNA controls. RNA markers, 100–1000 nt.
FIGURE 7.
mRPN1 down-regulation induced with a dsRNA construct induces accumulation of pre-gRNAs. (A) qPCR of mRPN1, REN, and mitochondrial RNA precursors at day 3 of induction. (B) Minicircles fragments (g1–g4) with two or three gRNAs and maxicircle fragments (m1–m4) with gene clusters from both strands (dashed lines). We confirmed the size and sequence of all amplicons. The assays were normalized as in Figure 5.
Together, these data suggest that mRPN1 is involved in the metabolism of gRNAs, presumably during processing of nascent precursors, since gRNAs decrease whereas extended unprocessed transcripts accumulate after induction of mRPN1 down-regulation.
mRPN1 associates with a subset of proteins typically found in relatively large MRB-related complexes
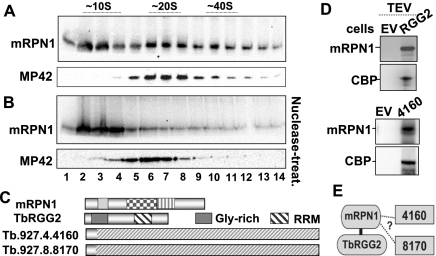
We found that native and tagged mRPN1 in mitochondrial lysates exhibit equivalent broad sedimentation, with clear accumulation in low density (top fractions) and a shallow peak in fractions containing about 20 S editing complexes. However, most mRPN1 shifted to low-density fractions, away from editing complexes, after nuclease treatment or affinity-purification (Fig. 8A,B; data not shown). We performed mass spectrometric analyses of TAP mRPN1 using about 20 S fractions as the starting extract. This purification helps remove most of the protein in the extract, which lies near the top of the gradient. As explained above, the tag has no discernible effect on sedimentation, and most mRPN1 associations in the extract break down upon purification. We found three nuclease-resistant associations, including TbRGG2, which has an RNA-binding domain, and the ∼80% identical Tb927.8.8170 and Tb927.4.4160, which have no recognizable conserved domains (termed here 8170 and 4160) (Fig. 8C; Supplemental Table S1). Reciprocal purifications confirmed the mRPN1 association with both TbRGG2 and 4160 (Fig. 8D). 8170 was not tested. Notably, these three proteins, but not mRPN1, are components of relatively large MRB-related complexes (Supplemental Table S1), which are thought to represent a group of protein assemblies with a core scaffold of at least six to seven subunits and multiple variable interactions (Hernandez et al. 2010; Ammerman et al. 2011). We performed a yeast two-hybrid analysis of all four proteins tested in fusions with Gal4 activation and DNA binding domains. Interestingly, mRPN1 bound to TbRGG2 in one orientation but not to the other two proteins in either orientation (depicted in Fig. 8E; Supplemental Fig. S5). As expected, mRPN1 did not interact with several components of MRB-related complexes included as the controls (Supplemental Fig. S5). Finally, tagged mRPN1 copurified with editing subunits and mitochondrial ribosomal proteins via unidentified RNA-dependent associations (data not shown), as has been observed with several reported mitochondrial factors (Weng et al. 2008; Hernandez et al. 2010). However, the biological relevance of these interactions was not addressed.
FIGURE 8.
mRPN1 nuclease-resistant associations, and yeast two-hybrid analysis. Western blots of endogenous mRPN1 and MP42 (KREPA3), which is a core subunit of RECC at about 20 S, in glycerol gradients of mitochondrial extracts, either untreated (A) or treated (B) with RNases A, T1, and V1 and micrococcal nuclease (nuclease treat.). Narrow slices of the same blot were used for MP42, which migrates just below mRPN1. Fraction 1 is the top of the gradient with sedimentation standards indicated. (C) Protein factors identified in nuclease-resistant TAP-purified mRPN1. (D) Reciprocal purifications of tagged RGG2 and tagged 4160. TEV eluates examined in Western blots with mRPN1 and anti-CBP antibodies. (E) Cartoon indicating a direct interaction of mRPN1 with RGG2 in a yeast two-hybrid analysis (Supplemental Fig. S5). The study did not show mRPN1 interaction with the other two protein fusions.
In summary, the endonuclease mRPN1 directly binds with TbRGG2 and exhibits a nuclease-resistant association with two more proteins, 4160 and 8170, previously found in reported MRB-related complexes. Since mRPN1 is not present in the latter complexes, these observations imply that TbRGG2, 4160, and 8170 may exhibit a specialized association with mRPN1, as well as alternative interactions with protein arrays in MRB-related complexes.
Discussion
This study characterized mRPN1, the first identified mitochondrial endonuclease in a mitochondrial RNA processing pathway other than RNA editing. Several lines of evidence indicate that mRPN1 is an RNase III endonuclease involved in the gRNA metabolism in T. brucei. First, mRPN1 has all the invariant catalytic residues of RNase III enzymes in a cluster mimicking a typical catalytic domain in a model structure. Second, recombinant mRPN1 cleavage requires Mg2+ and a conserved side-chain (D218). Moreover, its bacterial RNase III–like specificity toward duplexed RNA, including the generation of 2-nt 3′ overhangs, and size exclusion are indicative of a dimeric structure, as expected for RNase III enzymes. Third, inducible knockdown results in the loss of gRNAs and the accumulation of precursor sequences (pre-gRNAs), consistent with a role in processing. Interestingly, the apparent absence of mRPN1 in Leishmania may be tied to the fact that minicircles in these organisms encode a single gRNA (Sturm and Simpson 1991). In such case, Trypanosoma may have evolved specialized functions, including mRPN1, in the metabolism of gRNA polycistrons. Nothing is known about how gRNA biogenesis in Leishmania takes place.
In previous studies, the repression of RET1, which controls RNA stability via 3′ uridylylation, caused a loss of mature transcripts and significant accumulation of gRNA, rRNA, and select mRNA precursors (Aphasizheva and Aphasizhev 2010). Also, repression of GRBCs (GAPs) and REH2, proposed to modulate gRNA stability in MRB-related complexes, resulted in the loss of gRNA (Weng et al. 2008; Hashimi et al. 2009; Hernandez et al. 2010). Compared to the RET1-associated phenotype, mRPN1 down-regulation decreased the steady-state level but not the size of mature gRNA transcripts. This suggests that 3′ uridylylation is not affected by mRPN1. In line with this notion, the accumulation of precursors appeared modest compared to the observed reduction in gRNA. Accordingly, most previous blotting analyses of gRNA, including some of our own, have failed to detect pre-gRNA species (Blum and Simpson 1990; Riley et al. 1994; Clement et al. 2004; Aphasizheva and Aphasizhev 2010). Importantly, qPCR analysis overcomes this limitation as we showed for several gRNA precursor fragments. Thus, pre-gRNAs seem short-lived irrespective of mRPN1 expression. In an early report, metabolic labeling studies showed a fast turnover of mitochondrial rRNA precursors (Michelotti et al. 1992). This is most likely true for most, if not all, nascent precursor transcripts in mitochondria.
Although mRPN1 is a close homolog of the REN endonucleases in RNA editing, it failed to cleave a single/double-stranded junction and is not a subunit of RECC, consistent with this novel protein exhibiting a very different biological role. Since the mRPN1 cleavage specificity within a duplex region is reminiscent of bacterial RNase III processing mechanisms, this enzyme may participate in gRNA biogenesis through the specific recognition and cleavage of dsRNA-rich determinants in endogenous pre-gRNAs. Our current studies did not address the sequence specificity of mRPN1 on native transcripts. However further analyses of mRPN1 and cell-purified mRPN1, which exhibits detectable activity in our basic cleavage assays (data not shown), using additional model substrates will be important. A previous study reported that the T. brucei mitochondrial extract accurately processes the 5′-most gRNA in synthetic polycistronic pre-gRNA through 3′ cleavage, and proposed that the downstream sequence is degraded (Grams et al. 2000). It is possible that mRPN1 is associated with this processing, although additional cleavage and 3′ trimming may involve other nucleases. In line with the dsRNA specificity of mRPN1, targeted dsRNA-rich determinants may be provided through fold-back configurations of long pre-gRNAs. Sequence, structural determinants, or protein binding (i.e., anti-determinants) may protect the mature transcript from further cleavage, as seen with RNase III enzymes in other systems (Dasgupta et al. 1998). Alternatively, previously observed endogenous antisense RNA in minicircles could anneal with pre-gRNAs to direct the processing (Aphasizheva and Aphasizhev 2010). The minicircle and maxicircle gRNAs examined were similarly reduced by mRPN1 repression, and their precursors appear to have similar sizes (Aphasizheva and Aphasizhev 2010). This implies that gRNAs of different genomic origin are processed by similar mechanisms involving mRPN1. Also, since gRNAs are presumed to be primary transcripts, an intriguing question is whether their 5′ triphosphate is a determinant specifically recognized by mRPN1 during cleavage or by other factors involved in downstream steps in gRNA expression such as 3′ remodeling, turnover, or utilization by editing complexes.
We found a stable association of mRPN1 with three proteins previously found in MRB-related complexes, which do not include mRPN1. Particularly interesting is the direct mRPN1 interaction with one of these factors, TbRGG2. This protein is thought to play a role in the control of gRNA utilization by (or possibly entry into) RECC through undefined mechanisms that may involve annealing and unwinding (Ammerman et al. 2010). Another study proposed that its repression leads to accumulation of unprocessed mRNA precursors (Acestor et al. 2009), implying that this protein may be multifunctional. Other subunits of MRB complexes have been linked with the binding and stability of gRNAs and the processing of some mRNA and rRNA precursors (Hashimi et al. 2008; Panigrahi et al. 2008; Weng et al. 2008; Acestor et al. 2009; Hernandez et al. 2010). Since MRB complexes seem to represent a collection of compositionally related ribonucleoprotein particles (Hernandez et al. 2010; Ammerman et al. 2011), it is possible that TbRGG2, 4160, and 8170 alternatively interact with mRPN1 (i.e., in mRPN1 complexes) or with variable protein arrays in MRB complexes. Moreover, since mRPN1 but not TbRGG2 is involved in gRNA biogenesis, gRNAs may be shuttled by TbRGG2, 4160, and 8170, between mRPN1 complexes and downstream MRB complexes. In this “handover” model, depicted in Supplemental Figure S5C, the interactions of MRB complexes with RECC could in turn regulate the subsequent entry of gRNA into the editing pathway. Additional mitochondrial factors in a higher-order network of RNA-mediated interactions including RECC and MRB complexes (RECC* or L*) have been proposed to control 3′ remodeling and annealing of gRNAs with unedited mRNAs (Osato et al. 2009; Aphasizheva and Aphasizhev 2010; Hernandez et al. 2010; Li et al. 2011). An association with RECC* may explain the heterogeneous sedimentation of mRPN1 in mitochondrial lysates (Fig. 8A). Alternative models can be also proposed, including the possibility that TbRGG2 (and 4160 or 8170) may be involved in the metabolism of mitochondrial transcripts not affected by mRPN1 (e.g., mRNA and rRNA) or putative mRPN1-responsive RNAs other than gRNAs.
Besides mRPN1, only recombinant REN1 was shown to be processive in vitro (Kang et al. 2006), although substitutions of RNase III signature residues in each REN, like D218A in mRPN1, hindered the activity of editing complexes in vitro and in vivo, indicating that RENs are also processive (Carnes et al. 2005; Trotter et al. 2005). Class I RNase III family members typically contain a well-conserved dsRBD just downstream from their single RNase III domain. Importantly, rare exceptions where this dsRBD conservation is undetected or weak include the known cytosolic, nuclear (Shi et al. 2006; Patrick et al. 2009), and mitochondrial RNase III family members in kinetoplastids (Stuart et al. 2005). Interestingly, the proposed ZnF domain of mRPN1 and RENs is also weakly conserved, although it contains the signature C2H2 residues (Supplemental Fig. S2). The highly divergent primary sequence of these domains may reflect functional specialization or the evolutionary history of these early-branched eukaryotes.
Materials and Methods
Computational methods
Multiple sequence alignments were performed using ProbCons (Do et al. 2005) between REN1, REN2, REN3, and mRPN1 on individual regions with manually determined domain boundaries and on full sequences. These alignments were formatted using Alscript (Barton 1993), with similarly shaded residue groups [A, G, I, L, P, V], [R, H, K], [N, Q, S, T], [D, C, E], [M], and [F, W, Y] and starting and ending positions shown for each protein. Conserved ungapped motifs of REN1, REN2, REN3, and mRPN1 were identified using MEME (Bailey and Elkan 1994) with an e-value cutoff of 0.01 on unaligned full sequences (instead of on individual regions), so that these motifs represent conservation without domain boundary restrictions. Sequence identity was computed from its pairwise alignment by counting the percentage of columns that have identical residues, and sequence identity of a region was computed from its multiple alignment by the percentage of aligned pairs of identical residues over all pairs of proteins (aligned pairs that contain only gap characters were ignored).
Plasmid constructs and expression in cells
To prepare a TAP-mRPN1 construct, we amplified the entire ORF, which has no introns, directly from genomic DNA (procyclic 29-13 cells) using a proofreading thermostable polymerase mix (AccuTaq-Sigma) and oligonucleotides mRPN1-for and mRPN1-rev1. This product was cloned into pSC-A (Stratagene), digested with HindIII and BglII, and subcloned into pLew79-ada-TAP (Hernandez et al. 2008). For RNAi studies, a C-terminal mRPN1-specific fragment was cloned into p2T7-177 to expresses a dsRNA product (Wickstead et al. 2002). Alternatively, inverted copies of an mRPN1 fragment were cloned into pLew100 to express a hairpin construct (Wirtz et al. 1999). For stable expression in trypanosome, all plasmids were linearized, typically with NotI, or EcoRV in the case of pLew100, and transfected in procyclic 29-13 cells (Wirtz et al. 1999). Expression in all transgenic trypanosomes was induced with tetracycline at 1 μg/mL. For antibody production, the mRPN1 ORF was cloned into pET30b and expressed in Escherichia coli BL21 (DE3) by induction with 1 mM IPTG. All plasmid constructs were confirmed by DNA sequencing.
Protein analysis and purification
TAP purifications were conducted essentially as reported (Rigaut et al. 1999) with some modifications. Sedimentation fractions were obtained from freshly made mitochondrial extracts in 10%–30% glycerol gradients (Cruz-Reyes and Sollner-Webb 1996). Mitochondrial extracts were prepared in presence of DNase I (Harris and Hajduk 1992). Catalase was used as 10 S marker in concurrently run gradients. REAP-1 was used as 40 S marker in Western blots of the extract fractions in Figure 8A, as in a previous study (Halbig et al. 2004). Rabbit polyclonal serum against the recombinant purified mRPN1 fragment was produced by the LARR animal facilities at Texas A&M University. Specific antibodies were conjugated to protein A–Dynabeads (Invitrogen) for immunoprecipitation (IP) of the endogenous and ectopic mRPN1. Pull-downs of ectopic mRPN1 used anti-rabbit IgG Dynabeads (Invitrogen). All IP washes were performed at 150 mM KCl. For mass spectrometry analyses, the antibodies were cross-linked to the beads with 25 mM DMP (dimethylpimelimidate) in 0.2 M triethanolamine (pH 8.2), and protein identification was conducted as previously reported (Wohlschlegel 2009; Hernandez et al. 2010). Proteins were considered present in a sample if at least two peptides were identified per protein using a peptide-level false-positive rate of 5% as determined using a decoy database strategy (Elias and Gygi 2007).
Northern blotting
Total RNA (20 μg) from trypanosomes was treated with DNase I, dissolved in loading dye (90% formamide, 10 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue), heated for 5 min to 65°C, and resolved by 8 M urea/9% PAGE. RNA was transferred to bright star plus membrane (Ambion) by electrotransfer in 0.5× TBE for 1.5 h at 100 V and UV cross-linked twice at 0.12 J/Cm2. DNA oligonucleotide probes (10 pmol) were labeled with T4 kinase and 50 mCi of [γ-32P] ATP (6000 Ci/mmol). Overnight hybridizations (30°C) were performed in ULTRAhyb-oligo solution (Ambion) with 6 × 106 cpm/mL of radiolabeled probe. The membrane was washed three times with 50 mL of 4× SSPE, 0.5%SDS at 25°C and was exposed to phosphor screen. gRNA-labeling assays of total RNA were performed as reported (Hernandez et al. 2008). Protein samples coupled to Dynabeads were treated with the following nuclease mix at the given final concentration: RNase A (0.1 U/mL), T1 (0.125 U/mL), V1 (0.001 U/mL), and micrococcal nuclease (0.03 U/mL) for 60 min in ice. Quantitation of the density of bands in Northern blots was performed with the software Quantity one (Bio-Rad).
Structure-modeling studies
The homology modeling of the mRPN1 endonuclease domain was done using the SWISS-MODEL protein structure homology-modeling server (http://swissmodel.expasy.org/) in the alignment mode (sequence alignment was provided) with the Aa–RNase III structure (Protein Data Bank [PDB] entry 1JFZ) as the starting model (Blaszczyk et al. 2001). The resulting model from the server was used to construct the mRPN1–dsRNA complex based on the Aa–RNase III–dsRNA structure (PDB entries 2EZ6, 2NUF) (Gan et al. 2008). Manual adjustments, using program COOT, were carried out to remove steric clashes and the adjusted model was subject to energy minimization using the CNS program suite (Brunger and Rice 1997; Emsley and Cowtan 2004). The stereochemical quality of the final model was checked using PROCHECK (Laskowski et al. 1993) and WHATIF (Vriend 1990).
Endonuclease cleavage assay in vitro
The synthesized top and bottom strands of the dsRNA substrate were annealed and the duplex isolated from a native gel as described (Hernandez et al. 2008, 2010). The dephosphorylated top strand was 5′-end labeled with [γ-32P]ATP (6000 Ci/mmol). Individual reactions of 20 μL were set up with ∼50 cps (∼10 fmol) of dsRNA; reaction buffer (final concentration: 10 mM KCl, 1 mM EDTA at pH 8, 25 mM Tris-HCl at pH 8, and 5% glycerol); 1 U of SUPERase, with or without 2 mM MgCl2; and the indicated amount of recombinant mRPN1. One hundred nanograms of protein was estimated to contain ∼1.2 pmol of mRPN1 based on a molecular weight of 54 kDa and at least 70% purity. After incubation for 60 min at 26°C, the mixture was deproteinized and the RNA resolved in 9% acrylamide, 7 M urea gels. Each panel in the figures corresponds to one of two replica series performed simultaneously (i.e., one experiment). At least two independent experiments were performed for each figure, and the data shown are representative.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article. Detailed procedures for real-time RT-PCR, site-directed mutagenesis, the preparation of recombinant mRPN1 and anti-mRPN1 antibodies, yeast two-hybrid and oligonucleotide sequences used in this study are described in the Supplemental Methods section.
ACKNOWLEDGMENTS
We thank Damian Terry and Vikas Kumar in our laboratory for careful comments and discussion on the manuscript. Ricardo Zavala provided technical assistance with the preparation of plasmids. Craig Kaplan in our department provided detailed advice and access to equipment for real-time PCR. Ken Stuart and Paul Michels kindly provided monoclonal antibodies against core subunits of editing complexes and anti-serum against enolase, respectively. This work was supported by grants from the National Institutes of Health to J.C.-R. (GM067130, AI081529) and J.A.W. (GM089778) and by funding from the Jonsson Cancer Center at UCLA to J.A.W. and from the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to X.J.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2815911.
REFERENCES
- Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD 2009. The MRB1 complex functions in kinetoplastid RNA processing. RNA 15: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK 2010. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16: 2239–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Hashimi H, Novotna L, Cicova Z, McEvoy SM, Lukes J, Read LK 2011. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA 17: 865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648 [DOI] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R 2010. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol Cell Biol 30: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan CP 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Sys Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Barton GJ 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X 2001. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9: 1225–1236 [DOI] [PubMed] [Google Scholar]
- Blum B, Simpson L 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Rice LM 1997. Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol 277: 243–269 [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci 102: 16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement SL, Mingler MK, Koslowsky DJ 2004. An intragenic guide RNA location suggests a complex mechanism for mitochondrial gene expression in Trypanosoma brucei. Eukaryot Cell 3: 862–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Sollner-Webb B 1996. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci 93: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Fernandez L, Kameyama L, Inada T, Nakamura Y, Pappas A, Court DL 1998. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III—the effect of dsRNA binding on gene expression. Mol Microbiol 28: 629–640 [DOI] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15: 330–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R 2008. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J 27: 1596–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Ammerman ML, Presnyak V, Read LK 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem 28: 23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X 2008. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol 67: 143–154 [DOI] [PubMed] [Google Scholar]
- Grams J, McManus MT, Hajduk SL 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J 19: 5525–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbig K, De Nova-Ocampo M, Cruz-Reyes J 2004. Complete cycles of bloodstream trypanosome RNA editing in vitro. RNA 10: 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Hajduk SL 1992. Kinetoplastid RNA editing: in vitro formation of cytochrome b gRNA-mRNA chimeras from synthetic substrate RNAs. Cell 68: 1091–1099 [DOI] [PubMed] [Google Scholar]
- Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J 2009. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Panigrahi A, Cifuentes-Rojas C, Sacharidou A, Stuart K, Cruz-Reyes J 2008. Determinants for association and guide RNA-directed endonuclease cleavage by purified RNA editing complexes from Trypanosoma brucei. J Mol Biol 381: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J 2010. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem 285: 1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Simpson L 2003. Genomic organization of Trypanosoma brucei kinetoplast DNA minicircles. Protist 154: 265–279 [DOI] [PubMed] [Google Scholar]
- Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L 2006. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc Natl Acad Sci 103: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky DJ 2009. Complex interactions in the regulation of trypanosome mitochondrial gene expression. Trends Parasitol 25: 252–255 [DOI] [PubMed] [Google Scholar]
- Koslowsky DJ, Riley GR, Feagin JE, Stuart K 1992. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol Cell Biol 12: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM 1993. PROCHECK: a program to check stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L 2009. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci 106: 12306–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Herrera J, Zhou S, Maslov DA, Simpson L 2011. Trypanosome REH1 is an RNA helicase involved with the 3′-5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci 108: 3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Harris ME, Adler B, Torri AF, Hajduk SL 1992. Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processing and developmentally regulated expression. Mol Biochem Parasitol 54: 31–41 [DOI] [PubMed] [Google Scholar]
- Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ 2006. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol Biochem Parasitol 150: 37–45 [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T, Cipriano M, Hajduk SL 2007. KISS: the kinetoplastid RNA editing sequence search tool. RNA 13: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato D, Rogers K, Guo Q, Li F, Richmond G, Klug F, Simpson L 2009. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: in search of the editosome. RNA 15: 1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12: 1038–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Stuart KD 2007. Isolation and compositional analysis of trypanosomatid editosomes. Methods Enzymol 424: 3–24 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics 7: 534–545 [DOI] [PubMed] [Google Scholar]
- Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E 2009. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci 106: 17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Read LK 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertzev AV, Nicholson AW 2006. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res 34: 3708–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Riley GR, Corell RA, Stuart K 1994. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J Biol Chem 269: 6101–6108 [PubMed] [Google Scholar]
- Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16: 4069–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tschudi C, Ullu E 2006. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA 12: 2063–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Thieman OH, Savill NJ, Alfonzo JD, Maslov DA 2000. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci 97: 6986–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J 2010. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist 161: 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK 2005. Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30: 97–105 [DOI] [PubMed] [Google Scholar]
- Sturm NR, Simpson L 1991. Leishmania tarentolae minicircles of different sequence classes encode single guide RNAs located in the variable region approximately 150 bp from the conserved region. Nucleic Acids Res 19: 6277–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K 2005. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell 20: 403–412 [DOI] [PubMed] [Google Scholar]
- Vriend G 1990. WHAT IF: a molecular modeling and drug design program. J Mol Graph 8: 52–56 [DOI] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell 32: 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol 125: 211–216 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA 2009. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol 497: 33–49 [DOI] [PubMed] [Google Scholar]
- Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R 2003. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res 31: 6392–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zimmer SL, McEvoy SM, Li J, Qu J, Read LK 2011. A novel member of the RNase D exoribonuclease family functions in mitochondrial guide RNA metabolism in Trypanosoma brucei. J Biol Chem 286: 10329–10340 [DOI] [PMC free article] [PubMed] [Google Scholar]