Editing of mitochondrial pre-mRNAs in kinetoplastid protozoa involves the insertion or deletion of uridine residues. These reactions are dictated by guide RNAs that are partially complementary to the pre-mRNA. The complementary sequence is known as the anchor and it is composed of uridine residues that form canonical or wobble pairs with the pre-mRNA. Using X-ray crystallography, it is shown that the anchor duplex has a structure distinct from a helix with all canonical base pairs, results that suggest that the altered duplex may be an entry site for RNA editing proteins.
Keywords: trypanosome mRNA editing, guide RNA, trans-acting RNA, poly(U), wobble base pairs
Abstract
Guide RNAs bind antiparallel to their target pre-mRNAs to form editing substrates in reaction cycles that insert or delete uridylates (Us) in most mitochondrial transcripts of trypanosomes. The 5′ end of each guide RNA has an anchor sequence that binds to the pre-mRNA by base-pair complementarity. The template sequence in the middle of the guide RNA directs the editing reactions. The 3′ ends of most guide RNAs have ∼15 contiguous Us that bind to the purine-rich unedited pre-mRNA upstream of the editing site. The resulting U-helix is rich in G·U wobble base pairs. To gain insights into the structure of the U-helix, we crystallized 8 bp of the U-helix in one editing substrate for the A6 mRNA of Trypanosoma brucei. The fragment provides three samples of the 5′-AGA-3′/5′-UUU-3′ base-pair triple. The fusion of two identical U-helices head-to-head promoted crystallization. We obtained X-ray diffraction data with a resolution limit of 1.37 Å. The U-helix had low and high twist angles before and after each G·U wobble base pair; this variation was partly due to shearing of the wobble base pairs as revealed in comparisons with a crystal structure of a 16-nt RNA with all Watson–Crick base pairs. Both crystal structures had wider major grooves at the junction between the poly(U) and polypurine tracts. This junction mimics the junction between the template helix and the U-helix in RNA-editing substrates and may be a site of major groove invasion by RNA editing proteins.
INTRODUCTION
Insect-borne parasites from the genera Leishmania and Trypanosoma threaten over 500 million people with debilitating and sometimes fatal infections (St. Georgiev 2009). Current drugs have toxic side effects and drug resistance is emerging (Wilkinson et al. 2008; Rijal et al. 2010). Safer and more effective drugs are needed. These parasites share a RNA editing pathway (Benne et al. 1986) that is absent in humans, essential for parasite survival (Schnaufer et al. 2005), and a potential drug target (Amaro et al. 2008; Liang and Connell 2010; Moshiri et al. 2011). Large (∼20S) ribonucleoprotein editing complexes (Simpson et al. 2004; Carnes et al. 2008)—similar in dimensions and complexity to the ribosome and spliceosome (Golas et al. 2009; Li et al. 2009)—insert or delete uridylates (Us) through enzyme cascades (Seiwert and Stuart 1994; Adler and Hajduk 1997) that are under the direction of hundreds of different guide RNAs (gRNAs) (Blum et al. 1990; Pollard et al. 1990; Ochsenreiter et al. 2007) that bind antiparallel to the pre-mRNA at specific sites. The gRNAs have three functional domains: a 5′ anchor sequence that finds the editing site by forming mostly complementary base pairs and a few G·U wobble base pairs at the anchor-binding site (ABS) in the pre-mRNA, a template domain that directs the editing of often more than one site by mismatched base-pairing, and a 3′ oligo(U) tail (or U-tail) of about five to 24 Us (Blum and Simpson 1990) that a 3′ terminal uridyl transferase (Aphasizhev et al. 2003) adds to the gRNA after transcription. The U-tail binds nonspecifically to the purine-rich pre-mRNA upstream of the editing site (Leung and Koslowsky 2001) to form a double helix (the U-helix) (called the “5′ anchor” by Blum and Simpson [1990] because the duplex forms 5′ to the editing site in the unedited region of the mRNA) that is rich in G·U wobble base pairs.
Some editing substrates initially form a three-helical structure with the editing site at or near the three-way helical junction (Reifur and Koslowsky 2008). The U-helix and the anchor helix flank the editing site, and the template sequence of the gRNA forms a stem–loop opposite the editing site that serves as the third helix of the three-way junction. The formation of the U-helix often enhances the association of the gRNA with the target mRNA (Reifur et al. 2010), improves the stability of some editing substrates (Koslowsky et al. 2004), and protects the U-tail from the editing complex's exonuclease activity (McManus et al. 2000). The ABS in some mRNAs is initially inaccessible to binding by the gRNA because it forms part of a stem–loop structure in the mRNA (Reifur et al. 2010). The anchor helix and U-helix may keep the gRNA's template sequence bound to the pre-mRNA after cleavage of the pre-mRNA during editing, and thereby keep the two internal ends of the cleaved mRNA in close proximity for religation (Koslowsky et al. 2004). After the editosome has finished using the template domain of a particular gRNA, a RNA editing-specific helicase (Li et al. 2011) displaces the gRNA strand from the mRNA, a different gRNA binds to the next ABS 5′ to the last editing site on the mRNA, and the editing reaction cycles continue.
The U-helix is of interest because it is found in almost all editing substrates. The U-tail pairs with the purine-rich unedited mRNA, so the U-helix is rich in G·U wobble base pairs. The structural consequences of the G·U base pairs on the U-helix are unknown, but the asymmetry of the wobble base pairs is expected to add structural variation and distinctive electrostatic surfaces to the major and minor grooves of the U-helix (Masquida and Westhof 2000; Varani and McClain 2000; Xu et al. 2007). In other systems, G·U base pairs sometimes play functional roles by forming cation binding sites (Fan et al. 2005), anion binding sites (Masquida et al. 1999), ligand binding sites (Chow and Barton 1992), RNA tertiary interactions sites (Gagnon and Steinberg 2002; Mokdad et al. 2006), or protein recognition sites (Batey and Williamson 1996). G·U base pairs generally lower the thermal stability of the double helix in a manner that depends on the sequence context and their positions relative to the helical ends (Sugimoto et al. 1986; Mathews et al. 1999).
To gain insight into the structure of RNA editing substrates, we did crystallographic studies of the U-helix. We present a high-resolution (1.37 Å) crystal structure of a RNA 16-nt long with eight consecutive Us. Comparison of this structure to the structure of a 16-nt RNA with the G·U mismatches replaced with G–C Watson–Crick base pairs (the WC-helix) showed G·U-dependent changes in base-pair slide (translation along the long axis of the base step) and roll (rotation about the long axes of a base step) that are consistent with the conformational flexibility often ascribed to G·U wobble base pairs. This conformational flexibility may help modulate the changes in the structure of the gRNA/mRNA duplex during editing, as the editing complex advances from one editing site to the next along the pre-mRNA. In addition, the base step between the poly(U) and polypurine tracts may mimic the junction between the template helix and the U-helix in the RNA editing substrate. This base step was wide enough to allow protein side chains access to the base atoms in the major groove and may be a site of major groove invasion by RNA editing proteins.
RESULTS AND DISCUSSION
Construct design, crystallization, and structure determination
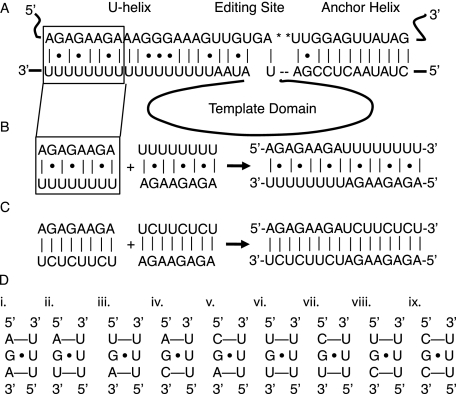
We fused two identical U-helices head-to-head to promote crystallization by (1) reducing the formation of out-of-register duplexes often formed by repeating sequences, (2) overcoming the tendency of poly(U) to form random coils at room temperature (Inners and Felsenfeld 1970), (3) avoiding solubility issues with long purine tracts, (4) preventing the formation of the triple-stranded structures favored by polypurines and polypyrimidines (Hoyne et al. 2000), and (5) increasing the stability of the duplex by doubling the number of base pairs. We made a 16-nt RNA (5′-AGAGAAGAUUUUUUUU-3′) (Fig. 1B, the U-helix) consisting of an 8-nt U-tail fused to the 3′-end of an 8-nt fragment of pre-mRNA. The fusion RNA self-annealed to form two copies of a fragment of the U-helix from the gA6-14 guide RNA/gA6 mRNA editing substrate (Fig. 1A; Seiwert et al. 1996). This fragment was located 16–23 nt 3′ to the second editing site in the gA6 mRNA. The fusion RNA was designed to self-anneal to form a 16-nt duplex with 10 standard A–U Watson–Crick base pairs and six G·U wobble base pairs. As a control on the effects of G·U wobble base pairs in the structure of the U-helix, we made a second RNA (5′-AGAGAAGAUCUUCUCU-3′) (Fig. 1C, the WC-helix) that formed only Watson–Crick base pairs upon self-annealing. The nucleotides were numbered 1–16 starting at the 5′ end of one strand. The residues in the opposing strand were denoted with an asterisk because they were related by crystallographic symmetry.
FIGURE 1.
(A) Base-pairing between the A6 mRNA (top strand) and the gA6[14] guide RNA (bottom strand). The asterisks mark the uridylates that are deleted by editing. The anchor helix is to the right of the editing site, and the U-helix is to the left. The template sequence in the middle of the guide RNA is represented by a loop. The box outlines the fragment of the U-helix that was crystallized. (B) The fragment was fused head-to-head with a duplicate fragment to give the RNA hexadecamer on the right. (C) The G·U base pairs in the U-helix were replaced with G–C base pairs to give the WC-helix on the left. Two copies of the WC-helix were fused head-to-head to give the 16-nt RNA on the right. (D) Triple base-pair motifs possible in U-helices. The first motif (i) occurred in the crystal structure of the U-helix.
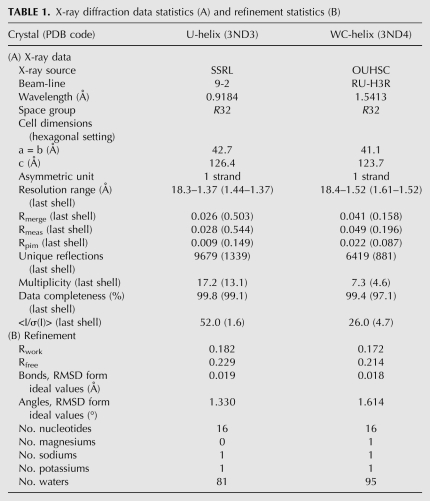
Crystals of both RNAs were grown from solutions containing 1–2 M Li2SO4. Satisfactory cryoprotection was achieved by passing the crystals rapidly through 1.9, 2.4, and 2.9 M Na malonate (pH 6.0) prior to flash cooling in a nitrogen cryostream. A single crystal of the U-helix gave 1.37 Å X-ray diffraction data with synchrotron radiation at SSRL beamline 9-2, and a single crystal of the WC-helix gave 1.52 Å X-ray data with in-house generated X-rays (Table 1). Both RNAs formed rod-shaped crystals with the same crystallographic symmetry (R32) (Table 1). The X-ray data were indexed in the hexagonal setting (H32). The lengths of the c edges of the two H32 unit cells differed by 2.2%, so they were not quite isomorphic. The volumes of the asymmetric units suggested that each contained one RNA strand (Table 1). The unit cell volumes per base pair (U-helix: 1,386 Å3/bp and WC-helix: 1,117 Å3/bp) were similar to values reported for the crystal structures of other double-stranded RNAs (dsRNAs) (e.g., 1350 Å3/bp for a 16-nt RNA in H3) (Pan et al. 1998).
TABLE 1.
X-ray diffraction data statistics (A) and refinement statistics (B)
Molecular replacement trials with the H32 X-ray data and one strand of RNA from a fiber diffraction-derived model failed. Trials succeeded with H3 X-ray data and a dsRNA search model. The best trial solution placed the search model's molecular dyad coincident with the crystallographic (y,x,−z) dyad in the H32 unit cell, suggesting that H32 was the correct space group. However, this dyad was parallel to the twinning operator (k,h,−l) in the H3 unit cell. We had three possible situations: H3 symmetry with the RNA's molecular dyad generating higher apparent symmetry in the diffraction pattern, H3 symmetry with twinning generating higher apparent symmetry, or H32 symmetry. Extensive refinements of dsRNA models with H3 X-ray data and the twinning operator failed to lower the Rfree significantly compared with refinements in H3 without the twinning operator. These results left H32 as the most appropriate choice for the space group. The final crystallographic Rfree values (Table 1) fell at or below the median Rfree value (median: 0.23, lower quartile: 0.21, upper quartile: 0.26 for a non-Gaussian distribution) for the 18 crystal structures of RNA hairpins or double-stranded duplexes in the resolution range of 1.37 to 1.53 Å (Protein Data Bank searched 7 June 2011).
Overall structures
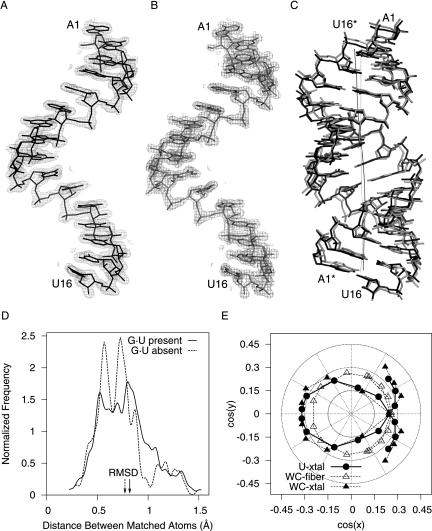
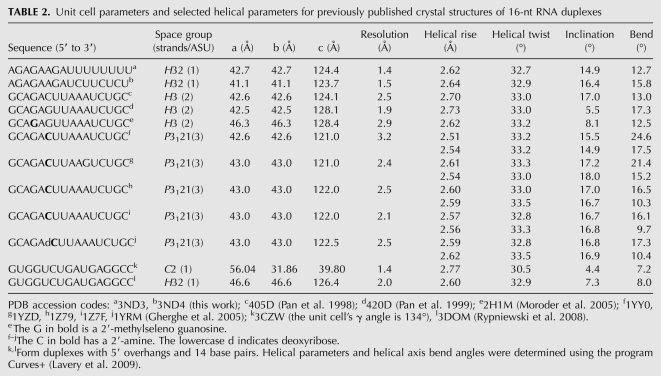
Both RNAs crystallized as right-handed, A-form, double-stranded helices with 16 bp and blunt ends (Fig. 2C). All of the nucleotides (including all of the backbone atoms) were clearly visible in the electron density (Fig. 2A,B). The U-helix adopted had an average helical rise (2.54 Å) that was within the range observed for single crystal structures of duplex RNAs (Table 2) and shorter than that for A-form fiber RNA (2.8 Å) (Arnott et al. 1973). The helical repeat of the U-helix was 10.9 base pairs per helical turn, which classifies it as a member of the A-form family (11 bp/turn) rather than the A′-form family (12 bp/turn) (Arnott et al. 1972). The average inclination angle of the U-helix was 15.3°, which was close to 16.1° expected for the A-form as opposed to 10.3° for the alternate A′-form, which has a wider major groove that is accessible to proteins.
FIGURE 2.
Comparisons of the U-helix and WC-helix RNA structures. Electron density (2m|Fo| – D|Fc|, 1.5 σ contour level) around the single-strand in the asymmetric unit of the crystal structures of the U-helix (A) and the WC-helix (B). (C) The crystal structure of the U-helix in black superposed on the crystal structure of the WC-helix in gray (RMSD = 0.78 Å when the G·U base pairs are excluded). The dark curve denotes the helical axis of the U-helix. The light curve denotes the helical axis of the WC-helix. (D) Probability distributions of the distances between corresponding atoms in the two superposed structures when all nucleotides (666 atoms) are used in the superposition (solid line) and when the six G·U base pairs are excluded (414 atoms) from the superposition (dashed line). (E) The direction cosines of the base-pair plane normal vector when the helical axis is aligned along the z-axis of the coordinate system.
TABLE 2.
Unit cell parameters and selected helical parameters for previously published crystal structures of 16-nt RNA duplexes
The U-helix was similar but not identical to the Watson–Crick helix (Fig. 2C). The coordinate uncertainty for each structure was ∼0.1 Å, so the uncertainty in the distances between corresponding atoms was ∼0.2 Å. The distribution of the paired distances started at a value larger than 0.2 Å (Fig. 2D), so all of the paired distances were significant. When the G·U base pairs were excluded from the comparison of the two structures, the distribution of the distances between corresponding atoms was approximately Gaussian (Fig. 2D). When the G·U base pairs were included in this comparison, the number of longer distances increased, the number of shorter distances decreased, and the distribution became more asymmetric, which reflected the systematic differences in structure introduced by the six G·U wobble base pairs that had a different shape from the remaining 12 A–U Watson–Crick base pairs. The average values of the helical parameters for both RNAs were similar to each other and to those of other 16-nt RNA crystal structures (Table 2).
The 5′-AGA-3′/5′-UUU-3′ motif
The presence of only Us in one strand of the U-helix limits to nine the possible nearest-neighbor sequence motifs that have a central G·U wobble base pair (Fig. 1D). Isolated G·U base pairs in U-helices are flanked by Watson–Crick A–U base pairs on both sides (Fig. 1D, i), on one side (Fig. 1D, ii–v), or by pyrimidine mismatches on both sides (Fig. 1D, vi–ix). Similar sequence considerations limit tandem G·U wobble base pair motifs in U-helices to the nonsymmetrical 5′-GG-3′/5′-UU-3′ motif (Gautheret et al. 1995). The crystal structure of the U-helix fragment provides three views of the only motif with two flanking A–U base pairs: 5′-AGA-3′/5′-UUU-3′ (Fig. 1D, i). The sequence 5′-AGA-3′ occurred in 37 RNA crystal structures with double helical fragments (Nucleic Acid Database accessed 25 May 2011), but none of these structures had 5′-UUU-3′ in the strand opposing 5′-AGA-3′.
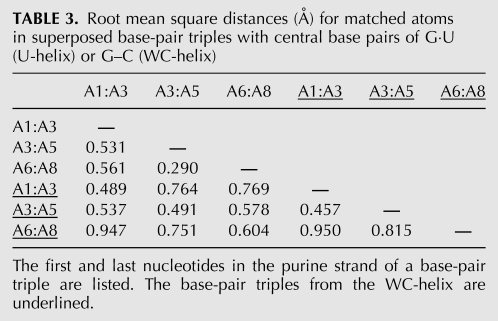
Pairs of base-pair triples were superposed, and the root-mean-square deviations (RMSDs) were measured (Table 3). These measurements were repeated with the corresponding base-pair triples in the crystal structure of the WC-helix and then between all pairs of base-pair triples in the two crystal structures (Table 3). The second and third base-pair triple in the U-helix were the most similar base-pair triples; their RMSD approached the experimental error in the distances between corresponding atoms. Crystal packing effects at the helical ends caused the first base-pair triple in the U-helix to differ more from the other two base-pair triples in the U-helix. The triples in the U-helix were more similar to each other than the corresponding base-pair triples in the WC-helix. The discrepancies between triples in the WC-helix were similar in magnitude to the discrepancies between triples from the U-helix and the WC-helix. The extended backbone of G7 in the WC-helix (described below) contributed to the large discrepancies measured in comparisons that included the third base-pair triple.
TABLE 3.
Root mean square distances (Å) for matched atoms in superposed base-pair triples with central base pairs of G·U (U-helix) or G–C (WC-helix)
Helical axis bending
The paths of the helical axes were determined with CURVES+ (Fig. 2C; Lavery et al. 2009). The total bending angles of both RNAs were in the range found for other 16-nt RNAs (Table 2).
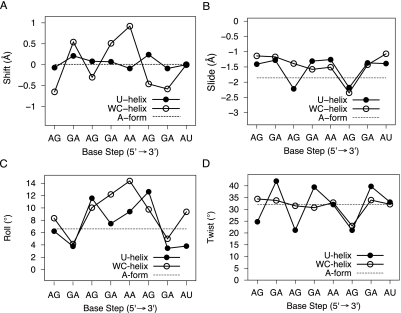
Base pair normal plots (Fig. 2E) were made with FREEHELIX (Dickerson 1998) and gave an independent analysis of helical axis bending. The straight helical axis of each RNA was calculated and aligned along the z-axis with FREEHELIX. The base pair normal vectors pointed along the helical axis, but the inclination and tilting of the base pairs in the A-form gave the base pair normal vectors significant x and y components that formed a large circle in the base pair normal plot (Fig. 2E). In contrast, both crystal structures gave noncircular plots that were symmetrical due to the molecular dyad. The angle between the base-pair normal vectors of the first and twelfth base pairs was 0° in the A-form model as expected for a double helix with a straight axis. This angle was 11° in the U-helix and 12° in the WC-helix; these values correspond well to the total bending angles reported in Table 2. The largest angles between adjacent base pair normal vectors in the U-helix occurred before each of the three unique G·U wobble base pairs (11°), which agreed with the large roll angles (positive rotation angles about long axes of base steps that indicated bending toward the major groove) at third and sixth base steps (Fig. 5C, below). The largest angles between adjacent base pair normal vectors in the WC-helix occurred at the fourth (12°) and fifth (14°) base steps; these values correlated with the large roll angles at these base steps (Fig. 6B, below).
FIGURE 5.
Base-step parameters for the first eight base steps (values for the last seven base steps are identical to the first seven due to symmetry): (A) shift, (B) slide, (C) roll, and (D) twist.
FIGURE 6.
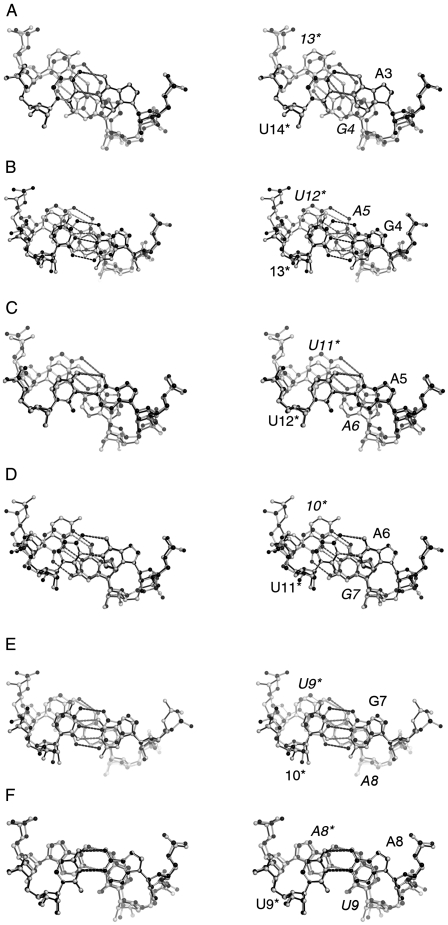
Base stacking in the U-helix and WC-helix. Stereo views made with PyMOL of the third to eighth base steps: (A) A3pG4, (B) G4pA5, (C) A5pA6, (D) A6pU7, (E) U7pA8, and (F) A8pU9. Corresponding base steps from each structure are superposed using the C1′ and N1 or N9 atoms of the base pairs closest to the viewer. Residues in the opposing strand have asterisks. The residues numbers for the base pairs far from the viewer are in italics. The single letter residue code is not given when either a C or U occupy a site. The bonds of the U-helix structure are colored black. The bonds of the WC-helix structure are colored light gray. The right-handed screws of the helical axes project out of the plane of the page.
Backbone torsion angles
All of the ribose rings were puckered in the C3′-endo conformation, and all of the bases were in the anti conformation about the glycosidic bond. The backbone torsion angles α (O5′–P) and γ (C4′–C5′) of all nucleotides were in the preferred gauche–, gauche+ conformation except for G7 in the WC-helix, which adopted the extended trans, trans-conformation (Fig. 6E, below). This extended backbone conformation was associated with a decrease in helical twist (Fig. 5D, below), a longer distance between phosphates (6.7 Å vs. 6.0 Å at the corresponding base step in the U-helix), and magnesium binding (Fig. 8A, below). The trans, trans-backbone conformation was previously associated with helix axis bending (Shi et al. 1999), was found with guanines in the syn conformation about the glycosidic bond (Pan et al. 1999; Kiliszek et al. 2011), and was thought to be common in G·U wobble base pairs in isolation or in tandem (Pan et al. 1998). However, none of the three isolated G·U wobble base pairs in the U-helix had the extended trans, trans-backbone conformation.
FIGURE 8.
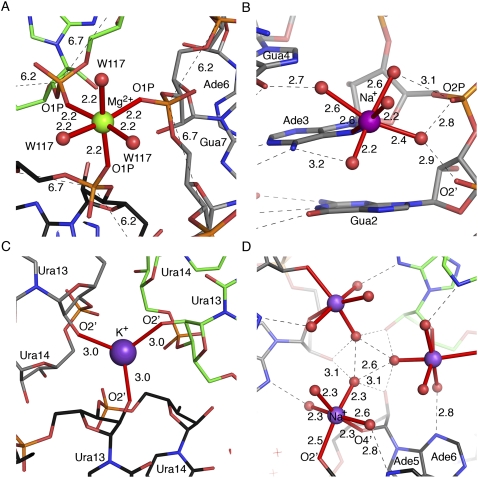
Metal binding sites in the crystal structures of the WC-helix (A,B) and U-helix (C,D). Distances are in Ångstroms. The figure in B is on a larger scale. The carbon atoms of symmetry-related molecules are colored green, black, and gray. The magnesium cation is colored green, and sodium and potassium cations are colored purple. (A) A magnesium-water complex [Mg2+(H2O)3] on a threefold crystallographic axis. The magnesium bound oxygen atoms of the phosphate of G7 and water 117. The distances between adjacent phosphates are shown. The typical distance in A-form RNA is 5.7 Å. (B) A sodium cation binds in the major groove with inner sphere coordination to the N7 nitrogen atom of A3. (C) A potassium cation sits on a crystallographic threefold axis and bridges three symmetry-related U13 phosphate O1P oxygen atoms. (D) A sodium–water complex [Na+(H2O)4] bound to the ribose O2′ hydroxyl of A5 and O4′ ring oxygen of A6 in the minor groove near a crystallographic threefold axis; the symmetry-related distances and labels are not shown.
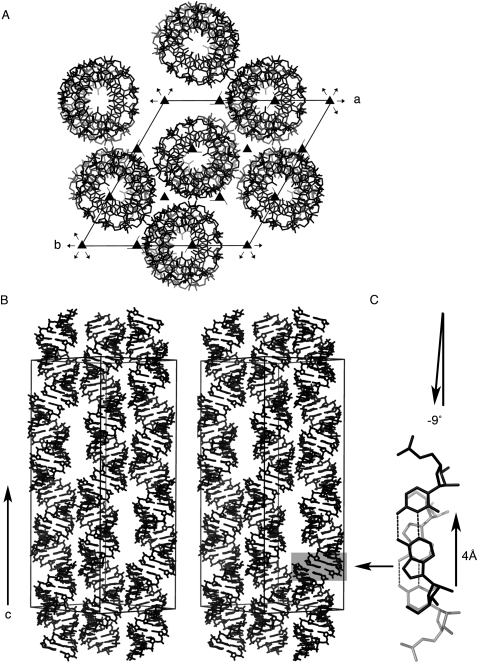
Crystal packing
Contacts with neighboring RNA helices in the crystal lattice determine the external forces acting on the double helix through contacts with neighboring RNAs. In both crystal structures, the helical axis of the RNA aligned parallel to the c-axis of the unit cell, and the molecular dyad superimposed on a crystallographic dyad, so that each strand was exposed to identical crystal packing forces. The double helices stacked parallel to the c-axis with their helical axes offset by a 4 Å translation along the long axis of the terminal base pair (Fig. 3C) and with a left-handed twist (−9°) between the terminal base pairs rather than the right-handed twist of ∼33° required for a pseudocontinuous helix. The translation along the long axis of the terminal base pair led to extreme cross-strand base stacking and positioned the O4′ oxygen atoms of the ribose rings over the six-membered rings of the uridines. Similar staggered stacking of helices was also found in the crystal structures of 16-nt RNAs that crystallized in H3 with two strands in the asymmetric unit (Pan et al. 1998, 1999).
FIGURE 3.
Crystal packing of the U-helix RNA. (A) View down the c-axis of the R32 unit cell in the hexagonal setting (H32). The molecular dyad (data not shown) of the RNA duplex in the center of the unit cell lies on the twofold rotation axis that runs along the short diagonal of the a × b face of the unit cell. (B) Stereo view along the short diagonal in the a × b plane. (C) Stick model of two adjacent terminal base pairs in a stack of helices.
The neighboring double helices packed side-by-side in a pseudo-hexagonal fashion aligned in layers normal to the c-axis of the unit cell. Direct contacts occurred via backbones packing in the minor grooves. In the crystal structure of the U-helix, eight of the 16 nucleotides in a strand formed 15 contacts—10 of which were hydrogen bonds (Table 4). In the crystal structure of the WC-helix, nine of the 16 nucleotides in a strand formed 21 contacts—nine of which were conventional hydrogen bonds. The two crystal structures shared only six contacts—four of which were hydrogen bonds. Two hydrogen bonds involved the free N2 nitrogen atom of G2 in the minor groove in the U-helix—an interaction that was not possible in the WC-helix because the N2 nitrogen atom was involved in base-pair formation with C13. In contrast, the 16-nt RNA r(GCAGACUUAAAUCUGC) that crystallized with H3 symmetry formed 11 contacts (five conventional hydrogen bonds), so the H32 crystal structures had three to four times as many hydrogen bonds. Additional indirect interactions between backbones were mediated by cations on or near the crystallographic threefold axes (Fig. 8, below) as discussed below under metal binding.
TABLE 4.
Contacts between double helices (distances <3.3 Å)
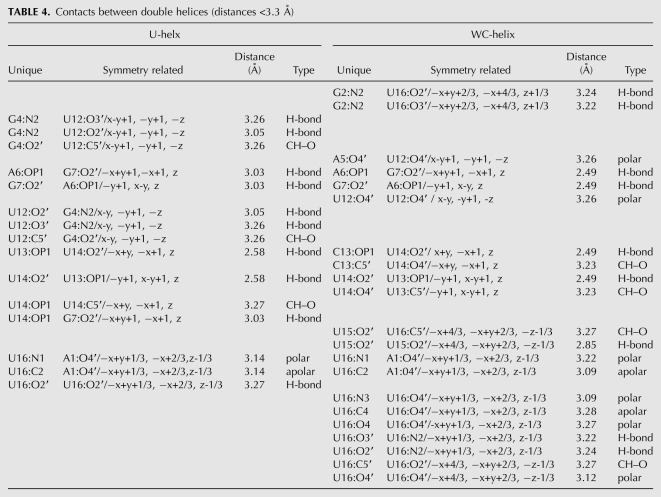
Base-pair parameters
The base-pair reference frame in 3DNA is taken with the x-axis along the short axis of the base pair and the positive direction as pointing away from the minor groove, the y-axis along the long axis of the base pair, and the z-axis along a normal to the base-pair plane. The orientations of the individual bases in a base pair are described by six base-pair parameters (i.e., translations and rotations about x, y, and z relative to the base-pair axial system: buckle, propeller, opening, stretch, shear, and stagger). Contact with the terminal base pair of a neighboring duplex caused a large buckle angle (6.35°) in the terminal base pair of the U-helix compared with the magnitudes (0.25° to 3.30°) for the other base pairs, whereas a slight difference in this interaction in the WC-helix crystal structure did not lead to significant buckle in the terminal base pairs of the WC-helix (Fig. 4A). Instead, the A6–U13 base pair in the WC-helix had a large buckle. This base pair was part of a base step that was also associated by helix axis bending. The two central A–U base pairs at the junction between the polypurine and the polypyrimidine tracts had large propeller twist in both structures (U-helix: −15.7°, WC-helix: −14.5°) that suppressed the roll angles and prevented the helical axes from bending at this base step (Fig. 4B). The G·U base pairs had large shear values that reflected the translation of the G toward the minor groove and the U toward the major groove (Fig. 4C). The base pairs in the WC-helix had insignificant base-pair shear values (Fig. 4C). The remaining base-pair parameters (opening, stretch and stagger) did not have significantly large values in both crystal structures.
FIGURE 4.
Base-pair parameters in the crystal structures of the U-helix (•) and the WC-helix (○): (A) Buckle, (B) propeller twist, (C) shear, (D) the relation between the change in base-pair shear and twist angle between adjacent base pairs. The line was fit to the U-helix data (R2 = 0.97). The base sequence (5′ to 3′) is along the x-axis for the U-helix (top) and WC-helix (bottom). Buckle and shear are inverted in sign about the molecular dyad in the center of the double helix (A,C).
Base-step parameters
The base-step parameters (translations and rotations about x, y, and z: shift, slide, rise, tilt, roll, twist, respectively) describe the orientation of two adjacent base pairs relative to their common base-step reference frame. The values of these parameters depend on the base-pair reference frame used to define the base-pair parameters described in the previous section (Olson et al. 2001). 3DNA's Watson–Crick base-pair reference frame was used for all of the base pairs in spite of the asymmetry of the G·U wobble base pairs. The application of this reference frame to wobble base pairs led to larger and smaller twist angles for motions of the wobble base pairs that may be better described by the base-step parameters slide and shift (Lu and Olson 2003). In the U-helix, the apparent twist values were low in the ApG base steps and high in the GpA base steps; in the WC-helix, the twist values were close to the average for A-form RNA, except at the sixth base step (A6pG7), where it was low. The change in shear between adjacent base pairs was strongly correlated with twist in the U-helix; a similar relation over a smaller range of shear values was reported for a group of A-DNA and B-DNA crystal structures (Olson et al. 2001).
Slide describes the translation of one base pair relative to the other base pair along the long axis of the base step. The A3pG4 base step in the U-helix had a larger slide than the corresponding base step in the WC-helix while the A6pG7 base step in both structures had similar slide and twist (Fig. 5B,D). The A6pG7 base step in WC-helix had an extended backbone associated with the large slide and low twist.
Shift describes the translation along the short axis of the base-pair step, and the signs of its values were symmetric about the molecular dyad. Both structures had mean shift values of 0.0 Å, because both halves of the duplexes were identical. The U-helix had a standard deviation of 0.14 Å and the WC-helix had a standard deviation of 0.59 Å. The largest values of shift in the U-helix were at GpA base steps (0.07–0.24 Å), whereas the ApA base step in the WC-helix had the largest value (0.92 Å).
Roll is the angle between base pairs about the long axis of the base step, with positive values indicating opening of the base step toward the minor groove. Large positive roll values also indicate bending of the helical axis toward the major groove. The A-form model had average roll angle values of 6.6° as a consequence of the base pairs spiraling around a straight helical axis. Larger roll angles were associated with bending of the helical axes toward the major groove. The variation in the roll angles values in the U-helix depended on the presence of G·U base pairs (Fig. 5C). The high-twist GpA base steps in the U-helix had low roll values, and the low-twist, large-slide ApG base steps had high roll values. The G2pA3 and G7pA8 base steps in the WC-helix had low roll values. The different patterns of roll angle values reflected the different patterns of bends in the helical axes.
Base stacking
Although the side view of the superposition of the U-helix and WC-helix (Fig. 2C) suggested that the structures were similar, views of the corresponding superposed base steps revealed conformation differences in base steps both with and without G·U base pairs. The first and second base steps were excluded from analysis due to end effects. The central A8pU9 base steps were the most similar (Fig. 6F). There were only some minor differences in the positions of the phosphates. This lone purine–pyrimidine base step had only intrastrand stacking of the bases due to the large propeller twist of the individual base pairs that prevented sliding of one base pair across the adjacent base pair.
The A5pA6 base step was the only other base step that did not include a G·U wobble base pair in the U-helix (Fig. 6C). This step had an almost exact match between the A5–U12 base pairs of both structures but not between the A6–U11 base pairs. The largest roll angle (14°) in the WC-helix occurred at this base step (Fig. 5C) and led to the difference in the positions of the A6–U11 base pairs. This difference in roll angle was a sequence dependent feature.
The A–U base pairs in the two ApG base steps (Fig. 6A,D) superposed very well, as expected. The largest discrepancies between the base steps were in the positions of the 5′ pyrimidine (Fig. 6A, 13* and Fig. 6D, 10*). The discrepancies around the 5′ pyrimidine were larger in the A6pG7 base step because the extended backbone of G7 in the WC-helix brought into alignment the ribose rings and bases of G7 in both structures (Fig. 6D), so residues in positions 6, 7, and 11* superposed well, while those in position 10* did not. In position 10*, the pyrimidine rings and backbone atoms had very little overlap between the two structures. In the A3pG4 base step, the discrepancies were evenly distributed across residues 4 and 13* and were larger than the ones seen when G·U base pairs were superposed on G–C base pairs (Fig. 6B). (The G·U wobble and Watson–Crick base pairs have similar C1′–C1′ distances, but different λ angles that subtend the glycosidic bond and the C1′–C1′ vector.) The larger differences in the positions of residues 4 and 13* in the two structures reflected the large difference in twist angles at this base step (Fig. 5D). The A3pG4 base step in the U-helix had less intrastrand base overlap, as expected in low twist base steps and as seen in the low twist A6pG7 base step of the WC-helix.
The base stacking in the two analyzed GpA base steps (Fig. 6B,E) were similar. The base-steps from the U-helix had larger apparent helical twist and larger intrastrand base overlap than the corresponding base steps in the WC-helix.
The large discrepancies in the position of the backbones of the 5′ pyrimidine in base steps with G·U or G–C base pair suggest significant twist motion about the helical axis.
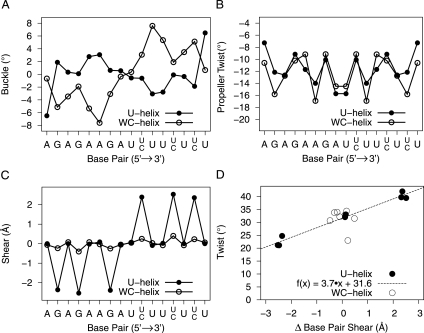
Major groove width
A-form RNA (11 bp/helical turn) 6 bp or longer has a major groove width of 3.8 Å that is too narrow to allow an α helix or a β strand to enter the major groove (Weeks and Crothers 1993). In comparison, A′-form RNA (12 bp/helical turn) has a major groove width of 8 Å (Tanaka et al. 1999), and B-DNA has a major groove width of 11.4 Å. Both of these latter grooves are wide enough for protein access. We used the method of El Hassan and Calladine (1998) as implemented in 3DNA to measure the refined phosphate–phosphate distance across the major grooves of the U-helix and WC-helix. This method takes into account the directions of the sugar-phosphates backbone. We subtracted 5.8 Å from each major groove width to allow for the van der Waals surface of the phosphates. The widths of the major groove of the first three base steps at each end of the 16-nt RNA crystal structures were impossible to measure directly due to end effects. The groove widths of the fourth through seventh base steps were related to the groove widths of the ninth through twelfth base steps by twofold rotational symmetry. The groove widths of the U-helix and the WC-helix increased toward the middle of the helix and exceeded 8 Å at the central and flanking base steps (Fig. 7C–E). The widest parts of the major grooves were similar in width to those parts of the K10 TLS RNA associated with protein binding (Bullock et al. 2010).
FIGURE 7.
Electrostatic potential mapped onto solvent accessible surface (probe radius 1.4 Å) of the U-helix (A,C) and the WC-helix (B,D). The electrostatic potential ranges from −5 kbT/e to 1 kbT/e− because the negative potential of the major groove is dominant. The N2 nitrogen atoms of the guanines are represented by blue spheres scaled by 0.7 of their van der Waals radius. (A,B) Views toward the shallow groove (minor groove) in the center. (C,D) Views toward the deep groove (major groove) in the center. (E) Major groove widths—measured with 3DNA—of the U-Helix and the WC-helix by base step.
Electrostatics
The electrostatic potential was calculated and mapped onto the solvent accessible surface (Fig. 7A–D) of each RNA. Both helices had high negative electrostatic potential in the major groove and along the backbones. The minor groove of the U-helix differed from that of the WC-helix by having patches of low positive potential (Fig. 7A,C) in the vicinity of the six guanine N2 nitrogen atoms. Different U-helices will have different patterns of positive potential patches in the minor groove. A subset of these patches may play roles in protein recognition.
Cation binding sites
Cation-binding sites were identified by the reliable distances and geometry of the ligands provided by the high-resolution X-ray diffraction data (Harding 2002). Sodium and potassium binding sites were identified in both crystal structures, but a magnesium-binding site was identified only in the WC-helix. One sodium–water complex [Na+(H2O)5] was found in the major groove of the WC-helix. The sodium atom bound directly to the N7 nitrogen atom of A3, and the water ligands bridged three adjacent purines (G2, A3, and G4) with five hydrogen bonds (Fig. 8B). The sodium cation in a sodium–water complex [Na+(H2O)4] bound directly to the O2′ hydroxyl of A5 and the O4′ of A6 on the minor groove side of the backbone without changing the backbone torsion angles (Fig. 8D). The sodium–water complex bridged two double helices by joining their minor grooves with hydrogen bonds to the N3 nitrogen atoms of A6 and A5*. Three copies of the sodium–water complex surrounded a crystallographic threefold rotation axis and were involved in direct hydrogen bonds with each other and the O2′ hydroxyl of A6 and a symmetry-related A6. The sodium–water complexes were probably introduced during cryoprotection with 3 M sodium malonate. A potassium cation on a crystallographic threefold axis bound to the O2′ hydroxyl of U13 in three symmetry-related molecules in both crystal structures (Fig. 8C). The crystallographic threefold axes of both structures appeared to have disordered sulfate anions. Refinement of sulfate ions on these axes failed to converge, so we deleted the sulfates long before the refinements of both crystal structures were completed.
A magnesium–water complex [Mg2+(H2O)3] sat on a crystallographic threefold rotation axis and bridged the outer phosphate O1P oxygen atoms of G7 in three symmetry-related strands in the WC-helix structure (Fig. 8A). The G7 backbone was extended in the trans, trans-conformation with a distance of 6.7 Å between the phosphates of G7 and A8. The phosphate–phosphate distance between A6 and G7 was 6.2 Å, whereas the average phosphate–phosphate distances of the remaining base steps was 5.7 Å. The geometry of the corresponding site in the crystal structure of the U-helix did not support the presence of a magnesium cation, and the backbone was not extended. Instead, a water molecule occupied this site.
Solvent structure
We compared the unique water molecules (waters nearest the one-strand of RNA in the asymmetric unit had been identified earlier) of the two RNAs after superimposing the WC-helix on the U-helix using all of the RNA atoms. Water molecules closer than 2.4 Å were treated as the same water molecule and were assigned the same residue number in the final coordinate files. There are 50 matched pairs of water molecules with a RMSD of 1.21 Å, 34 unmatched waters in the U-helix structure, and 44 unmatched waters in the WC-helix structure.
The most striking differences in the hydration patterns occurred around the G–C and G·U base pairs. Waters bridged both strands in both the major and minor grooves of the G·U base pairs in the U-helix. In the minor groove, a water bridged the guanine N2 nitrogen atom and the uridine O2′ hydroxyl oxygen atom. In the major groove, a water molecule bridged the guanine O6 and uracil O4 oxygen atoms. The corresponding G–C base pairs in the WC-helix lacked waters that bridged both strands in the minor or major grooves.
The widening of the major groove by 6 Å from the fourth base step to the eighth base step (Fig. 6E) did not change the number of first shell waters in the major groove. Overall, the hydration patterns of the base pairs were similar to the average pattern found in an analysis of crystal structures of dsRNA (Auffinger and Westhof 1998).
Biological implications of the crystal structure of the U-helix
The A-form conformation of the U-helix sequesters the U-tail in a squat helix in which all Us form two-hydrogen bonds with the purines of the pre-mRNA strand, are well-stacked, and have low solvent accessibilities. The compact conformation of the U-helix protects the U-tail from editing exonucleases that target single-stranded RNAs (McManus et al. 2000). Likewise, a U-helix that contains a nascent ABS may be able to protect it from premature binding by the next gRNA.
The U-helix may stabilize the editing substrate by binding the pre-mRNA downstream to the editing site and holding the 3′-end of the gRNA's template domain in position during the editing reaction, while the anchor domain holds the 5′-end of the template domain in position (Koslowsky et al. 2004). As mismatches between the template sequence and the mRNA are removed by editing, the duplex that started as the anchor helix grows in length and increases in stability relative to the U-helix, which probably remains constant in length. Large differences in stability between the anchor helix and the U-helix may be a means by which RNA binding proteins distinguish the U-helix from the anchor helix.
U-helix may also play a role in alternative RNA editing in which gRNAs with the same anchor sequence, but different template sequences, make transcripts that encode different proteins (Ochsenreiter and Hajduk 2006; Ochsenreiter et al. 2008). Some alternate gRNAs may form less-stable editing substrates than their canonical counterparts due to a greater number of mismatches with the pre-mRNA. The binding of the mRNAs by U-helices may make larger positive contributions to the stability of less stable RNA-editing substrates.
CONCLUSIONS
The crystal structure of the U-helix showed that the U-tail forms standard Watson–Crick A–U and wobble G·U base pairs with the pre-edited mRNA. The crystal structure of the U-helix contained three samples of the 5′-AGA-3′/5′-UUU-3′ sequence motif—the only single G·U base-pair motif possible in the U-helix that has two flanking Watson–Crick base pairs. The asymmetry of the wobble base pairs introduced sequence-dependent base-pair shear and patches of positive electrostatic potential in the shallow groove that may be important in protein recognition. The junction between the two fused U-helices may mimic the junction between the U-helix and the template domain. The major groove at this junction was wide enough to allow protein access.
MATERIALS AND METHODS
RNA purification and crystallization
RNA oligonucleotides were designed to self-hybridize to form two U-helices fused head-to-head (5′-AGAGAAGAUUUUUUUU-3′) and two WC-helices fused head-to-head (5′-AGAGAAGAUCUUCUCU-3′). The RNAs were synthesized, gel-purified, and desalted by Dharmacon Research (Lafayette) and then resuspended at a concentration of 0.33 mM in 10 mM sodium cacodylate (pH 6.5). To promote proper duplex formation, the RNAs were heated to 85°C for 3 min and cooled in 90 min to 22°C. When the temperature dropped to 40°C, MgCl2 was added to a final concentration of 10 mM. The RNA solutions were passed through 0.22-μm filters prior to crystallization experiments.
Crystals grew at 22°C by vapor diffusion using the hanging drop method. We mixed 2 μL of RNA in annealing buffer with 2 μL of reservoir solution to make the crystallization drops. The U-helix RNA crystallized as triangular blocks from 50 mM Na cacodylate (pH 6.0), 10 mM MgCl2, and 1 M Li2SO4. The WC-helix RNA crystallized as triangular blocks from solutions containing RNA at 0.25 mM, 50 mM sodium cacodylate (pH 6.5), 15 mM MgCl2, 10 mM spermine, and 2 M Li2SO4. Crystals appeared in 1 wk. Crystals were moved sequentially through drops of 1.9, 2.4, and 2.9 M Na malonate (pH 6.0) prior to vitrification at –170°C in a nitrogen cryostream.
X-ray diffraction data collection and processing
X-ray data for the U-helix RNA were collected from a single crystal at 100 K with a Mar 325 CCD detector at beamline 9-2 of the Stanford Synchrotron Radiation Lightsource (SSRL). X-ray data were collected at a temperature of –170°C and at a long distance with the X-ray beam attenuated 99% to properly measure the intense low-resolution reflections. The detector was moved closer to the crystal to collect the high-resolution data using the nonattenuated X-ray beam. The X-ray data from the crystal of the WC-helix were collected in-house at –170°C with a Rigaku RU-H3R X-ray generator operating at 50 kV and 96 mAmps, a Varimax-HF confocal optical system, a Raxis-IV image plate, and a X-stream cryosystem. The X-ray diffraction data were indexed and integrated with XDS (Kabsch 1988) and scaled with SCALA (Evans 2006). PHENIX was used to aid space group assignment, detect translational pseudo-symmetry, test for the presence of twinning operators, and assign 10% of the reflections to a test set to monitor Rfree, while taking into account lattice symmetry and noncrystallographic symmetry-related reflections (Adams et al. 2010).
Structure determination
The structure of the U-helix RNA was determined by molecular replacement searches using MOLREP (Vagin and Teplyakov 1997). A 16-bp A-RNA search model was built with COOT (Emsley et al. 2010) and used with X-ray data reduced in space group H3 (R3 in the hexagonal setting). The best solution had the molecular dyad of the RNA placed coincident with a crystallographic twofold in H32 (R32 in the hexagonal setting). A single strand from the correct solution was used as the starting model for refinement against the H32 data. The model was refined initially using simulated annealing with PHENIX (Adams et al. 2010). The models were iteratively rebuilt using COOT, and the coordinates were refined further with PHENIX. Automated water picking was done with PHENIX using m|Fo| – D|Fc| maps. Each water was inspected for fit to 2m|Fo| – D|Fc| electron density and hydrogen bonds with the RNA. The anisotropic temperature factors were introduced at the midpoint in refinement and significantly decreased Rfree. MOLPROBITY (Davis et al. 2004) was used to check for bad contacts. RNABC (Wang et al. 2008) was used to check for bad backbone stereochemistry.
The structure of the WC-helix RNA was determined by molecular replacement using the same protocol as for the U-helix and refined using the above protocol. Refinement trials with anisotropic temperature factors lowered Rwork but not Rfree, so isotropic temperature factors were retained. The structure factors and models were deposited in the Protein Data Bank (accession codes: 3ND3, 3ND4).
Structure analysis
Helical parameters were calculated using 3DNA (Lu and Olson 2008). CURVES+ (Lavery et al. 2009) was used to find the helical axes. FREEHELIX (Dickerson 1998) was used to calculate the base pair normal vectors. The probability distributions of the paired atom distances were made with the kernel density estimation method using gnuplot (Janert 2010) after bandwidth optimization for Gaussian kernels (Shimazaki and Shinomoto 2009). Contacts to symmetry-related RNAs were found with the CCP4 (Winn et al. 2011) program NCONT. To prepare the coordinates for electrostatic calculations, hydrogen atoms, atomic charges, and atomic radii were assigned with PDB2PQR (Dolinsky et al. 2004) using the AMBER94 force-field parameters (Cornell et al. 1995). Electrostatic potentials were calculated with the Adaptive Poisson-Boltzmann Solver (APBS) (Baker et al. 2001) using the Non-linear Poisson–Boltzmann (NLPB) equation. The NLPB equation was solved for a solution with 150 mM Na, 150 mM Cl, a solute dielectric of 2.0, and a solvent dielectric of 78.5. The electrostatic maps were essentially unchanged when recalculated with 3.0 M Na malonate (the cryoprotectant) and a solvent dielectric of 65. Molecular images were prepared with PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
ACKNOWLEDGMENTS
We thank Drs. Tzanko Doukov, Peter Dunten, and Irimpan Mathews for help with data collection at Stanford Synchrotron Radiation Lightsource (SSRL) beam line 9-2. The Structural Molecular Biology Program at SSRL is supported by DOE-OBER, NIH-NCRR (P41RR001209), and NIH-NIGMS. We thank Dr. Michael Quillin for permission to use his program RESOLVE. This work was supported by grants to B.M. from the OUHSC College of Medicine Alumni Association, the Presbyterian Health Foundation (PHF#1545-Mooers), and the Oklahoma Center for the Advancement of Science and Technology (OCAST HR08-138).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2880311.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler BK, Hajduk SL 1997. Guide RNA requirement for editing-site-specific endonucleolytic cleavage of preedited mRNA by mitochondrial ribonucleoprotein particles in Trypanosoma brucei. Mol Cell Biol 17: 5377–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA 2008. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci 105: 17278–17283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L 2003. A tale of two TUTases. Proc Natl Acad Sci 100: 10617–10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S, Hukins DW, Dover SD 1972. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun 48: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Arnott S, Hukins DW, Dover SD, Fuller W, Hodgson AR 1973. Structures of synthetic polynucleotides in the A-RNA and A′-RNA conformations: x-ray diffraction analyses of the molecular conformations of polyadenylic acid·polyuridylic acid and polyinosinic acid·polycytidylic acid. J Mol Biol 81: 107–122 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E 1998. Hydration of RNA base pairs. J Biomol Struct Dyn 16: 693–707 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Williamson JR 1996. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: II. Specificity determinants of RNA-protein recognition. J Mol Biol 261: 550–567 [DOI] [PubMed] [Google Scholar]
- Benne R, Van Den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826 [DOI] [PubMed] [Google Scholar]
- Blum B, Simpson L 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397 [DOI] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198 [DOI] [PubMed] [Google Scholar]
- Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ 2010. A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol 17: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K 2008. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol 28: 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CS, Barton JK 1992. Recognition of G-U mismatches by tris(4,7-diphenyl-1,10-phenanthroline)rhodium(III). Biochemistry 31: 5423–5429 [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117: 5179–5197 [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson RE 1998. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res 26: 1906–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan MA, Calladine CR 1998. Two distinct modes of protein-induced bending in DNA. J Mol Biol 282: 331–343 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Fan Y, Gaffney BL, Jones RA 2005. RNA GG·UU motif binds K+ but not Mg2+. J Am Chem Soc 127: 17588–17589 [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Steinberg SV 2002. GU receptors of double helices mediate tRNA movement in the ribosome. RNA 8: 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Konings D, Gutell RR 1995. G · U base pairing motifs in ribosomal RNA. RNA 1: 807–814 [PMC free article] [PubMed] [Google Scholar]
- Gherghe CM, Krahn JM, Weeks KM 2005. Crystal structures, reactivity and inferred acylation transition states for 2′-amine substituted RNA. J Am Chem Soc 127: 13622–13628 [DOI] [PubMed] [Google Scholar]
- Golas MM, Böhm C, Sander B, Effenberger K, Brecht M, Stark H, Göringer HU 2009. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J 28: 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MM 2002. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr 58: 872–874 [DOI] [PubMed] [Google Scholar]
- Hoyne PR, Gacy AM, McMurray CT, Maher LJ 2000. Stabilities of intrastrand pyrimidine motif DNA and RNA triple helices. Nucleic Acids Res 28: 770–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inners LD, Felsenfeld G 1970. Conformation of polyribouridylic acid in solution. J Mol Biol 50: 373–389 [DOI] [PubMed] [Google Scholar]
- Janert PK 2010. Gnuplot in action: understanding data with graphs. Manning Publications Co, Greenwich, CT [Google Scholar]
- Kabsch W 1988. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Crystallogr 21: 916–924 [Google Scholar]
- Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W 2011. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res doi: 10.1093/nar/gkr368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky DJ, Reifur L, Yu LE, Chen W 2004. Evidence for U-tail stabilization of gRNA/mRNA interactions in kinetoplastid RNA editing. RNA Biol 1: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K 2009. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res 37: 5917–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SS, Koslowsky DJ 2001. Interactions of mRNAs and gRNAs involved in trypanosome mitochondrial RNA editing: Structure probing of an mRNA bound to its cognate gRNA. RNA 7: 1803–1816 [PMC free article] [PubMed] [Google Scholar]
- Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L 2009. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci 106: 12306–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Herrera J, Zhou S, Maslov DA, Simpson L 2011. Trypanosome REH1 is an RNA helicase involved with the 3′-5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci 108: 3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Connell GJ 2010. Identification of specific inhibitors for a trypanosomatid RNA editing reaction. RNA 16: 2435–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK 2003. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31: 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK 2008. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc 3: 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Westhof E 2000. On the wobble GoU and related pairs. RNA 6: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B, Sauter C, Westhof E 1999. A sulfate pocket formed by three GoU pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA 5: 1384–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- McManus MT, Adler BK, Pollard VW, Hajduk SL 2000. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol Cell Biol 20: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokdad A, Krasovska MV, Sponer J, Leontis NB 2006. Structural and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic Acids Res 34: 1326–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroder H, Kreutz C, Lang K, Serganov A, Micura R 2006. Synthesis, oxidation behavior, crystallization and structure of 2′-methylseleno guanosine containing RNAs. J Am Chem Soc 128: 9909–9918 [DOI] [PubMed] [Google Scholar]
- Moshiri H, Acoca S, Kala S, Najafabadi H, Hogues H, Purisima EO, Salavati R 2011. Naphthalene-based RNA editing inhibitor blocks RNA editing activities and editosome assembly in Trypanosoma brucei. J Biol Chem 286: 14178–14189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Hajduk SL 2006. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep 7: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Cipriano M, Hajduk SL 2007. KISS: the kinetoplastid RNA editing sequence search tool. RNA 13: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Anderson S, Wood ZA, Hajduk SL 2008. Alternative RNA editing produces a novel protein involved in mitochondrial DNA maintenance in trypanosomes. Mol Cell Biol 28: 5595–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, et al. 2001. A standard reference frame for the description of nucleic acid base-pair geometry. J Mol Biol 313: 229–237 [DOI] [PubMed] [Google Scholar]
- Pan B, Mitra SN, Sundaralingam M 1998. Structure of a 16-mer RNA duplex r(GCAGACUUAAAUCUGC)2 with wobble C·A+ mismatches. J Mol Biol 283: 977–984 [DOI] [PubMed] [Google Scholar]
- Pan B, Mitra SN, Sundaralingam M 1999. Crystal structure of an RNA 16-mer duplex r(GCAGAGUUAAAUCUGC)2 with nonadjacent G(syn)·A+(anti) mispairs. Biochemistry 38: 2826–2831 [DOI] [PubMed] [Google Scholar]
- Pollard VW, Rohrer SP, Michelotti EF, Hancock K, Hajduk SL 1990. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell 63: 783–790 [DOI] [PubMed] [Google Scholar]
- Reifur L, Koslowsky DJ 2008. Trypanosoma brucei ATPase subunit 6 mRNA bound to gA6-14 forms a conserved three-helical structure. RNA 14: 2195–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifur L, Yu LE, Cruz-Reyes J, Vanhartesvelt M, Koslowsky DJ 2010. The impact of mRNA structure on guide RNA targeting in kinetoplastid RNA editing. PLoS ONE 5: e12235 doi: 10.1371/journal.pone.0012235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal S, Bhandari S, Koirala S, Singh R, Khanal B, Loutan L, Dujardin JC, Boelaert M, Chappuis F 2010. Clinical risk factors for therapeutic failure in kala-azar patients treated with pentavalent antimonials in Nepal. Trans R Soc Trop Med Hyg 104: 225–229 [DOI] [PubMed] [Google Scholar]
- Rypniewski W, Adamiak DA, Milecki J, Adamiak RW 2008. Noncanonical G(syn)–G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex. RNA 14: 1845–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K 2005. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J 24: 4029–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiwert SD, Stuart K 1994. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266: 114–117 [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Heidmann S, Stuart K 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84: 831–841 [DOI] [PubMed] [Google Scholar]
- Shi K, Wahl M, Sundaralingam M 1999. Crystal structure of an RNA duplex r(GGGCGCUCC)2 with non-adjacent G·U base pairs. Nucleic Acids Res 27: 2196–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki H, Shinomoto S 2009. Kernel bandwidth optimization in spike rate estimation. J Comput Neurosci 29: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Gao G, Kang X 2004. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA 10: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Georgiev V 2009. Tropical medicine and parasitic diseases. In National Institute of Allergy and Infectious Diseases, NIH: Impact on global health. Vol. 2, Chapter 6 (ed. St. Georgiev V), p. 57–64 Humana Press, Totowa, NJ [Google Scholar]
- Sugimoto N, Kierzek R, Freier SM, Turner DH 1986. Energetics of internal GU mismatches in ribooligonucleotide helices. Biochemistry 25: 5755–5759 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Fujii S, Hiroaki H, Sakata T, Tanaka T, Uesugi S, Tomita K, Kyogoku Y 1999. A′-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC). Nucleic Acids Res 27: 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Varani G, McClain WH 2000. The G·U wobble base pair: a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kapral G, Murray L, Richardson D, Richardson J, Snoeyink J 2008. RNABC: forward kinematics to reduce all-atom steric clashes in RNA backbone. J Math Biol 56: 253–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Crothers DM 1993. Major groove accessibility of RNA. Science 261: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Wilkinson SR, Taylor MC, Horn D, Kelly JM, Cheeseman I 2008. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc Natl Acad Sci 105: 5022–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M, Ballard C, Cowtan K, Dodson E, Emsley P, Evans P, Keegan R, Krissinel E, Leslie AG, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Landon T, Greenbaum NL, Fenley MO 2007. The electrostatic characteristics of G·U wobble base pairs. Nucleic Acids Res 35: 3836–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]