It has been established that alternative and constitutive splicing decisions are dictated in part by cis-acting RNA sequences known as splicing enhancers or inhibitors. Both types of elements can be found either in exons or introns. Here, the authors have used computational methods to identify predicted intronic and exonic splicing enhancers in Drosophila. Many of these were previously shown to be enhancers of vertebrates. The evolutionary implications of these findings are discussed.
Keywords: Drosophila, splicing, splicing regulatory elements, ESE, ISE
Abstract
RNA sequence elements involved in the regulation of pre-mRNA splicing have previously been identified in vertebrate genomes by computational methods. Here, we apply such approaches to predict splicing regulatory elements in Drosophila melanogaster and compare them with elements previously found in the human, mouse, and pufferfish genomes. We identified 99 putative exonic splicing enhancers (ESEs) and 231 putative intronic splicing enhancers (ISEs) enriched near weak 5′ and 3′ splice sites of constitutively spliced introns, distinguishing between those found near short and long introns. We found that a significant proportion (58%) of fly enhancer sequences were previously reported in at least one of the vertebrates. Furthermore, 20% of putative fly ESEs were previously identified as ESEs in human, mouse, and pufferfish; while only two fly ISEs, CTCTCT and TTATAA, were identified as ISEs in all three vertebrate species. Several putative enhancer sequences are similar to characterized binding-site motifs for Drosophila and mammalian splicing regulators. To provide additional evidence for the function of putative ISEs, we separately identified 298 intronic hexamers significantly enriched within sequences phylogenetically conserved among 15 insect species. We found that 73 putative ISEs were among those enriched in conserved regions of the D. melanogaster genome. The functions of nine enhancer sequences were verified in a heterologous splicing reporter, demonstrating that these sequences are sufficient to enhance splicing in vivo. Taken together, these data identify a set of predicted positive-acting splicing regulatory motifs in the Drosophila genome and reveal regulatory sequences that are present in distant metazoan genomes.
INTRODUCTION
The splicing of pre-mRNAs is an important level in the regulation in metazoan gene expression. The precise excision of introns and the joining of flanking exons is essential for accurate protein synthesis. Introns contain several sequence elements required for pre-mRNA splicing: 5′ and 3′ splice sites (5′ss, 3′ss), branch point, and polypyrimidine tract. Splice sites can be classified as “weak” or “strong” according to their similarity to consensus motifs. The degree to which a splice site is used is thought to increase as its strength increases (Lim and Burge 2001; Roca et al. 2005), exemplified by the fact that consitutively spliced introns have stronger splice sites than alternatively spliced introns (Koren et al. 2007). There are also splicing regulatory elements (SREs) within the pre-mRNA, which influence splicing efficiency (Lim and Burge 2001). SREs are named according to their function and location: exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), or intronic splicing silencers (ISSs). ESEs are thought to most often be recognized by serine–arginine-rich proteins (SRs), ESSs, and ISSs most often recognized by heterogeneous nuclear ribonucleoproteins (hnRNPs) (Chen and Manley 2009). Some hnRNPs and other RNA-binding proteins, such as Nova and Fox, have been shown to recognize ISEs (Chen and Manley 2009). The specific combination of SREs and their distances from splice junctions contributes to splicing outcome (Zhang et al. 2009). In addition, current models for splice-site selection suggest that splice sites are recognized through interactions of the spliceosome across exons, termed “exon definition,” when exons are flanked by long introns and across introns, termed “intron definition,” when introns are short (Robberson et al. 1990; Romfo et al. 2000; Lim and Burge 2001; Yeo et al. 2004). Therefore, the relative size of introns is important for splice-site selection.
Putative SREs have previously been identified with in vitro SELEX experiments, as well as in vivo functional selection of minigene reporter libraries (Shi et al. 1997; Liu et al. 1998; Amarasinghe et al. 2001; Wang et al. 2004; Smith et al. 2006; Blanchette et al. 2009). Previous computational approaches have also been successful at identifying SREs. Since RNA-binding domains typically bind to six to eight nucleotides, computational searches focus on finding enriched RNA elements of this size in functionally relevant locations (Fedorov et al. 2001). One such approach is the Relative Enhancer and Silencer Classification by Unanimous Enrichment (RESCUE) method (Fairbrother et al. 2002), which has been applied to numerous genomes including mammals, fish, and plants (Fairbrother et al. 2002; Yeo et al. 2004; Zhang and Chasin 2004; Pertea et al. 2007). The RESCUE method detects motifs enriched near weak splice sites of constitutively spliced introns based on the principle that neighboring sequences act as enhancers to compensate for poor splice-site recognition. Other approaches use the premise that functional splicing regulatory elements are under stringent evolutionary constraint (Goren et al. 2006; Kabat et al. 2006; Voelker and Berglund 2007; Yeo et al. 2007; Churbanov et al. 2009).
Here we used a combination of these two methods, the RESCUE approach and a statistical model to define genomic regions under evolutionary sequence constraint, to predict SREs in Drosophila melanogaster. To define constrained sequences, we used 15 highly diverged insect species to identify phylogenetically conserved intronic elements. By comparing our set of fly-splicing regulatory sequences with those found in vertebrates, we have identified sequence elements whose function is conserved across distant animal species. Interestingly, 58% of the putative enhancer elements identified here in Drosophila have also been identified in vertebrates. Several of the motifs are predicted binding sites of both Drosophila and mammalian RNA-binding proteins. Compared with vertebrate genomes with characterized splicing regulatory elements, the D. melanogaster genome has the unique feature of a large proportion of short introns. We have taken advantage of this feature to ask whether there are different regulatory sequences present near short and long introns. A selection of putative SREs was tested for functionality in vivo in a minigene reporter. The majority of sequences examined had significant effects on the level of splicing, indicating the robustness of the computational approach used in this study.
RESULTS
Long and short introns have different distributions of splice-site strengths
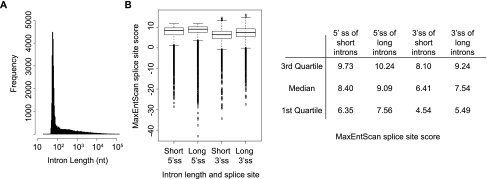
The number and type of regulatory elements near an intron is dependent upon intron length and splice-site strength (Lim and Burge 2001; Yeo et al. 2004; Xiao et al. 2007); therefore, we looked for potential biases in SREs arising from intron length. The length distribution of constitutively spliced introns in D. melanogaster consists of a peak with a mode at 69 nt and a long tail (Fig. 1A). This length distribution is different from that of human introns, with a mode of 1500 nt (Lim and Burge 2001; Yeo et al. 2004). Given the intron-length distribution, we divided constitutively spliced introns into two categories: short (≤80 nt; 22,329 introns) and long (>80 nt; 15,474 introns).
FIGURE 1.
Splice sites of short constitutively spliced introns are weaker than long constitutively spliced introns in Drosophila. (A) Length distribution of constitutively spliced introns in Drosophila. (B) A significant difference in the distribution of MaxEntScan splice-site scores (Yeo and Burge 2004) of short and long introns (P < 2.26e-16, Wilcoxon rank sum test). The first quartile, median, and third quartile of splice-sites scores are given in the table. Higher MaxEntScan scores correspond to a stronger splice-site sequence.
Using MaxEntScan (Yeo and Burge 2004) to score splice-site strengths, we found that longer introns have significantly stronger 5′ and 3′ splice-site strengths than shorter introns (P < 2.26e-16 for 5′ and 3′ splice sites, Wilcoxon rank sum test) (Fig. 1B). Although MaxEntScan's scores are derived from human splice sites, Drosophila splice-site motifs are highly similar to human, and many spliceosomal components involved in splice-site recognition are highly conserved (Barbosa-Morais et al. 2006; Schwartz et al. 2008). Dividing the data into two length groups accounts for effects of intron length and permits the identification of motifs that may be specific to each length class.
Identification of exonic and intronic splicing enhancers in D. melanogaster
To identify ESEs and ISEs in Drosophila, we implemented the RESCUE method (Fairbrother et al. 2002), which allows direct comparisons between the Drosophila motifs and those that were previously identified in other species by RESCUE.
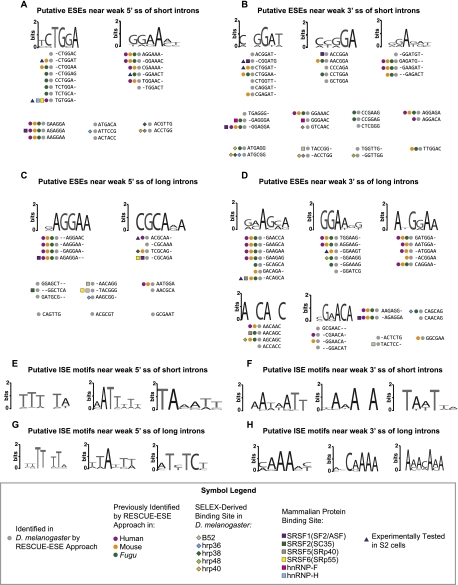
We applied the RESCUE method to constitutive splicing events to identify potential ESEs, characterized as hexamers significantly enriched in exons compared with introns and significantly enriched near either a weak 5′ or weak 3′ splice site, compared with strong splice sites (see Materials and Methods; Supplemental Fig. S1A–D). ESEs were identified near short and long introns, separately. Putative ESE sequences were identified up to 100 bp upstream of 5′ splice sites and up to 100 bp downstream from 3′ splice sites, excluding the splice-site sequences. If an exon was shorter than 100 bp, sequence from the entire exon was used and was not extended into the next intron. We used the distribution of splice-site strengths in both data sets to define cutoffs for weak and strong splice sites (Fig. 1B): Weak splice sites were defined as the first quartile of scores and strong splice sites in the fourth quartile. This analysis identified 22 hexamers near 5′ss and 34 hexamers near 3′ss of short introns as putative ESEs (Fig. 2A,B). Five sequences were enriched near both splice sites (CTGGAG, CTGGAT, CTGGAA, CCTGGA, GGAAAC). Although enhancers are predicted to act at the RNA level, we include thymines when reporting their hexamers, since they were discovered using the genomic DNA sequence. Near long introns, 19 hexamers were found to be enriched in exons at the 5′ss and 33 hexamers at the 3′ss (Fig. 2C,D). Two sequences were found in exons near long introns at both splice sites (AGAGGA, AATGGA). Interestingly, just two hexamers (AAGGAA, AGAGGA) were shared between the ESEs identified near short introns and near long introns for either splice site. This suggests that largely distinct regulatory sequences are present in exons proximal to introns of different sizes.
FIGURE 2.
Hexamers and motifs enriched in exons and introns near weak splice sites of constitutive introns. Putative exonic splicing enhancers (ESEs) enriched near short introns of (A) weak 5′ splice sites and (B) weak 3′ splice sites, and long introns near (C) weak 5′ splice sites and near (D) weak 3′ splice sites. Representative putative intronic enhancer motifs (ISEs) enriched near (E) weak 5′ splice sites and (F) weak 3′ splice sites of short introns, and near (G) weak 5′ splice sites and (H) weak 3′ splice sites of long introns. Hexamers identifed using the RESCUE-ESE method in this study and in human (Fairbrother et al. 2002), mouse, or Fugu (Yeo et al. 2004) are indicated by colored circles. Hexamers containing high-affinity binding sites of D. melanogaster splicing regulators (Shi et al. 1997; Amarasinghe et al. 2001; Blanchette et al. 2009) are indicated by colored diamonds, while binding sites for mammalian proteins are indicated by colored squares (Cartegni et al. 2002; Goren et al. 2006; Smith et al. 2006). The seven hexamers we tested for enhancer activity in an in vitro splicing reporter are indicated by blue triangles.
Closely related hexamers were clustered according to their edit distances (Supplemental Fig. S2A–D) (Fairbrother et al. 2002; Böckenhauer and Bongartz 2007) and sequence logos were created to identify general motifs (Schneider and Stephens 1990; Crooks et al. 2004). Binding sites for many splicing regulators are known to be highly degenerate (Chen and Manley 2009); therefore, such motifs might correspond to sequence elements bound by proteins. Upon inspecting hexamer clusters, we found GGAA-containing ESE motifs present near both 5′ss and 3′ss, regardless of intron length (Fig. 2). This 4-mer is part of the binding site of multiple hnRNP and SR proteins, including hnRNP-H, hnRNP-F, and SRSF6 (SRp55) (Goren et al. 2006). We observed a motif unique to short introns, CTGGA, as well as motifs unique to long introns, CGCA and A[A/G/C]CA[A/G/C]C (Fig. 2). By clustering similar hexamers, we identified motifs that are shared and distinct between short and long introns.
Potential ISEs were identified using the RESCUE method, by seeking hexamers over-represented in introns relative to exons and enriched near weak splice sites relative to strong splice sites (Supplemental Fig. S1E–H). As with ESEs, short and long introns were analyzed separately and sequences were identified within 100 bp of each splice site. For introns shorter than 100 bp, the entire intron sequence was used. A total of 96 hexamers were identified in short introns at 5′ss and 76 hexamers at 3′ss (Supplemental Fig. S3A,B). Fifteen sequences found in short introns were located both near 5′ss and 3′ss. Seventy-eight hexamers were identified in long introns at 5′ss and 43 hexamers at 3′ss, seven of which were enriched at both splice sites (Supplemental Fig. S3C,D). Twenty-four putative ISEs were found near 5′ss in both long and short introns and 10 ISEs were found near 3′ss of both intron length classes. There is a greater overlap of ISEs found in short and long introns than ESEs found near both intron lengths.
Similar ISEs were clustered into motifs (Fig. 2E–H; Supplemental Fig. S3). Most ISE clusters were found to be AT-rich and many were present near both short and long introns. CAA motifs were preferentially found near weak 3′ splice site of long introns. Although some clustered hexamers revealed motifs preferentially found near short or long introns (e.g., TAAT and T[T/C]TC, respectively), sequences containing these motifs could be identified near both length classes (Supplemental Fig. S3).
We next looked to see whether these enhancer sequences were enriched in positions closer to the splice sites, farther away from the splice sites, or evenly distributed across the search space—up to 100 bp from a splice site. Given that the search space length varied by the length of the exon or intron, we divided the search space into four equally sized bins to indicate a proximal, distal, or intermediate distance from each splice site. As a control, we also compared the distribution of SREs observed near weak splice sites to those observed near strong splice sites. As expected, there was a greater enrichment of enhancer sequences across the entire search space near weak splice sites than near strong splice sites (Supplemental Fig. S4). ESEs appear to be evenly distributed across the search space, while ISEs show some positional biases (Supplemental Fig. S4). ISEs identified near weak 5′ splice sites of short introns tend to be more distal to the 5′ splice site. This trend is also observed in shuffled control sequences, indicating that the trend may be due to a general occurrence of A and T nucleotides, as the ISEs are AT-rich (Supplemental Fig. S4A, black lines). We also observe a trend for ISEs identified near weak 3′ splice sites of short introns to occur more proximal to the 3′ splice site (Supplemental Fig. S4A).
58% of RESCUE-identified D. melanogaster hexamers are identical to those found in vertebrates
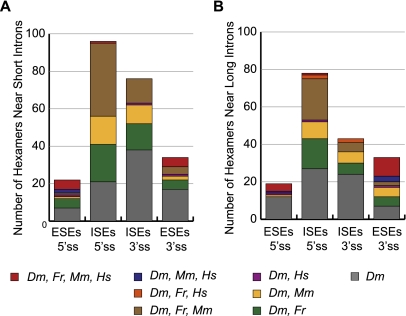
We compared the putative ESEs and ISEs we found in Drosophila with those identified in other vertebrate species using the same method, and we found a significant overlap with human (Homo sapiens), mouse (Mus musculus), and pufferfish (Fugu rubripes) sequences (Fisher's exact test, P < 2.2e-16) (Fairbrother et al. 2002; Yeo et al. 2004). A total of 57 of 99 putative Drosophila ESEs and 136 of 231 putative Drosophila ISEs were previously identified in one or more of the vertebrates (Figs. 2, 3; Supplemental Fig. S3). We found that of the three species, Drosophila had the most overlap with Fugu (Fig. 3), which may arise from the similar intron length distributions between the two species (Fig. 1; Lim and Burge 2001; Yeo et al. 2004). Among Drosophila ISEs, 45% (104 of 231) are identical to Fugu ISEs. A large proportion of ISEs are also identical to mouse; however, very few are shared with human (Fig. 3; Supplemental S3).
FIGURE 3.
A majority of D. melanogaster RESCUE-identified ESEs and ISEs are identical to those found in vertebrates. The number of hexamers found near (A) short introns or (B) long introns that are shared with pufferfish (Fugu rubripes, Fr), mouse (Mus musculus, Mm), and/or human (Homo sapiens, Hs) is shown along with the number of hexamers that were uniquely identified in fly (Drosophila melanogaster, Dm).
We observed that ESEs were more conserved across all four species than ISEs, perhaps due to added evolutionary constraint from the protein-coding sequence of exons (Fig. 3). A total of 20% of predicted fly ESE hexamers (20 of 99) are shared with human, mouse, and pufferfish predicted enhancers, with a greater proportion of shared ESEs near 3′ splice sites of long Drosophila introns (30%, 10 of 33) (Fig. 3). In contrast, only two putative ISEs are conserved between all four species: CTCTCT and TTATAA. CTCTCT is predicted to be a binding site for the splicing regulator PTB (Chen and Manley 2009; Robida et al. 2010), while no cognate binding protein for TTATAA was found through literature searches. In HeLa cells, PTB has been shown to activate splicing of exons containing PTB binding sites in the downstream intron (Llorian et al. 2010). The CTCTCT ISE we identified in our study was found in long introns near weak 5′ splice sites, consistent with the location of PTB-bound enhancer sequences.
In addition to the shared ESEs and ISEs between fly and vertebrates, there were five fly ESEs that were identified as vertebrate ISEs and 14 fly ISEs identified as vertebrate ESEs (Supplemental Data set). These may be bound by proteins that can act both from exonic and intronic locations, but are preferentially enriched in exons or introns due to inherent differences in genome composition of different organisms. All but one of these 19 enhancers were shared specifically with human and/or mouse. These sequences are identified as splicing enhancers in both vertebrates and fly, despite the difference in their enrichment in exonic sequences versus intronic sequence. Differences in exonic sequences may be the result of different codon usages in different organisms.
Overlap with known RNA protein-binding sites
To identify potential cognate proteins for the putative ESEs and ISEs, we compared the hexamers against SELEX-derived binding sites of six D. melanogaster proteins (Shi et al. 1997; Amarasinghe et al. 2001; Blanchette et al. 2009) and against multiple mammalian RNA-binding proteins defined in ESRSearch (Goren et al. 2006) and ESEfinder (Fig. 2; Cartegni et al. 2002; Smith et al. 2006). Though there may be differences in the RNA-binding specificities of mammalian and Drosophila RNA-binding proteins, there are examples of proteins whose binding motif is well conserved, such as PTB/Hephaestus and Nova/Pasilla (Pérez et al. 1997; Jensen et al. 2000; Robida et al. 2010; Brooks et al. 2011). We found one putative ESE, ATGCGG, to be a high-affinity binding site for Drosophila hrp36, hrp38, and hrp40, despite the fact that their SELEX-derived consensus motifs are distinct. There are regions of the transcriptome that are known to be bound by these three hnRNPs, though not necessarily simultaneously (Blanchette et al. 2009). We identified several SR and hnRNP recognition motifs in both ESEs and ISEs (Fig. 2; Supplemental Fig. S3), supporting the observation that SR proteins do not exclusively bind to exonic sequences and hnRNPs do not exclusively bind to intronic sequences or act as splicing silencers (Sanford et al. 2008, 2009; Blanchette et al. 2009; Llorian et al. 2010). In addition, the putative ISE TCTATC, found near weak 3′ splice sites of long introns, was recently identified in Caenorhabditis elegans as a binding site for HRP-2, a homolog to hnRNPs Q and R (Kabat et al. 2009).
Hexamers enriched in conserved regions of constitutively spliced introns
Because stringent evolutionary constraints on intronic sequences have been used to indicate function (Kabat et al. 2006; Voelker and Berglund 2007; Yeo et al. 2007), we identified hexamers over-represented in conserved regions within the set of constitutive introns. Introns are more amenable to the identification of conserved sequence elements than exons, because they are free from protein-coding constraints. This search identified additional SREs in introns and indicated which RESCUE-identified ISEs tend to be conserved. The phastCons model was used to define genomic regions under evolutionary constraint between D. melanogaster, 11 other Drosophila species, and three additional divergent insects (Siepel et al. 2005; Fujita et al. 2011) (http://genome.ucsc.edu). This statistical model allowed us to identify conserved sequences across evolutionary distances greater than those used to identify conserved intronic sequences in vertebrates or in worm (Siepel et al. 2005; Kabat et al. 2006; Voelker and Berglund 2007; Yeo et al. 2007). As before, we separated constitutively spliced introns into short and long introns. We identified hexamers that were over-represented in conserved regions of introns relative to nonconserved regions (Bonferroni-corrected P-value <0.001 for 4096 tested hexamers).
In general, short introns did not overlap with phastCons regions. Only 1% of all possible 4096 hexamers overlapped with at least one phastCons region in short introns compared with 12% of hexamers in long introns. One hexamer was significantly enriched in conserved regions of short introns, CTAATT. Although branch-point sequences have not been extensively studied in Drosophila, the 5-mer CTAAT (branch-point A, underlined) matches well with the consensus branch-point sequence of human introns (Gao et al. 2008; Pastuszak et al. 2011). It is not surprising that one of the most conserved motifs in short introns is a key feature of introns in general, given that the branch point is located in the window we are searching. This putative branch-point sequence is also a RESCUE-identified ISE; thus, a consensus branch-point sequence may work analogously to ISEs to promote splicing.
We identified 298 hexamers in long introns that were significantly enriched in conserved regions (Supplemental Fig. S5). CTAATT was also found in conserved regions of long introns. Similar hexamers were clustered to identify common motifs shared by multiple sequences and most clusters were found to be markedly AT-rich in nature (Supplemental Fig. S5A). Within the conserved elements in long introns, we identified previously reported high-affinity binding sites for multiple D. melanogaster and mammalian proteins (Supplemental Fig. S5).
A total of 73 conserved hexamers overlapped with the 231 hexamers identified via RESCUE-ISE analysis, indicating which RESCUE-identified ISEs are also more likely to be phylogenetically conserved among insects (Supplemental Fig. S5). The hexamers CTCTCT and TTATAA identified as ISEs in fly, pufferfish, mouse, and human are also enriched in conserved regions of the fly genome, further supporting their functional role in splicing. We compared Drosophila conserved intronic hexamers to those identified in conserved regions of human introns (Yeo et al. 2007) and to 40 hexamers reported as conserved in regions near alternative introns in C. elegans (Kabat et al. 2006; Supplemental Fig. S5). A total of 35% (105 of 298) of conserved intronic hexamers in fly are also conserved intronic hexamers in human and 10 fly hexamers are also conserved intronic hexamers in worm (Supplemental Fig. S5). Most conserved intronic sequences shared between fly and human are AT-rich (Supplemental Fig. S5A). However, the significance of this is uncertain, as it is possible that the sequence overlap between fly and human is an artifact resulting from the AT-rich nature of introns in general.
Computationally predicted ESEs and ISEs stimulate cassette exon inclusion in vivo
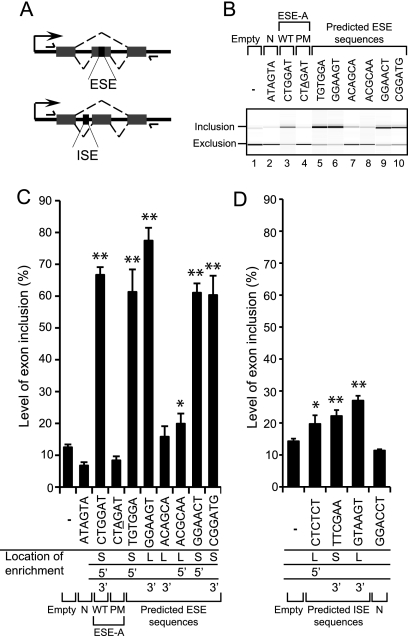
To test whether putative ESEs and ISEs that we identified are sufficient for splicing enhancer activity in vivo, their ability to stimulate splicing in a minigene reporter was examined. SREs were tested in this minigene assay in order to validate our computational approach. The minigene consisted of an alternatively spliced cassette exon event from a Drosophila endogenous gene (pep); therefore, the activity of putative ESEs and ISEs was monitored in a different context from the constitutive splicing events where they were identified. ESEs were inserted into the 101-nt cassette exon at a location that is within 100 nt of both 3′ and 5′ splice sites (Fig. 4A). ISEs were tested in the long (811 nt) upstream intron within 100 nt of the 3′ splice site (Fig. 4A). The downstream intron of the cassette exon was also long (252 nt). Representative putative ESE and ISEs from long and short introns, and from both 5′ and 3′ splice-site locations were tested.
FIGURE 4.
Predicted ESEs and ISEs exhibit stimulatory activity in minigene reporter assay. (A) Schematic of minigene reporter with an alternative cassette exon. ESEs and ISEs were cloned into the cassette exon (101 nt) and upstream intron (811 nt), respectively, as indicated (downstream intron is 252 nt). RT–PCR primers designed to the plasmid backbone are shown as arrows. (B) Semiquantitive RT–PCR Bioanalyzer 2100 image indicates that ESEs stimulate cassette exon inclusion compared with empty vector (−) and negative control hexamers (PM and N). Wild-type (WT) ESE-A is shown alongside a single-point mutant (PM), which is underlined. A neutral hexamer control is also shown (N). (C,D) RT–PCR quantitation of cassette exon inclusion levels with minigenes containing putative ESEs (C) and ISEs (D). Error bars represent mean ± SD of three independent experiments. Asterisks indicate significant differences from empty vector control (t-test; [*] P < 0.05 and [**] P < 0.005). The location at which each SRE was identified is indicated under the bar chart, from long (L) or short (S) introns, at 5′ or 3′ splice sites.
The activities of seven ESEs were examined alongside the minigene reporter without any inserted sequence (−) (Fig. 4B, lane 1). To assess whether the effect of inserting hexamers was specific and not just the result of inserting additional sequence within the cassette exon, a neutral control hexamer sequence was also tested (ATAGTA, N). This hexamer was selected based on its distinct sequence composition from our predicted ESEs. The neutral hexamer showed exon inclusion levels similar to the empty vector (Fig. 4B, lane 2). Among hexamers selected to test for enhancer activity were sequences previously identified in other organisms, for example CTGGAT (ESE-A), which stimulated cassette exon inclusion from 12% with no inserted sequence (−) to 66% (Fig. 4B, lane 3; Fig. 4C). We also tested several hexamers predicted to be bound by splicing factors. TGTGGA is recognized by mammalian hnRNP H, the ortholog of Drosophila Glorund (Barbosa-Morais et al. 2006), and it exhibited a strong enhancing effect on cassette exon inclusion (Fig. 4B, lane 5). A novel fly-specific ESE, CGGATG, also showed a stimulatory effect (Fig. 4B, lane 10).
A single-point mutation (PM) to ESE-A resulted in a hexamer that was not enriched near weak splice sites and not predicted to possess enhancer activity. We found that the point mutation exhibited exon inclusion levels similar to background (Fig. 4B, lane 4). We would not necessarily assume that point mutants of the ESE and ISE hexamers would all exhibit activity close to background. One base change to RNA-binding motifs may not ablate protein binding. Also, RESCUE scores of point mutants of our tested hexamers do not give significantly under-represented hexamers, which would be expected to not have enhancement activity. In fact, other point mutants that we tested ranged in activity from levels similar to their wild-type hexamer or to the empty vector background (data not shown).
Out of seven ESEs tested, six exerted a statistically significant stimulatory effect on cassette exon inclusion (t-test, P < 0.05) (Fig. 4C). ESEs identified near short and long introns showed no difference in activity in the splicing reporter, even though the introns surrounding the cassette exon are both long.
The activities of three ISEs were tested when inserted in the intron near the 3′ splice site of the cassette exon (Fig. 4A). All had small but significant effects on exon inclusion (t-test, P < 0.05) (Fig. 4D). A neutral hexamer (N), having a different sequence composition from identified ISEs, exhibited background levels of exon inclusion, indicating that ISE effects are specific. One tested ISE matches the consensus 5′ splice-site sequence, GTAAGT, for D. melanogaster (Schwartz et al. 2008). Addition of this splice-site sequence did not introduce a cryptic splice site. It has been shown that, in some circumstances, neighboring splice sites can assist in splice-site recognition (Chiara and Reed 1995; Hicks et al. 2010). One of the exceptionally conserved ISEs, CTCTCT, was also tested and had significant enhancement activity. Interestingly, TTCGAA, which was identified from short introns, was just as active as predicted ISEs from long introns, even though the minigene has long introns. Two of the ISEs tested were found near 3′ splice sites but were still active near the 5′ splice site in the reporter. Two other putative ISEs, identified from earlier iterations of RESCUE analysis, but not above the final cut-off, were also tested in the reporter and stimulated cassette exon inclusion (Supplemental Fig. S5). The difference in the magnitude of enhancement between the tested ESEs and ISEs may be due to effects from local sequence context or relative position (Fig. 4D; Goren et al. 2006; Zhang et al. 2009).
DISCUSSION
We used the RESCUE method (Fairbrother et al. 2002) to predict 99 ESEs and 231 ISEs that were over-represented near weak splice sites of constitutive introns in D. melanogaster. Within our set of computationally predicted SREs, we identified binding sites of multiple Drosophila and mammalian splicing regulators, implicating putative cognate binding proteins for our sequences. We found many SR and hnRNP binding sites within our set of ESEs and ISEs, giving further evidence that these proteins can act as enhancers and bind to both exons and introns. Seven ESEs and three ISEs were tested in vivo for enhancer activity when introduced in a minigene reporter assay, and all but one showed a statistically significant enhancement of splicing.
We identified putative SREs separately near short and long introns and found that the majority of enhancer sequences were specific to each intron class. This suggests, on average, genome-wide differences in splicing regulation that correlate with intron length. Splice-site recognition is thought to occur through the definition of introns or exons, depending on intron length. Perhaps the distinct regulatory sequences found near different length introns are associated with factors preferentially used for intron or exon definition; however, our genome-wide approach cannot implicate intron or exon definition modes of regulation for specific splicing events. When putative SREs were tested for their ability to stimulate cassette exon inclusion of a minigene reporter where the introns were long, there was no difference in activity between those SREs found near long and short introns. This may be the result of the different context in which the SREs were tested from where they were found.
A previous study of SREs cautions that many computational predictions have been “too successful,” because now at least 75% of a typical human exon sequence can be shown to influence splicing (Zhang et al. 2009). Our study indicates which of these many SREs are particularly relevant by identifying SREs that overlap between Drosophila and vertebrates. We found that a significant portion (58%) of fly putative enhancer sequences were identical to human, mouse, or pufferfish enhancer sequences. Moreover, a substantial fraction (20%) of fly ESEs were identical to ESEs found in all three vertebrate species, highlighting enhancer sequences whose function has been maintained throughout evolution.
In addition to RESCUE-identified ISEs, we also made use of 15 insect species to report a set of intronic sequence elements phylogenetically conserved at a greater evolutionary depth than previous analyses (Siepel et al. 2005; Kabat et al. 2006; Voelker and Berglund 2007; Yeo et al. 2007). Some of these conserved intronic hexamers may not be involved in splicing; however, 73 of these sequences were also identified as ISEs using the RESCUE approach. The hexamers CTCTCT and TTATAA were highlighted as exceptionally conserved, since they were identified as ISEs through the RESCUE method in fly and three vertebrates, and were also enriched in conserved intronic regions of the Drosophila genome. The hexamer CTCTCT is predicted to be recognized by the splicing regulator PTB, which is itself conserved between fly and vertebrates (Barbosa-Morais et al. 2006). We did not find a previously reported cognate binding protein for TTATAA, yet this orphan putative regulatory sequence appears important due to its conservation. Given that the sequence is a palindrome, perhaps it is acting through RNA secondary structure. The sequence may also act through splicing regulation at the DNA level by affecting transcription rates (Kornblihtt 2006) or chromatin states (Schwartz and Ast 2010). Most identified SREs are likely binding sites for trans-acting regulatory proteins; however, some may regulate splicing through these alternative mechanisms.
This study presents the most comprehensive computational analysis of splicing enhancer sequences in Drosophila melanogaster to date, and it has revealed splicing regulatory elements whose function is conserved across metazoan evolution. Since splicing patterns can differ between tissue type and developmental stages, it is also necessary to study splicing regulation in diverse cellular contexts (Matlin et al. 2005; Zhang et al. 2009), taking into account the SREs' role in the pantheon of splice affecters.
MATERIALS AND METHODS
Intron coordinates in the D. melanogaster genome
The D. melanogaster genome sequence and annotations from FlyBase release 5.4 (Tweedie et al. 2009), which includes 37,803 constitutively spliced introns, were used for the RESCUE method. Using modified scripts from the Drosophila Exon Database (Lee et al. 2004), constitutively spliced introns were identified as intron coordinates that are present in all isoforms of the same gene.
Measurements of splice-site strength with MaxEntScan
MaxEntScan was used to assess how well a sequence conforms to the well-established 5′ss or 3′ss consensus motif (Yeo and Burge 2004; Schwartz et al. 2008). This score was taken as an indication of splice-site strength. The 5′ss sequence is defined as position (−3, +6) and the 3′ss sequence at position (−20, +3), relative to the exon–intron junction. MaxEntScan models short sequence motifs and accounts for relationships between adjacent and nonadjacent nucleotide positions.
Defining short and long introns in D. melanogaster
A histogram was created from the lengths of all constitutive introns in FlyBase r5.4. By visual inspection, we identified a sharp peak in the distribution of lengths with most introns ≤80 nt in length (“short”) and a tail corresponding to introns longer than 80 nt (“long”) (Fig. 1).
RESCUE-ESE and RESCUE-ISE method
The frequency of all 4096 possible hexamers was determined, using a sliding 6-bp window and allowing overlaps in each of the following locations: exonic sequence, intronic sequence, near a weak 5′ss or 3′ss, and near a strong 5′ss or 3′ss. Sequences within 100 bp of the exon–intron boundary, excluding the nucleotides (−3, +6) relative to the 5′ ss and (−20, +3) relative to the 3 ss, were used for the RESCUE method (Fairbrother et al. 2002). If the intron or exon was <100 bp, then the entire intron sequence, excluding splice sites, was used. Hexamer frequencies were calculated separately for exonic sequences near short and long introns and intronic sequences within short and long introns. When identifying ESEs near 5′ss, the length of the downstream intron was used to separate between short and long introns, while the length of the upstream intron was used to separate by length when identifying ESEs near 3′ss.
ΔEI, Δ5WS, and Δ3WS scores were calculated for each hexamer using the formula as described in the Supporting Online Material for Fairbrother et al. (2002). Hexamers with ΔEI and Δ5WS, or Δ3WS, scores above 2.5 (P < 0.01), were selected as potential 5′ and potential 3′ ESEs, respectively. Therefore, the P-value for significance of each putative enhancer is <10−4, given that the significance threshold for the two independent scores has a P-value of 0.01. A similar procedure was performed for identifying ISEs, where a ΔIE, Δ5WS, and Δ3WS was calculated for each hexamer found in intron sequences.
Positional biases of enhancers near weak splice sites
To determine whether enhancer sequences had a positional bias relative to the splice sites, we counted the frequency of each set of enhancers at positions near the splice sites, distal to the splice sites, and at intermediate distances to the splice sites.
Exon and intron sequence within 100 bp of the intron–exon boundary was used for the analysis. If the exon or intron was <100 bp, the entire sequence was used. Due to the varying lengths caused by shorter introns or exons, each region was divided into four equal length windows (proximal to the splice site, distal to the splice site, and two intermediate windows), and the frequency of enhancers was divided by the length of each window to get a proportion of enhancers found in each window. As a control, the nucleotides in each sequence window were randomly shuffled and enhancer frequencies were calculated from these shuffled sequences. Enhancer frequencies were obtained separately in regions near weak splice sites and strong splice sites. Frequency of enhancers were only determined in the sequence region from which they were identified as enhancers. For example, in exonic sequences 100 bp upstream of 5′ splice sites, with a short downstream intron, the frequency of ESEs found near 5′ splice sites near short introns was determined.
Clustering hexamers
The following manipulations were done separately for hexamers in the different intron length and splice-site groups. An edit distance (number of insertions, deletions, and substitutions) was calculated between all possible pairs of hexamers, using the Levenshtein distance algorithm (Schneider and Stephens 1990; Crooks et al. 2004; Böckenhauer and Bongartz 2007). The MATLAB “linkage” function was used to generate a hierarchical cluster tree from the unweighted average distances, such that sequences with the lowest edit distances would fall into the same cluster. The tree was then visualized using the MATLAB “dendrogram” function with the “colorthreshold” parameter. In the original RESCUE-ESE study, a dissimilarity cutoff of 2.7 was used to select clusters of hexamers (Fairbrother et al. 2002). We increased the cutoff up to 3.0 whenever it led to the inclusion of several additional hexamers to any cluster. Clusters composed of four or more hexamers were aligned using ClustalW, and a sequence logo was generated for each cluster using WebLogo (Schneider and Stephens 1990; Crooks et al. 2004; Larkin et al. 2007).
Identifying high-affinity binding sites from SELEX-derived binding matrices
The SELEX-derived binding affinities for B52, PSI (Amarasinghe et al. 2001), hrp36, hrp38, hrp40, and hrp48 (Blanchette et al. 2009) in D. melanogaster and SRSF1 (SF2/ASF), SRSF2 (SC35), SRSF5 (SRp40), and SRSF6 (SRp55) (Cartegni et al. 2002; Smith et al. 2006) in human were used to determine high-affinity binding sites for each protein. The SRSF1-binding motif from functional SELEX of the IgM- and BRCA1-derived minigene was used. The average and standard deviation of position weight matrix (pwm) scores against all exons and introns in FlyBase r5.4 were used to calculate a Z-score for a given hexamer. When comparing against hrp36, hrp38, hrp40, hrp48, and SRSF6 pwms, hexamers with a Z-score ≥2 (P ≤ 0.05; two-tailed) were considered high-affinity binding sites. The binding site for PSI, SRSF1, SRSF2, and SRSF5 is >6 nt; therefore, hexamers were compared against all subhexamer windows within the matrix. A Bonferroni-correction for the multiple subhexamer windows was used to maintain an overall P-value of ≤0.05 for matches to these binding sites. The exact binding sequences for B52 are reported in Shi et al. (1997); therefore, exact matches to the conserved 17-nt core of the B52 binding site was used to identify binding sites.
Identifying previously published enhancer sequences in vertebrates
Hexamers identified as ESEs and ISEs in human, mouse, or Fugu were identified through queries to the ACESCAN2 web server (http://genes.mit.edu/acescan2/index.html).
Identification of conserved intronic hexamers
Conserved regions of the genome were defined by phastCons conserved elements that had a transformed log-odds score greater than 0 (Siepel et al. 2005), and coordinates (dm3) were downloaded from the UCSC Genome Browser public MySQL server (Fujita et al. 2011) (http://genome.ucsc.edu). The frequency of hexamers within 100 nt of either splice site of constitutive introns was calculated. A χ2 statistic with Yates' continuity correction was computed for each hexamer using a two-by-two contingency table as performed with conserved sequences in humans (Yeo et al. 2007). The table for each hexamer compared (1) the number of times the hexamer occurred in phastCons elements within 100 bp of a splice site versus the number of times all other hexamers occurred in conserved elements, (2) the number of times the hexamer occurred within 100 bp of a splice site versus the number of times all other hexamers occurred within 100 bp of a splice site. Only hexamers with counts greater than 10 were selected for testing. Hexamers with Bonferroni-corrected P-values <0.001 were selected as significantly enriched in conserved intronic regions.
Reporter plasmid construction
The minigene reporter (obtained from M. Blanchette, Stowers Institute) was prepared from exons 1, 2, and 3 of the Drosophila pep gene (CG6143) fused to EGFP in pMT/V5 (Invitrogen). Oligo pairs containing ESEs were ligated into NheI-digested plasmid, 17 bp into exon 4, resulting in the addition of 12 bp, 6 bp of which corresponded to ESEs. ISEs were inserted into a BglII site 84 bp upstream of the cassette exon.
Tissue culture, DNA transfections, RNA purification, and RT–PCR
Drosophila Schneider (S2) cells were grown in standard tissue culture conditions at 26°C with M3 supplemented with 5% fetal bovine serum. Plasmid DNA was transfected using Effectene (Qiagen), according to manufacturer's instructions. After 24 h, CuSO4 was added to a final concentration of 0.5 mM. After a further 48 h, cells were harvested and total RNA purified. Samples were DNaseI treated, followed by nucleic acid purification. RNA was subjected to reverse transcription using SuperScript II Reverse Transcriptase (Invitrogen) with oligo d(T)15. PCR was performed on the resulting cDNA with HotStar polymerase (Qiagen), and primers were designed to anneal to the vector backbone (forward: cgtagaatcgagaccgagg, reverse: gctcctcgcccttgctca). PCR products were examined using a 2100 Bioanalyzer (Agilent) and subsequently quantitated using 2100 Expert Software.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank K. Hansen and S. Dudoit for helpful discussions on the study and Marco Blanchette for the minigene reporter. We thank the UC Berkeley Undergraduate Research Apprentice Program. This work was funded by a contract from the National Human Genome Research Institute modENCODE project, U01-HG004271 to S.E. Celniker. Additional support was provided by National Instituties of Health grants R01-GM071655 (S.E.B.), R01-GM61987 (J.L.A. and D.C.R.), and a National Science Foundation Graduate Research Fellowship (A.N.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2696311.
REFERENCES
- Amarasinghe AK, MacDiarmid R, Adams MD, Rio DC 2001. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA 7: 1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Carmo-Fonseca M, Aparício S 2006. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell 33: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckenhauer HJ, Bongartz D 2007. Algorithmic aspects of bioinformatics. Springer-Verlag, New York [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Reed R 1995. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature 375: 510–513 [DOI] [PubMed] [Google Scholar]
- Churbanov A, Vorechovsky I, Hicks C 2009. Computational prediction of splicing regulatory elements shared by Tetrapoda organisms. BMC Genomics 10: 508 doi: 10.1186/1471-2164-10-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Fedorov A, Saxonov S, Fedorova L, Daizadeh I 2001. Comparison of intron-containing and intron-lacking human genes elucidates putative exonic splicing enhancers. Nucleic Acids Res 29: 1464–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36: 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G 2006. Comparative analysis identifies exonic splicing regulatory sequences—The complex definition of enhancers and silencers. Mol Cell 22: 769–781 [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ 2010. Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol Cell Biol 30: 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci 97: 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM 2006. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput Biol 2: e86 doi: 10.1371/journal.pcbi.0020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, Zahler AM 2009. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J Biol Chem 284: 28490–28497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren E, Lev-Maor G, Ast G 2007. The emergence of alternative 39 and 59 splice site exons from constitutive exons. PLoS Comput Biol 3: e95 doi: 10.1371/journal.pcbi.0030095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2006. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol 13: 5–7 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee BT, Tan TW, Ranganathan S 2004. DEDB: a database of Drosophila melanogaster exons in splicing graph form. BMC Bioinformatics 5: 189 doi: 10.1186/1471-2105-5-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB 2001. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci 98: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev 12: 1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW 2005. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Pastuszak AW, Joachimiak MP, Blanchette M, Rio DC, Brenner SE, Frankel AD 2011. An SF1 affinity model to identify branch point sequences in human introns. Nucleic Acids Res 39: 2344–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez I, Lin CH, McAfee JG, Patton JG 1997. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778 [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Mount SM, Salzberg SL 2007. A computational survey of candidate exonic splicing enhancer motifs in the model plant Arabidopsis thaliana. BMC Bioinformatics 8: 159 doi: 10.1186/1471-2105-8-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson BL, Cote GJ, Berget SM 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol 10: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida M, Sridharan V, Morgan S, Rao T, Singh R 2010. Drosophila polypyrimidine tract-binding protein is necessary for spermatid individualization. Proc Natl Acad Sci 107: 12570–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Sachidanandam R, Krainer AR 2005. Determinants of the inherent strength of human 5′ splice sites. RNA 11: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfo CM, Alvarez CJ, van Heeckeren WJ, Webb CJ, Wise JA 2000. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol Cell Biol 20: 7955–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF 2008. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS ONE 3: e3369 doi: 10.1371/journal.pone.0003369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Ast G 2010. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J 29: 1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G 2008. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res 18: 88–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hoffman BE, Lis JT 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol 17: 2649–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR 2006. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15: 2490–2508 [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker RB, Berglund JA 2007. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res 17: 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845 [DOI] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Burge CB 2007. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci 104: 18583–18588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yeo G, Hoon S, Venkatesh B, Burge CB 2004. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci 101: 15700–15705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand EL, Nostrand EL, Liang TY 2007. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet 3: e85 doi: 10.1371/journal.pgen.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Arias MA, Ke S, Chasin LA 2009. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA 15: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]