Hepatitis C is a plus-strand RNA virus that replicates in the cytoplasm. It has previously been shown that viral replication is enhanced by the miRNA miR-122. Here it is demonstrated that Ago2 and miR-122 are recruited to lipid droplets to facilitate HCV replication. Conversely, suppression of HCV by siRNAs (and Ago2) appears to involve P-bodies. These results indicate that opposing activities of small RNAs take place in distinct cytoplasmic compartments.
Keywords: Argonaute 2, Dicer, RNAi, hepatitis C, miRNA-122, P-bodies
Abstract
The antiviral role of RNA interference (RNAi) in humans remains to be better understood. In RNAi, Ago2 proteins and microRNAs (miRNAs) or small interfering RNAs (siRNAs) form endonucleolytically active complexes which down-regulate expression of target mRNAs. P-bodies, cytoplasmic centers of mRNA decay, are involved in these pathways. Evidence exists that hepatitis C virus (HCV) utilizes host cellular RNAi machinery, including miRNA-122, Ago1-4, and Dicer proteins for replication and viral genome translation in Huh7 cells by, so far, nebulous mechanisms. Conversely, synthetic siRNAs have been used to suppress HCV replication. Here, using a combination of biochemical, transfection, confocal imaging, and digital image analysis approaches, we reveal that replication of HCV RNA depends on recruitment of Ago2 and miRNA-122 to lipid droplets, while suppression of HCV RNA by siRNA and Ago2 involves interaction with P-bodies. Such partitioning of Ago2 proteins into different complexes and separate subcellular domains likely results in modulation of their activity by different reaction partners. We propose a model in which partitioning of host RNAi and viral factors into physically and functionally distinct subcellular compartments emerges as a mechanism regulating the dual interaction of cellular RNAi with HCV RNA.
INTRODUCTION
Small noncoding RNAs are increasingly at the forefront of active research across biology, virology, and biomedical and drug discovery sciences. These RNAs are key regulators in a variety of cellular processes which, in a broader context, are referred to as RNA interference (RNAi). Through the action of genetically encoded microRNA (miRNAs), a distinct class of biological regulators with conserved functions, a cell modulates gene expression typically resulting in silencing of specific genes. RNAi is also known as a fundamental defense mechanism against viruses in plants and lower invertebrates. The question of whether the antiviral arm of RNA silencing was conserved in mammals—as in plants and invertebrates—has been intriguing researchers since the discovery of RNAi in mammalian systems and remains enigmatic (Cullen 2006; 2010; Lin and Cullen 2007; Umbach and Cullen 2009). The hepatitis C virus (HCV), a human pathogen classified within positive-strand RNA viruses propagating primarily in the liver, has emerged as an interesting case in terms of its interaction with the host cellular RNAi system. On the one hand, the HCV apparently evolved to utilize a highly abundant, well-conserved liver-specific miRNA-122 and key RNAi proteins, including Argonaute 1-4 and Dicer, for replication (Jopling et al. 2005), translation of the viral genome (Henke et al. 2008; Jangra et al. 2010b), and infectious virus production (Randall et al. 2007; Jangra et al. 2010b). On the other hand, the cellular RNAi machinery is capable of potently suppressing replication of HCV RNA after being seeded with the exogenous “anti-HCV” small interfering RNA (siRNAs) (Kapadia et al. 2003; Randall et al. 2003; Supekova et al. 2008). The exact mechanisms of these complex interactions remain to be better explained.
The current understanding of the RNAi molecular mechanisms suggests that Argonaute 2 (Ago2) protein and small, 20- to 24-nucleotide (nt) noncoding double-stranded miRNAs or siRNAs form endonucleolytically active complexes which down-regulate expression of target mRNAs. During formation of such RNA-induced silencing complexes (RISCs), one strand of siRNAs or miRNAs gets loaded into the Ago2 enzyme with the help of TAR RNA binding proteins (TRBPs) and Dicer proteins (Chendrimada et al. 2005; Meister et al. 2005; Joshua-Tor 2006; MacRae et al. 2008). This guide RNA strand targets RISC binding to mRNA, typically but not invariably in its 3′ UTR region, bearing a complementary sequence. Depending on the degree of a base pair match, RISC binding either initiates the endonucleolytic cleavage of mRNA by Ago2 (siRISC) or results in translational repression of the target transcript (miRISC). The RNAi repression pathway has been functionally linked to specialized cytoplasmic compartments known as P-bodies. P-bodies are nonmembrane-bound dynamic cytoplasmic centers highly enriched in mRNA turnover proteins (Eulalio et al. 2007; Kulkarni et al. 2010). Accumulation of Ago2-miRNA (Liu et al. 2005a,b; Bhattacharyya et al. 2006) and Ago2-siRNA (Jakymiw et al. 2005; Ikeda et al. 2006; Lian et al. 2007) complexes, together with their target transcripts in the P-bodies, is considered as a possible mechanism of temporal translational repression or final decay of impaired mRNAs. Impaired mRNA in P-bodies interacts with a host of proteins including Dcp1a, Dcp2, GW182, RCK/p54, and others. These interactions determine mRNA fate, usually resulting in its degradation.
In normal biology of liver cells, miRNA-122 was reported to regulate the mevalonate pathway and expression of genes involved in hepatic lipids and cholesterol metabolism (Krutzfeldt et al. 2005; Esau et al. 2006; Elmen et al. 2008; Norman and Sarnow 2010). In addition, miRNA-122 temporally represses host cationic amino acid transporter CAT-1 mRNA in cultured Huh7 cells (Chang et al. 2004), and this function was linked to sequestering of CAT-1 mRNA by miRNA-122 and Ago2 in P-bodies (Bhattacharyya et al. 2006). Whether P-bodies also play a role in regulation of HCV RNA by miRNA-122 is not understood. However, recent studies have shown that P-body proteins Rck/p54 (or DDX6 helicase) (Scheller et al. 2009; Jangra et al. 2010a), Lsm1-7, and PatL1 (Scheller et al. 2009) are required for the optimal replication of HCV RNA and translation of the viral genome. An existing model proposes that assembly of translationally repressed RNA-protein complexes containing the HCV RNA in P-bodies might be a mechanism sequestering and directing translationally inactive HCV RNA to allow a competing replication process (Beckham and Parker 2008). To gain insights into the mechanisms behind the dual regulation of HCV RNA by the cellular RNAi system and to understand P-bodies' roles in these processes, we examine here how key components of the RNAi machinery—Ago2, Dicer, miRNA-122, and synthetic anti-viral siRNA—distribute and interact inside Huh7 cells bearing replication of HCV RNA. We find that up-regulation and repression functions correlate with differential localization of these RNAi components into separate cellular compartments, P-bodies and lipid droplets, which likely results in modulation of activity by different spatial localization relative to other reaction partners.
RESULTS
HCV RNA replication in Huh7 cells recruits Ago2 and Dicer to lipid droplets
We used an unmodified human hepatocarcinoma Huh7 cell line and Huh7 cell lines stably expressing two different modifications of genotype 1b autonomously replicating full-length (Pietschmann et al. 2002) and subgenomic pFK-1398neo/luc/NS3-3′/5.1 (Lohmann et al. 1999; Supekova et al. 2008) HCV replicons (Fig. 1, constructs I and II, respectively). In a first set of experiments, we examined how RNAi proteins Ago2 and Dicer distribute in the Huh7 cells in the absence and presence of a self-replicating HCV RNA, using immunofluorescence and confocal imaging (methodological details are provided in the Materials and Methods section). In these experiments, in addition to looking at localization of key RNAi proteins with respect to P-bodies, our attention was also focused on another type of intracellular compartment, lipid droplets (LDs). LDs are known as intracellular platforms of HCV replication and infectious virus production (Moradpour et al. 1996, 2004; Boulant et al. 2007; Miyanari et al. 2007; Shavinskaya et al. 2007).
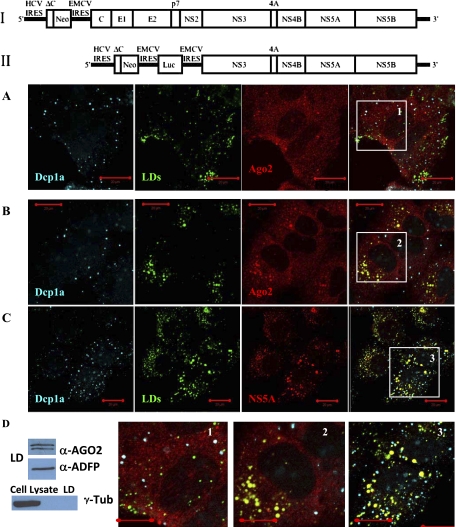
FIGURE 1.
Argonaute 2 is enriched at and around lipid droplets (LDs) in Huh7 cells with full-length HCV replicon. (I) Schematic of genotype 1b autonomously replicating full-length HCV replicon stably expressed in Huh7 cells. (II) Schematic of subgenomic pFK-1398neo/luc/NS3-3′/5.1 HCV replicon. (A) Ago2 does not localize at lipid droplets in native Huh7 cells (enlargement 1). (B) Ago2 is enriched in foci located at and around LDs in Huh7 HCV cells with the full-length HCV replicon (enlargement 2). (C) Nonstructural viral protein NS5A localizes at and around LDs (enlargement 3). (A–C) P-body protein Dcp1a, unlike Ago2, is always seen separate from LDs. (D) Western blot analysis of the lipid droplet fraction from HCV replicating cells confirms that Ago2 associates with LDs. LD protein ADRP was used as LD marker. Whole cell lysates and LD fraction were also probed with cytoplasmic protein γ-Tubulin to verify purity of LD separation. Scale bar corresponds to 20 μm.
The obtained imaging data show that most of Ago2 in Huh7 and Huh7 HCV cells is, as expected (Leung et al. 2006), diffusely distributed in the cytoplasm (Fig. 1A,B). Strikingly, though, in the presence of replicating HCV RNA, Ago2 becomes distinctly enriched in the perinuclear weblike structures and at the LDs (Fig. 1B). In contrast, no such Ago2 accumulation at LDs is detectable in the Huh7 cells without HCV RNA. Instead, in the cells having no HCV RNA, Ago2 is seen accumulating in the P-bodies (Fig. 1A). P-bodies are seen spatially separated from LDs, with the P-body marker protein Dcp1a neither assembling into web like structures nor being enriched at LDs in Huh7 cells with HCV RNA (Fig. 1B). Ago2 localization in the P-bodies of Huh7 cells is not surprising and was observed also in other cell lines (Jakymiw et al. 2005; Chu and Rana 2006; Leung et al. 2006). However, Ago2 docking at LDs in the presence of HCV RNA in these cells is a new and intriguing finding, which we explore in more detail.
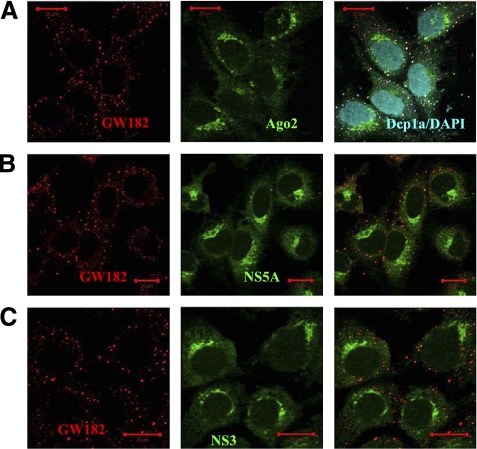
Because HCV uses LDs as the intracellular platforms for virus production and assembles viral RNA, replicative dsRNA intermediates, viral Core, nonstructural NS3, NS5A, and envelope proteins at these compartments, we speculated that Ago2 accumulation at LDs might enable or be a result of its interaction with viral protein machinery. In support of this hypothesis, we observed that viral nonstructural NS5A protein is strongly enriched at LDs (Fig. 1C). At the same time, NS5A spots are clearly seen separated from P-bodies (Fig. 1C). To get more detail about P-body and LD distribution in Huh7 cells with HCV RNA, we analyzed intracellular localization of GW182 protein, another Ago2 binding partner in the RNAi pathway which is known to concentrate in the P-bodies (Lian et al. 2007; Eulalio et al. 2009). In obtained images, GW182 clearly colocalizes with Dcp1a in the joint foci, and none of these proteins accumulate into the dot- and weblike structures enriched with viral NS5A (Supplemental Fig. S1). Thus, unlike Ago2, Dcp1a and GW182 proteins do not get condensed at LDs in the Huh7 cells with HCV RNA, indicating that P-bodies stay apart from the sites of viral RNA synthesis. Western blot analysis of the LD fraction from the Huh7 HCV replicating cells provides biochemical evidence that Ago2 proteins associate with LDs (Fig. 1D). A lipid droplets fraction was prepared according to previously described protocols (Miyanari et al. 2007; Herker et al. 2010). Lipid droplet-associated adipose differentiation-related protein (ADRP) was used as a marker of the lipid droplet fraction and γ-Tubulin was used to verify the purity of the fractionation.
In the Huh7 cells without HCV RNA, a small fraction, up to ∼10%, of all visible Ago2 foci localize separate from P-bodies (Fig. 1A). Because we hypothesize that Ago2 redistribution from its predominant diffusive cytoplasmic localization into a condensed state in the cellular subdomains might have functional implication, we tested if these “non-P-body” Ago2 foci are specific for the Huh7 cell line. Arguing against specificity of this observation only for Huh7 cells, we detected a similar type of “non-P-body” Ago2 foci (residing separately from Dcp1a and GW182 joint foci) in HEK293 cells (Supplemental Fig. S2). The same observation was also noted by others in the HeLa cells (Azuma-Mukai et al. 2008). Therefore, formation of Ago2 foci separate from P-bodies appears to be more general and indicates additional layers of heterogeneity in the Ago2 distribution, whose details and functional implications need to be better understood in future studies.
We next investigated how Ago2 distribute in the Huh7 cell line with a subgenomic HCV replicon that lacks expression of Core protein (Fig. 1, construct II). The HCV Core protein is understood to be a key factor targeting viral machinery to LDs (Miyanari et al. 2007). In the absence of Core, viral RNA synthesis occurs in the perinuclear membranous lipidic “web,” hosting NS3 and NS5A proteins (Gosert et al. 2003). Indeed, we observed that in the cells with the subgenomic HCV replicon, viral nonstructural proteins NS3 and NS5A (Fig. 2B,C) assemble into the characteristic perinuclear weblike patterns. Ago2 localization in this cell line very strongly resembles these dot- and weblike perinuclear structures (Fig. 2A; nuclear marker DAPI was applied to counter-stain nuclei). Notably, NS3-NS5A-Ago2 enriched spots are clearly devoid of P-body proteins Dcp1a and GW182. Therefore, in both cases, with full and subgenomic HCV replicons, we observed that Ago2 became enriched at intracellular modified lipid enclaves hosting HCV RNA replication, but P-bodies were spatially separated from HCV replication sites.
FIGURE 2.
Argonaute 2 localizes into perinuclear weblike structures in Huh7 cells with subgenomic HCV replicon. (A) In Huh7 cells with subgenomic HCV replicon, Ago2 accumulates into a weblike perinuclear pattern, which is devoid of GW182 and Dcp1a proteins. Nuclei are visualized with DAPI. (B,C) Perinuclear structures enriched in nonstructural viral NS5A and NS3 proteins are devoid of GW182. Scale bar corresponds to 20 μm.
To assess observed distributions more quantitatively, all images were processed using an image colocalization analysis algorithm home-written in MATLAB 9 software (MathWorks). To quantify colocalization, we applied an earlier described approach based on the Pearson's correlation coefficient, r (Bolte and Cordelieres 2006; French et al. 2008). In particular, we implemented a version of the algorithm that uses image masks. An example of digitally generated image masks using original images for LDs and NS5A protein is provided in Supplemental Figure S3. The colocalization algorithm produces r values in the range [−1, 1], with 0 indicating that there is no discernable correlation and −1 and +1 meaning strong negative and positive correlations, respectively (French et al. 2008). The obtained r values for Ago2 and LDs, NS5A and LDs, Dcp1a and LDs in Huh7 HCV cells were respectively r = 0.8, 0.9, and 0.09, with 0.5 < r < 1.0 corresponding to a high correlation and 0 < r < 0.1 regarded as none (Supplemental Table S1). Taken together, these observations indicate that, in the presence of HCV RNA in the Huh7 cells, Ago2 protein gets enriched at the LDs where viral protein machinery is assembled, but Ago2 partners in the RNAi pathway—Dcp1a and GW182 proteins—do not localize at LDs.
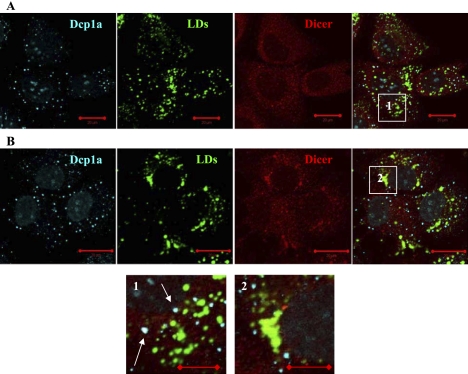
We then explored how Dicer, the other key protein partner of Ago2 in the RNAi pathway, distributes in the presence and the absence of HCV RNA in the Huh7 cell line. The images show that Dicer in the cells with replicating HCV RNA localizes at and around LDs (Fig. 3). P-bodies (Dcp1a protein) are seen spatially separated from LDs in these cells. In contrast, in the Huh7 cells without HCV RNA, Dicer is seen enriched in P-bodies. Quantitative image analysis indicates that, in the absence of replicating HCV RNA, up to 75% of all visible Dicer foci in Huh7 cells colocalize with P-bodies. An analogous observation of Dicer localization in P-bodies was reported earlier for another cell line (Moser et al. 2007) and was linked to its role in the RNAi down-regulation pathway. Thus, the intracellular distribution of Dicer is changed in the cells bearing replication of HCV RNA, and this protein, similar to Ago2, is seen docking at LDs.
FIGURE 3.
Intracellular localization of Dicer is altered in Huh7 cells with replicating HCV RNA. (A) In native Huh7 cells, Dicer can be found in P-bodies (enlargement 1, arrows). P-bodies are spatially separated from lipid droplets. (B) In the Huh7 cell with the full length HCV replicon, Dicer accumulates at perinuclear weblike structures and at lipid droplets (enlargement 2). P-body protein Dcp1a does not localize to these structures. Nuclei were counter-stained with DAPI. Scale bars correspond to 20 μm in full images and 10 μm in enlargements.
Summarizing this set of experiments, we report that replication of HCV RNA in Huh7 cells alters the intracellular distributions of two key RNAi proteins, Ago2 and Dicer, which both get locally concentrated at the sites of viral RNA synthesis at LDs. P-body proteins Dcp1a and GW182 showed no accumulation at LDs, and the two types of intracellular subdomains, P-bodies and LDs, were seen spatially separated.
miRNA-122 and Ago2 regulation of HCV RNA replication does not involve P-bodies
miRNA-122 and Ago2 were reported to repress CAT-1 mRNA in Huh7 cells by withholding it in P-bodies (Bhattacharyya et al. 2006). Our results presented above showed that in the Huh7 cells with replicating HCV RNA, Ago2 accumulates at LDs. Consequently, we decided to examine the miRNA-122 distribution in the Huh7 cells bearing HCV RNA replication. For visualization of cellular miRNA-122, we used a labeled miRNA-122 LNA probe (Exiqon) and performed in situ RNA-RNA hybridization (methodological details and specificity controls are described in the Materials and Methods section), following previously reported protocols (Bhattacharyya et al. 2006).
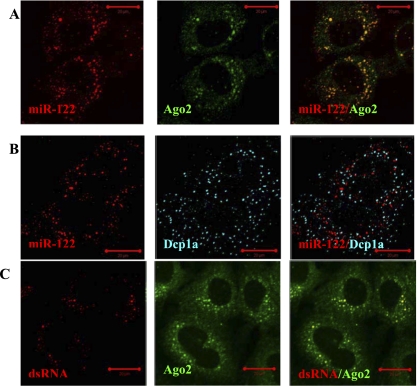
We observed that in Huh7 cells with the full-length HCV replicon, miRNA-122 is seen as multiple foci scattered in the cytoplasm (Fig. 4A,B, left panel). Intriguingly, we found that these miRNA-122 spots do not colocalize with P-bodies as was observed for miRNA-122 and Ago2 in the Huh7 cells without HCV RNA (Bhattacharyya et al. 2006). Quantitative image analysis identified that <5% of all visible miRNA-122 foci colocalized with P-bodies in the cells with HCV RNA (Fig. 4B). Although not enriched in P-bodies, miRNA-122, nevertheless, clearly associates with Ago2 in the joint foci (Fig. 4A). Taking this information along with our imaging data showing that Ago2 colocalizes with LDs in Huh7 cells with HCV RNA (Fig. 1B), we infer that miRNA-122 and Ago2 seen in the joint foci are located at LDs. The presence of replicating viral dsRNA in these cells was confirmed by immunostaining with monoclonal K1 antibody, known to recognize double-stranded RNA with a length larger than 40 bp (Schonborn et al. 1991; Targett-Adams et al. 2008), which showed punctuated staining in the presence of the HCV replicon (no such staining was observed in native Huh7 cells). Interestingly, visible dsRNA foci demonstrated colocalization with Ago2 foci (Fig. 4C). We also note that, in the hybridization experiments using miRNA-122 fluorescence probes in the Huh7 cells bearing no HCV RNA replication, we did not detect visible miRNA-122 spots beyond an elevated fluorescence signal distributed throughout the cytoplasm. Therefore, quantification of the miRNA-122 spots distribution with respect to P-bodies and Ago2 was not feasible. Based on the above-described observations, we speculate that the docking of miRNA-122 and Ago2 to LDs instead of P-bodies in Huh7 cells with HCV RNA is linked to their up-regulating function in replication of HCV RNA. On the other hand, whether P-bodies play a role in regulation of HCV RNA replication is unclear.
FIGURE 4.
Argonaute 2 foci associate with miRNA-122 and dsRNA in Huh7 cells with replicating HCV RNA. (A) In Huh7 cells with full-length HCV replicon, miR-122 and Ago2 distribute into joint spots localized in the perinuclear area. (B) P-bodies (visualized using Dcp1a protein) and miRNA-122 spots are seen separated. (C) dsRNA (visualized using K1 antibody) shows punctate staining and exhibits foci overlapping with Ago2 foci. Scale bars correspond to 20 μm.
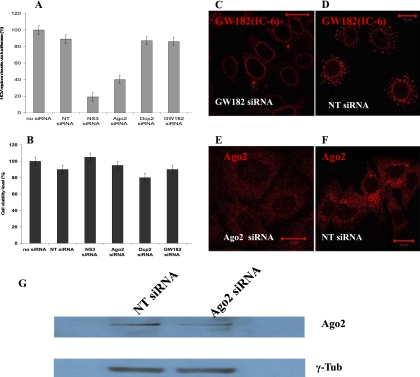
To explore a role for P-bodies in the regulation of HCV replication, we performed a series of siRNA silencing experiments using a Huh7 cell line stably expressing the subgenomic firefly luciferase reporter HCV replicon (Fig. 1, construct II). Using two well-characterized anti-Dcp2 and anti-GW182 siRNAs, we down-regulated expression of these P-body proteins in the Huh7 cells with the subgenomic HCV replicon (Jakymiw et al. 2005; Liu et al. 2005a,b). For comparison, we also down-regulated Ago2 using a cocktail of two anti-Ago2 siRNAs as described previously (Meister et al. 2004; Aleman et al. 2007). In addition, we used a well-defined siRNA targeting nonstructural NS3 protein of HCV genome as a positive control (Supekova et al. 2008) and several nontargeting (NT) siRNAs as a nonspecific control (Dharmacon). Relative HCV replicon levels in response to the above protein silencing were measured using the luciferase assay 48 h post-siRNA transfection (Fig. 5A).
FIGURE 5.
P-body disruption has no effect on HCV RNA replication in Huh7 cells. (A) Down-regulation of P-body proteins GW182 and Dcp2 had no effect on HCV RNA level in Huh7 cells as measured by subgenomic HCV replicon with firefly luciferase reporter. For comparison, silencing of Ago2 protein in the same cell line reduced HCV replicon level by ∼55%. Nontargeting (NT) siRNA was used as a negative control. NS3 siRNA was used as a positive control and inhibited HCV RNA by ∼80%. All siRNAs were transfected for 48 h. (B) Cell viability was not affected after 48 h post-siRNAs transfection (measured via Alamar Blue). All values are average of two sets of triplicate ± SD. (C,D) P-bodies are seen dispersed in Huh7 cells with subgenomic HCV replicon after 48 h down-regulation of GW182 protein. In control experiment with NT siRNA, P-bodies remain intact. (E,F) Ago2 foci are seen disrupted in the Huh7 cells after 48 h down-regulation of Ago2, while in the cells treated with NT siRNA, distribution of Ago2 into foci remained unaffected. Scale bar corresponds to 20 μm. (G) Western blot data show visible down-regulation of Ago2 expression after 48 h as identified by monoclonal 4F9 anti-Ago2. γ-Tubulin results are from the same reprobed membrane.
We found that NS3 siRNA reduces replicon levels by nearly 80%, similar to previous reports (Supekova et al. 2008). Ago2 siRNAs inhibit HCV replication by ∼55%, in agreement with data by others (Randall et al. 2007). However, HCV replicon levels remain largely unaffected by Dcp2 and GW182 siRNAs (Fig. 5A) and are approximately the same as in the cells transected with NT control siRNAs. We also noted, by using several different nontargeting siRNAs as nonspecific controls, that NT siRNAs slightly decreased HCV replicon levels as compared to mock treated cells using only the transfection reagent. This might be due to the fact that any siRNA can trigger formation of RISC complexes and in this way depletes RNAi protein machinery, lessening its function in HCV RNA replication. All applied siRNAs were checked for toxicity and showed no effect on cell viability (Fig. 5B).
Disruption of P-bodies and Ago2 foci in response to down-regulation of the respective proteins was tested by immunofluorescence imaging. In cells transfected with a cocktail of Ago2 siRNA1, and siRNA2, the total fluorescence signal in the cytoplasm was significantly lower as compared to cells transfected with nonspecific NT siRNA, and cells lost virtually all Ago2 foci (Fig. 5E,F). The down-regulation of Ago2 expression was also confirmed by Western blot data (Fig. 5G), showing a decreased amount of the protein in the cells transfected with the cocktail of two anti-Ago2 siRNAs as proposed earlier (Meister et al. 2004). The cells transfected with GW182 siRNA showed decreased cytoplasmic staining of GW182 protein and nearly full depletion of P-bodies (GW182 was visualized using IC-6 serum) (Fig. 5C,D). A cross-control of P-body dispersal after down-regulation of GW182 was performed using Dcp1a marker and showed a similar result of nearly complete P-body loss (Supplemental Fig. S4). We also noted that dispersal of Ago2 foci after Ago2 down-regulation has no effect on the integrity of P-bodies. Therefore, the results of the above series of combined experiments designed to investigate P-body function in miRNA-122 regulation of HCV RNA replication reveal that P-body disruption neither enhances nor inhibits HCV RNA replication.
siRNA and Ago2 repression of HCV RNA replication is facilitated by P-bodies
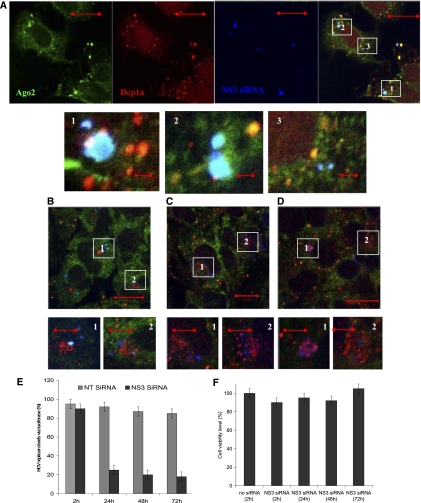
Our next question was whether P-bodies have a functional role in siRNA and Ago2 repression of HCV RNA replication. To answer this question, we performed a series of imaging, silencing, and biochemical experiments in which intracellular localization of siRNA, Ago2, and P-bodies could be linked to their functions. To visualize intracellular distribution of “anti-HCV” NS3 siRNA, we used siRNA molecules labeled at the 3′ end of a guide strand with Alexa 647 (Berezhna et al. 2006). Huh7 cells with a subgenomic HCV replicon were transfected with these labeled NS3 siRNAs for different time intervals. At selected time points, the cells were fixed, immunostained using Ago2 and Dcp1a antibodies, and observed under the microscope.
At the first time point, ∼2 h post-transfection, NS3 siRNA molecules were visible in condensed variable-size spots in the cytoplasm (Fig. 6A, blue). These siRNA spots also contained a locally increased concentration of Ago2 (Fig. 6A, green). The joint siRNA-Ago2 foci (Fig. 6A, enlargements, cyan overlay) did not contain a detectable amount of Dcp1a (Fig. 6A, red). However, in some locations, small Dcp1a dots were seen adjacent to siRNA-Ago2 foci (Fig. 6A, enlargements 1–3). The cells also had P-bodies enriched in Ago2 (Fig. 6A, yellow overlay) that did not contain any detectable amount of labeled siRNAs. Based on these observations, we speculate that siRNA molecules, being endocytosed by the cells and released into the cytoplasm, interacted with Ago2 molecules diffusing in the cytoplasm and formed siRISC complexes. Formation of such complexes might be enhanced at the sites of siRNA release from the endosomes, where local concentration of these molecules is temporally elevated. Emergence of small P-bodies around a siRNA-Ago2 spot might indicate the next step in the RNAi pathway and corresponds to “in situ” P-body formation at sites of siRNA-Ago2 interaction with their targets. The P-bodies enriched in Ago2 and not showing fluorescent siRNA molecules are, instead, pre-existing formations in the cells and are not involved in interaction with exogenously introduced NS3 siRNAs. A siRNA accumulation in the dot-type structures was also observed for the labeled siRNA being directly micro-injected into the Huh7 HCV cells that were fixed and observed under the microscope at different time intervals post-injection. These siRNA molecules entered the cells, bypassing endosome-mediated entry, and indicate that the observed siRNA spots are not a result of endosome-trapped accumulations.
FIGURE 6.
Intracellular localization and activity of NS3 siRNA in Huh7 HCV cells at different intervals post-transfection. (A) In 2 h post-transfection, a fluorescently labeled guide strand of NS3 siRNA (blue) and Ago2 (green) are seen in the colocalizing spots in the cytoplasm. Small Dcp1a (red) spots can be found adjacent (enlargements 1–3). (B–D) In the course of the next 72 h, siRNA and Ago2 spots dispersed into fragmented pattern in the perinuclear area, with increase of Dcp1a punctuate staining at siRNAs sites (B) 24 h; (C) 48 h; (D) 72 h (enlargements 1,2). (E) Inhibition activity of labeled NS3 siRNA was not compromised by labeling. Nontargeting siRNA was used as an unspecific control. (F) Cell viability (measured via Alamar Blue) remained unaffected after transfection and prolonged incubation with labeled NS3 siRNA. Scale bars correspond to 20 μm in full images and 5 μm in enlargements.
In support of this interpretation of the imaging data, the HCV RNA luciferase reporter assay showed that, after 2 h, NS3 siRNA decreased the HCV RNA level by ∼2% (a normalized value versus NT siRNA) (Fig. 6E). Although this down-regulation is small, it indicates that siRISC started to silence their target HCV RNAs. We also performed an indirect test of siRISC formation using NS3 siRNA, fluorescently labeled at both strands (Alexa 647 at the 3′ end of the guide strand and Alexa 488 at the 3′ end of the passenger strand) which allowed us to assess the passenger strand distribution with respect to “Ago2-siRNA guide strand” spots. We did not detect a visible fluorescence signal from passenger strand in the Ago2 foci. This result suggests that a siRNA passenger strand got separated and was likely cleaved during RISC formation. Dual-labeled NS3 siRNA used in these experiments was tested with luciferase subgenomic HCV RNA reporter in Huh7 cells and determined to be active (Fig. 6E).
The next imaging time points were selected at 24, 48, and 72 h after transfection (Fig. 6B–D, respectively). The siRNA-Ago2 foci got fragmented and after 24 h are seen as multiple smaller spots typically concentrated in the perinuclear area. Initially only insignificantly enriched at siRNA-Ago2 foci, Dcp1a protein accumulated at siRNA locations by 24 h, and the amount of Dcp1a-siRNA overlaying spots increased over the next time intervals (Fig. 6B–D). By 72 h, ample P-bodies were visible in the cells, some of which were still enriched in siRNA (Fig. 6C, enlargements). Luciferase-based measurement of HCV RNA inhibition by NS3 siRNA showed that major suppression of HCV RNA replication, by ∼70%, occurred within the first 24h (Fig. 6E) and only slightly increased, up to ∼80%, after 48 h. Incubation for a longer interval, up to 72 h, did not increase inhibition above ∼80%. Controls showed that transfection reagents and all applied siRNAs did not affect cell viability throughout the experiment (Fig. 6F). Therefore, we observed that NS3 siRNA initially interacted with Ago2 in the cytoplasm without Dcp1a, but within a time interval needed for efficient suppression of HCV RNA replication (∼24 h), Dcp1a became enriched at siRNA spots and remained associated together for a prolonged interval (∼72 h). Taken together, these observations show a correlation between P-body enrichment at siRNA sites and the degree of HCV RNA inhibition and suggest that P-bodies might be involved in the regulation of HCV RNA by NS3 siRNA.
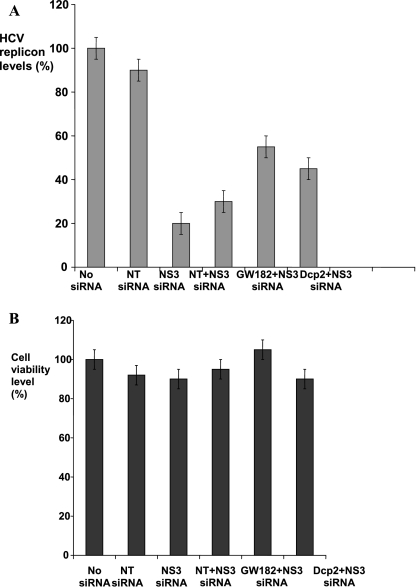
To verify whether P-bodies play a functional role in siRNA down-regulation of HCV RNA, we tested how depletion of P-bodies from cells would affect NS3 siRNA inhibition of HCV RNA. To disrupt P-bodies, we individually down-regulated Dcp2 and GW182 proteins using corresponding siRNAs. We observed that, in the cells transfected with either of the siRNAs that disrupted P-bodies, NS3 siRNA inhibition of HCV RNA was less efficient compared to the cells transfected with the control NT siRNA (Fig. 7A). The effect was more pronounced when GW182 protein was knocked down, resulting in ∼25% reduction of NS3 siRNA activity (Fig. 7A, column 5). Down-regulation of Dcp2 protein reduced NS3 siRNA activity by ∼15% (Fig. 7A, column 6). All applied siRNAs were checked for toxicity and did not affect cell viability (Fig. 7B). P-body dispersals in response to Dcp2 and GW182 down-regulation were individually verified using immunofluorescence imaging of control and down-regulated cells. Imaging data obtained in these experiments confirmed P-body disruption in the cells treated with Dcp2 and GW182 siRNAs. Thus, we conclude that P-body disruption does not abolish NS3 siRNA inhibition of HCV RNA replication but does reduce its efficiency.
FIGURE 7.
P-body disruption impairs NS3 siRNA inhibition of HCV RNA replication. (A) NS3 siRNA inhibition of subgenomic HCV replicon is compromised respectively by ∼25% in the cells with silenced GW182 (column 5) and by ∼15% in the cells with silenced Dcp2 (column 6). All values are normalized to HCV RNA level in the cells transfected with NT siRNA (column 4). HCV replicon levels measured in “no siRNA” (column 1), NT siRNA (24 h, column 2), and “NS3 siRNA only” (24 h, column 3) Huh7 cells are provided as controls. GW182, Dcp2, and NT siRNAs were transected for 48 h and subsequently, the cells were cotransfected with NS3 siRNA for additional 24 h. (B) Cell viability level via Alamar Blue measured after treatment with corresponding siRNAs as identified for plot A. Error bars correspond to a standard mean deviation (SD) in three independent experiments, each done in triplicate.
DISCUSSION
A human host responds to viral infection in multiple ways, including a key defense mechanism via the interferon pathway. Whether RNAi in mammals also functions in the antiviral defense remains open and is being actively investigated (Cullen 2010; Pfeffer 2010). The hepatitis C virus shows a very complex interaction with the human host RNAi system. Because HCV replication occurs through a minus-strand, producing long dsRNA intermediates (Chisari 2005), the prediction holds that Ago2 and Dicer can target and slice HCV replicative intermediates into short 21–23-nt siRNA fragments, potentially suppressing HCV propagation. Such siRNAs produced by slicing of the HCV genome have been actively sought for in the HCV-infected cells (Pfeffer et al. 2005) and not found until very recently (Parameswaran et al. 2010). This latest study, by using multiplexed high-throughput sequencing, identified small RNAs derived from a viral genome (vsRNAs) in the cells carrying a hepatitis C replicon and strand-selective loading of these “viral” siRNAs into Argonaute complexes (Parameswaran et al. 2010). These findings define vsRNAs as possible direct evidence of the antiviral RNAi defense in the HCV-infected cells. At the same time, compelling data suggest that HCV can counteract the host antiviral RNAi system and even use its components for efficient propagation. It was demonstrated that, in the absence of the HCV protein machinery, the Dicer enzyme is capable of completely digesting the HCV IRES and the HCV RNA replicative intermediate into short 22-nt pieces (Wang et al. 2006). However, addition of the HCV Core protein blocks Dicer activity via direct binding and rescues HCV RNA replication (Wang et al. 2006). Moreover, host RNAi molecular machinery, including miRNA-122, Ago2, and Dicer, appears to positively regulate HCV propagation in cultured Huh7 cells (Jopling et al. 2005; Randall et al. 2007; Henke et al. 2008; Jangra et al. 2010b). How exactly these multiple activities of RNAi machinery are orchestrated in the Huh7 cells bearing HCV replication and whether any of these functions are correlated with and perhaps controlled by localization in the intracellular subdomains is not well-understood. We addressed these questions by a combined approach using several methodologies and gained new insights about this complex interplay.
We observed that miRNA-122 and Ago2 in Huh7 cells with replicating HCV RNA were enriched together in multiple cytoplasmic foci. However, these spots do not colocalize with P-bodies, and a detectable amount of miRNA-122 was only found in <5% of P-bodies. Instead, joint miRNA-122 and Ago2 foci were seen accumulating at and around LDs. Depletion of P-bodies in cells by silencing GW182 and Dcp2 proteins showed no impact on HCV RNA level. In contrast, down-regulation of the Ago2 protein reduced the HCV RNA level by >50%, consistent with earlier data (Randall et al. 2007). Ago2 down-regulation was accompanied by dispersal of Ago2 foci but did not cause dispersal of P-bodies. Our observation that miRNA-122 does not accumulate in the P-bodies of Huh7 cells bearing replication of HCV RNA is complemented by recent data from another group showing that HCV replicating dsRNA in FT3-7 cells (a clonal derivative of Huh7 cells) is seen in the cytoplasm as multiple scattered foci which do not localize to P-bodies (Jangra et al. 2010b). In addition to Ago2 and miRNA-122, Dicer also became enriched at lipid droplets in the cells with replicating HCV RNA. Based on the fact that LDs and associated membranous web structures are used by the virus as cellular platforms for replication, we speculate that Ago2, Dicer, and miRNA-122 accumulation at these sites is due to or facilitates their interaction with the replicating HCV RNA. Thus, while P-bodies play an essential role in miRNA-122 repression of CAT-1 mRNA, their dispersal appears to be unimportant for miRNA-122 up-regulation of HCV RNA, and instead, lipids droplets come into action in this process.
The enrichment of the core RNAi components at LDs in the cells with replicating HCV RNA may indicate a development in which miRISC complexes bind the HCV genome and/or replicative intermediate but are not able to direct the bound HCV RNA to the decay pathway (Eulalio et al. 2007; Kulkarni et al. 2010) due to counteraction by the HCV proteins residing at LDs. The possible subversion of the RNAi function by viral protein machinery at LDs is consistent with the fact that Dicer endonucleolytic capacity is neutralized by Core binding (Wang et al. 2006), which likely occurs at LDs, and is also congruent with Dicer docking at these subdomains, as we observed here. Furthermore, Core and envelope protein 2 (E2) were shown to form complexes with RISC components resulting in E2 binding to Ago2 and blocking its activity (Ji et al. 2008). These RISC-blocking actions are likely facilitated by trapping Ago2 and miRNA-122 at LDs. Thus, we postulate that retention of Ago2, Dicer, and miRNA-122 at LDs and associated membranous structures emerges as a possible mechanism by which HCV protein machinery gets hold of and modulates cellular RNAi response.
Our observations with an exogenous NS3 siRNA transfected into cells to repress replication of HCV RNA provide more insight about HCV interaction with cellular RNAi. The siRNA data further support our hypothesis that condensing and/or withholding of active RNAi components in intracellular subdomains is an important regulatory mechanism in RNAi function. In this case, such spatially restricted activity enclaves are P-bodies. The imaging experiments with fluorescently labeled NS3 siRNA demonstrated that siRNA molecules associated with cytoplasmically distributed Ago2 molecules, and the siRNA-Ago2 spots were eventually enriched with Dcp1a molecules. Thus, unlike miRNA-122, which stays largely outside P-bodies in Huh7 cells bearing HCV RNA, NS3 siRNA instead appears to initiate formation of P-bodies at the sites of its activity. A functional involvement of P-bodies in NS3 siRNA inhibition of HCV RNA was confirmed by the luciferase reporter data showing that disruption of P-bodies compromised NS3 siRNA capacity to suppress replication. Based on these observations, we hypothesize that binding of siRISC complexes to their targeted HCV RNA molecules is followed by recruitment of Dcp1a proteins from the cytoplasm and on-site P-body formation, which facilitates degradation of the impaired HCV RNA. Dcp1a proteins are known to be dynamic components of P-bodies which continuously exchange with the cytoplasm (Aizer et al. 2008). Therefore, an elevated local concentration of stalled RISC-impaired HCV RNA likely drives assembly of the complexes with soluble Dcp1a proteins into microscopically visible P-bodies. Hence, the NS3 siRNA data provide more evidence in favor of our postulation that partitioning of key RNAi components into specialized spatially restricted intracellular subdomains has a regulatory role in determining RNAi outcome.
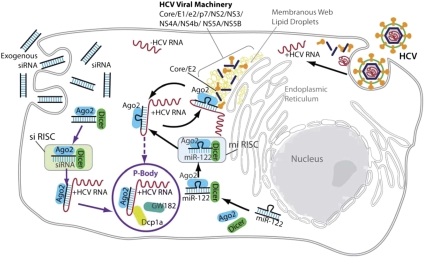
Throughout the viral lifecycle, HCV RNA is involved in multiple processes. It constantly replicates on positive and negative templates, serves as a messenger for translation of viral proteins, and gets packaged into progeny virus particles. These events are mutually competing and proceed in a tightly regulated mode, most of which remains nebulous. It has been proposed that contending events in the HCV life cycle are regulated by partitioning of different viral and host factors into functionally and physically distinct subcellular compartments (Randall et al. 2007; Tellinghuisen et al. 2007). An emerging view suggests that, in addition to lipid droplets, P-bodies might also serve as such intracellular enclaves, playing distinct roles in the viral life cycle (Beckham and Parker 2008). One putative mechanism proposes that transition of the HCV genome from active translation to a translationally inactive state that enables replication and/or packaging is mediated by withholding of translationally repressed HCV RNA in the P-bodies (Beckham and Parker 2008; Cullen 2010). We did not observe this proposed role of P-bodies in the facilitation of HCV RNA replication by host miRNA-122. Instead, we identified that P-bodies are important for inhibition of viral replication by siRNA complexes. At the same time, we found that retention of key RNAi components including miRNA-122, Ago2, and Dicer at lipid droplets may act as a possible regulatory mechanism by which HCV hijacks RNAi molecular complexes and reverses their functions. Overall, taking together our results and findings from other groups we propose a compartmentation model for cellular RNAi interaction with the HCV genome, in which the fate of HCV RNA molecules bound to RISC complexes is modulated by docking of these complexes at physically distinct intracellular compartments such as P-bodies and lipid droplets, where different sets of proteins augment or counteract antiviral RNAi function (Fig. 8).
FIGURE 8.
A model of functional compartments regulating dual interaction of cellular RNAi with HCV RNA. Ago2 and Dicer proteins interact with exogenous siRNA or endogenous miRNA-122 in the cytoplasm to form RISC complexes. Active siRISC or miRISC complexes can recognize and bind HCV RNA molecules released into the cytoplasm from the viral particles. siRISC binding to HCV RNA induces cleavage of the bound HCV RNA strand and eventual stalling of impaired HCV RNA molecules in the cytoplasm. Subsequently, degradation mechanisms either orchestrate recruitment of GW182 and Dcp1a proteins from the cytoplasm and “in situ” P-body formation at the sites of stalled HCV RNA fragments, or the impaired HCV RNA molecules with bound siRISC complexes can be shuttled to the pre-existing P-body. Segregation of siRISC complexes and their targeted HCV RNA molecules in the P-bodies might reflect a cellular condition of the efficient degradation of incompetent RNA. Whether binding of miRNA-122 and Ago2 to HCV RNA at its 5′ end would result in translocation of the viral strand into P-bodies remains open. At the same time, HCV viral proteins Core and E2 can bind to Ago2 and Dicer that formed complexes with miRNA-122 and HCV RNA molecules, and this binding ultimately affects interaction of miRISC complexes with HCV RNA. Apparently, functional actions of HCV proteins might be facilitated by sequestering Ago2, Dicer, and miRNA-122 complexes with HCV RNA at the enclaves formed by lipid droplets.
Finally, the idea that concentration of key RNAi molecular components in separate intracellular compartments is an important cellular lever regulating a dual interaction between RNAi machinery and HCV RNA likely reflects a more general cellular strategy of formation of cellular subdomains to promote the rate and specificity of reactions (Strzelecka et al. 2010; Yoon and Parker 2010). Nonmembrane subcellular bodies in the cytoplasm (P-bodies, stress granules) and the nucleus (Cajal bodies, nucleoli, paraspeckles) are being actively investigated as such cellular platforms serving to increase the rates of assembly of multicomponent cellular machines (Yoon and Parker 2010). Another example of intracellular subdomains involved in RNAi biology was observed for miRNA-function oscillating between mRNA repression and activation throughout the cell cycle, when Ago2 and miRNAs were seen distributed into different types of condensed foci (Vasudevan and Steitz 2007; Vasudevan et al. 2007). Lipid rafts are a type of condensed microdomain in the plasma membrane; these are widely understood to be specialized dynamic structures that compartmentalize cellular processes by serving as platforms for the assembly of signaling molecules, influencing membrane fluidity and membrane protein trafficking, and regulating neurotransmission and receptor trafficking (Simons and Vaz 2004). Continued study of the human RNAi system and careful comparison of its multiple aspects including interaction with viruses, in vitro, with cellular systems and in human subjects, promises to further unravel the complexity of RNAi mechanisms and functions.
MATERIALS AND METHODS
siRNAs
All siRNAs were purchased from Dharmacon as single strands and prepared according to manufacturer's instructions. Sequences of siRNAs (antisense strand) used in this study are as follows:
NS3: 5′-ACAUUGGUGUACAUUUGGG dTdT-3′
GW182: 5′-UAGCGGACCAGAGCAUUUCUUdTdT-3′
Dcp1: 5′-UCUAUGACAAGGCAAUUUCUUdTdT-3′
NT control: 5′-UUGAUGUGUUUAGUCGCUAUUdTdT-3′
hAgo2 #1 (sense strand): 5′-GCACGGAAGUCCAUCUGAAUU-3′
hAgo2 #2 (sense strand): 5′-GCAGGACAAAGAUGUAUUAUU-3′
Cell culture, transfection, and siRNA visualization
The Huh-7 cell line stably expressing the full-length HCV replicon (Pietschmann et al. 2002) was received as a gift from Dr. Bartenschlager's laboratory (University of Heidelberg, Germany). The Huh7 cell line harboring the subgenomic replicon was generated by us as described elsewhere (Supekova et al. 2008). Unmodified Huh7 and Huh7 HCV, HeLa, HeLa EGFP-Ago2 and HEK293 cell lines were cultured in DMEM with 10% FBS in a 37°C incubator with 5% CO2. Huh7 HCV and HeLa EGFP-Ago2 cells were grown in the presence of 500 μg/mL Geneticin G418 (Invitrogen). For fluorescence imaging experiments, the cells were plated overnight at ∼50% confluency in 2-well chambered coverglass plates (LabTec, Nalge Nunc International). Luciferase assay experiments were performed in standard 6- or 96-well plates. For transfection experiments, Lipofectamine 2000 (Invitrogen) was diluted 50-fold with Opti-MEM (Invitrogen), and 25 μL of this mixture were added per well to 96-well plates.
In the fluorescence experiments investigating NS3 siRNA distribution in Huh7 HCV cells, live cells were transfected with 60 nM labeled siRNA and monitored periodically under the microscope to look at siRNA intracellular uptake. After 2 h, the cells were washed three times with PBS to remove external siRNAs and kept in normal growth medium in a 37°C incubator, allowing for siRNA inhibiting activity. The transfected cells were fixed and immunostained with Ago2 and Dcp1a antibodies at different time intervals as indicated in the Results section.
In double down-regulation experiments, the cells plated at ∼40% confluency in 6-well plates were first transfected with either 100 nm or 50 nM anti-GW182, anti-Dcp2, and control nontargeting siRNAs in serum-free OptiMEM using Lipofectamine 2000. Four h post-transfection, 10% FBS was added to the transfection medium, and cells were incubated with siRNAs for, respectively, ∼24–72 h in the 37°C incubator with 5% CO2. Next, the cells were washed with PBS, resuspended, transferred to 96-well plates, and transfected with either 100 nm or 50 nM anti-NS3 siRNA for 24 h. We detected that ∼72 h incubation with anti-GW182 siRNA provided the strongest disruption of P-bodies as observed by immunofluorescence imaging. Therefore, for NS3 siRNA activity tests in P-body-depleted cells, we performed NS3 siRNA transfection after ∼72 h incubation with anti-GW182 and anti-Dcp2 siRNAs, respectively. The luciferase assay was performed as directed by the manufacturer (Promega). For fluorescent studies of P-body distribution during down-regulation of GW182 and Dcp2, the cells were plated in glass-bottom 6-well plates (MatTek) and transfected with siRNAs using the same transfection protocol. At different post-transfection times, the cells were fixed, permeabilized, immunostained with corresponding antibodies, and observed under the microscope. Similarly, in the fluorescent experiments exploring labeled NS3 siRNA distribution with respect to Ago2 foci and P-bodies, the cells were cultured overnight at ∼50% confluency in 2-well chambered cover glass plates (LabTec, Nalge Nunc International), transfected with siRNAs for 2 h in serum-free OptiMEM. Two h post-transfection, the cells were PBS-washed and kept in normal cell growth medium until fixation and immunostaining. All presented data are results of at least three independent experiments. For each presented imaging data set, at least 200 cells were examined in three independent experiments. Double transfection experiments were repeated three times.
Cell viability, luciferase activity
Cell viability was determined by incubating cells in 96-well plates with 10% Alamar Blue solution (Trek Diagnostic System) for 2–4 h prior to data collecting time points and measuring fluorescence. To quantify firefly luciferase activity, the Bright-Glo luciferase assay system (Promega) was used according to the manufacturer's instructions. Fluorescence and luminescence signals were measured using an Analyst AD reader (Molecular Device).
Immunofluorescence and antibodies
For immunofluorescence, cells were fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized in 0.5% Triton–×100 in PBS for 5 min at 4°C. Incubations with antibodies were performed in PBS containing 1.5% BAF as blocking agent at 4°C for 1 h. Standard controls of using only secondary and no primary antibodies were performed for each combination of antibodies to ensure no articulate staining. The primary antibodies and their respective dilution were: mouse monoclonal anti-Ago2 4F9 (1:100, 1:200) (sc-53521, Santa Cruz Biotechnology), rabbit polyclonal anti-Ago2 (1:100, 1:200) (Upstate Biotechnology), rabbit anti-Dcp1a (1:200) (a gift from J. Lykke-Andersen, University of Colorado), IC-6 and 18033 sera (1:1000) (a gift from T. Eystathioy and M. Fritzler, University of Calgary, Canada), mouse monoclonal anti-GW182 (1:100) (Abcam), mouse monoclonal anti-Dicer (1:100) (ab14601, Abcam), mouse monoclonal anti-NS3 and anti-NS5A (1:50) (ViroStat). The secondary antibodies, goat anti-human Alexa 488, Alexa 555, donkey anti-rabbit Alexa 405, Alexa 488, Alexa 555, goat anti-mouse Alexa 488, Alexa 555, chicken anti-goat Alexa 488 were all purchased from Invitrogen and used in 1:250 dilution. Anti-dsRNA monoclonal antibody K1 (Scicons) was used in 1:500 dilution. For lipid droplet visualization, Bodipy 493/503 (Invitrogen) was diluted in ethanol as a saturated solution and added into the secondary antibodies incubation solution at final concentration 20 μg/mL.
Western blot analysis of the lipid droplet fraction was performed using polyclonal anti-ADFP (Abcam), polyclonal anti-Ago2 (Upstate Biotechnology), and monoclonal anti-Tubulin (Sigma) antibodies.
Specificity controls for Ago2 antibodies
Following earlier reports, for Ago2 visualization we used two different well-established Ago2 antibodies, mouse monoclonal (4F9) from Santa Cruz Biotechnology and rabbit polyclonal from Upstate Biotechnology. However, considering potential cross-reactivity issues of these Ago2 antibodies raised before, we validated their specificity in a HeLa cell line stably expressing EGFP-Ago2 (Leung et al. 2006). Results obtained in a combination of several different controls (Supplemental Fig. S5) confirmed that both tested Ago2 antibodies provide a consistent visualization of endogenous Ago2 in the cytoplasm and in P-bodies. In addition, rabbit polyclonal Ago2 antibody also recognizes nuclear distributed Ago2, similar to a newly developed multifunctional monoclonal 11A9 Ago2 (Weinmann et al. 2009) antibody, which was also tested in a separate control. We also tested the rabbit polyclonal Ago2 alongside 11A9 Ago2 and observed that both antibodies equally well-identify Ago2 in the nucleus and in P-bodies. We note that, during protein overexpression, EGFP-Ago2 becomes strongly enriched in P-bodies and, therefore, creates a strong gradient of rather bright EGFP fluorophore in the foci comparing to its averaged concentration in the cytoplasm. In order to avoid signal saturation in the foci, we had to tune excitation power of a 488-nm laser typically below 1 mW and under such conditions, cytoplasmic signal became fairly faint. If one increases excitation power, EGFP-Ago2 fluorescence is clearly seen throughout the cytoplasm, but the signal from foci get oversaturated, and therefore, such images are distorted and cannot be used for comparison of EGFP-Ago2 distribution. For visualization of P-bodies, we used, respectively, two key protein markers, Dcp1a and GW182. The corresponding antibodies were rabbit anti-Dcp1a serum from the J. Lykke-Andersen laboratory (University of Colorado) and two types of GW182 human autoimmune sera 18033, IC-6 from the M. Fritzler laboratory (University of Calgary, Canada). Lipid droplets were visualized using Bodipy 493/503.
Preparation of the lipid droplets
Cells at a confluency of ∼80% on a dish with a diameter of 14 cm were scraped in PBS. The cells were pelleted by centrifugation at 1500 rpm. The pellet was resuspended in 500 μL of hypotonic buffer (50 mM HEPES, 1 mM EDTA, and 2 mM MgCl2 at pH 7.4) supplemented with Complete protease inhibitors tablet (Roche Diagnostics) and was incubated for 10 min at 4°C. The suspension was homogenized with 30 strokes of a glass Dounce homogenizer using a tight-fitting pestle. Then, 50 μL of 10× sucrose buffer (0.2 M HEPES, 1.2 M KoAc, 40 mM Mg(oAc)2, and 50 mM DTT at pH 7.4) was added to the homogenate. The nuclei were removed by centrifugation at 2000 rpm for 10 min at 4°C. The supernatant was collected and centrifuged at 16,000g for 10 min at 4°C. The supernatant was mixed with an equal volume of 1.04 M sucrose in isotonic buffer (50 mM HEPES, 100 mM KCl, 2 mM MgCl2, and protease inhibitors). The solution was set at the bottom of 2.00-mL ultracentrifuge tube (Beckman Coulter). One milliliter of isotonic buffer was loaded onto the sucrose mixture. The tube was centrifuged at 100,000g in a TLA-100.3 rotor (Beckman Coulter) for 60 min at 4°C. After the centrifugation, the LD fraction on the top of the gradient solution was recovered in isotonic buffer. The suspension was mixed with 1.04 M sucrose and centrifuged again at 100,000g, as described above, to eliminate possible contamination with other organelles. The collected LD fraction was used for Western blotting.
Western blot
Western blot experiments to verify down-regulation of Ago2 protein in the Huh7 cells with a full-length HCV replicon were performed using cells treated with nontargeting control NT siRNA and a cocktail of two anti-Ago2 siRNA1 and siRNA2 applied at 55 nM concentration of total siRNA in 6-well plates for 48 h. The cell lysates were subjected to SDS-PAGE and transferred to a nitrocellulose membrane (Invitrogen). The blots were probed with monoclonal antibodies against Ago2 protein (Santa Cruz Biotechnology). Immunoreactive proteins were detected with the SuperSignal West Dura detection System (Thermo Scientific). A Western blot of the LD fraction was performed using polyclonal anti-ADFP (Abcam), polyclonal anti-Ago2 (Upstate Biotechnology), and monoclonal anti-Tubulin (Sigma) antibodies.
In situ miRNA-122 hybridization and visualization
Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and permeabilized in 0.5% Triton–×100 in PBS for 5 min at 4°C. For hybridization, each well was covered with 500 μL of hybridization buffer containing 2xSSC, 30% formamide, 1g/10ml dextran sulfate, 0.2mg/mL BSA, 250μg/mL yeast tRNA, RNaseOUT (Invitrogen) at 20unit/10μL of reaction mixture and 90 ng of a DIG-labeled LNA probe for miRNA-122 (Exiqon). After 18 h of hybridization at 49°C under the humidified conditions, cells were washed three times for 30 min at 37°C in 2×SSC and 30% formamide. Following in situ hybridization, cells were stained using a fluorescent antibody enhancer set for DIG detection with fluorescein anti-DIG Fab fragments (Roche Diagnostics) according to the manufacturer's protocol.
We note that, although the visualization method as applied here does not have the sensitivity to resolve individual miRNA molecules and observed fluorescent spots likely contain dozens of miRNA molecules (based on the integrated intensity of the spot), we roughly estimated that the number of spots per cell ranging from ∼50 to ∼150 is consistent with the reported number of miRNA-122 copies per cell exceeding 1000 (Sarasin-Filipowicz et al. 2009). In addition, our specificity controls as described below ruled out the possibility of nonspecific staining, which gives us further confidence that fluorescent signal comes from miRNA-122 hybridized probes. For quantification of miRNA-122 fluorescent spots in the individual cells, cell boundaries were approximately drawn based on the digital overlay of fluorescence and phase contrast images. A pale fluorescent signal was detectable in cell nuclei, which is likely due to the weak hybridization of the LNA probe to pre-miRNA-122 in the nuclei.
As a specificity control, EGFP-HeLa cells, which do not express miRNA-122 were microplated next to Huh7 HCV cells into adjacent quadrants of the same grid cover glass plate (LabTec, Nalge Nunc International). These two different cell lines were hybridized side by side and subsequently stained by rhodamine-labeled anti-DIG Fab fragments. We did not observe miRNA-122-related fluorescence in HeLa cells that were located next to Huh7 HCV cells and easily discriminated by clearly visible EGFP-Ago2 foci. Therefore, this control confirms specificity of miRNA-122 staining in Huh7 HCV cells. After hybridization of miRNA-122, Huh7 HCV cells were immunostained for Ago2 or P-bodies using either rabbit anti-Ago2 or rabbit anti-Dcp1a primary and corresponding secondary fluorescent antibodies.
Confocal imaging and quantitative data analysis
Confocal imaging was done on a BioRad-Zeiss 2100 LSM with a 60× 1.4 NA oil immersion objective. Fluorophores were excited, using a blue diode at 406 nm and Argon-ion, green HeNe, and red diode lasers at 488 nm, 543 nm, and 647 nm, respectively. Fluorescence emission was separated by corresponding dichroic mirrors 500DCLPXR, 560DCLPXR, and a set of blocking HQ420LP, HQ500LP, HQ570LP, and emission HQ 450SP/30 nm, HQ530SP/30 nm, and HQ610SP/40 nm filters, respectively. Optical slicing corresponded to 0.9 μm. Quantification of colocalization of Ago2, NS5A, Dcp1a proteins and LDs was performed using a house-made digital image correlation general approach described in detail elsewhere (French et al. 2008). After subtracting fixed values of background signals coming from cytoplasmic staining in both channels, corresponding image masks were generated, and total numbers of all visible Ago2 foci, miRNA-122 foci, and P-bodies were calculated for all cells in the image. Next, numbers of overlapping Ago2 foci and P-bodies or miRNA-122 foci and P-bodies were determined and respective percentage rates calculated. Two foci were considered as colocalized if their corresponding pixel areas were overlapping by >50%. The foci were separate if their pixel areas have zero overlap. Otherwise, foci were considered as partially overlapping and were not included in calculations. In total, 200 cells were selected from randomly chosen images in four independent experiments for each of data sets.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Drs. J. Lykke-Andersen (University of Colorado), T. Eystathioy and M. Fritzler (University of Calgary, Canada), M. Kiledjian (Rutgers University), R. Parker (University of Arizona), and G. Meister (Max Planck Institute for Biochemistry, Marttinsried) for sharing valuable antibodies, Dr. R. Bartenschlager (University of Heidelberg, Germany) for providing the pFK-1398neo/NS3-3′/5.plasmid and Huh7 cell line harboring the full-length HCV replicon, and Drs. A. Leung and P. Sharp (MIT) for the HeLa EGFP-Ago2 cell line. We also thank Dr. T. L. Tellinghuisen (The Scripps Research Institute, Scripps Florida) for careful reading of the manuscript and critical comments, and Ms. Katya Kadyshevskaya for help with Figure 8. This work was supported by the National Institute of General Medical Sciences, NIH (Grant Nos. RO1 GM073104 and GM066833 to A.A.D.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2523911.
REFERENCES
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y 2008. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell 19: 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman LM, Doench J, Sharp PA 2007. Comparison of siRNA-induced off-target RNA and protein effects. RNA 13: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhna SY, Supekova L, Supek F, Schultz PG, Deniz AA 2006. siRNA in human cells selectively localizes to target RNA sites. Proc Natl Acad Sci 103: 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Boulant S, Targett-Adams P, McLauchlan J 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88: 2204–2213 [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology 1: 106–113 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV 2005. Unscrambling hepatitis C virus-host interactions. Nature 436: 930–932 [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210 doi: 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol 7: 563–567 [DOI] [PubMed] [Google Scholar]
- Cullen BR 2010. Five questions about viruses and microRNAs. PLoS Pathog 6: e1000787 doi: 10.1371/journal.ppat.1000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. 2008. LNA-mediated microRNA silencing in nonhuman primates. Nature 452: 896–899 [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Ott M 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16: 1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Satoh M, Pauley KM, Fritzler MJ, Reeves WH, Chan EK 2006. Detection of the argonaute protein Ago2 and microRNAs in the RNA-induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods 317: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM 2010a. DDX6 (Rck/p54) Is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol 84: 6810–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM 2010b. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84: 6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Glaser A, Wernli M, Berke JM, Moradpour D, Erb P 2008. Suppression of short interfering RNA-mediated gene silencing by the structural proteins of hepatitis C virus. J Gen Virol 89: 2761–2766 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L 2006. The Argonautes. Cold Spring Harb Symp Quant Biol 71: 67–72 [DOI] [PubMed] [Google Scholar]
- Kapadia SB, Brideau-Andersen A, Chisari FV 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci 100: 2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with “antagomirs.” Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G 2010. On track with P-bodies. Biochem Soc Trans 38: 242–251 [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci 103: 18125–18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S, Fritzler MJ, Katz J, Hamazaki T, Terada N, Satoh M, Chan EK 2007. Small interfering RNA-mediated silencing induces target-dependent assembly of GW/P bodies. Mol Biol Cell 18: 3375–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Cullen BR 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 81: 12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR III, Parker R, Hannon GJ 2005a. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005b. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–113 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T 2005. Identification of novel argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Moradpour D, Englert C, Wakita T, Wands JR 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222: 51–63 [DOI] [PubMed] [Google Scholar]
- Moradpour D, Brass V, Bieck E, Friebe P, Gosert R, Blum HE, Bartenschlager R, Penin F, Lohmann V 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol 78: 13278–13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser JJ, Eystathioy T, Chan EKL, Fritzler MJ 2007. Markers of mRNA stabilization and degradation, and RNAi within astrocytoma GW bodies. J Neurosci Res 85: 3619–3631 [DOI] [PubMed] [Google Scholar]
- Norman KL, Sarnow P 2010. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol 84: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. 2010. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 6: e1000764 doi: 10.1371/journal.ppat.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S 2010. RNA silencing as a natural antiviral defense system in mammals: Where are we now? Mol Ther 18: 871–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Gra¨sser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol 76: 4008–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Grakoui A, Rice CM 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci 100: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci 104: 12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W 2009. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med 15: 31–33 [DOI] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Diez J 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci 106: 13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res 19: 2993–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282: 37158–37169 [DOI] [PubMed] [Google Scholar]
- Simons K, Vaz WL 2004. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33: 269–295 [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM 2010. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17: 403–409 [DOI] [PubMed] [Google Scholar]
- Supekova L, Supek F, Lee J, Chen S, Gray N, Pezacki JP, Schlapbach A, Schultz PG 2008. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J Biol Chem 283: 29–36 [DOI] [PubMed] [Google Scholar]
- Targett-Adams P, Boulant S, McLauchlan J 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol 82: 2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Evans MJ, von Hahn T, You S, Rice CM 2007. Studying hepatitis C virus: Making the best of a bad virus. J Virol 81: 8853–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23: 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA 2007. AU-rich-element-mediated up-regulation of translation by FXR1 and Argonaute 2. Cell 128: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2007. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M 2006. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology 130: 883–892 [DOI] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets Argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Yoon JH, Parker R 2010. Coil-in-to snRNP assembly and Cajal bodies. Nat Struct Mol Biol 17: 391–393 [DOI] [PubMed] [Google Scholar]