Eukaryotic RNases P and MRP function as RNPs and are closely related but have different substrate specificities. RNase P participates in the processing of pre-tRNAs while RNase MRP is involved in pre-rRNA processing. Here, it is shown that two protein constituents of both RNase P and RNase MRP form a heterodimer that binds directly to the presumed catalytic site of RNase MRP. Intriguingly, this site corresponds to the binding site of bacterial protein that binds to prokaryotic RNase P RNA.
Keywords: ribonuclease MRP, ribonuclease P, Saccharomyces cerevisiae, Pop5, Rpp1, RNA–protein interactions
Abstract
Ribonuclease (RNase) MRP is a multicomponent ribonucleoprotein complex closely related to RNase P. RNase MRP and eukaryotic RNase P share most of their protein components, as well as multiple features of their catalytic RNA moieties, but have distinct substrate specificities. While RNase P is practically universally found in all three domains of life, RNase MRP is essential in eukaryotes. The structural organizations of eukaryotic RNase P and RNase MRP are poorly understood. Here, we show that Pop5 and Rpp1, protein components found in both RNase P and RNase MRP, form a heterodimer that binds directly to the conserved area of the putative catalytic domain of RNase MRP RNA. The Pop5/Rpp1 binding site corresponds to the protein binding site in bacterial RNase P RNA. Structural and evolutionary roles of the Pop5/Rpp1 heterodimer in RNases P and MRP are discussed.
INTRODUCTION
The ribonucleoprotein complex RNase MRP (Chang and Clayton 1987a,b; Karwan et al. 1991) is a site-specific endoribonuclease found in practically all eukaryotes (Piccinelli et al. 2005; Rosenblad et al. 2006). RNase MRP is essential for the viability of eukaryotes (Schmitt and Clayton 1992; Schneider et al. 2010). Though it was originally identified as a mitochondrial enzyme, the vast majority of RNase MRP is found in the nucleolus and, transiently, in the cytoplasm (Chang and Clayton 1987a; Karwan et al. 1991; Gill et al. 2006, and references therein). Mitochondrial RNase MRP has a distinct protein composition which is yet to be fully characterized (Lu et al. 2010) and will not be discussed in this study.
Outside of the mitochondria, RNase MRP is shown to be involved in the maturation of 5.8S rRNA (Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996) and apparently plays additional, not yet characterized role(s) in early steps of rRNA maturation (Lindahl et al. 2009; Schneider et al. 2010). Yeast RNase MRP is also involved in the regulation of the cell cycle by cleaving the 5′ UTR of cyclin B2 mRNA (Cai et al. 1999, 2002; Gill et al. 2006). Defects in RNase MRP function result in a variety of recessive pleiotropic diseases typically characterized by dwarfism (Ridanpaa et al. 2001; Thiel et al. 2005; Martin and Li 2007; Glazov et al. 2011); the mechanisms responsible for the development of these diseases are not known.
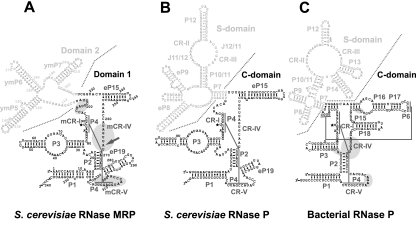
RNase MRP is closely related to RNase P, a universal RNA-based enzyme primarily responsible for the maturation of the 5′-end of tRNA (Altman 2010). Yeast RNase MRP shares eight of its 10 protein components with RNase P (Chamberlain et al. 1998; Walker et al. 2010). The RNA component of RNase MRP (Lopez et al. 2009) also shows clear similarities to the catalytic RNA component of RNase P; particularly, the secondary structure of the putative catalytic domain of RNase MRP closely resembles that of the catalytic domain of RNase P (Fig. 1A,B; for a recent review, see Esakova and Krasilnikov 2010). Footprinting studies of RNase MRP and RNase P holoenzymes suggest that the catalytic domain of RNase P and the corresponding domain of RNase MRP have similar structural organizations (Esakova et al. 2008). The differences in the substrate specificities of RNases MRP and P (Esakova et al. 2011) are likely to be due to the divergence in parts of their RNA components (Fig. 1A,B) and the presence of unique proteins.
FIGURE 1.
RNA components of yeast RNase MRP (A), yeast RNase P (B), and bacterial RNase P (Thermotoga maritima) (C). Catalytic (C-) domains of RNase P and Domain 1 of RNase MRP are shown in black; specificity (S-) domains of RNase P and Domain 2 of RNase MRP are shown in gray. (A) Areas of RNase MRP RNA that are protected in the presence of the Pop5/Rpp1 heterodimer are highlighted in gray; the location of the Pop5–RNA UV cross-link in the RNase MRP holoenzyme is indicated by an arrow. (C) Areas of bacterial RNase P RNA that are within 5 Å of the protein component (Reiter et al. 2010) are highlighted in gray. The diagrams are based on Esakova and Krasilnikov (2010).
The structural organization of bacterial RNase P, which consists of a phylogenetically conserved catalytic RNA and a small protein (Altman 2010) has been established, and several structures of the protein (Stams et al. 1998; Spitzfaden et al. 2000; Kazantsev et al. 2003), RNA (Krasilnikov et al. 2003, 2004; Kazantsev et al. 2005; Torres-Larios et al. 2005), and holoenzymes (Reiter et al. 2010; Kazantsev et al. 2011) are available. At the same time, the structural organizations of the much more complex eukaryotic RNases MRP and P are not clear (Esakova and Krasilnikov 2010). The only available crystal structure is that of the P3 RNA subdomain (Fig. 1A,B) in a complex with protein components Pop6 and Pop7 (Perederina and Krasilnikov 2010; Perederina et al. 2010a,b). Two- and three-hybrid studies, UV-cross-linking studies, as well as pull-down assays have been performed on yeast and human RNases MRP/P (Pluk et al. 1999; Jiang and Altman 2001; Jiang et al. 2001; Houser-Scott et al. 2002; Welting et al. 2004; Aspinall et al. 2007; Reiner et al. 2011), but how most of the multiple protein components interact with the catalytic RNA moiety and what are the roles of the proteins, remain unclear.
One of the major challenges in studying eukaryotic RNases MRP/P is that most of the proteins from Saccharomyces cerevisiae RNases MRP/P are not soluble or tend to aggregate when individually expressed in Escherichia coli (Aspinall et al. 2007, and references therein), requiring the presence of other components for their proper folding. A coexpression approach to studying “difficult” eukaryotic RNase MRP/P proteins has been previously applied to the structural characterization of protein components Pop6 and Pop7 (Perederina et al. 2007, 2010a,b).
Our results from screening for suitable combinations of yeast RNase MRP/P proteins have demonstrated that Pop5 (Chamberlain et al. 1998) and Rpp1 (Stolc and Altman 1997), which are found in both RNase MRP and RNase P (Chamberlain et al. 1998), form a stable soluble complex, whereas, individually, these proteins are not soluble. The formation of a complex between Pop5 and Rpp1 is consistent with the available results of two-hybrid studies and pull-down experiments performed for yeast RNases MRP/P (Houser-Scott et al. 2002; Aspinall et al. 2007) as well as with the results obtained for archaeal homologs of Pop5 and Rpp1 (Tsai et al. 2006; Pulukkunat and Gopalan 2008; Honda et al. 2010).
To better understand the structural organization of RNase MRP, we studied interactions of phylogenetically conserved RNase MRP/P proteins Pop5/Rpp1 (Stolc and Altman 1997; Chamberlain et al. 1998) with the RNA component of RNase MRP using holoenzymes purified from yeast as well as partially reconstituted complexes.
Results
Pop5 and Rpp1 form a heterodimer that interacts with RNase MRP RNA
Yeast Pop5 and Rpp1 were not soluble when individually expressed in E. coli (data not shown). However, coexpression of Pop5 and Rpp1 resulted in a soluble complex that could easily be purified and analyzed.
The complex of Pop5 and Rpp1 was purified using a His6 tag attached to the carboxyl terminus of Rpp1 (see Materials and Methods). The complex was stable at NaCl concentrations of up to 1 M and could be concentrated to 6 mg/mL in 100 mM NaCl/ 50 mM KCl without aggregation (data not shown).
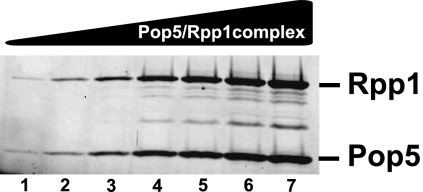
Dynamic light scattering (DLS) experiments indicated that the complex was homogeneous with a particle size of 3.19 nm, suggesting that the complex was a heterodimer; the mobility of the complex in a gel-filtration column was also consistent with it being a heterodimer (data not shown). To verify the Pop5 to Rpp1 ratio, the complex was analyzed on SYPRO-stained SDS–polyacrylamide gels (Fig. 2). The quantification of the Pop5 and Rpp1 protein bands showed that the proteins were present in a 1.14 ± 0.10 to 1 Pop5:Rpp1 molar ratio. Thus, it was concluded that Pop5 and Rpp1 do, indeed, form a heterodimer.
FIGURE 2.
Varying amounts (75–1600 ng) of the Pop5/Rpp1 complex analyzed on a SYPRO Orange-stained 15% SDS–polyacrylamide gel. Quantification of the intensities of the protein bands shows a 1:1 Pop5:Rpp1 molar ratio. Combined with the Dynamic Light Scattering data (particle size 3.19 nm), this indicates that Pop5 and Rpp1 form a heterodimer.
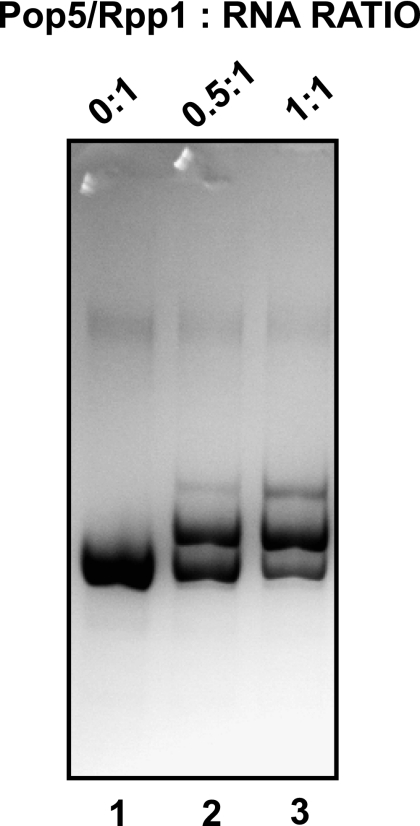
Incubation of the Pop5/Rpp1 heterodimer with in vitro-transcribed RNase MRP RNA resulted in the formation of an RNA–protein complex (Fig. 3), where one Pop5/Rpp1 heterodimer bound one RNA molecule (the complex was formed at concentrations of RNA and proteins that were more than an order of magnitude higher than the dissociation constant [below], allowing us to estimate the stoichiometry of the RNA–protein complex). We did not observe the binding of the Pop5/Rpp1 heterodimer to control yeast tRNA or to 140-nt-long (mostly single-stranded) RNA containing a fragment of the internal transcribed spacer 1 (ITS1), a known substrate for RNase MRP (Esakova et al. 2011, and references therein; data not shown). Our attempts to perform similar experiments using yeast RNase P RNA did not yield interpretable results, apparently due to misfolding of RNA; the same problem was encountered previously (Perederina et al. 2007).
FIGURE 3.
Binding of the Pop5/Rpp1 heterodimer to RNase MRP RNA. A total of 2 μg (final concentration 1.8 μM) of the full-length RNase MRP RNA was incubated with varying amounts of the Pop5/Rpp1 heterodimer and analyzed on a 5% native polyacrylamide gel stained with Toluidine Blue. (Lane 1) no Pop5/Rpp1; (lane 2) 0.9 μM Pop5/Rpp1; (lane 3) 1.8 μM Pop5/Rpp1. The concentrations of the components of the RNA–protein complex were at least an order of magnitude higher than the dissociation constant for the complex (∼60 nM).
An estimation of the dissociation constant of the Pop5/Rpp1–RNase MRP RNA complex, performed in the presence of a 100-fold excess of competitor yeast tRNA using filter-binding assays (Materials and Methods), yielded Kd = 60 ± 20 nM. This value is comparable to the one obtained for another heterodimer known to bind RNase MRP RNA, Pop6/Pop7 (Kd = 110 ± 40 nM) (Perederina et al. 2007). It was suggested that the presence of proteins already bound to RNA, or a simultaneous cooperative binding of several protein subcomplexes, may improve Pop6/Pop7 binding during RNase MRP assembly (Perederina et al. 2007); it is likely that this is the case for the Pop5/Rpp1 heterodimer as well.
Footprinting studies of interactions between the Pop5/Rpp1 heterodimer and RNase MRP RNA
To determine the localization of the Pop5/Rpp1 binding site on RNase MRP RNA, we performed RNA footprinting analysis using RNase V1 (cleaving accessible double-stranded or stacked RNA) and RNase A (cleaving RNA at accessible single-stranded pyrimidines) as probes; to cover the entire length of RNase MRP RNA, both 5′-end and 3′-end-labeled RNA was used (Materials and Methods).
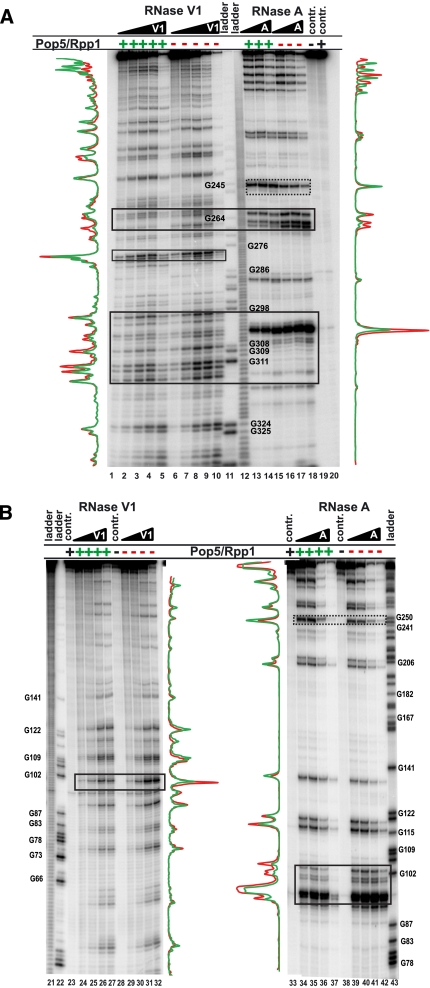
The results of the footprinting analysis (Fig. 4) showed that the binding of the Pop5/Rpp1 heterodimer affected several regions of RNase MRP RNA (Fig. 1A). All of the affected regions belonged to the putative catalytic domain of RNase MRP RNA. While the protected regions are spread over the RNA component, an analysis taking into account the available crystal structures of the corresponding phylogenetically conserved parts of bacterial RNase P (Kazantsev et al. 2005; Torres-Larios et al. 2005; Reiter et al. 2010) suggests that these regions are confined to a relatively small three-dimensional area (see Discussion).
FIGURE 4.
Footprinting analysis of the Pop5/Rpp1–RNase MRP RNA complex. Pop5/Rpp1 was mixed with an equimolar amount of refolded in vitro-transcribed RNA. (A) 3′-end 32P-labeled RNase MRP RNA; (B) 5′-end 32P-labeled RNase MRP RNA. (Lanes 1–5,24–27) increasing concentrations of RNase V1 in the presence of the Pop5/Rpp1 heterodimer; (lanes 6–10,29–32) increasing concentrations of RNase V1 without Pop5/Rpp1; (lanes 11,22,43) RNase T1 digestion of RNase MRP RNA (sequence ladders); (lanes 12,21) alkaline hydrolysis of RNase MRP RNA (ladders); (lanes 13–15,34–37) increasing concentrations of RNase A in the presence of the Pop5/Rpp1 heterodimer; (lanes 16–18,39–42) increasing concentrations of RNase A without Pop5/Rpp1; (lanes 19,28,38) undigested RNase MRP RNA (controls); (lanes 20,23,33) undigested complex of RNase MRP with Pop5/Rpp1 (controls). Regions protected in the presence of Pop5/Rpp1 are traced with solid lines; the region displaying hypersensitivity in the presence of proteins is traced with a dotted line. RNase MRP RNA nucleotide numbering (shown next to sequence ladders) matches that in Figure 1A. Graphs next to the gels represent bands’ intensities in the presence of Pop5/Rpp1 (shown in green; lanes 3,13,27,35 were quantified) and for RNA alone (shown in red; lanes 8,16,32,40 were quantified).
One region of RNase MRP RNA, the terminal loop of the eP15 stem (Fig. 1A), became more sensitive to RNase A cleavage in the presence of the Pop5/Rpp1 heterodimer (Fig. 4A, around G245). It was previously observed (Perederina et al. 2007) that the binding of the Pop6/Pop7 heterodimer to the P3 subdomain of RNase MRP had a similar effect on the sensitivity of the closest terminal loop (the P3 terminal loop). The Pop6/Pop7 heterodimer did not interact with the P3 terminal loop (Perederina et al. 2010b), and the increased sensitivity of this region apparently resulted from a more complete folding of RNA in the presence of protein (misfolding in a fraction of RNA molecules can make the terminal region unsuitable for RNase A cleavage, whereas protein binding can force RNA into the proper conformation, providing additional molecules with the proper single-stranded conformation in the loop region). We suggest that the increased sensitivity of the eP15 loop in the presence of Pop5/Rpp1 is similarly caused by a more complete RNA folding when proteins are present. Consistent with this explanation are the results of footprinting analysis performed on the RNase MRP holoenzyme (Esakova et al. 2008), which indicate that the eP15 loop is not involved in interactions with proteins, as are the results of mutational studies (Shadel et al. 2000; Li et al. 2004), which indicate that the terminal part of the eP15 stem is not essential for the function of RNase MRP.
UV cross-linking of Pop5 and Rpp1 with the RNA component of the RNase MRP holoenzyme
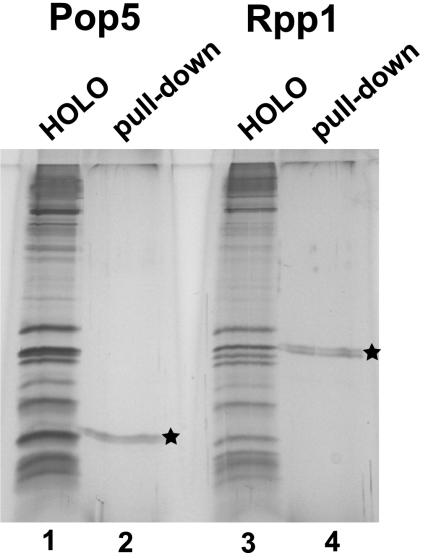
To verify the results obtained for the reconstituted complex of RNase MRP RNA with the Pop5/Rpp1 heterodimer (above), we performed UV cross-linking studies using RNase MRP holoenzymes purified from S. cerevisiae. We generated two yeast strains, each containing a TAP tag (Rigaut et al. 1999) attached to the carboxyl terminus of RNase MRP/P protein component Pop4 (Salinas et al. 2005; Esakova et al. 2008) plus a His6 tag attached to the carboxyl terminus of either Pop5 or Rpp1. Active RNase MRP was purified from both yeast strains using established protocols (Salinas et al. 2005; Esakova et al. 2008); the TAP tag was used as the purification handle. Following purification, RNase MRP was subjected to UV irradiation. Irradiated RNase MRP was transferred to a denaturing buffer containing high concentrations of SDS, then His6-tagged proteins (Pop5 or Rpp1) were pulled down using Ni2+ affinity resin and washed using a denaturing buffer. This procedure resulted in the removal of all protein components except for Pop5 or Rpp1 (depending on the strain used) (Fig. 5) and allowed for the isolation of the RNA molecules that were specifically cross-linked to these proteins. Cross-linked RNA was extracted by Proteinase K digestion, followed by phenol extraction, and analyzed for the location of cross-links using primer extension with a reverse transcriptase. For more details see the Materials and Methods section.
FIGURE 5.
Isolation of His6-tagged proteins from purified RNase MRP/P holoenzymes. (Lane 1) Proteins in the RNase MRP/P holoenzyme mix purified from yeast strain EK-Pop5 (TAP-tagged Pop4 and His6-tagged Pop5); (lane 2) Pop5 isolated from holoenzymes under denaturing conditions; a star indicates the Pop5 protein band; (lane 3) proteins in the RNase MRP/P holoenzyme mix purified from yeast strain EK-Rpp1 (TAP-tagged Pop4 and His6-tagged Rpp1); (lane 4) Rpp1 isolated from holoenzymes under denaturing conditions; a star indicates the Rpp1 protein band. Silver-stained 15% SDS–polyacrylamide gel.
The results of UV cross-linking studies show that, consistent with the results of footprinting studies, Pop5 binds to the conserved area mCR-IV of Domain 1 (the putative catalytic domain) (Figs. 1A, 6). No other cross-linking sites were detected for Pop5; UV cross-linking of Rpp1 did not reveal any cross-linking sites on RNase MRP RNA (data not shown). It should be noted that while the presence of a cross-link allows for reliable identification of an RNA–protein interaction, the absence of a cross-link does not by itself necessarily mean that there is no close RNA–protein contact. Thus, the absence of observed Rpp1–RNA cross-links does not automatically mean that the two do not interact.
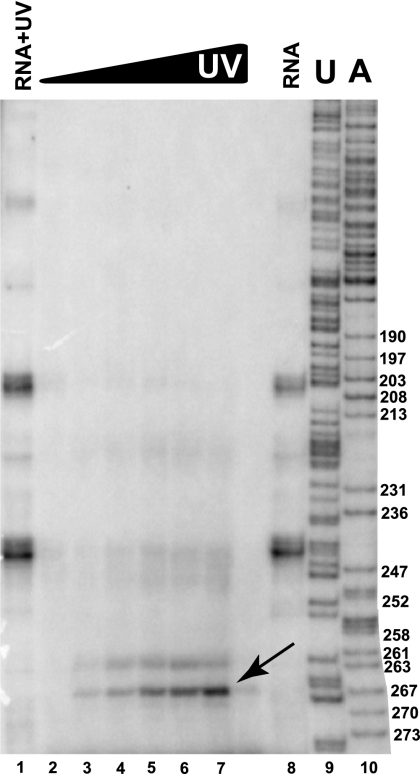
FIGURE 6.
UV cross-linking analysis of interactions of Pop5 with RNA in the RNase MRP holoenzyme purified from yeast. RNase MRP holoenzyme was purified from yeast, subjected to UV cross-linking, and disassembled under denaturing conditions; then, His6-tagged Pop5 with cross-linked RNA was isolated, and RNA cross-linked to Pop5 was extracted and finally subjected to primer extension analysis. (Lane 1) RNase MRP RNA extracted from purified holoenzyme and subjected to UV irradiation at 1.28 J/cm2 (control for RNA–RNA cross-links); (lanes 2–7) UV-irradiated RNase MRP holoenzyme (0, 0.04, 0.08, 0.16, 0.32, and 0.64 J/cm2, respectively); (lane 8) RNase MRP RNA extracted from the purified holoenzyme (control, no irradiation); (lanes 9,10) sequence ladder. 5% denaturing (8 M urea) polyacrylamide gel.
DISCUSSION
We have demonstrated that Pop5 and Rpp1, protein components of yeast RNases MRP/P, form a heterodimer that binds to the putative catalytic domain (Domain 1) of RNase MRP (Fig. 1A).
Archaeal homologs of Pop5 and Rpp1 were shown to act as a pair, binding to the catalytic domain of archaeal RNase P, a phylogenetically conserved counterpart of RNase MRP Domain 1 (Tsai et al. 2006; Pulukkunat and Gopalan 2008; Chen et al. 2010; Honda et al. 2010; Jarrous and Gopalan 2010), matching the behavior of eukaryotic Pop5, Rpp1, and suggesting a degree of similarity between archaeal RNase P and yeast RNase MRP (and, by inference, yeast RNase P as well). Indeed, footprinting analysis of interactions of archaeal RNase P RNA with archaeal homologs of Pop5 and Rpp1 (Xu et al. 2009) yielded results similar to our results obtained for yeast RNase MRP.
In the course of evolution, RNase P, an apparent ancestor of RNase MRP (Piccinelli et al. 2005; Rosenblad et al. 2006; Zhu et al. 2006; Woodhams et al. 2007), underwent a transformation from an RNA-based enzyme with a single small protein component (∼10% of the holoenzyme mass in bacterial RNase P) to an RNA-based enzyme with much more protein than RNA (about three-quarters of the holoenzyme mass in yeast RNase P) (Esakova and Krasilnikov 2010). While this transformation was accompanied by changes in the catalytic RNA component, several key regions of RNA remained well conserved (Conserved Regions [CR] I through V) (Fig. 1B,C; Chen and Pace 1997; Brown 1999; Li et al. 2011).
The recent crystal structure of bacterial RNase P holoenzyme (Reiter et al. 2010) has revealed that its single protein component interacts mostly with RNA regions that are conserved in RNases P throughout the three domains of life (Fig. 1C). Surprisingly, neither archaeal RNase P nor eukaryotic RNases P/MRP have a protein component that would show any homology with the bacterial RNase P protein (for a recent review, see Esakova and Krasilnikov 2010), and it is not clear how unrelated archaeal/eukaryotic protein(s) replace the bacterial protein when the binding site seems to remain similar. The results presented in this work position the Pop5/Rpp1 heterodimer in the area of RNase MRP RNA that corresponds to the protein binding site in bacterial RNase P (Fig. 1A,C), suggesting that in RNase MRP it is the Pop5/Rpp1 heterodimer that serves to substitute (in terms of its interactions with the RNA component, at least) for the bacterial protein.
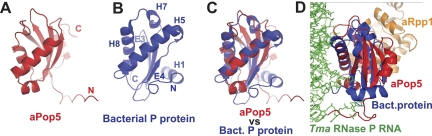
Crystal structures of archaeal homologs of Pop5 and Rpp1 have been determined (Takagi et al. 2004; Kawano et al. 2006; Wilson et al. 2006). Neither protein is related to the bacterial RNase P protein, and their structural organizations are distinct. However, Pop5 adopts an α/β sandwich fold (Fig. 7A), where some of the distinctly connected secondary structure elements are positioned in a way that is similar to that in the bacterial RNase P protein (Fig. 7A–C; for review, see Esakova and Krasilnikov 2010). An analysis of the recent crystal structure of the bacterial RNase P holoenzyme (Reiter et al. 2010) shows that the area of similarity between the bacterial protein and archaeal Pop5 is the one that forms the RNA-binding interface in bacterial RNase P (Fig. 7D).
FIGURE 7.
Structural elements in the archaeal homolog of Pop5 (A) and in the protein component of bacterial RNase P (B) have distinct connectivities, but adopt similar overall mutual orientations (C). (A) Crystal structure of an archaeal homolog of Pop5 (PDB ID 2CZV) shown in red; N and C termini are indicated. (B) Crystal structure of T. maritima RNase P protein (PDB ID 3OKB) shown in blue; N and C termini are indicated. Secondary structure elements (H: α-helices, E: β-strands) are marked according to the designation in Figure 8 and Supplemental Figure S1. (C) Archaeal homolog of Pop5 (PDB ID 2CZV, shown in red) superposed on the protein component of bacterial RNase P (PDB ID 3OKB, shown in blue). (D) An archaeal aPop5/aRpp1 heterodimer (PDB ID 2CZV; aPop5 is in red, aRpp1 is in gold) is shown superposed on the crystal structure of T. maritima RNase P (PDB ID 3OKB; the protein component is in blue, the RNA component is in green).
A structure-based alignment of eukaryotic Pop5 sequences and their archaeal homologs strongly suggests that the area resembling bacterial RNase P protein is structurally conserved from archaea to eukaryotes (Supplemental Fig. S1). This conservation, combined with the observed similarity between the protein binding site in bacterial RNase P RNA and the Pop5/Rpp1 heterodimer binding site in yeast RNase MRP RNA (Fig. 1A,C), makes it tempting to suggest that yeast Pop5 interacts with RNase MRP RNA in a manner resembling that of the protein component in bacterial RNase P (Fig. 7D); a similar suggestion has been put forward for archaeal RNase P (Chen et al. 2010).
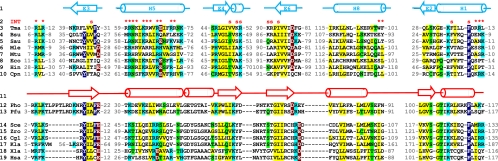
If eukaryotic Pop5 were to interact with the generally conserved area of RNA using the suggested interface (Fig. 7D), one would expect to observe a phylogenetic conservation of key amino acids involved in this interaction; moreover, since the RNA interface involved in this interaction is well conserved from bacteria to eukaryotes (Chen and Pace 1997; Lopez et al. 2009; Li et al. 2011), one should expect a degree of similarity between phylogenetically conserved amino acids involved in interactions with RNA in bacterial RNase P and amino acids occupying equivalent spatial positions in eukaryotic (as well as in archaeal) Pop5. Indeed, a phylogenetic comparison of the sequences of the protein's regions involved in interactions with RNA in bacteria with the corresponding regions of archaeal/eukaryotic Pop5 shows similarities between the two (Fig. 8; Supplemental Fig. S1C), consistent with the suggested mode of Pop5–RNA interaction (Fig. 7D).
FIGURE 8.
Spatially superposable elements in Pop5 and bacterial RNase P protein are connected differently, but show resembling patterns of phylogenetic conservation in areas corresponding to the protein–RNA interface in bacterial RNase P. Sequences of major secondary structure elements (line 1) in bacterial RNase P proteins (lines 3–10) shown against corresponding (in space) elements (line 11) in archaeal (lines 12,13) and eukaryotic (lines 14–19) Pop5 proteins. The bacterial and archaeal/eukaryotic structure elements are superposed as shown in Figure 7C. The complete alignment is shown in Supplemental Figure S1C. (Line 1) Secondary structure elements in bacterial RNase P protein (PDB ID 3OKB) marked according to Figure 7B and Supplemental Figure S1, A and B. α-helices are shown by cylinders; β-strands are shown by arrows. (Line 2) Amino acids of the bacterial RNase P protein that are involved in interactions with the RNA component of T. maritima RNase P (Reiter et al. 2010) are indicated by red asterisks; amino acids involved in interactions with bacterial RNase P substrate (Reiter et al. 2010) are indicated by red letters (s). (Line 3) T. maritima; (line 4) Bacillus subtilis; (line 5) Staphylococcus aureus; (line 6) Mycobacterium leprae; (line 7) Mycobacterium tuberculosis; (line 8) E. coli; (line 9) Haemophilus influenzae; (line 10) Chlamydophila pneumoniae. (Line 11) Secondary structure elements in the archaeal homolog of Pop5 (PDB ID 2CZV); eukaryotic Pop5 is expected to be similar. (Line 12) Pyrococcus horikoshii OT3; (line 13) Pyrococcus furiosus; (line 14) S. cerevisiae; (line 15) Zygosaccharomyces rouxii; (line 16) Candida glabrata; (line 17) Kluyveromyces lactis; (line 18) Xenopus laevis; (line 19) human. Residues showing similar patterns of conservation in bacterial RNase P protein versus archaeal/eukaryotic Pop5 are highlighted as follows: nonpolar aliphatic (GAPVLIM, yellow), aromatic (FYW, dark blue), polar uncharged (STCNQ, green), positively charged (KHR, light blue), negatively charged (DE, brown).
Eukaryotic Pop5 forms a heterodimer with Rpp1 (above); the archaeal homolog of Pop5 clearly acts in a pair with the archaeal homolog of Rpp1 (Kawano et al. 2006; Tsai et al. 2006; Pulukkunat and Gopalan 2008; Honda et al. 2010), although the stoichiometry of the archaeal pair is not clear and the archaeal complex crystallizes as a heterotetramer (Kawano et al. 2006). This heterotetramer can be broken down into two possible dimers; based on the similarity between eukaryotic Pop5, Rpp1, and their archaeal counterparts, we assume that one of the dimers observed in the crystal structure is relevant to the observed eukaryotic Pop5/Rpp1 heterodimer. The dimer that does not result in steric clashes is presumed to be relevant and is used in Figure 7D. The absence of Rpp1–RNA contacts in this model is consistent with the observed absence of cross-links between Rpp1 and RNA in RNase MRP holoenzyme (above).
The analysis of the results of footprinting and cross-linking studies supports the idea that in RNase MRP (and, by inference, in eukaryotic RNase P as well) the protein component Pop5, acting in a heterodimeric complex with Rpp1, performs a role similar to that of the single protein component in bacterial RNase P.
MATERIALS AND METHODS
Purification of RNase MRP holoenzyme
RNase MRP (as a ∼1:1 mix with RNase P) was purified from S. cerevisiae strains EK-Pop5 and EK-Rpp1 using tandem affinity purification (Rigaut et al. 1999) and following a protocol described in Salinas et al. (2005) and Esakova et al. (2008). Strains EK-Pop5 and EK-Rpp1 were based on yeast strain YSW1 (Salinas et al. 2005), a generous gift from Mark Schmitt. His6 tags were added to the carboxyl termini of Pop5 and Rpp1, respectively, using standard PCR-based methods; kanMX4 was used as the selectable marker (Brachmann et al. 1998). The Pop5 and Rpp1 genes in EK-Pop5 and EK-Rpp1 were PCR amplified and sequenced to verify the proper insertion of the tags.
UV cross-linking analysis of interactions of Pop5 and Rpp1 with the RNA component of RNase MRP holoenzyme
RNase MRP samples (15-μL drops of a 0.5 μM solution in 20 mM Tris-HCl at pH 7.8, 150 mM NaCl, 2.5 mM MgCl2, 0.05 mM EDTA, 5% [v/v] glycerol) were irradiated with UV light (λ = 254 nm, the dosage varied from 0.04 to 0.64 J/cm2) in a UV cross-linker SpectroLinker XL-1500 (Spectronics) on ice. After irradiation, 20 μL of a denaturing buffer (100 mM Tris-HCl at pH 7.8, 4% SDS, 20% [v:v] glycerol) and DTT to the final concentration of 30 mM were added, and the samples were incubated for 10 min at 65°C. Following the first round of incubation, an additional 30 mM of DTT was added, and incubation was continued for another 10 min; then, the DTT addition/incubation cycle was repeated three more times. Following initial denaturation, 1 mL of a binding buffer (15 mM Tris-HCl at pH 7.8, 8 M urea, 0.5 M NaCl, 10 mM Na-imidazole at pH 7.4, 1% [v:v] Tween 20) was added, and the samples were incubated with 50 μL of Ni-IDA resin (Macherey-Nagel) for 2 h at room temperature, with intensive agitation. After incubation, supernatant was carefully removed, and the resin was washed with 1 mL of the binding buffer (above) five times. Bound proteins were eluted with 100 μL of an elution buffer (15 mM Tris-HCl at pH 7.8, 4 M urea, 0.25 M NaCl, 300 mM Na-imidazole at pH 7.4, 0.5% [v:v] Tween 20). The quality of protein isolation was monitored using silver-stained SDS–polyacrylamide gels.
Samples containing His6-tagged proteins with covalently attached cross-linked RNA were treated with 300 μg of Proteinase K for 20 min at 37°C; then, RNA was extracted with phenol, precipitated with ethanol in the presence of acrylic carrier (MRC), dissolved in a buffer containing 5 mM Tris-HCl (pH 8.0), 10 mM KCl, and 0.1 mM EDTA, and stored at −70°C.
RNA samples were analyzed using primer extension with a reverse transcriptase following a previously described protocol (Esakova et al. 2008) and using previously described primers RTP1AL, RTP1B, RTP15A, RTP2A, RTP21, RTP25, RTP3B, and RTP4 (Esakova et al. 2008) that were specific to RNase MRP RNA. This set of primers allowed for reliable coverage of the entire length of RNase MRP RNA except for the last 25 nucleotides at the 3′-end of RNA. UV-irradiated (1.28 J/cm2) and not irradiated RNase MRP RNA that was phenol-extracted from purified RNase MRP holoenzyme preparations was used as controls. DNA sequencing reactions obtained using RNase MRP RNA plasmid p31/51 (Perederina et al. 2007) and corresponding primers were used as ladders. 32P-labeled primer extension products were resolved on 5% denaturing (8 M urea) polyacrylamide gels and visualized using a PhosphorImager (Molecular Dynamics).
Purification of the Pop5/Rpp1 heterodimer
The Pop5/Rpp1 heterodimer was expressed in E. coli from a polycistronic plasmid 5Rpp1C. 5Rpp1C plasmid was constructed by the insertion of the S. cerevisiae Pop5 gene, followed downstream by the Rpp1 gene with a fused C-terminal His6 tag, into pET21b plasmid (Novagen); the sequence of the insert was verified. The protein complex was expressed in BL21 (DE3, cotransformed with pRARE plasmid [Novagen]) strain of E. coli at 30°C by overnight autoinduction.
For the purification of the RNase-free Pop5/Rpp1 complex, cells were harvested by centrifugation at 4000g at 4°C and resuspended in Buffer A containing 50 mM Tris-HCl (pH 8.3), 250 mM NaCl, 50 mM KCl, 2 mM MgCl2, 0.1 mM PMSF, 5 mM β−mercaptoethanol, and 10 mM imidazole (pH 9.0). The cells were disrupted by sonication. The lysate was cleared by centrifugation for 30 min at 17,000g (4°C) and incubated with 2 mL of Ni-NTA agarose (Qiagen) for 40 min at 4°C with light agitation. The Ni-NTA resin was washed with 20 vol of Buffer A, then with 20 vol of Buffer A supplemented with 30 mM imidazole (pH 9.0). The complex was eluted with a buffer containing 50 mM MES-NaOH (pH 6.5), 50 mM KCl, 250 mM NaCl, 10 mM β−mercaptoethanol, 0.1 mM PMSF, and 400 mM imidazole (pH 6.5). Fractions containing the Pop5/Rpp1 complex were combined, diluted twofold with Buffer B (50 mM MES-NaOH at pH 6.5, 50 mM KCl, 5 mM NaCl, 10 mM β−mercaptoethanol, 0.1 mM PMSF, 0.5 mM EDTA), and loaded onto a Hi-Trap Heparin HP column (Amersham) pre-equilibrated with Buffer B. The complex was eluted with a linear gradient of 0.250–1.5 M NaCl in the starting buffer; fractions of interest were combined, concentrated using an Amicon Ultra-15 concentrator (10,000 MWCO, Millipore), and fractionalized through a Superdex 75 gel-filtration column (Amersham) that was pre-equilibrated with Buffer C (10 mM Tris-HCl at pH 8.0, 200 mM NaCl, 50 mM KCl, 10 mM DTT, 0.1 mM PMSF). The fractions containing the purified Pop5/Rpp1 complex were pooled and concentrated with an Amicon Ultra-15 concentrator (10,000 MWCO). The concentration of the complex was determined using a NanoDrop ND-1000 spectrophotometer. This procedure yielded ∼4 mg of RNase-free Pop5/Rpp1 complex per liter of culture.
Estimation of Pop5:Rpp1 molar ratio in the Pop5/Rpp1 complex
To estimate the molar ratio of Pop5 to Rpp1 in the Pop5/Rpp1 complex, varying amounts of the complex (75–1600 ng) were separated on a 15% denaturing SDS–polyacrylamide gel and stained with SYPRO Orange dye. The intensities of the bands were quantified using a PhosphorImager (Molecular Dynamics). The relative intensities of the bands corresponding to Pop5 and Rpp1 were normalized by the molecular weights of the proteins.
Production of the RNA component of RNase MRP by in vitro transcription
RNA was produced by run-off in vitro transcription with T7 RNA polymerase following a standard protocol as previously described (Perederina et al. 2007). For gel mobility-shift experiments and footprinting assays involving 5′-end 32P-labeled RNA, plasmid p31/51 (Perederina et al. 2007) was used as a template; for footprinting assays involving 3′-end 32P-labeled RNA, plasmid pHST7*NME1* (a generous gift from Johanna Avis) containing RNase MRP RNA sequence flanked by 5′-hammerhead and 3′-hepatitis delta virus ribozymes (Walker and Avis 2004) was used to avoid the heterogeneity of the 3′-end of the transcription product. In vitro-transcribed RNase MRP RNA was purified using 6% denaturing polyacrylamide gels.
Gel mobility-shift assays
RNase MRP RNA was incubated at 75°C for 10 min in 20 mM Tris-HCl (pH 7.5), followed by a 30-min cooling to room temperature in a styrofoam rack. RNA–protein complexes were formed by incubation of 2 μg of RNA (final concentration 1.8 μM) with varying amounts of protein in a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, and 0.5 mM MgCl2 (Buffer D) for 15 min at 28°C, followed by 15 min at room temperature. Under those conditions, the concentrations of the components of the RNA–protein complex were at least an order of magnitude higher than the dissociation constant for the complex (∼60 nM). The samples were loaded on a 5% native polyacrylamide gel and fractionated at 4°C. The gels were stained with Toluidine Blue.
Filter-binding assays
Filter-binding assays were performed according to a previously published protocol (Perederina et al. 2007). RNase MRP RNA was 5′-end labeled with 32P, gel purified, and RNA–protein complexes were formed as described above. The final concentration of labeled RNA was 2 nM; the protein concentrations ranged from 20 to 800 nM. In experiments involving competitor RNA, 100-fold molar excess of unlabeled yeast tRNA was added to the labeled RNase MRP RNA prior to the formation of RNA–protein complexes. The complexes were filtered through a stack of BA83 nitrocellulose (Whatman), followed by Hybond-N+ (Amersham) membranes. The membranes were washed with Buffer D (above) and dried. The intensities of the spots on the nitrocellulose membrane (corresponding to the amount of loaded labeled RNA that formed complexes with proteins) and the intensities of the spots on the Hybond-N+ membrane (corresponding to the amount of loaded labeled RNA that did not form complexes with proteins; the sum of the intensities on the two membranes corresponded to the total amount of labeled RNA loaded) were quantified using a PhosphorImager (Molecular Dynamics); the experiments were repeated five times. To obtain an estimate of the dissociation constant, we used a single-site binding model. The proportion of the RNA that formed complexes with Pop5/Rpp1 was plotted as a function of the Pop5/Rpp1 concentration, and the graph was interpolated to the point corresponding to 50%, which under the conditions of the experiment (Pop5/Rpp1 were always present in high excess) corresponded to the estimated Kd.
Footprinting analysis
For the experiments involving 5′-end-labeled RNase MRP RNA, in vitro-transcribed RNA (above) was dephosphorylated with calf intestinal alkaline phosphatase and labeled using [γ-32P]ATP and T4 polynucleotide kinase. For the experiments involving 3′-end-labeled RNase MRP RNA, in vitro-transcribed RNA (above) was labeled using 32P pCp and RNA ligase 1.
For footprinting analysis, 1 pmol of labeled RNA was mixed with 9 pmol of unlabeled gel-purified RNase MRP RNA and subjected to folding by incubation at 75°C for 10 min in 20 mM Tris-HCl (pH 7.5), followed by 30 min of cooling to room temperature in a styrofoam rack. KCl and MgCl2 were added to the folded RNA to 100 mM and 0.5 mM, respectively. RNA–protein complexes were formed by the incubation of RNA with 10 pmol (1 μM, a 1:1 RNA:protein molar ratio) of Pop5/Rpp1 complex for 15 min at 28°C followed by 15 min at room temperature. Partial digestions with RNases A and V1 were performed for 10 min at 0°C and 15 min at 25°C, respectively, using varying concentrations of the enzymes. Following digestion, RNA was extracted with a phenol/chloroform mix, precipitated with ethanol, and analyzed on 6% denaturing (8 M urea) polyacrylamide gels.
The reference ladders were produced by alkaline hydrolysis in a buffer containing 5 mM glycine (pH 9.5), 2 mM MgSO4 (1 min at 95°C), and by digestion with RNase T1 (2 min at 65°C). The radioactive bands were visualized using a PhosphorImager (Molecular Dynamics) and quantified using ImageQuant TL (GE Healthcare).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Joe Reese and Mark Schmitt for valuable comments and suggestions, Johanna Avis and Mark Schmitt for generous gifts of yeast strains and plasmids, and Lydia Krasilnikova for help with the manuscript preparation. This work was supported by NIH Grant GM085149 to A.S.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2855511.
REFERENCES
- Altman S 2010. History of RNase P and overview of its catalytic activity. In Ribonuclease P, protein reviews 10 (ed. Liu F. and Altman S.), pp. 1–15 Springer, New York [Google Scholar]
- Aspinall TV, Gordon JMB, Bennett HJ, Karahalios P, Bukowski J-P, Walker SC, Engelke DR, Avis JM 2007. Interactions between subunits of Saccharomyces cerevisiae RNase MRP support a conserved eukaryotic RNase P/MRP architecture. Nucleic Acids Res 35: 6439–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davis A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Brown JW 1999. The ribonuclease P database. Nucleic Acids Res 27: 314–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Reilly TR, Cerio M, Schmitt ME 1999. Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol Cell Biol 19: 7857–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME 2002. The Saccharomyces cerevisiae RNase Mitochondrial RNA Processing is critical for cell cycle progression at the end of mitosis. Genetics 161: 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA 1987a. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J 6: 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA 1987b. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 235: 1178–1184 [DOI] [PubMed] [Google Scholar]
- Chen J-L, Pace NR 1997. Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560 [PMC free article] [PubMed] [Google Scholar]
- Chen W-Y, Pulukkunat DK, Cho I-M, Tsai H-Y, Gopalan V 2010. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res 38: 8316–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci 91: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS 2010. Of proteins and RNA: The RNase P/MRP family. RNA 16: 1725–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Quan C, Schmitt ME, Krasilnikov AS 2008. Footprinting analysis demonstrates extensive similarity between eukaryotic RNase P and RNase MRP holoenzymes. RNA 14: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Quan C, Berezin I, Krasilnikov AS 2011. Substrate recognition by ribonucleoprotein ribonuclease MRP. RNA 17: 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Aulds J, Schmitt ME 2006. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J Cell Biol 173: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Zankl A, Donskoi M, Kenna TJ, Thomas GP, Clark GR, Duncan EL, Brown MA 2011. Whole-exome re-sequencing in a family quartet identifies POP1 mutations as the cause of a novel skeletal dysplasia. PLoS Genet 7: e1002027 doi: 10.1371/journal.pgen.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Hara T, Nan J, Zhang X, Kimura M 2010. Archaeal homologs of human RNase P protein pairs Pop5 with Rpp30 and Rpp21 with Rpp29 work on distinct functional domains of the RNA subunit. Biosci Biotechnol Biochem 74: 266–273 [DOI] [PubMed] [Google Scholar]
- Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR 2002. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc Natl Acad Sci 99: 2684–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Gopalan V 2010. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res 38: 7885–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Altman S 2001. Protein–protein interactions with subunits of human nuclear RNase P. Proc Natl Acad Sci 98: 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Guerrier-Takada C, Altman S 2001. Protein–RNA interactions in the subunits of human nuclear RNase P. RNA 7: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwan R, Bennett JL, Clayton DA 1991. Nuclear RNase MRP processes RNA at multiple discrete sites: Interaction with an upstream G box is required for subsequent downstream cleavages. Genes Dev 5: 1264–1276 [DOI] [PubMed] [Google Scholar]
- Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M 2006. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: Implication of dimer formation of the holoenzyme. J Mol Biol 357: 583–591 [DOI] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Carter RJ, Holbrook SR, Adams PD, Pace NR 2003. High-resolution structure of RNase P protein from Thermotoga maritima. Proc Natl Acad Sci 100: 7497–7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci 102: 13392–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Rambo RP, Karimpour S, Santalucia J, Tainer JA, Pace NR 2011. Solution structure of RNase P RNA. RNA 17: 1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764 [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107 [DOI] [PubMed] [Google Scholar]
- Li X, Zaman S, Langdon Y, Zengel JM, Lindahl L 2004. Identification of a functional core in the RNA component of RNase MRP of budding yeast. Nucleic Acids Res 32: 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Gößringer M, Hartmann RK 2011. Archaeal-bacterial chimeric RNase P RNAs: Towards understanding RNA's architecture, function and evolution. ChemBioChem 12: 1536–1543 [DOI] [PubMed] [Google Scholar]
- Lindahl L, Bommankanti A, Li X, Hayden L, Jones A, Khan M, Oni T, Zengel JM 2009. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA 15: 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MD, Rosenblad MA, Samuelsson T 2009. Conserved and variable domains of RNase MRP RNA. RNA Biol 6: 208–220 [DOI] [PubMed] [Google Scholar]
- Lu Q, Wierzbicki S, Krasilnikov AS, Schmitt ME 2010. Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components. RNA 16: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by Ribonuclease MRP in vitro. Science 272: 268–270 [DOI] [PubMed] [Google Scholar]
- Martin AN, Li Y 2007. RNase MRP RNA and human genetic diseases. Cell Res 17: 219–226 [DOI] [PubMed] [Google Scholar]
- Perederina A, Krasilnikov AS 2010. The P3 domain of eukaryotic RNases P/MRP: Making a protein-rich RNA-based enzyme. RNA Biol 7: 534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS 2007. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA 13: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS 2010a. Crystallization and preliminary X-ray diffraction analysis of the P3 RNA domain of yeast ribonuclease MRP in a complex with RNase P/MRP protein components Pop6 and Pop7. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS 2010b. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J 29: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli P, Rosenblad MA, Samuelsson T 2005. Identification and analysis of Ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res 33: 4485–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJM, van Venrooij WJ 1999. RNA–protein interactions in the human RNase MRP ribonucleoprotein complex. RNA 5: 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulukkunat DK, Gopalan V 2008. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res 36: 4172–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Alfiya-Mor N, Berrebi-Demma M, Wesolowski D, Altman S, Jarrous N 2011. RNA binding properties of conserved protein subunits of human RNase P. Nucleic Acids Res. doi: 10.1093/nar/gkr126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpaa M, Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. 2001. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, Cartilage-Hair Hypoplasia. Cell 104: 195–203 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rosenblad MA, Lopez MD, Piccinelli P, Samuelsson T 2006. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res 34: 5145–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas K, Wierzbicki S, Zhou L, Schmitt ME 2005. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem 280: 11352–11360 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1992. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev 6: 1975–1985 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Bains AK, Rajendra TK, Dominski Z, Matera AG, Simmonds AJ 2010. Functional characterization of Drosophila MRP (mitochondrial RNA processing) RNA gene. RNA 16: 2120–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME 2000. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene 245: 175–184 [DOI] [PubMed] [Google Scholar]
- Spitzfaden C, Nicholson N, Jones JJ, Guth S, Lehr R, Prescott CD, Hegg LA, Eggleston DS 2000. The structure of Ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J Mol Biol 295: 105–115 [DOI] [PubMed] [Google Scholar]
- Stams T, Niranjanakumari S, Fierke CA, Christianson DW 1998. Ribonuclease P protein structure: Evolutionary origins in the translational apparatus. Science 280: 752–755 [DOI] [PubMed] [Google Scholar]
- Stolc V, Altman S 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev 11: 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Watanabe M, Kakuta Y, Kamachi R, Numata T, Tanaka I, Kimura M 2004. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem Biophys Res Commun 319: 787–794 [DOI] [PubMed] [Google Scholar]
- Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Rüschendorf F, Sticht H, Spranger J, Müller D, et al. 2005. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet 77: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A 2005. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437: 584–587 [DOI] [PubMed] [Google Scholar]
- Tsai H-Y, Pulukkunat DK, Woznick WK, Gopalan V 2006. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci 103: 16147–16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Avis JM 2004. A conserved element in the yeast RNase MRP RNA subunit can participate in a long-range base-pairing interaction. J Mol Biol 341: 375–388 [DOI] [PubMed] [Google Scholar]
- Walker SC, Marvin MC, Engelke DR 2010. Eukaryote RNase P and RNase MRP. In Ribonuclease P, protein reviews 10 (ed. Liu F. and Altman S.), pp. 173–202 Springer, New York [Google Scholar]
- Welting TJM, van Venrooij WJ, Pruijn GJM 2004. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res 32: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Bohlen CJ, Foster MP, Bell CE 2006. Structure of Pfu Pop5, an archaeal RNase P protein. Proc Natl Acad Sci 103: 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams MD, Stadler PF, Penny D, Collins LJ 2007. RNase MRP and the RNA processing cascade in the eukaryotic ancestor. BMC Evol Biol 7: S13 doi: 10.1186/1471-2148-7-S1-S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP 2009. Solution structure of an archaeal RNase P binary protein complex: Formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J Mol Biol 393: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Stribinskis V, Ramos KS, Li Y 2006. Sequence analysis of RNase MRP RNA reveals its origination from eukaryotic RNase P RNA. RNA 12: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]