The mechanism(s) by which unsegmented negative-strand RNA viruses initiate genome replication is poorly understood. Using respiratory syncytial virus as an example, this paper shows that RSV RNA-dependent RNA polymerase can correctly initiate antigenome synthesis even if the first two nucleotides of the template are deleted. The nontemplated addition of the nucleotides suggests a model for how the RSV polymerase initiates antigenome synthesis.
Keywords: RNA-dependent RNA polymerase, replication, encapsidation, respiratory syncytial virus, nonsegmented negative-strand RNA virus
Abstract
There is limited knowledge regarding how the RNA-dependent RNA polymerases of the nonsegmented negative-strand RNA viruses initiate genome replication. In a previous study of respiratory syncytial virus (RSV) RNA replication, we found evidence that the polymerase could select the 5′-ATP residue of the genome RNA independently of the 3′ nucleotide of the template. To investigate if a similar mechanism is used during antigenome synthesis, a study of initiation from the RSV leader (Le) promoter was performed using an intracellular minigenome assay in which RNA replication was restricted to a single step, so that the products examined were derived only from input mutant templates. Templates in which Le nucleotides 1U, or 1U and 2G, were deleted directed efficient replication, and in both cases, the replication products were initiated at the wild-type position, at position −1 or −2 relative to the template, respectively. Sequence analysis of the RNA products showed that they contained ATP and CTP at the −1 and −2 positions, respectively, thus restoring the mini-antigenome RNA to wild-type sequence. These data indicate that the RSV polymerase is able to select the first two nucleotides of the antigenome and initiate at the correct position, even if the 3′-terminal two nucleotides of the template are missing. Substitution of positions +1 and +2 of the template reduced RNA replication and resulted in increased initiation at positions +3 and +5. Together these data suggest a model for how the RSV polymerase initiates antigenome synthesis.
INTRODUCTION
A challenge that viruses with linear genomes face is that they must be able to form an initiation complex that can replicate the terminal nucleotides of their templates, or they may yield truncated defective RNAs. RNA viruses have evolved a variety of RNA replication initiation mechanisms to allow them to accurately position their polymerases opposite the terminal nucleotide, or otherwise generate their terminal nucleotides (for review, see Kao et al. 2001). In the case of RNA-dependent RNA polymerases that initiate RNA synthesis de novo (i.e., without a primer), the initiation complex assembles in a way to ensure accurate initiation. For example, the phi6 bacteriophage polymerase binds the template RNA such that the first nucleotide of the template is 1 nt away from the catalytic site. The initiating NTP, GTP, becomes bound to the initiation site of the complex by forming specific contacts with the polymerase and hydrogen-bonding with the penultimate nucleotide of the template. The position of the template is then adjusted to allow base-pairing between the terminal nucleotide and GTP, and initiation of RNA synthesis can ensue (Butcher et al. 2001). A similar situation occurs with rotavirus polymerase VP1, which recognizes the sequence at the 3′ end of the plus-strand RNA but binds to it out of register and then undergoes structural changes to reposition the RNA relative to the active site (Lu et al. 2008). In the Flaviviridae, GTP is required at high concentration for de novo initiation, irrespective of the sequence at the 3′ end of the template (Kao et al. 1999; Luo et al. 2000). Structural analysis of the bovine viral diarrhea virus polymerase showed that the GTP is positioned within the polymerase at the −1 position relative to the product and acts as a platform that helps orient the initiating NTP to allow phosphodiester bond formation with the second NTP (Choi et al. 2004; Choi and Rossmann 2009). In the case of reovirus lambda 3 and rotavirus, the polymerase contains a priming loop, which fulfills the same function as the GTP of bovine viral diarrhea virus (Tao et al. 2002; Lu et al. 2008).
Other RNA viruses use a primer to initiate replication. Picornaviruses use uridylylated VPg protein as a primer, and the polymerase initiation complex accommodates and stabilizes the primer, allowing the uridine bases to base-pair with the template for initiation of RNA synthesis (Paul et al. 1998; Ferrer-Orta et al. 2009 and references therein). Members of the segmented negative-strand viruses can initiate replication using a prime-realign mechanism, in which the polymerase initiates internally on the template to generate a short oligonucleotide primer and then repositions so that the primer anneals to the 3′ terminus of the template (Garcin et al. 1995; Deng et al. 2006). The polymerase can then incorporate nucleotides on the 3′ end of the primer to continue RNA synthesis. In these cases, accurate polymerase repositioning is facilitated by repetitive terminal sequences or by secondary structures in the template RNA.
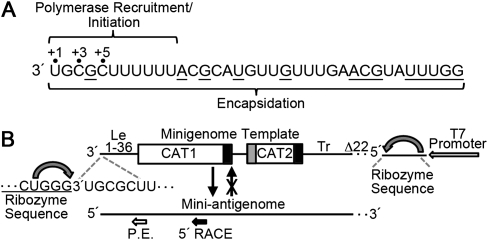
In contrast to the viruses described above, the replication initiation mechanism used by the nonsegmented negative-strand (NNS) RNA viruses is poorly understood. Respiratory syncytial virus (RSV) is a member of the Paramyxoviridae, one of four virus families with an NNS RNA virus genome. The NNS RNA viruses replicate via a positive-strand RNA replication intermediate called the antigenome, and both the antigenome and genome RNAs are encapsidated with viral nucleoprotein (N) as they are synthesized to form a helical nucleocapsid (Whelan et al. 2004; Albertini et al. 2006; Green et al. 2006; Tawar et al. 2009). Concurrent encapsidation of the nascent replication product is believed to be necessary for polymerase processivity and so is a key feature of RNA replication, although it is not yet clear if it is a component of the replication initiation complex or becomes associated after initiation has occurred (Vidal and Kolakofsky 1989; Gubbay et al. 2001; Qanungo et al. 2004; McGivern et al. 2005). In RSV, as in the other NNS viruses, there are two promoter regions for RNA replication: the leader (Le) region at the 3′ end of the genome and the trailer-complement (TrC) region at the 3′ end of the antigenome (Mink et al. 1991). In their natural contexts, the two promoter regions have functional differences, with the Le region being able to signal both transcription and RNA replication (Fearns et al. 2002), while the TrC region signals a higher level of RNA replication than the Le (Fearns et al. 2000; Hanley et al. 2010). The RSV Le and TrC promoter regions are identical for 10 of the first 11 nucleotides and share significant similarity for the first 36 nucleotides, but then diverge (Mink et al. 1991). Functional mapping of the Le region showed that the first 11 nucleotides are sufficient to recruit the RSV polymerase and direct initiation of RNA synthesis (Fearns et al. 2002; Cowton and Fearns 2005), while nucleotides up to position 36 are required in addition for encapsidation of the replication product (Fig. 1A; McGivern et al. 2005). The first 36 nt of the TrC promoter are also sufficient to signal synthesis of an encapsidated replication product (Fearns et al. 2000).
FIGURE 1.
(A) Diagram showing the sequence of the RSV Le nucleotides 1–36, with the regions required for polymerase recruitment and initiation, and encapsidation indicated. Positions +1, +3, and +5, which were found to be used as initiation sites in this study, are marked. Nucleotides that are different from those in the TrC promoter are underlined. (B) Schematic diagram (not to scale) depicting the dicistronic minigenome template. The 3′ end of the minigenome consists of the first 36 nt of Le sequence, which directly abuts a CAT-specific sequence that has the RSV N-P gene junction inserted, resulting in two segments, CAT 1 (552 nt) and CAT 2 (190 nt). The 3′ terminus of the Le sequence is generated by a delta ribozyme (curved arrow), which creates a 2′,3′ cyclic phosphate group at the end of the template RNA, while the 5′ terminus of the minigenome is created by a hammerhead ribozyme. Note that CAT 1 lacks a transcription-specific gene start signal but possesses a gene end signal, as indicated by a black box. CAT 2 is separated from CAT 1 by a short intergenic region and is flanked by gene start (gray box) and gene end signals. The 5′ terminus of the minigenome consists of the RSV Tr, with a 22-nt deletion at the 5′ end, as indicated by a dotted line. This deletion removes the promoter from the mini-antigenome replication product such that it is unable to act as a template for new minigenomes, as depicted by the arrow with the cross. The positions on the replication product where reverse transcription primers for primer extension (P.E.) and 5′ rapid amplification of cDNA ends (5′-RACE) hybridize are shown.
Studies of RSV RNA replication initiation have been greatly aided using a minigenome system in which RNA replication is reconstituted in cells using plasmid-expressed RNA and polymerase proteins (Grosfeld et al. 1995; Yu et al. 1995; Fearns et al. 1997, 2000, 2002; Peeples and Collins 2000; Marriott et al. 2001; Cowton and Fearns 2005; McGivern et al. 2005; Noton et al. 2010). Of particular significance is the use of a minigenome (or mini-antigenome) in which the template RNA provided to the polymerase is limited to a single step of replication so that the products reflect what is produced from mutant templates, rather than a template that is selected over multiple replication cycles (Fearns et al. 2002; Cowton and Fearns 2005; McGivern et al. 2005; Noton et al. 2010). This system has allowed analysis of replication initiation in an intracellular setting using defined mutant templates. Using this approach, a study to examine initiation from the RSV Le promoter region indicated that the promoter sequence, rather than the 3′ terminus of the template, is responsible for positioning the polymerase and determining the site of replication initiation (Cowton and Fearns 2005). A subsequent study using the TrC promoter gave a similar result, showing that the polymerase was highly constrained to initiate at the correct position, even if substitutions were introduced into nucleotide 1 of the TrC promoter (Noton et al. 2010). Remarkably, this study also showed that if the first nucleotide of the TrC promoter was deleted, the polymerase was still able to initiate at the correct wild-type (wt) position with the wt nucleotide (A). The wt A residue was also introduced at high frequency if there was a substitution at position +1. Together, these data suggest that RSV replication initiation involves a mechanism in which the polymerase is positioned by sequences within the promoter and is able to select the first nucleotide of the product independently of the 3′-terminal nucleotide of the template.
The high level of identity of the Le and TrC promoter regions within the 3′-terminal 11 nt, and their similarity over the first 36 nt suggests that they direct replication initiation in an identical manner. However, mutagenesis studies have indicated that some nucleotides that share identity between Le and TrC differ in their abilities to tolerate mutations, indicating that this might not be the case (Peeples and Collins 2000; Fearns et al. 2002; Noton et al. 2010). For this reason, in this study we performed mutation analysis of the Le promoter to determine if the terminal nucleotide of the antigenome is selected independently of the 3′-terminal nucleotide of the Le promoter, similarly to genome initiation from the TrC promoter, and to investigate what differences exist between the initiation properties of the Le and TrC promoters.
RESULTS
RSV Le sequence containing 3′-terminal deletions can direct efficient RNA replication
As described in the Introduction, a study investigating the mechanism of RSV genome initiation from the TrC promoter showed that the polymerase could efficiently initiate RNA synthesis on a template containing a deletion at the first position of the promoter to yield an RNA product in which the terminal nucleotide was restored to wt sequence. In contrast, deletion of the first two positions of the TrC promoter inhibited RNA replication (Noton et al. 2010). To determine if initiation of antigenome synthesis from the Le promoter follows a similar pattern, we investigated RNA synthesis from a minigenome template that was mutated to delete the first one, two, or three nucleotides from the 3′ terminus of the Le region.
The template used for these studies is shown in Figure 1B. This template was based on a dicistronic minigenome similar to that used for a number of studies of RSV transcription and replication (Fearns et al. 2002; Cowton and Fearns 2005; McGivern et al. 2005; Noton et al. 2010). The minigenome is generated from a T7 promoter, and its 3′ terminus is generated by ribozyme cleavage. The minigenome was modified compared to that used in previous studies such that its 3′ end contained the first 36 nt of Le sequence directly adjoining heterologous chloramphenicol acetyl transferase (CAT) sequence (Noton et al. 2010). The first 36 nt of Le contain all the signals necessary for initiation of RNA replication and encapsidation of the replication product. The transcription-specific signals that include nucleotides 37–44 of Le and the first gene start signal were omitted to increase the strength of the Le replication promoter, to facilitate RNA detection, and to ensure that the probes used in the experiments described below would specifically detect products generated from signals at the 3′ terminus of the template and not mRNA generated from the first gene start signal. The minigenome was also engineered so that it was limited to a single step of replication by introducing a 22-nt deletion at the 5′ end of the Tr region. This deletion removed the TrC promoter from the 3′ end of the mini-antigenome RNA produced from the minigenome template, so that the mini-antigenome was unable to be recognized as a template by the RSV polymerase. Importantly, this modification allowed analysis of replication products generated from a defined input template, rather than a template selected for “fitness” over multiple replication cycles.
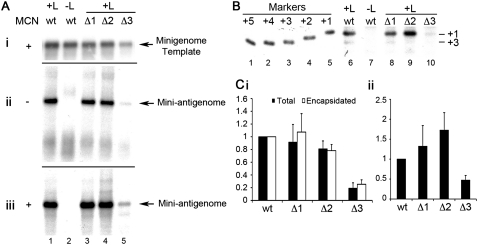
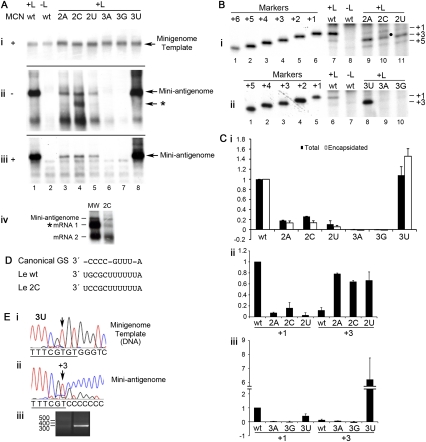
RSV antigenome synthesis was reconstituted intracellularly by transfecting each minigenome plasmid individually into cells together with plasmids expressing the RSV polymerase proteins. The levels of each input template and replication product were then determined by Northern blotting with strand-specific probes. The levels of input minigenome template RNA were examined by harvesting the cell lysates and treating them with nuclease before isolating the RNA. The nuclease treatment degrades unencapsidated RNA, allowing detection of encapsidated template specifically. This analysis was used to determine the levels of input minigenome template (Fig. 2Ai). Analysis of the levels of antigenome replication products showed that the template containing a single nucleotide deletion directed efficient antigenome synthesis, similar to the results observed previously for the TrC promoter (Fig. 2Aii, cf. lanes 1,3; Fig. 2Ci). Interestingly, a template containing a 2-nt deletion also directed efficient RNA replication, producing RNA at ∼80% of wt levels (Fig. 2Aii, cf. lanes 1,4; Fig. 2Ci). The Δ3 template generated a small amount of RNA (Fig. 2Aii, lane 5), demonstrating that nucleotide 3 of the Le plays a key role in RNA replication, but even in its absence the Le promoter retains some functionality. Analysis of RNA isolated following treatment of cell lysates with nuclease confirmed that the RNA products detected in the total intracellular RNA samples were nuclease-resistant (Fig. 2Aiii; Fig. 2Ci), indicating that they were encapsidated RNAs.
FIGURE 2.
Impact of deleting the 3′ terminal 1–3 nt of the minigenome template on RSV RNA replication in HEp-2 cells. (A) Northern blot analysis of minigenome RNAs: nuclease-resistant negative-sense minigenome templates expressed by MVA-T7 polymerase (i); positive-sense replication products expressed by the RSV polymerase from the minigenome templates in total RNA samples (ii); and positive-sense, nuclease-resistant replication products (iii). (Lane 2) A negative control in which the L plasmid, which encodes the enzymatic activity of the RSV polymerase complex, was omitted from the wild-type (wt) minigenome transfection. (B) Primer extension analysis of RNA generated from wt, Δ1, Δ2, and Δ3 templates (lanes 6,8–10). (Lane 7) A negative control in which L was omitted from the wt minigenome transfection. The band that can be detected at the top of the gel in these lanes (and in subsequent figures) is a nonspecific background band. Molecular weight markers in lanes 1–5 are end-labeled oligonucleotides, representing products initiated from position +5 to +1 of the wt template, respectively. (C) Quantitation of total (black bars) and nuclease-resistant RNA (white bars) expressed from each minigenome, as determined by Northern blotting (i); and quantitation of RNA initiated at the +1 position (relative to the wt minigenome) from each minigenome, as determined by primer extension (ii). Each value was calculated relative to the wt value, and each error bar represents standard error of the mean of at least three independent experiments.
These results showed that the RSV Le could still direct RNA replication if the first position was deleted, similar to the findings with the RSV TrC promoter. However, in contrast to the TrC promoter, a Le promoter containing deletion of the first two positions was also highly efficient for RNA replication, demonstrating that nucleotides 1U and 2G of the Le promoter are not essential for antigenome initiation to occur.
Terminal deletions of 1 or 2 nt from the 3′ end of Le were repaired in the antigenome product
Studies of the RSV TrC promoter showed that the genome RNA product was initiated correctly at the wt position +1, even if the first nucleotide of the template was deleted. To determine what effects the Le deletion mutations had on the replication initiation site, RNA products present in the total intracellular RNA samples were examined by primer extension analysis. The primer that was used for this analysis was designed to hybridize close to the 5′ end of the transcripts, at positions 24–50 relative to the wt replication product, to allow precise size resolution of the primer extension products. Because this primer hybridized close to the initiation site, it had the potential to detect prematurely terminated replication products, which might not be detected on a Northern blot, in addition to full-length antigenome.
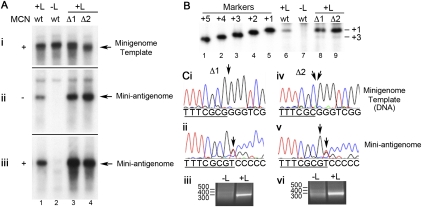
Examination of RNA produced from the Δ1 template showed that it was initiated at the same site as RNA generated from a wt RNA template (Fig. 2B, cf. lanes 6,8). This result indicates that the antigenome was initiated at the +1 position relative to a wt template, and at the −1 position relative to the Δ1 mutant template. Thus, initiation from the Δ1 Le template was similar to initiation from a Δ1 TrC promoter, with the polymerase apparently being able to introduce the first nucleotide of the RNA product without base-pairing with the terminal nucleotide of the template. Interestingly, analysis of RNA generated from the Δ2 Le promoter showed that it was also initiated at the same position as RNA initiated from a wt promoter (Fig. 2B, lane 9), indicating that in this case, the polymerase could initiate at the −2 position relative to the truncated template. Quantitation of the levels of the RNA initiated at the +1 position indicated that the Δ1 and Δ2 mutant templates actually generated slightly higher levels of RNA than the wt promoter, indicating that these mutant promoters were highly efficient, despite the terminal deletions (Fig. 2Cii). Remarkably, RNA derived from the Δ3 mutant template was also initiated at the wt +1 position, although in this case, only a very low level of RNA could be detected, suggesting that initiation from this template was highly inefficient (Fig. 2B, lane 10; Fig. 2Cii). It should be noted that a low level of initiation from the +3 position, the next pyrimidine in the template, could also be detected even on the wt template, suggesting that this is an alternative initiation site for the RSV polymerase (note that throughout the paper, numbering is relative to the wt template). Initiation from the +3 position was also observed for the TrC promoter (Noton et al. 2010).
Repair of the antigenome 5′ termini occurred in the absence of vaccinia virus
The minigenome system used for the experiments described above (and throughout the rest of the study) used recombinant modified vaccinia virus Ankara (MVA-T7) to generate T7 RNA polymerase to drive plasmid expression. Because vaccinia virus is a cytoplasmic virus that expresses several nucleic acid–modifying enzymes, this raised the possibility that the MVA-T7 was providing an enzyme activity that allowed repair of the minigenome or mini-antigenome RNA termini. To eliminate this possibility, the Δ1 and Δ2 mutants were examined by reconstituting RNA synthesis in BSR-T7 cells, a BHK-21 cell line that has been engineered to express T7 RNA polymerase (Buchholz et al. 1999). The Northern blot and primer extension results obtained using BSR-T7 cells were identical to those obtained from the MVA-T7 system, demonstrating that neither the RNA templates nor their products were being modified by vaccinia virus (Fig. 3A,B).
FIGURE 3.
Impact of deleting nucleotides 1 and 2 of the minigenome template on RSV RNA replication in BSR-T7 cells. (A,B) The panels are arranged as in Figure 2. (C) 5′-RACE sequence analysis of the RNA products generated from the Δ1 and Δ2 mutant templates: sequence analysis of the input minigenome plasmid DNA isolated from Δ1 (i) and Δ2 (iv) transfected cells. 5′-RACE sequence trace of the corresponding Δ1 (ii) and Δ2 (v) replication products (tailed with dCTP). All sequence traces are shown as template sense DNA. The Le sequence (underlined); sites of mutation in the templates (arrows); and the reintroduced nucleotides in the replication products. Note that the minor sequence that can be detected (beneath the 5′-RACE sequence trace) corresponds to the sequence of input plasmid DNA. Agarose gel electrophoresis of the 5′-RACE products generated from Δ1 and Δ2 mutant RNA samples from transfections that were lacking or containing the L polymerase plasmid, as indicated (iii and vi).
To determine which bases were incorporated at positions 1 and 2 during replication initiation from the Δ1 and Δ2 templates, the RSV replication products generated in BSR-T7 cells were subjected to 5′ rapid amplification of cDNA ends (5′-RACE) and sequenced. Sequence analysis of DNA isolated from the transfected cells confirmed that the plasmid templates contained the Δ1 (Fig. 3Ci) and Δ2 (Fig. 3Civ) deletions. In contrast, sequence analysis of antigenome replication products isolated from the same cells showed that for both the Δ1 (Fig. 3Cii) and Δ2 (Fig. 3Cv) Le templates, it was restored to wt sequence. This confirmed the results obtained by primer extension showing that the RNA was initiated at the correct wt position, rather than opposite the first nucleotide of the template, and in addition indicated that the 5′-terminal nucleotides of the replication product were selected with high specificity, even though there was no template base-pairing to govern their selection. Figure 3C, panels iii and vi, shows agarose gel electrophoresis analysis of the 5′-RACE products and demonstrate that intense specific bands were only generated in the case of transfections including the RSV L polymerase subunit, confirming that the RNA being sequenced was largely a product of the RSV polymerase. Faint bands could also be detected in the −L lanes. These are likely to be derived from specific and nonspecific primer binding to the input plasmid DNA, which can be detected as a minor sequence on the sequencing trace (Fig. 3Cii,v).
Nucleotide substitutions at Le position +1 increase initiation at positions +3 and +5
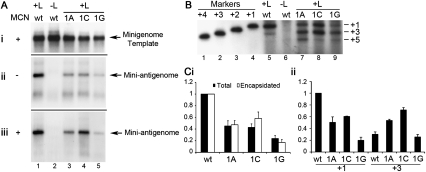
Having found that deletion of nucleotide 1U did not affect the site of replication initiation, it was of interest to determine what effect substitution at this position had. This nucleotide was substituted with the three alternative residues, and the RNAs produced were examined by Northern blotting, to determine the effect of the mutation on the accumulation of full-length replication product, and primer extension analysis, to determine the site at which RNA products were initiated. Northern blot analysis showed that each of the substitutions reduced mini-antigenome accumulation, with the 1G substitution resulting in replication at <20% of wt levels, for example (Fig. 4Aii,iii; Fig. 4Ci). Primer extension analysis of the RNA products showed a significantly different pattern of initiation compared to the wt template (Fig. 4B; Fig. 4Cii). As described above, the RNA from the wt template was predominantly initiated at nucleotide +1, with a lower level of initiation detected at position +3. In contrast, the products from the 1A substitution template showed a reduced level of initiation at position +1, but an increased level of initiation at position +3. There was also clear evidence of initiation at position +5 (Fig. 4B, lane 7). Similarly, the 1C substitution resulted in a slight decrease in the level of initiation at position +1 and a significant increase in initiation at position +3, with detectable initiation at position +5 (Fig. 3B, lane 8). The 1G substitution caused an overall decrease in initiation levels and a change in the relative abundances of the +1, +3, and +5 initiations (Fig. 3B, lane 9). These findings indicate that substitution at Le position +1 inhibits initiation at this site and promotes initiation at downstream pyrimidines (Fig. 1A), suggesting that the presence of a mutant base at position +1 interferes with replication initiation.
FIGURE 4.
Impact of substituting nucleotide 1U of the minigenome template on RSV RNA replication. Minigenome replication was reconstituted in HEp-2 cells simultaneously infected with MVA-T7, and the resulting RNA was analyzed. Panels are as described for Figure 2, except that panel Cii shows quantitation of RNA initiated at positions +1 and +3; each value was calculated relative to the wt +1 initiation value, and each error bar represents range of the mean of two independent primer extension analyses.
Nucleotide substitutions at Le position +2 inhibit initiation at position +1
Similar analysis to that described above was performed for mutant Le templates containing substitutions at positions +2 and +3 (Fig. 5). Although deletion of nucleotides 1 and 2 was well tolerated, each of the three possible nucleotide substitutions at position +2 significantly inhibited replication, with mini-antigenome levels being reduced to ∼20% of wt levels in each case (Fig. 5Aii,iii; Fig. 5Ci). Primer extension analysis of the RNA indicated that in each case, initiation at position +1 was significantly reduced, whereas initiation at position +3 was increased, such that the total levels of detectable initiation were similar for the mutants as for the wt template (Fig. 5Bi, lanes 9–11; Fig. 5Cii). These data show that the identity of the nucleotide at position +2 of the template can affect the ability of the polymerase to initiate at position +1 and suggests that the nucleotide at position +2 affects the accuracy of replication initiation, without necessarily affecting the ability of the Le promoter to direct initiation of RNA synthesis.
FIGURE 5.
Impact of substituting nucleotides 2G and 3C of the minigenome template on RSV RNA replication. Minigenome replication was reconstituted in HEp-2 cells simultaneously infected with MVA-T7, and the resulting RNA was analyzed. (A–C) The panels are arranged similarly to Figure 2 except that in B, the primer extensions of the RNA produced from the position 2 and position 3 substitution mutants are separated into two panels, i and ii, respectively, and (C) quantitation of the RNA initiated at +1 and +3 from the position 2 and position 3 substitution mutant templates are separated into two panels, ii and iii, respectively. (Aiv) Northern blot analysis of positive-sense RNA generated from a transcription-competent minigenome, to act as a molecular weight marker (MW), and the 2C mutant template, as indicated. The positions of the antigenome, CAT 1, and CAT 2 mRNAs are indicated, and the subgenomic RNA generated from the 2C minigenome is indicated by an asterisk in panels Aii and Aiv. In Bi the position of the +2 band detected from the 2C minigenome is indicated with a dot. (D) Sequence alignment showing the similarity between Le nucleotides 3–12 and the gene start (GS) signal sequence. (E) 5′-RACE sequence analysis of the RNA products generated from the 3U mutant template; arranged as Figure 3C.
Surprisingly, substitution of position 2G with a C residue also resulted in production of a nuclease-sensitive, subgenomic RNA (indicated with an asterisk), reminiscent of an mRNA transcription product (Fig. 5Aii, lane 4). As described above, the minigenome template did not contain a promoter-proximal gene start signal to direct mRNA initiation but did contain an internal gene junction (Fig. 1B). Therefore, it seemed likely that the subgenomic RNA was initiated within the Le as a consequence of the 2C mutation and terminated at the internal gene end signal. To examine this possibility, the size of the subgenomic RNA was compared to CAT 1 mRNA generated from a transcription-competent minigenome template that contained the complete wt 44-nt Le and promoter-proximal gene start signal. Northern blot analysis of the RNA produced from this minigenome showed that it generated an antigenome and two subgenomic mRNAs, corresponding to CAT 1 and CAT 2 mRNAs. The subgenomic RNA generated from the 2C minigenome template was similar in size to CAT 1 mRNA, consistent with it being terminated at the internal gene end signal (Fig. 5Aiv). The 2C minigenome also generated a smaller subgenomic RNA, which was the appropriate size to be CAT 2 mRNA, suggesting that the polymerase had terminated and reinitiated RNA synthesis at the gene junction, as is typical during transcription. However, this smaller RNA was also generated from several other templates and often comigrated with a nonspecific background band (e.g., see Fig. 2), and so its significance could not be interpreted.
Primer extension analysis of the RNA generated from the 2C mutant showed that an additional faint band was detectable that corresponded with initiation at position +2 (Fig. 5Bi, lane 10, marked with a dot). Together with the Northern blot data, this finding suggests that the 2G-to-C substitution caused initiation at position +2, which resulted in production of subgenomic RNA, terminated at the internal gene end signal. Inspection of the wt Le sequence shows that there is a gene-start-like sequence lying from positions 3 to 12. The 2G-to-C mutation increased the similarity of this sequence to the canonical RSV gene start signal (Fig. 5D). Thus, it seems likely that this change allowed transcription to be initiated at position +2, accounting for the production of subgenomic RNA.
A 3C-to-U substitution allows efficient replication to occur
Substitution analysis was also performed for nucleotide 3C. Substitution with an A or G residue completely inhibited antigenome production and ablated initiation as detected by primer extension (Fig. 5Aii,iii, lanes 6,7; Fig. 5Bii, lanes 9,10). This finding suggests that these two substitutions either inactivated the Le promoter, such that initiation of RNA synthesis could not occur, or that the products containing these substitutions were so unstable as to be undetectable. Interestingly, substitution of position +3 with a U residue resulted in highly abundant levels of replication product, which was resistant to nuclease digestion, indicating that it was encapsidated (Fig. 5Aii,iii, lane 8). In addition, a significant amount of RNA produced from the 3U template migrated more slowly than mini-antigenome RNA on the Northern blot. Attempts to determine if this was due to inaccurate termination at the end of the template showed no evidence that this is the case (data not shown). Therefore, it is currently unclear why this RNA migrated abnormally. Primer extension analysis of the RNA generated from this mutant showed that there was only a very low level of detectable initiation from position +1, but abundant initiation from position +3 (Fig. 5Bii, lane 8; Fig. 5Ciii). 5′-RACE analysis of the RNA confirmed this result, showing that it was initiated at position +3 and was the complement of the input template (Fig. 5E). Thus, the data suggest that, in this case, the RNA initiated at position +3 was encapsidated and elongated. This was different from what was observed with the other templates in the study, in which production of full-length encapsidated antigenome correlated with the levels of initiation at position +1, rather than +3 (e.g., cf. the graphs in Fig. 5Ci,ii). It should be noted that the 3U substitution creates a 3′-UGC sequence at positions 3–5 of the Le, which is the same sequence as at positions 1–3 of the wt Le (Fig. 1A). Thus, it seems likely that the 3U substitution not only allowed efficient initiation of RNA synthesis from the +3 site, but also encapsidation of the RNA to generate a full-length replication product.
DISCUSSION
In a previous study, we showed that a mutant TrC promoter in which the first position was deleted could still direct RNA replication with high efficiency (60% of wt levels) and generate a product in which the terminal nucleotide was restored in 100% of sequenced transcripts (Noton et al. 2010). The study described here extends this observation, demonstrating that in the case of the Le promoter, the RSV polymerase can tolerate a 2-nt deletion and produce an RNA that is restored to wt sequence. Analysis of templates containing substitutions at positions +1 to +3 indicated that mutations could affect the preferred site of initiation and also the outcome of the initiation event. These findings provide information on the mechanism of replication initiation, as discussed below.
“Template-independent” initiation
Remarkably, deletion of either one or two nucleotides from the 3′ end of the Le promoter resulted in RNA that was initiated at the wt +1 position, in a negative register relative to the input template (Figs. 2, 3). In addition, 5′-RACE analysis of the RNA produced from the Δ1 and Δ2 templates showed that it was restored to wt sequence (Fig. 3). The efficiency with which the restoration occurred was high: The Δ2 Le template generated replication product at 80% of wt levels, and consensus sequencing showed that almost all of the detectable RNA was initiated at the wt +1 position with 5′-AC, the wt terminal nucleotides. This high efficiency suggests that the mechanism by which deletion mutations were repaired in this study is a normal event in RSV replication, and not a rare, chance occurrence.
The mechanism by which the deletions were repaired is not known. We considered the possibility that the template RNA did not contain the intended 3′-terminal deletions due to events occurring following template synthesis in the transfected cells. For example, one possibility is that the template generated by T7 RNA polymerase was restored to wt sequence by a cellular (or viral) terminal transferase activity. However, this seems unlikely since the 3′ end of the template was created by a ribozyme and so contained a 2′,3′-cyclic phosphate at the terminus, rather than a 3′-hydroxyl group (Tomar et al. 2006). In addition, it would be surprising if a terminal transferase were to restore the length and sequence of the Δ2 template so efficiently that the product yields were similar to those generated when a wt template was transfected into cells. Another possibility is that replication products were generated from templates containing the entire ribozyme, or a few nucleotides of ribozyme sequence, as a consequence of inefficient or inaccurate cleavage, respectively. However, both these scenarios would result in a G residue being present at nucleotide 1, which was found to be deleterious to replication and to result in an altered initiation pattern (Fig. 4). In addition, we have previously shown that addition of six or more nucleotides to the 3′ end of the RSV Le significantly inhibits promoter activity, indicating that a template retaining the ribozyme would not be viable (Cowton and Fearns 2005). Thus, it is unlikely that the replication products were generated from longer-than-expected templates, indicating that the restoration to wt sequence occurred during replication initiation. Finally, it is worth noting that the minigenome assay cannot completely mimic the conditions that exist during the different stages of RSV infection. It is possible that the high rate of repair that we observed was dependent on a particular polymerase: template ratio, which might only occur transiently or rarely during viral infection. Even if this were the case, the results obtained describe a novel aspect of RSV molecular biology, which could play an important role in virus replication.
Based on the arguments outlined above and the efficiency of the repair of the RNAs generated from the Δ1 and Δ2 mutant templates, we propose that the mechanism by which the RNA products were restored to wt sequence is related to events that occur during normal replication initiation, and that the results described here provide insight into this process. The finding that the polymerase was able to initiate at the correct position, even if the 3′-terminal one or two nucleotides of the promoter were deleted, suggests that the site of initiation is not determined by the location of the 3′ terminus of the template, but rather by the nucleotide sequence of the promoter. This finding is consistent with results obtained in a previous study of the RSV Le region, which showed that if a small number of nucleotides were added to the 3′ end of the Le promoter, the replication product was still initiated at the correct wt position, even if the additional nucleotides represented an optimal initiation sequence (Cowton and Fearns 2005). Together these data indicate that the 3′ terminus of the template provides little or no guidance to the RSV polymerase during initiation, but rather that the polymerase is precisely positioned by the promoter sequence to ensure accurate initiation. In addition, it was previously shown that substitutions at nucleotides 3, 5, 8, 9, and 10 of the Le significantly inhibit all positive-sense RSV RNA synthesis (Fearns et al. 2002). This finding, in combination with the data regarding the Δ1 and Δ2 mutants described here, suggests that it is Le nucleotides 3, 5, 8, 9, and/or 10 that anchor the polymerase during initiation of RNA synthesis.
Whereas the deletion mutants yielded high levels of antigenome, which was initiated correctly, substitution of nucleotides 1 or 2 reduced the yield of antigenome RNA and altered the site of RNA synthesis initiation, leading to an increase in RNA initiated from positions +3 and +5 (Figs. 4, 5). These data show that although the first and second nucleotides of the Le promoter were not required for initiation of replication (because the Δ2 mutant was functional), having an incorrect nucleotide at either of these positions affected the site of RNA synthesis initiation and significantly inhibited the ability of the polymerase to generate replication product from position +1. These findings are consistent with a model in which the 5′-terminal nucleotides of the RSV antigenome are selected independently of the 3′-terminal nucleotides of the genome template. This could occur if RSV replication initiation involved recruitment or synthesis of a sequence-specific primer. According to this hypothesis, deletion of the terminal nucleotides of the template might have no significant effect on the ability of the primer to be used, but substitution of nucleotides 1 and 2 could sterically hinder formation of the template/primer/polymerase complex at the +1 initiation site, resulting in increased use of the +3 and +5 initiation sites.
As described in the Introduction, several RNA viruses use an RNA primer to initiate RNA replication (Garcin et al. 1995; Paul et al. 1998; Deng et al. 2006; Tomar et al. 2006; Ferrer-Orta et al. 2009). If this were the case for RSV, it raises the question of how a sequence-specific primer could be generated. Viruses that are known to initiate RNA replication with a nucleic acid primer use a prime-realign mechanism for initiation, and in these cases, there is repetitive sequence at the termini to allow the prime-realign process to occur (Garcin et al. 1995; Deng et al. 2006). However, this is not the case for RSV in which the next UG sequence in the template is at Le positions 17 and 18, which can be substituted with only a minor effect on replication efficiency (Fearns et al. 2002). Therefore, if a prime-realign mechanism were used for RSV antigenome initiation, it would involve a distinct mechanism from what has been described previously. An alternative possibility is that RSV has an internal cis-acting replication element to signal primer synthesis. However, there are relatively few RSV-specific sequences in the minigenome construct, and it would be expected that encapsidation of the template would obscure any internal initiation site. A third possibility is that the RSV polymerase might have the capacity to use any RNA as a template, similarly to the polymerase of phage Qβ (Hill and Blumenthal 1983; Chetverin et al. 1991; Ugarov et al. 2003), allowing it to initiate abortive RNA synthesis at a UG sequence to create a dinucleotide primer that it can then use to initiate RNA replication at the 3′ end of the genome. A fourth possibility is that the RSV polymerase is able to recruit oligonucleotides from the cytoplasmic milieu to act as primers. There are several examples in which short oligonucleotides have been used as primers to initiate viral RNA synthesis (Kawakami et al. 1981; Garcin and Kolakofsky 1992; Kao and Sun 1996; Nagy et al. 1997; Chen and Patton 2000; Nomaguchi et al. 2003). In some of these cases, oligonucleotides of appropriate sequence or length were selectively used, indicating that RNA virus polymerases might have the ability to bind specifically to oligonucleotides that would be suitable for initiation. Finally, it is possible that the RSV polymerase has the capacity to self-generate a primer in a template-independent fashion. Although this seems an extraordinary possibility, it is not without precedent. For example, the cellular enzyme, tRNA nucleotidyltransferase, can generate a sequence-specific polyribonucleotide in a template-independent manner to modify the 3′ termini of tRNA molecules (Weiner 2004), and a study with rotavirus polymerase showed that it was able to polymerize a G-G dinucleotide in a reaction in which the template had been diluted to the extent that no template function was detectable (Chen and Patton 2000). Further genetic, biochemical, and high-resolution structural studies will be necessary to investigate these possibilities and determine the mechanism underlying RSV RNA replication initiation. Whichever initiation mechanism is used by the RSV polymerase, the data suggest that the process is highly coordinated to ensure accurate initiation, consistent with what has been shown for other viruses with linear genomes. Moreover, the first two nucleotides of the RSV replication products (5′-AC) are conserved throughout all sequenced paramyxoviruses, despite considerable diversity in the organization and sequences of their promoters. This suggests that the mechanism that is used by RSV is conserved throughout the Paramyxoviridae.
Comparison of the Le and TrC promoters
We previously showed that the RSV polymerase could efficiently and accurately select the first nucleotide of the genome product even if the 3′-terminal nucleotide of the TrC promoter was mutated (Noton et al. 2010). Thus, the results described for Le and TrC are similar, and in both cases are consistent with a mechanism involving loading of the polymerase with the first nucleotide(s) of the product independently of the sequence at the 3′ terminus of the template. However, the data also revealed differences between the Le and TrC promoters. Whereas a 2-nt deletion was tolerated in the context of the Le promoter, deletion of the first two nucleotides of the TrC promoter ablated all detectable RNA synthesis. In addition, substitutions at positions +1 and +2 were generally tolerated better in the context of the Le, than in the TrC promoter. These differences are surprising given that the Le and TrC promoter sequences are very similar, sharing identity at nucleotides 1–11, except for position +4. It is possible that there are differences in the mechanisms of initiation on the Le and TrC promoters, as has been shown for influenza virus (Deng et al. 2006). However, an alternative explanation is that the same mechanism is used in both cases, but that the sequence of the promoter at nucleotides 4, 12, and downstream affects the stability of the initiation complex, resulting in differences in the way in which the promoters respond to mutation analysis.
It should be noted that some of the substitution mutants behaved differently than in a previous study of the Le promoter (Fearns et al. 2002). There are two differences between the templates used in the two studies. First, the template used in the previous study contained all the sequences required for transcription initiation at the first gene. We have found that the presence of the gene start signal at position 45 significantly reduces the level of detectable replication product (DR McGivern, C Zack, and R Fearns, unpubl.), indicating that there is competition between the cis-acting elements. Second, in the previous study, the parental Le sequence contained a C residue at position +4, rather than a wt G residue. The 4C Le variant has been observed to arise spontaneously in vaccine strains of RSV and is more efficient for RNA replication than the wt Le. Therefore, in the past, it was used to allow accurate quantitation of the effects of mutations on RNA replication in a situation in which transcription was also being analyzed (and in which RNA replication from a wt Le would be virtually undetectable). Thus, either of these two differences, or their combination, had the potential to have an impact on the effects of mutations in the terminal nucleotides.
Initiation at +3
Primer extension analysis of the RNA generated from a wt Le sequence indicated that in addition to RNA initiated at the +1 position, which had the potential to be extended into full-length antigenome, a much smaller amount of RNA initiated at +3 was also detected. The significance of this RNA is unknown. Initiation at +3 was also observed in experiments with the TrC promoter; in this case, the RNA initiated at position +3 was extended between 50 and 100 nt, suggesting that this initiation event results in abortive RNA synthesis and the production of truncated RNA (Noton et al. 2010). It should be noted that because the experiments performed in this study monitored steady-state levels of RNA in a cellular environment, the +3 initiation event might be considerably more frequent than is apparent from the primer extension analyses presented here, particularly if this RNA were uncapped and unencapsidated. Inspection of the Le sequence shows that nucleotides 3–12 of the Le share strong similarity with the canonical RSV gene start signal (Fig. 5D) and even greater similarity with the slightly different RSV L gene start signal, with 8/10 nucleotides being identical. Thus, initiation at +3 might occur as a result of polymerase interacting directly with this signal. Substitution at position +1 or +2 resulted in a significant increase in +3 initiations (Figs. 4, 5). Even if nucleotide 1U was replaced with a C residue, there was decreased initiation at +1C and increased initiation at +3C, compared to the wt template, suggesting that the increase in initiation at position +3 was not simply due to the polymerase attempting to initiate at the first pyrimidine in the sequence (Fig. 4). This finding suggests that initiations at positions +1 and +3 occur by distinct mechanisms and are competitive, such that failure of the polymerase to form a +1 initiation complex creates more opportunity for the +3 initiation complex to form. It is not known if the RNA initiated at +3 is significant for viral infection, and further studies are ongoing to explore this possibility.
Production of subgenomic RNA
The data obtained with the 2C mutant template were intriguing as they showed that this minigenome generated subgenomic RNA, in addition to full-length mini-antigenome. Primer extension analysis indicated that there was a faint band at the +2 site, which was not detected for RNA produced from other minigenome templates, indicating that this was the initiation site for the subgenomic RNA (Fig. 5Bi). The size of the subgenomic RNA was consistent with it having been terminated at the internal gene end signal in the minigenome (Fig. 5Aiv), making it reminiscent of an mRNA transcription product. Unfortunately, the levels of the subgenomic RNA were too low for us to determine if it had other characteristics of an mRNA, such as a 5′ cap and 3′-poly(A) tail (data not shown). (Note that if RNA initiated at position +2 were capped, at least some of the primer extension product would contain an additional nucleotide at the 5′ end and would migrate at position +1, accounting for the apparent discrepancy between the intensity of the +2 initiation and the abundance of subgenomic RNA.) However, the 2G-to-C substitution increased the similarity of the sequence near the 3′ end of the Le to the RSV gene start sequence, and so it seems likely that this allowed an mRNA transcript to be initiated from this site. If this were the case, it suggests that the transcriptase form of the RSV polymerase is not constrained to initiate at the gene start signal that usually lies at position 45 on the genome, but can be engaged to initiate at the 3′ end of Le. Whether this has relevance during an authentic infection is not known, but this finding might have significance for understanding the mechanism of RSV transcription initiation.
Encapsidation
Production of antigenome RNA not only depends on initiation of RNA replication, but also concurrent encapsidation of the nascent RNA. In this study, with the exception of the 3U template (discussed below), the levels of full-length, encapsidated replication product correlated with levels of RNA initiated at position +1, rather than RNA initiated at +3 and +5, suggesting that RNA initiated at position +1 was efficiently encapsidated, whereas RNA initiated at +3 or +5 was not. This finding suggests that either the polymerase able to initiate at +1 was a replicase complex poised to initiate encapsidation, as has been suggested for the VSV replicase (Qanungo et al. 2004), whereas polymerase that initiated at positions +3 or +5 was not; or that the 5′-AC sequence at the end of the +1 product is an important factor in initiating encapsidation. These findings suggest that incorrectly initiated replication products would not be encapsidated, and likely not elongated, allowing the virus to avoid replicating defective templates. These findings also help to explain the unusual result obtained with the 3U mutant template. Whereas the 3A and 3C mutations significantly inhibited replication, the 3U mutant yielded more encapsidated replication product than the wt template. In the case of the 3U mutant, there was very little detectable initiation at position +1 and a considerable amount of RNA initiated from position +3, indicating that in this case, the +3 initiation product was encapsidated. It seems likely that the substitution of position 3G with a U residue either allowed replicase to initiate from position +3 to generate encapidated RNA, or re-created a 5′-AC motif at the 5′ end of the RNA initiated at position +3, and that this sequence (in concert with other downstream sequences) was able to direct encapsidation.
CONCLUSIONS
In summary, we have found that nucleotides 1 and 2 of the antigenome replication product can be selected independently of the 3′-terminal nucleotides of the genome template, suggesting the possibility that RSV RNA replication initiation occurs via a primer-mediated initiation mechanism. Such a mechanism could be a means of allowing the RSV polymerase to form a stable complex that enables efficient initiation on a linear RNA template. In addition, we have found evidence that the 5′-AC sequence at the terminus of the replication product correlates with efficient encapsidation, suggesting a link between initiation of RSV RNA replication and encapsidation of the nascent RNA.
MATERIALS AND METHODS
Plasmid construction and mutagenesis
The plasmid encoding the minigenome template was derived from a previously described plasmid, MP-28 (Fearns et al. 2002). MP-28 contains a dicistronic CAT-containing minigenome that is flanked at its 5′ end by a T7 promoter and at its 3′ end by the hepatitis delta virus ribozyme to create the 3′ terminus of the RNA template. The CAT open reading frame contains the RSV N-P gene junction inserted within it, which creates two genes, CAT 1 and CAT 2. MP-28 was modified as follows: RSV-specific sequence from nucleotide 37 of Le to the end of the NS1 untranslated region was deleted such that nucleotides 1–36 of Le were directly adjacent to the nonspecific CAT 1 sequence. This modification removed transcription-specific signal sequences from the 3′ end of the minigenome such that the template was optimized for RNA replication. In addition, 22 nt was deleted from the 5′ terminus of the Tr, and a hammerhead ribozyme was inserted between the remaining Tr region and the T7 promoter. Position +4 of the MP-28 was mutated from 4C to 4G to reflect the wt A2 Le sequence. The resulting plasmid was the backbone into which all mutations were introduced and is referred to as wt throughout this study. Deletions and single-nucleotide substitutions were introduced at the 3′ terminus of the minigenome by PCR mutagenesis, and the 3′- and 5′-terminal sequences of each minigenome template were confirmed by sequencing. The minigenome used to size subgenomic RNAs (MW) (Fig. 5Aiv) was identical to that described above, except the RSV-specific sequence from nucleotide 37 to the end of the NS1 untranslated region in the MP-28 backbone was retained such that CAT 1 transcription signals were present.
Intracellular minigenome RNA synthesis assay
RNA synthesis was reconstituted in HEp-2 cells, as described previously (Fearns et al. 1997). Briefly, HEp-2 cells were transfected with 0.2 μg of minigenome DNA, 0.4 μg of pTM1 N, 0.2 μg of pTM1 P, 0.1 μg of pTM1 M2-1, and 0.1 μg of pTM1 L plasmids using lipofectin (Invitrogen) according to the manufacturer's instructions. HEp-2 cells were simultaneously infected with MVA-T7 (Wyatt et al. 1995) to express T7 RNA polymerase and drive plasmid expression. After ∼18 h, the transfection mixture was replaced with Opti-MEM containing 2% fetal bovine serum (FBS) and actinomycin D at 2 μg/mL (Calbiochem) for 2 h, aspirated and Opti-MEM containing 2% FBS added. In addition, RSV RNA synthesis was reconstituted in BSR-T7 cells, which is a cell line that has been engineered to constitutively express T7 RNA polymerase (Buchholz et al. 1999). BSR-T7 cell monolayers in six-well dishes were transfected as follows. Eight microliters of Lipofectamine 2000 (Invitrogen) was vortexed briefly in 100 μL of Opti-MEM (GIBCO), incubated for 5 min at room temperature, and then mixed with 150 μL of Opti-MEM containing 0.2 μg of minigenome DNA, 0.4 μg of pTM1 N, 0.2 μg of pTM1 P, 0.1 μg of pTM1 M2-1, and 0.1 μg of pTM1 L plasmids. After 20 min at room temperature, the mixture was applied to the BSR-T7 cells previously washed with serum-free Opti-MEM. Cells were incubated for 6 h before FBS was added to a final concentration of 3%. After 18 h, the media were replaced with 2% FBS and actinomycin D at 2 μg/mL for 2 h and aspirated, and Opti-MEM containing 2% FBS added. For both HEp-2 and BSR-T7 RNA synthesis assays, at 48 h post-transfection, RNA was either directly harvested to isolate total intracellular RNA, or cells were incubated with a nonionic detergent containing micrococcal nuclease prior to RNA extraction, as previously described (Fearns et al. 1997). An RNeasy kit (QIAGEN) was used to isolate RNA samples, according to the manufacturer's instructions.
RNA analysis
Northern blot analysis was performed as described previously, using CAT-specific riboprobes to detect the RNA (Fearns et al. 1997; McGivern et al. 2005). Phosphorimage quantitation was carried out using a Molecular Dynamics PhosphorImager. Primer extension analysis was performed by combining RNA representing one-tenth of a well of a six-well dish with 0.2 pmol of 32P-end-labeled primer that hybridized at positions 24–50 of the wt replication product (5′-TTCTCCATTCTAGAGGTTTATGCAAGT). The reaction was heated for 3 min to 80°C and cooled on ice, and the RNA was reverse-transcribed using Sensiscript (QIAGEN) according to the manufacturer's instructions. The reaction was stopped after 1.5 h by the addition of 95% deionized formamide containing 20 mM EDTA, bromophenol blue, and xylene cyanol. End-labeled DNA oligonucleotides corresponding to template sense nucleotides 1–50, 2–50, 3–50, 4–50, 5–50, and 6–50 were used as molecular weight markers. Samples were analyzed on 6% polyacrylamide gels containing 7 M urea in tris-borate-EDTA buffer. Products were visualized by autoradiography and quantified using either ImageJ software or by phosphorimage analysis (the two quantitation methods gave similar results). For RNA quantification, in cases in which examination of the Northern blots showed evidence of experimental error at the level of the transfection or RNA preparation steps, the levels of replication product were normalized to adjust for altered levels of template RNA. To examine the 5′-terminal consensus sequence of the mini-antigenome population, 5′-RACE analysis of total intracellular RNA was performed as follows. RNA was annealed to a template-sense CAT-specific primer (5′-CCAGCTCACCGTCTTT) and reverse-transcribed using Sensiscript, according to the manufacturer's instructions. Following purification, the cDNA was tailed with dCTP using terminal transferase (NEB). The tailed cDNA was PCR-amplified using a nested CAT-specific primer (5′-GCGTCTGCAGCGGAATTCCGGATGA) and a primer that annealed to the dCTP tail (e.g., 5′-GACCACGGTCGAGGGGGGGGGGGGGGG). The PCR reactions were carried out in triplicate, and the products were pooled and gel-purified, and analyzed by DNA sequencing.
ACKNOWLEDGMENTS
We thank Dr. Klaus Conzelmann (Ludwig-Maximilians-University Munich) for providing BSR-T7 cells, Dr. Bernard Moss (National Institutes of Allergy and Infectious Diseases, National Institutes of Health) for providing MVA-T7, and Dr. Peter Collins (National Institutes of Allergy and Infectious Diseases, National Institutes of Health) for providing plasmids. We also thank the Fearns laboratory group fors helpful discussion. The work was funded by grants R01AI074903 and R21AI079328 to R.F.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2813411.
REFERENCES
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW 2006. Crystal structure of the rabies virus nucleoprotein–RNA complex. Science 313: 360–363 [DOI] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240 [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT 2000. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin AB, Chetverina HV, Munishkin AV 1991. On the nature of spontaneous RNA synthesis by Qβ replicase. J Mol Biol 222: 3–9 [DOI] [PubMed] [Google Scholar]
- Choi KH, Rossmann MG 2009. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol 19: 746–751 [DOI] [PubMed] [Google Scholar]
- Choi KH, Groarke JM, Young DC, Kuhn RJ, Smith JL, Pevear DC, Rossmann MG 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc Natl Acad Sci 101: 4425–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowton VM, Fearns R 2005. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol 79: 11311–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Vreede FT, Brownlee GG 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol 80: 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearns R, Peeples ME, Collins PL 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236: 188–201 [DOI] [PubMed] [Google Scholar]
- Fearns R, Collins PL, Peeples ME 2000. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J Virol 74: 6006–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearns R, Peeples ME, Collins PL 2002. Mapping the transcription and replication promoters of respiratory syncytial virus. J Virol 76: 1663–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C, Agudo R, Domingo E, Verdaguer N 2009. Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase. Curr Opin Struct Biol 19: 752–758 [DOI] [PubMed] [Google Scholar]
- Garcin D, Kolakofsky D 1992. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol 66: 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol 69: 5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M 2006. Structure of the vesicular stomatitis virus nucleoprotein–RNA complex. Science 313: 357–360 [DOI] [PubMed] [Google Scholar]
- Grosfeld H, Hill MG, Collins PL 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol 69: 5677–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay O, Curran J, Kolakofsky D 2001. Sendai virus genome synthesis and assembly are coupled: A possible mechanism to promote viral RNA polymerase processivity. J Gen Virol 82: 2895–2903 [DOI] [PubMed] [Google Scholar]
- Hanley LL, McGivern DR, Teng MN, Djang R, Collins PL, Fearns R 2010. Roles of the respiratory syncytial virus trailer region: Effects of mutations on genome production and stress granule formation. Virology 406: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D, Blumenthal T 1983. Does Qβ replicase synthesize RNA in the absence of template? Nature 301: 350–352 [DOI] [PubMed] [Google Scholar]
- Kao CC, Sun JH 1996. Initiation of minus-strand RNA synthesis by the brome mosaicvirus RNA-dependent RNA polymerase: Use of oligoribonucleotide primers. J Virol 70: 6826–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Del Vecchio AM, Zhong W 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253: 1–7 [DOI] [PubMed] [Google Scholar]
- Kao CC, Singh P, Ecker DJ 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287: 251–260 [DOI] [PubMed] [Google Scholar]
- Kawakami K, Ishihama A, Ohtsuka E, Tanaka T, Takashima H, Ikehara M 1981. RNA polymerase of influenza virus. II. Influence of oligonucleotide chain length on the priming activity of RNA synthesis by virion-associated RNA polymerase. J Biochem 89: 1759–1768 [DOI] [PubMed] [Google Scholar]
- Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC 2008. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16: 1678–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott AC, Smith JM, Easton AJ 2001. Fidelity of leader and trailer sequence usage by the respiratory syncytial virus and avian pneumovirus replication complexes. J Virol 75: 6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern DR, Collins PL, Fearns R 2005. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol 79: 2449–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink MA, Stec DS, Collins PL 1991. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology 185: 615–624 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Carpenter CD, Simon AE 1997. A novel 3′-end repair mechanism in an RNA virus. Proc Natl Acad Sci 94: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R 2003. De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J Virol 77: 8831–8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton SL, Cowton VM, Zack CR, McGivern DR, Fearns R 2010. Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc Natl Acad Sci 107: 10226–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393: 280–284 [DOI] [PubMed] [Google Scholar]
- Peeples ME, Collins PL 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol 74: 146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanungo KR, Shaji D, Mathur M, Banerjee AK 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci 101: 5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Farsetta DL, Nibert ML, Harrison SC 2002. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111: 733–745 [DOI] [PubMed] [Google Scholar]
- Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, et al. 2009. Crystal structure of a nucleocapsid-like nucleoprotein–RNA complex of respiratory syncytial virus. Science 326: 1279–1283 [DOI] [PubMed] [Google Scholar]
- Tomar S, Hardy RW, Smith JL, Kuhn RJ 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80: 9962–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarov VI, Demidenko AA, Chetverin AB 2003. Qβ replicase discriminates between legitimate and illegitimate templates by having different mechanisms of initiation. J Biol Chem 278: 44139–44146 [DOI] [PubMed] [Google Scholar]
- Vidal S, Kolakofsky D 1989. Modified model for the switch from Sendai virus transcription to replication. J Virol 63: 1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM 2004. tRNA maturation: RNA polymerization without a nucleic acid template. Curr Biol 14: R883–R885 [DOI] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283: 61–119 [DOI] [PubMed] [Google Scholar]
- Wyatt LS, Moss B, Rozenblatt S 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210: 202–205 [DOI] [PubMed] [Google Scholar]
- Yu Q, Hardy RW, Wertz GW 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol 69: 2412–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]