A review of the current knowledge of the role of viral miRNAs in viral replication and pathogenesis and how viruses influence cellular miRNA expression.
Keywords: microRNAs, RNAi, viruses, herpesviruses
Abstract
Analyses of small RNA expression profiles have revealed that several DNA viruses—including particularly, herpesviruses—express high levels of multiple viral microRNAs (miRNAs) in infected cells. Here, I review our current understanding of how viral miRNAs influence viral replication and pathogenesis and discuss how viruses reshape the pattern of cellular miRNA expression. Indeed, viruses are now known to both activate and repress the expression of specific cellular miRNAs, and disrupting this process can perturb the ability of viruses to replicate normally. In addition, it is now clear that virally encoded miRNAs play a key role in inhibiting antiviral innate immune responses and can also promote cell transformation in culture. While our understanding of how viruses interact with miRNAs remains somewhat rudimentary, it is nevertheless already clear that these interactions can play a critical role in mediating viral pathogenesis and therefore may represent novel and highly specific targets for therapeutic intervention.
MicroRNAs (miRNAs) are ∼22-nucleotide (nt)-long regulatory RNAs that are expressed by all multicellular eukaryotes (Bartel 2009). miRNAs act as guide RNAs for the RNA-induced silencing complex (RISC), a protein complex consisting minimally of an argonaut (Ago) protein and a member of the GW182 protein family (Hammond et al. 2000). miRNAs repress specific mRNAs by guiding RISC to complementary target sites that are most commonly located in mRNA 3′ untranslated regions (3′ UTRs). Most functional interactions between RISC and an mRNA require full complementarity between the mRNA and nucleotides 2–7 or 8 of the miRNA, the so-called seed region, although ∼25% of functional mRNA target sites may have incomplete seed complementarity; e.g., a single nucleotide insertion or a G:U base pair, often combined with complementarity to other regions of the miRNA. Binding of RISC to partially complementary target sites generally results in inhibition of mRNA translation followed by retargeting of the mRNA to translationally inactive cytoplasmic processing bodies (P bodies), where the repressed mRNA may be deadenylated and degraded (Huntzinger and Izaurralde 2011). In contrast, binding of RISC to fully complementary target sites can result in endonucleolytic cleavage by the RISC component Ago2, leading to rapid mRNA degradation. Importantly, RISC functions enzymatically on highly complementary target sites but largely stoichiometrically on the far more common partially complementary sites. The ratio of expression of the mRNA to the miRNA is therefore important in determining the efficiency of repression of mRNAs bearing partially complementary target sites but is potentially less important in the case of fully complementary target sites, which RISC can bind, cleave, and release efficiently.
Over 1000 miRNAs are encoded by the human genome, and it is now apparent that miRNAs play a key role in many aspects of normal cellular development and function and, moreover, that dysregulation of miRNA expression or function is associated with numerous disease states, including, particularly, cancer (Croce 2009). It is believed that >50% of cellular mRNAs are subject to regulation by one or more miRNAs, and individual miRNAs have been found to repress >100 mRNAs in expressing cells, although it remains unclear how many of these are physiologically relevant targets (Bartel 2009).
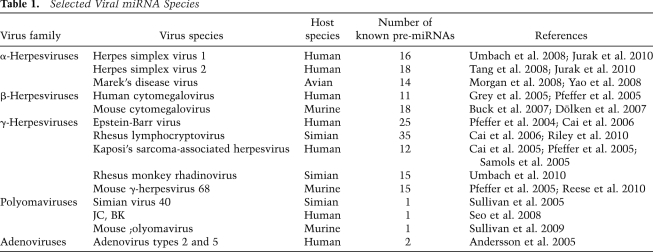
The small size of miRNAs, combined with their ability to specifically repress the expression of multiple mRNA targets, would seem to make them ideal tools for viruses to reshape the gene expression profile of an infected cell in ways that favor viral replication. The first viral miRNAs were discovered in 2004 in the human γ-herpesvirus Epstein-Barr virus (EBV) (Pfeffer et al. 2004), and subsequent work has identified multiple miRNAs expressed by members of all three herpesvirus subfamilies, including the α-herpesviruses herpes simplex virus 1 (HSV-1), HSV-2, and Marek's disease virus (MDV); the β-herpesviruses human cytomegalovirus (hCMV) and mouse cytomegalovirus (mCMV); and the γ-herpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV) and mouse γ-herpesvirus 68 (MHV68) (Table 1). While the expression of multiple viral miRNAs in infected cells appears to be almost a defining characteristic of herpesviruses, some other DNA viruses also express miRNAs. In particular, members of the polyomavirus family and the human adenoviruses have been reported to express one or two miRNAs in infected cells. In contrast, other DNA viruses, including papillomaviruses and poxviruses, do not encode any miRNAs, and investigation of a wide range of RNA viruses has yet to identify any viral miRNA species (Skalsky and Cullen 2010).
Table 1.
Selected Viral miRNA Species
Identification and expression of viral miRNAs
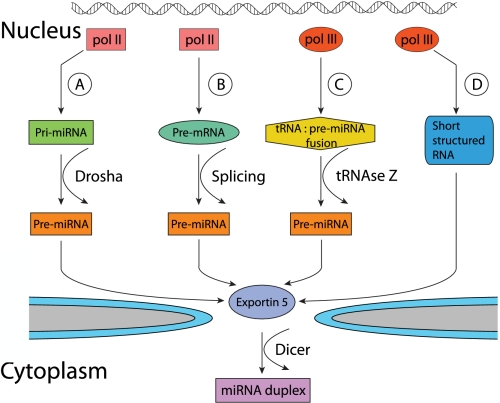
The canonical pathway of mRNA expression begins with the transcription of a capped, polyadenylated primary miRNA (pri-miRNA) precursor by RNA polymerase II (Pol II) (Fig. 1A; Cullen 2004). The pri-miRNA may be thousands of nucleotides in length and contains one or more ∼80-nt pri-miRNA stem–loop structures. The mature miRNA forms the upper part of the ∼32-base-pair (bp) imperfect stem, which is surmounted by a large terminal loop and flanked by single-stranded sequences (Zeng et al. 2005; Han et al. 2006). This stem–loop structure is recognized by the nuclear “microprocessor,” consisting of the RNase III enzyme Drosha and its cofactor, DGCR8, which cleave ∼22 bp down the stem to liberate an ∼60-nt RNA hairpin bearing a 2-nt 3′ overhang (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). This “pre-miRNA” hairpin is bound by the nuclear export factor Exportin 5 (Exp5), which transports the pre-miRNA to the cytoplasm (Yi et al. 2003; Lund et al. 2004), where a second protein complex, consisting of the RNase III enzyme Dicer and its cofactor, TRBP, removes the terminal loop, leaving a second 2-nt 3′ overhang (Chendrimada et al. 2005). One strand of the resultant miRNA duplex intermediate is then incorporated into RISC in a process that results in the degradation of the other strand, termed the miRNA passenger or star strand. This discrimination, which reflects the relative stability of base pairing at the 5′ end of each strand in the duplex (Khvorova et al. 2003; Schwarz et al. 2003), may be close to absolute or may be only partial, and several examples exist where a miRNA duplex gives rise to functionally relevant miRNAs from both strands (Yang et al. 2011).
Figure 1.
Schematic representation of the known miRNA biogenesis pathways in mammalian cells. (A) In the canonical miRNA processing pathway, Drosha liberates the pre-miRNA by direct cleavage of the pri-miRNA. (B) In the miRtron pathway, the pre-miRNA is generated by pre-mRNA splicing. (C) The miRNA expression pathway used by MHV68 requires processing of a tRNA:pre-miRNA fusion intermediate by tRNAseZ to liberate the pre-miRNA. (D) In this final pathway, short structured RNAs, generated by Pol III transcription, are directly exported to the cytoplasm, where they are cleaved by Dicer. As shown, export is mediated by Exp5, but this may not always be the case.
Analysis of mouse cells lacking Drosha, DGCR8, or Dicer has demonstrated that the vast majority of cellular miRNAs are dependent on DGCR8 and Drosha function, and all but one miRNA are dependent on Dicer function (Babiarz et al. 2008). The small number of cellular miRNAs that are still expressed in the absence of Drosha primarily derive from so-called miRtrons, short introns located in pre-miRNAs that, upon excision by the splicing machinery, are debranched and then fold to form RNA hairpins that are analogs of pre-miRNAs (Fig. 1B; Berezikov et al. 2007; Ruby et al. 2007). In a third miRNA expression pathway, short, highly structured RNA molecules transcribed by RNA polymerase III (Pol III) are directly transported to the cytoplasm (Fig. 1D), where they may be subject to direct cleavage by Dicer; e.g., the ∼102-nt-long “Y” RNA family or the ∼128-nt small nucleolar RNA (snoRNA) family (Babiarz et al. 2008; Ender et al. 2008; Meiri et al. 2010). Other small RNAs may undergo a structural rearrangement that renders them susceptible to Dicer cleavage; e.g., tRNA molecules that can collapse their normal cruciform structure to instead give an ∼70-nt RNA hairpin (Babiarz et al. 2008). Importantly, although the short RNAs that result from Dicer cleavage of tRNAs, Y RNAs, snoRNAs, or other highly structured small RNAs can function as miRNAs (Ender et al. 2008), processing appears to be far less efficient than seen with authentic pre-miRNAs. Although only a small percentage of a given tRNA, snoRNA, or Y RNA may therefore be cleaved by Dicer, the high level of expression of these RNAs nevertheless means that the resultant miRNAs can often be readily detected by deep sequencing. Whether these are physiologically relevant miRNAs or instead result from a low but physiologically irrelevant level of “off-target” Dicer cleavage is currently unclear.
Analysis of viral miRNA expression has revealed that the vast majority of known viral miRNAs are processed via the canonical miRNA pathway and, so far, no viral miRtrons have been identified. However, the processing pathway that requires short stem–loop RNA precursors transcribed by Pol III is used by adenovirus, which expresses an ∼160-nt, highly structured RNA called VA1. VA1, which normally functions to repress antiviral innate immune responses, is transcribed at very high levels by Pol III and then exported to the cytoplasm by Exp5. Efficient binding of RNAs by Exp5 is dependent on a short, terminal RNA stem bearing a 3′ overhang (Zeng and Cullen 2004), and this is similar to what is recognized by the Dicer:TRBP heterodimer, which preferentially binds the base of structured RNAs that have terminal stems that are blunt or contain short 3′ overhangs. As a result, Dicer can bind and process VA1 to yield functional miRNAs. However, this processing is very inefficient; i.e., <1% of VA1 is processed by Dicer in adenovirus-infected cells (Lu and Cullen 2004; Andersson et al. 2005). Moreover, VA1 actually competitively inhibits Dicer function, resulting in an inhibition in cellular pre-miRNA processing in adenovirus-infected cells. At present, it remains unclear whether the VA1-derived miRNAs seen in adenovirus-infected cells are important, or whether the low level of processing of VA1 by Dicer has instead not been evolutionarily selected against because the loss of <1% of total VA1 is insufficient to perturb their essential functions as repressors of innate immune responses. Resolution of this question will require the analysis of adenovirus VA1 mutants where the basal stem has been mutated to change the miRNA seed sequence while maintaining the integrity of the stem, which is required for VA1 nuclear export and function.
A fourth pathway of miRNA biogenesis was first identified in the murine γ-herpesvirus MHV68. MHV68 encodes eight tRNA:pre-miRNA fusion transcripts that each consist of an ∼70-nt tRNA-like molecule linked to one or two ∼60-nt pre-miRNA hairpins (Fig. 1C; Pfeffer et al. 2005). The initial, highly structured ∼190-nt RNAs are transcribed by Pol III using internal promoters located within each tRNA (Bogerd et al. 2010). This RNA structure is then cleaved by the enzyme tRNaseZ, which normally functions in the definition of mature tRNA 3′ ends, resulting in nonfunctional—i.e., nonaminoacylated—tRNA-like molecules and viral pre-miRNAs that are indistinguishable from normal pre-miRNAs. Indeed, there does not appear to be anything unique about the MHV68 tRNA:pre-miRNA fusions, as analogous expression cassettes consisting of an authentic cellular tRNA linked to a cellular pre-miRNA are also cleaved by tRNaseZ to liberate the pre-miRNA, which is then exported to the cytoplasm and processed by Dicer (Fig. 1C; Bogerd et al. 2010). It remains unclear whether an analogous Drosha-independent, tRNaseZ-dependent pathway is used by any cellular miRNA.
As noted above, it is now clear that many different herpesviruses express high levels of viral miRNAs, and polyomavirus- and adenovirus-encoded miRNAs are also readily detected in infected cells. Yet other viruses, especially RNA viruses, appear to entirely lack miRNAs. The reasons for this discrepancy are unclear, but one can speculate that excision of a miRNA from an RNA virus genome, which would result in the cleavage of that genome, would be disadvantageous. Moreover, the majority of RNA viruses (the major exceptions are retroviruses and orthomyxoviruses) replicate in the cytoplasm, and their transcripts would therefore be largely inaccessible to the nuclear microprocessor complex. Indeed, insertion of the EBV miR-BART2 pri-miRNA sequence into the genome of the flavivirus tick-borne encephalitis virus (TBEV) was found to generate only ∼500 copies of miR-BART2 per cell, even though infected cells on average contained ∼70,000 copies of the TBEV genome, a processing efficiency of ∼0.7% (Rouha et al. 2010). Interestingly, processing of this miRNA was nevertheless dependent on Drosha, thus suggesting either that a low percentage of TBEV genomic RNA enters the nucleus or that the TBEV genome and Drosha interact at a low level as a result of nuclear membrane breakdown during mitosis. Although the level of expression of this inserted miRNA in TBEV-infected cells was sufficient to down-regulate an indicator construct containing a perfect miR-BART2 3′ UTR target site, it is unclear whether this low level of expression would be sufficient to globally down-regulate mRNAs that are authentic miR-BART2 targets. In contrast, insertion of pri-miRNAs into the genomes of nuclear RNA viruses, such as retroviruses and influenza virus, gives rise to efficient pri-miRNA processing and, hence, high levels of miRNA expression (Gottwein et al. 2007; Varble et al. 2010).
While most efforts to identify miRNAs in RNA viruses have been unsuccessful, there are occasional reports that suggest that some RNA viruses, especially HIV-1, might encode a miRNA (Omoto et al. 2004; Ouellet et al. 2008). In contrast, published data from this laboratory and others (Pfeffer et al. 2005; Lin and Cullen 2007), as well as unpublished deep sequencing data from my group (H Bogerd, J Powers, and BR Cullen, unpubl.) have consistently failed to reveal any HIV-1-encoded miRNAs. In this context, it is worth briefly reviewing the characteristics that one would expect to observe for an authentic viral miRNA identified during deep sequencing of an RNA sample derived from virus-infected cells. This is important, as it is now routinely possible to obtain >5,000,000 short RNA sequence reads, yet the total number of miRNAs expressed by a given cell is generally ∼50,000 or so. Therefore, even if you obtain a particular short RNA sequence 100 times during deep sequencing, this actually represents only approximately one copy of that sequence per cell. As miRNAs bind to RNA target sites that are defined by as little as 6 nt or 7 nt of homology, and current evidence indicates that >100 mRNAs, many of which may be expressed at >10 or even >100 copies per cell, can be significantly down-regulated by a single cellular or viral miRNA (Bartel 2009), it is clear that miRNAs expressed at <50 copies per cell are likely to be of limited functional relevance; i.e., there are too many available binding sites on too many cellular transcripts for such a small number of miRNA copies to be able to repress any one mRNA effectively. Other criteria for the validation of authentic viral miRNAs include:
They should derive from a tightly defined region(s) of the viral genome, and their expression level should substantially exceed the “noise” generated by small viral RNA degradation products, which can be quite common in cells undergoing a viral cytopathic effect.
They should have a discrete size that falls within the 21- to 23-nt—maximally 19- to 25-nt—size range characteristic of miRNAs.
Given the importance of the miRNA seed region—i.e., nucleotides 2–8 from the 5′ end—in determining target identification, the 5′ end of the miRNA should be highly discrete.
As the miRNA duplex intermediate invariably gives rise to levels of both RNA strands, and these are readily detectable by deep sequencing, it should be possible to identify an ∼80-nt hairpin structure where each recovered RNA strand is located in the upper part of each side of the predicted ∼32-bp stem, offset by ∼2 nt.
If these criteria cannot be satisfied, and especially if the candidate miRNA is expressed at vanishingly low levels, then it is unlikely to be an authentic viral miRNA.
Viral regulation of viral miRNA expression
As discussed above, all viral miRNAs are generated through processing steps that rely on cellular factors, and, at present, no viral factors that specifically regulate viral miRNA processing are known. Nevertheless, it is clear that viral miRNA expression is regulated at both the transcriptional and post-transcriptional levels. At the post-transcriptional level, it is clear that there must be cis-acting sequences that regulate the efficiency of viral miRNA expression because viral miRNAs that derive from the same primary miRNA precursor are often expressed at quite different levels. For example, the KSHV miRNAs miR-K4 and miR-K7, which derive from a single primary miRNA, differ in their level of expression by ∼400-fold in latently KSHV-infected BC-3 cells (Umbach and Cullen 2010), and similar differences in expression levels have been noted for several other herpesvirus miRNAs (Pratt et al. 2009; Umbach et al. 2009). At the transcriptional level, there is a clear difference in the expression of the two EBV miRNA clusters that encode the three miR-BHRF1 miRNAs and the 22 miR-BART miRNAs, respectively, depending on the stage of viral latency (Cai et al. 2006; Xing and Kieff 2007; Cosmopoulos et al. 2009). Specifically, the miR-BHRF1 miRNA cluster is expressed during EBV latency stage III in infected B cells, but is not detectably expressed during latency stage II in epithelial cells, such as nasopharyngeal carcinoma (NPC) cells. Conversely, the miR-BART miRNA cluster is expressed at very high levels in NPCs but only at moderate levels in latency III B cells. Viral miRNA expression may also be temporally regulated, similar to the expression of proteins in cells infected by DNA tumor viruses. For example, the single miRNA expressed by polyomaviruses is transcribed by the viral “late” promoter (Sullivan et al. 2005) and the known CMV miRNAs also appear to be selectively expressed in the later stages of the viral replication cycle (Grey et al. 2005; Dölken et al. 2007; Murphy et al. 2008). In HSV-1, there are six miRNAs that are expressed primarily during latency and nine that are expressed exclusively during productive replication; a 16th miRNA, miR-H6, is expressed during both (Jurak et al. 2010). Presumably, this regulation directly relates to the roles played by viral miRNAs in different stages of the viral life cycle.
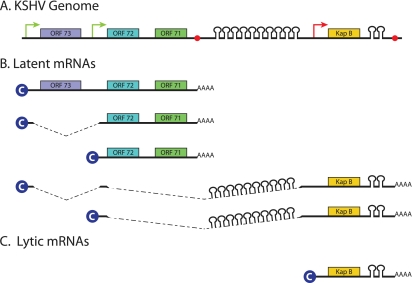
An interesting example of both transcriptional and post-transcriptional regulation of viral miRNA expression has recently been reported for KSHV, which encodes a single cluster of 12 viral pre-miRNAs that nevertheless shows a rather complex pattern of regulation (Fig. 2). During latency, all 12 KSHV pre-miRNAs are expressed from two latency-specific promoters that also direct the expression of four viral latent proteins: ORF71, ORF72, ORF73, and kaposin B (KapB) (Fig. 2B; Cai and Cullen 2006). Ten KSHV miRNAs are located within an ∼5-kb intron located between ORF71 and KapB, while the other two viral miRNAs (miR-K10 and miR-K12) are located in the KapB 3′ UTR. Clearly, because nuclear excision of miR-K10 or miR-K12 would result in the cleavage of the KapB mRNA, generation of KapB mRNA versus miR-K10/miR-K12 represents mutually exclusive fates for the initial transcript. However, as miR-K10 and miR-K12 excision by Drosha is fairly inefficient, all three KSHV gene products are nevertheless expressed in latently KSHV-infected cells (Lin et al. 2010; Umbach and Cullen 2010).
Figure 2.
Schematic of the miRNA locus in KSHV. (A) This figure shows a simplified schematic of the latency region of KSHV at the genomic level, including the four major latency-specific gene products and the 12 KSHV pre-miRNAs. (B) The two major latency-specific promoters (green arrows) give rise to a complex mixture of mRNAs, depending on alternative splicing and polyadenylation, that express all four latent proteins and all 12 viral pre-miRNAs. (C) Upon activation of lytic replication, a powerful lytic promoter (red arrow) is activated that transcribes RNAs encoding the KapB protein and two of the 12 viral miRNAs. As discussed in the text, lytic replication also results in inhibition of Drosha expression, which favors KapB mRNA production over miRNA biogenesis.
Interestingly, this pattern is perturbed upon activation of lytic KSHV replication. Specifically, there is a powerful lytic KSHV promoter located immediately 5′ to the KapB ORF, and this produces high levels of an mRNA that encompasses the KapB ORF as well as miR-K10 and miR-K12, but not any of the other, latency-specific KSHV miRNAs (Fig. 2C; Cai and Cullen 2006). However, analysis has revealed that KapB expression increases far more than does expression of miR-K10 and miR-K12 when productive KSHV replication is activated. It now appears that this results from a substantial inhibition of Drosha expression during the KSHV lytic replication cycle (Lin and Sullivan 2011). The mechanism underlying this effect is unclear, but it could simply reflect the global degradation of cellular mRNAs, including Drosha mRNAs, that occurs during the lytic replication of many herpesvirus species. In any event, this study reveals a nice example of viral regulation of viral miRNA expression via a combination of cis-acting DNA and RNA sequences and trans-acting proteins.
Cellular miRNAs can promote viral replication
An interesting question that arises is whether viruses are also able to regulate cellular miRNA transcription and processing in infected cells in ways that result in the loss of deleterious cellular miRNAs and/or in the expression of advantageous cellular miRNAs. In fact, a number of examples of cellular miRNA dysregulation in virally infected cells have now been reported (Triboulet et al. 2007; Cameron et al. 2008; Yin et al. 2008; Ho et al. 2011).
Viral infection is a highly traumatic event, and it is well established that cellular mRNA expression patterns can change dramatically. Similarly, because most miRNAs are also transcribed by Pol II, using many of the same cellular transcription factors, it is not surprising that viral infection also perturbs the pattern of miRNA expression. It is difficult, however, to discern a priori whether up-regulation of a specific miRNA represents part of the host innate immune response and therefore might inhibit viral replication, is specifically induced by the virus and therefore might enhance virus replication, or is simply irrelevant. However, several cellular miRNAs have now emerged that either repress or enhance virus replication and that show substantial changes in expression upon viral infection.
The best established cellular miRNA that promotes the replication of a specific virus is miR-122, a liver-specific miRNA that is essential for hepatitis C virus (HCV) genome replication and that, counterintuitively, may also enhance HCV mRNA translation (Jopling et al. 2005; Henke et al. 2008). Interestingly, miR-122 acts via two partially complementary “seed” targets located in the 5′ UTR (not the 3′ UTR) of the HCV RNA genome (Jopling et al. 2008). Importantly, inhibition of miR-122 by a locked nucleic acid (LNA) antisense drug injected into HCV-infected chimpanzees has been shown to dramatically reduce HCV viral loads in vivo (Lanford et al. 2010). This potential therapeutic is now in phase II clinical trials in humans, and represents a proof of the principle that specific inhibition of a miRNA that plays a critical role in viral replication is not only feasible, but also practicable.
Because miR-122 is expressed at high levels in uninfected hepatocytes and is not substantially induced upon HCV infection, this example of a cellular miRNA that facilitates viral replication is not directly relevant to the question of whether the induction of specific cellular miRNAs upon infection affects viral replication. However, a clear case where induction of a cellular miRNA is critical for establishment of virus infection is provided by the oncogenic γ-herpesvirus EBV and the cellular miRNA miR-155. EBV is able to induce the immortalization of primary human B cells infected in culture to generate so-called lymphoblastoid cell lines (LCLs). This process is induced and maintained by a handful of viral proteins and also requires the action of virally encoded miRNAs (see below). However, EBV also dramatically induces the expression of several cellular miRNAs, including miR-155, whose expression increases by >100-fold within days of infection, apparently due entirely to activation of pri-miR-155 transcription (Gatto et al. 2008; Yin et al. 2008). This is an intriguing observation, given that two other transforming herpesviruses, KSHV and the avian herpesvirus MDV, have been shown to encode viral orthologs of cellular miR-155 that down-regulate a similar set of cellular mRNAs (see below; Gottwein et al. 2007; Skalsky et al. 2007; Zhao et al. 2009). Moreover, overexpression of cellular miR-155 is associated with oncogenic transformation in several species, including humans (Croce 2009). In fact, inhibition of miR-155 function using a miRNA sponge—i.e., an overexpressed RNA containing multiple partially complementary target sites that specifically blocks miR-155 function (Ebert et al. 2007)—was found to result in the cell cycle arrest and apoptotic death of EBV-infected LCLs in culture (Linnstaedt et al. 2010).
Several herpesviruses, including KSHV and hCMV, have been shown to up-regulate another cellular miRNA, miR-132, by >10-fold after infection (Lagos et al. 2010). This miRNA appears to indirectly inhibit the expression of a number of interferon-stimulated genes by targeting the transcriptional coactivator p300. As a result, inhibition of miR-132 expression in cells infected by KSHV or hCMV leads to a significant, but modest, approximately twofold drop in viral replication. Many other cellular miRNAs are also activated after infection by a wide range of virus species, and it will be interesting to see how many of these inductions actually reflect viral evolutionary adaptations leading to enhanced replication. If the effect is profound, as seen with EBV and miR-155, then it seems possible that these cellular miRNAs could also emerge as targets for treatment with antisense drugs.
Another interesting example of a virus that induces the expression of a cellular miRNA that then facilitates viral gene expression is provided by enteroviruses (EVs) (Ho et al. 2011). EVs are RNA viruses whose positive sense genomes lack a 5′ cap and instead rely on an internal ribosome entry site (IRES) for translation. Infection with an EV was shown to strongly induce expression of cellular miR-141, primarily at the transcriptional level. This, in turn, directly represses the expression of mRNAs encoding the cellular translation factor eIF4E, which is required for cap-dependent, but not IRES-dependent, mRNA translation. Blocking miR-141 function was found to inhibit EV replication, while ectopic overexpression of miR-141 was found to selectively promote IRES-dependent mRNA expression. It therefore appears that EVs have evolved to favor the translation of viral mRNAs by activating the expression of a cellular miRNA that then selectively inhibits cap-dependent mRNA translation (Ho et al. 2011).
Cellular miRNAs can function as inhibitors of viral replication
In principle, there is no reason why cellular miRNAs should not be able to also bind to viral mRNA species and then inhibit their expression. Indeed, there have been several reports documenting the tissue-specific attenuation of viruses upon the insertion of artificial target sites for cellular miRNAs that are specifically expressed in that tissue (Barnes et al. 2008; Kelly et al. 2008; Cawood et al. 2009). This approach has considerable potential in the development of attenuated viruses for use in vaccines and in the design of oncolytic viral vectors that target transformed but not normal tissues. However, one concern with this approach is that these engineered viruses are under obvious selective pressure to delete the inserted miRNA target sites, and this generally occurs within days of infection. Nevertheless, if it takes less time to mount a protective immune response than it takes to select for virus variants lacking the inserted miRNA target sites, then this may be a manageable concern.
There have also been several reports documenting the inhibition of viral replication by endogenous cellular miRNA species. At first, this seems like an odd result. Viruses, especially RNA viruses, have quite plastic genome sequences and would be expected to rapidly evolve away from repression by specific miRNAs, especially as this could often be achieved by a single nucleotide mutation. In contrast, cellular miRNAs are generally highly conserved through evolution, with some miRNAs (e.g., let-7) being conserved all the way from humans to nematodes. Therefore, viruses have certainly had ample time to evolve in ways that preclude the binding of RISCs programmed by cellular miRNAs. The rapid loss of cognate RISC-binding sites is indeed exactly what is seen when rapidly evolving viruses such as HIV-1 are grown in the presence of artificial siRNAs, which function identically to miRNAs when they bind to perfectly complementary RNA targets (Boden et al. 2003; Westerhout et al. 2005). Further examination, however, reveals that many studies documenting inhibition of viral replication by endogenous miRNAs were performed in cell lines that may not be good models for the target tissues that are infected in vivo by the virus in question. For example, an early study clearly demonstrated inhibition of primate foamy virus (PFV) replication in the human embryonic kidney cell line 293T by the cellular miRNA miR-32 (Lecellier et al. 2005). However, PFV is thought to primarily replicate in salivary glands in vivo, so this inhibition is of unclear physiological relevance and PFV is unlikely to have been selected to shed miR-32-binding sites during PFV evolution. As miRNA expression patterns differ profoundly between different tissues in vivo, and given that many viruses show a high degree of tissue tropism, it is clearly important to carefully control for this issue before concluding that a given cellular miRNA is really repressing the replication of a particular virus in vivo. Nevertheless, there are reports that document inhibition of viral replication by a cellular miRNA expressed in the appropriate target tissue; e.g., inhibition of HIV-1 replication by cellular miR-29a in T cells via a conserved target site in the viral RNA genome (Nathans et al. 2009). This result is difficult to understand, unless miR-29a binding also has a beneficial effect on viral replication that exceeds the more obvious negative effect. For example, could miRNA binding to the HIV-1 genome retarget this RNA away from translation and into a pathway favoring packaging into progeny virions? Certainly, the issue of how cellular miRNAs expressed in the physiologically relevant target tissues affect the replication of HIV-1 and other RNA viruses deserves more scrutiny.
While miRNAs could obviously inhibit virus replication by directly targeting viral mRNA species, this is, as noted above, readily reversed by the selection of a viral variant lacking the target site in question. However, it also seems possible that cellular miRNAs may inhibit virus replication by, for example, repressing the expression of cellular factors required for efficient viral replication cycle or repressing inhibitors of protective innate immune responses. In this case, the virus has two possible paths to circumvent this problem. One is to globally or selectively inhibit miRNA biogenesis, while the second is to specifically block the action of the miRNA in question.
As noted above, the adenoviral noncoding RNA VA1 represents a good example of a virally induced global inhibition of miRNA biogenesis. VA1 is expressed at ≥108 copies per cell in adenovirus-infected cells, and this level of VA1 synthesis appears to saturate the function of Exp5, which mediates the nuclear export of both pre-miRNAs and VA1 (Lu and Cullen 2004). As a result, pre-miRNA nuclear export, and hence pre-miRNA processing, is blocked. Moreover, and as noted above, VA1 also is an efficient competitive inhibitor of Dicer function, possibly due to the fact that it can bind the Dicer:TRBP heterodimer effectively, yet subsequent cleavage is highly inefficient (Andersson et al. 2005).
Another viral gene product that has been proposed to globally inhibit miRNA biogenesis, again by inhibiting Dicer function, is the HIV-1 Tat protein (Bennasser et al. 2005). However, two other groups (Lin and Cullen 2007; Sanghvi and Steel 2011) have failed to reproduce this result, and the observed induction of specific cellular miRNAs in HIV-1-infected, Tat-expressing cells also seems inconsistent with this hypothesis (Triboulet et al. 2007). While viral gene products that block RISC function are common in plants and insects, where RNAi represents a major innate immune response to viral infections, it remains unclear whether viruses that infect mammals, which do not mount an RNAi response to viral infection of somatic cells, commonly encode inhibitors of miRNA biogenesis (Cullen 2006). Nevertheless, viruses that are known to block Pol II-mediated mRNA transcription in infected cells would certainly be expected to inhibit the production of most cellular miRNAs. It should be remembered that miRNAs are generally very stable, with half-lives measured in days, while most lytic viral replication cycles are over in a matter of hours. Whether the global inhibition of cellular miRNA biogenesis plays an important role in facilitating the replication of any mammalian virus is therefore currently unclear.
While global inhibition of miRNA expression upon viral infection may therefore be unusual in mammalian cells, a nice example of the specific inhibition of the expression of a cellular miRNA is provided by human papillomavirus (HPV), which establishes a latent infection in skin epithelial cells that switches to productive virus replication once these cells differentiate into suprabasal cells. Normally, this differentiation results in cell cycle arrest, a process that is in part mediated by the repression of the p63 family of transcription factors by cellular miR-203. However, the HPV oncoproteins E6 and E7 have been reported to inhibit miR-203 expression at the transcriptional level, thus increasing the expression of p63 and its target genes (McKenna et al. 2010; Melar-New and Laimins 2010). This, in turn, facilitates the continued replication of HPV-infected cells and formation of the hyperplasias that are characteristic of HPV infection. Moreover, recent data indicate that p63 plays a key role in mediating HPV DNA genome amplification and late gene expression, thus implicating the repression of miR-203 as a potentially critical step in the production of progeny virions (Mighty and Laimins 2011).
Given the issue of generally short viral life cycles and long miRNA half-lives, it seems clear that the best way for most viruses to block the function of deleterious cellular miRNAs would be not at the level of miRNA biosynthesis, but rather directly at the level of mRNA function and/or stability. One laboratory technique that not only blocks miRNA function but also induces miRNA destabilization is the miRNA sponge technique. As noted above, sponges are highly expressed transcripts that contain tandem sites that are partially complementary to a specific miRNA (Ebert et al. 2007). As a result, this miRNA binds the sponge RNA stoichiometrically and can be titrated away from its normal target mRNAs. Moreover, and for reasons that are not entirely clear, sponges often substantially reduce the stability of the miRNAs they bind.
Recently, it has been suggested that viruses also use natural miRNA sponges to attenuate miRNA function in vivo. The γ-herpesvirus herpesvirus saimiri (HVS), a simian virus closely related to KSHV, expresses high levels of a small nuclear noncoding RNA called HSUR1 in HVS-transformed, latently infected simian T cells (Cook et al. 2004). Despite its largely nuclear localization, HSUR1 appears able to bind miR-27 and thereby sequester it away from its normal cellular mRNA targets, which as a result show a significant increase in expression (Cazalla et al. 2010). This interaction also induces the degradation of miR-27 in a manner similar to what is seen with artificial miRNA sponges; i.e., without affecting pri-miR-27 transcription or processing (Cazalla et al. 2010). Interestingly, mCMV also induces the destabilization of miR-27 in infected cells, again without affecting pri-miR-27 transcription or expression (Buck et al. 2010). While this result is clearly suggestive of an mCMV-encoded, miR-27-specific sponge, this putative viral transcript has yet to be identified. However, forced expression of miR-27 has been shown to inhibit mCMV replication, consistent with the idea that the miR-27-mediated repression of specific cellular mRNAs that exert a positive effect on viral replication also exerts a repressive effect on the replication of mCMV—as well as HVS and possibly other virus species—that must be blocked for maximal virus replication to occur. Many viruses are known to encode noncoding RNAs of generally unknown function, and it will certainly be of interest to determine whether any of these viral RNAs also function as miRNA sponges.
Phenotypes of viral miRNA mutants
A major question that remains inadequately explored is how viral miRNAs affect viral replication and pathogenicity in vivo and in culture. The first study to analyze the effect of a viral miRNA on viral replication in vivo focused on mouse polyomavirus (mPy). Like all other polyomavirus family members, mPy encodes a single pre-miRNA that is expressed as a viral late gene product. This pre-miRNA is located antisense to the viral T antigen (TAg) transcripts and, because of their perfect complementarity, these miRNAs cleave TAg mRNAs and hence repress early mPy gene expression late in the viral life cycle (Sullivan et al. 2009). Because TAgs represent targets for cytotoxic T cells (CTLs), it was proposed that this inhibition reduced CTL killing of infected cells during the late phase of the viral life cycle (Sullivan et al. 2005). However, infection of immunocompetent mice with wild-type mPy or with an mPy mutant lacking the single viral pre-miRNA resulted in no detectable differences in viral load or immune response to viral infection (Sullivan et al. 2009). We are therefore left with the conundrum that a pre-miRNA that has been conserved in location (but not in sequence) throughout polyomavirus evolution has no obvious effect on virus replication in vivo!
A result more consistent with an important role for viral miRNAs in vivo was observed upon analysis of an mCMV mutant lacking two out of the 18 miRNAs encoded by this virus, which serves as a model system for the study of CMV replication in vivo (Dölken et al. 2010a). Infection of C57BL/6 mice (which are highly resistant to mCMV infection due to a strong NK cell response) versus BALB/C mice (which are more susceptible) failed to reveal any difference in viral load at 3 d post-infection (dpi). However, at 14 dpi, Dölken et al. (2010a) noted an ∼100-fold lower viral load in the salivary glands of C57BL/6 mice infected with the mutant mCMV, while in BALB/C mice, high and comparable titers were noted whether the miRNAs were expressed or not. In contrast, in both mouse strains, similar viral loads were observed in the lung. Interestingly, in C57BL/6 mice in which both the NK cell response and the CD4+ T-helper cell response to mCMV infection were blocked, mCMV viral loads in the salivary gland were high and comparable regardless of whether the viral miRNAs were present. Dölken et al. (2010a) concluded that these two viral miRNAs protect the virus from immune elimination specifically in the salivary glands, which play a key role in transmission by releasing mCMV virions into the saliva. This interesting finding raises the possibility that the miRNA encoded by mPy might also play a role in virus transmission and makes the point that the identification of in vivo phenotypes for specific viral miRNAs may require not only the analysis of viral loads in a range of different tissues, but also measurement of viral transmission efficiency.
An even more striking phenotype was observed upon inactivation of a miRNA, miR-M4, encoded by the avian α-herpesvirus MDV. miR-M4 functions as an ortholog of cellular miR-155, which, as noted above, plays a critical role in the transformation of B cells in culture by EBV (Linnstaedt et al. 2010; Zhao et al. 2011). Moreover, KSHV also encodes a functional analog of miR-155 called miR-K11 that shares a largely identical list of mRNA targets with both miR-M4 and miR-155 (Gottwein et al. 2007; Skalsky et al. 2007). Mutational inactivation of miR-M4 in MDV was found to not affect viral replication per se but did block the ability of MDV to induce T-cell lymphomas in infected chickens. It therefore appears that MDV, KSHV, and EBV all use either cellular or viral miR-155 to promote their replication in vivo, and that miR-155 activity is also likely to play a key role in the transformation of lymphoid cells by not only MDV, but also EBV and KSHV, which cause B-cell lymphomas in humans.
Another recently published study looked at the effect of mutagenesis of the viral miRNA miR-H6 on the ability of HSV-2 to replicate and establish latency in vivo in guinea pigs (Tang et al. 2011). HSV-2 encodes ∼17 miRNAs, of which eight, including miR-H6, have been reported to be expressed during viral latency (Jurak et al. 2010). Nevertheless, reduced miR-H6 expression did not affect the establishment of latency or viral reactivation in infected guinea pigs. Tang et al. (2011) did, however, note a significant reduction in neurological complications in the animals infected with the mutant virus.
Because human herpesviruses (the exceptions are HSV-1 and HSV-2) are unable to replicate in animals, this represents our current knowledge of how viral miRNAs affect herpesvirus replication and pathogenicity in vivo. Clearly, one can expect to see more information in the future about how animal herpesviruses that act as models for human herpesviruses behave in vivo when they have lost one or more of the miRNAs they encode. In the interim, it is possible to examine the replication of miRNA mutants of human herpesviruses in culture. As noted above, EBV is unique in that it is able to immortalize primary human B cells in culture and, after infection, EBV expresses both the viral miR-BHRF1 miRNA cluster and the miR-BART cluster (Cai et al. 2006). Perhaps surprisingly, mutational inactivation of the entire 22-pre-miRNA miR-BART cluster does not substantially affect B-cell transformation by EBV (Seto et al. 2010), although it should be recalled that in vitro replication assays would be unable to detect effects on the host immune response, among other things. However, mutational inactivation of the three miRNA miR-BHRF1 clusters does substantially reduce the ability of EBV to transform primary B cells in culture, and this effect reflects the reduced ability of cells infected by the mutant virus to proceed from the G1 to the S phase of the cell cycle (Seto et al. 2010; Feederle et al. 2011). These data represent the first observations indicating that viral miRNAs may play a key role in viral transformation in humans.
Identification of mRNA targets for viral miRNAs
Because several recent reviews have comprehensively discussed the previously published mRNA targets for virally encoded miRNAs (Skalsky and Cullen 2010; Grundhoff and Sullivan 2011; Plaisance-Bonstaff and Renne 2011), I will not attempt to do the same here, but rather present a selection of well-documented gene targets for miRNAs encoded by human herpesviruses, focusing primarily on mRNA targets for which phenotypic evidence also exists.
Human α-herpesviruses
As noted above, the clinically important, and closely related, human α-herpesviruses HSV-1 and HSV-2 both encode a number of miRNAs, several of which are expressed at high levels during latent infection (Tang et al. 2008; Umbach et al. 2008; Jurak et al. 2010). Unfortunately, both HSV-1 and HSV-2 only establish latency in primary, nondividing sensory neurons in vivo, which means that it is experimentally very difficult to identify repressed mRNA species. However, it does appear that HSV-1 and HSV-2 use miRNAs to down-regulate the expression of several viral genes. In particular, both HSV-1 and HSV-2 express a miRNA, miR-H2, that is located antisense to the mRNA encoding the viral immediate early transcription factor ICP0 and that represses ICP0 expression (Umbach et al. 2008; Tang et al. 2009). In addition, the HSV-1 miRNA miR-H6 bears full seed homology with a target sequence located in the mRNA encoding a second immediate early transcription factor, ICP4, and can repress ICP4 expression in vitro (Umbach et al. 2008). As ICP0 and ICP4 are thought to play a critical role in initiating the viral lytic replication cycle, these findings are consistent with the hypothesis that these viral miRNAs promote entry into latency and/or stabilize viral latency by inhibiting ICP0 and ICP4 expression. Two other miRNAs that are conserved between HSV-1 and HSV-2, miR-H3 and miR-H4, are located antisense to a viral late gene, ICP34.5, and have been shown to repress ICP34.5 expression (Tang et al. 2008, 2009). ICP34.5 is important for inhibition of cellular innate immune responses to viral infection and ICP34.5 expression enhances neurovirulence in HSV-1-infected animals. Why the virus has evolved miRNAs that repress ICP34.5 expression is currently unclear, but this may prolong the viability of HSV-1/HSV-2-infected neurons after reactivation.
Human β-herpesviruses
Unlike α-herpesviruses and γ-herpesviruses, where research has focused largely on miRNAs expressed during viral latency, research into the miRNAs expressed by hCMV has so far only identified viral miRNAs expressed during lytic replication in cultured fibroblasts. It would clearly be of considerable interest to characterize the hCMV miRNAs expressed during latency, which is thought to occur in infected monocytes and/or hematopoetic stem cells, to determine whether any of the known hCMV miRNAs or additional, currently unknown hCMV miRNAs are expressed during latency.
Analysis of hCMV miRNA function has demonstrated that the viral miR-UL112 miRNA specifically targets a site located in the 3′UTR of the mRNA encoding the major viral immediate early transcription factor IE1/IE72 (Grey et al. 2007; Murphy et al. 2008). As noted above, it is currently unclear whether miR-UL112 is also expressed during viral latency, and it is therefore unknown whether hCMV, by analogy to HSV-1 and HSV-2, uses miRNAs to stabilize viral latency. During lytic replication, miR-UL112 accumulates late in the viral replication cycle, at which point it acts to reduce IE1/IE72 protein expression. The purpose of this inhibition is unclear, but it may prevent the accumulation of toxic levels of IE1/IE72 or, by analogy to the proposed mechanism of action of the polyomavirus miRNAs, it may function to prevent CTL killing of hCMV-infected cells by reducing the presentation of epitopes derived from IE1/IE72.
Efforts to identify cellular mRNAs targeted by the hCMV miRNA miR-US25-1 using RISC immunoprecipitation (RIP) linked to microarray analysis (RIP-ChIP [chromatin immunoprecipitation]) in transfected 293 cells have identified a number of cell cycle control genes as targets for miR-US25-1, including the mRNA encoding cyclin E2. Moreover, Grey et al. (2010) noted that cyclin E2 was overexpressed in cells infected by an hCMV mutant lacking miR-US25-1. How this would benefit the virus is currently unclear.
A more obviously advantageous mRNA target for an hCMV miRNA was identified by Stern-Ginossar et al. (2007), who reported that hCMV miR-UL112 (the same viral mRNA that represses viral IE1/IE72 expression) represses the cellular gene product MICB. MICB is a stress-induced ligand for a receptor expressed on NK cells that plays an important role in NK cell killing of virus-infected cells. Interestingly, subsequent work has revealed that the KSHV miRNA miR-K7 and the EBV miRNA miR-BART2 also repress MICB and facilitate immunoevasion in vitro, indicating that MICB repression is advantageous for multiple herpesvirus species (Nachmani et al. 2009).
Human γ-herpesviruses
Unlike human α-herpesviruses and β-herpesviruses, human γ-herpesviruses are clearly associated with tumorigenesis in vivo. EBV, which infects B cells and epithelial cells, is linked to several varieties of B-cell lymphoma, including Burkitt's lymphoma (BL) and non-Hodgkin's lymphoma, as well as to NPC, which is of epithelial origin. KSHV infects B cells and endothelial cells and is the etiologic agent of primary effusion lymphoma (PEL) in B cells and Kaposi's sarcoma in endothelial cells. Both EBV and KSHV readily establish latent infections both in vivo and in vitro and both viruses express high levels of viral miRNAs during latent infection. As a result, work on the miRNAs encoded by EBV and KSHV has almost entirely focused on their role in latently infected cells.
Analysis of mRNA species targeted by EBV-encoded miRNAs has identified a substantial number of potential cellular mRNA targets using a RIP-ChIP approach, including two genes, TOMM22 and Importin 7, that play a role in intracellular transport (Dölken et al. 2010b). The phenotypic consequences of this down-regulation remain unclear. A more striking result was reported for miR-BART5, which has been shown to down-regulate the mRNA encoding the proapoptotic protein “p53 up-regulated modulator of apoptosis” (PUMA) in expressing cells (Choy et al. 2008). Strikingly, inhibition of miR-BART5 function in the latently EBV-infected NPC cell line C666-1 induced apoptosis, and this could be rescued by the simultaneous addition of an siRNA specific for PUMA. EBV infection is known to induce a cellular DNA damage response (Nikitin et al. 2010), and miR-BART5 may play a key role, at least in epithelial cells, by preventing the p53 activation that is often seen in EBV-infected cells from leading to cell death. The ability of the EBV miR-BART miRNAs to inhibit apoptosis in epithelial cells has also been reported to result in part from repression of a second proapoptotic cellular gene product called Bim, although in this case the combined action of several miR-BART miRNAs appeared to be required for efficient Bim repression (Marquitz et al. 2011).
Another interesting mRNA target that is down-regulated by an EBV-encoded miRNA is the T-cell-attracting chemokine CXCL-11 (Xia et al. 2008). Unusually, the EBV miRNA miR-BHRF1-3 shows complete complementarity to a target site located in the CXCL-11 mRNA, and CXCL-11 mRNA expression in EBV-infected BL cells was enhanced by inhibition of miR-BHRF1-3 function. Together with the work, noted above, identifying MICB as a target for EBV miR-BART2 (Nachmani et al. 2009), this adds up to four cellular factors known to play a role in the innate immune response to viral infection that have so far been identified as targets for repression by an EBV miRNA, and it is likely that many more examples will be identified in EBV and other herpesviruses.
In terms of viral mRNA targets for EBV-encoded miRNAs, the most fully validated example is represented by the EBV DNA polymerase gene BALF5, which lies antisense to the viral miRNA miR-BART2 (Pfeffer et al. 2004; Barth et al. 2008). As expected, miR-BART2 can induce the endonucleolytic cleavage of this mRNA at the predicted target site in EBV-infected cells. As BALF5 is not thought to play an important role in the activation of lytic EBV replication, the significance of this inhibition is not currently clear. Another EBV-encoded protein, LMP1, has been reported to be down-regulated by no less than three viral miRNAs: miR-BART1, miR-BART16, and miR-BART17-5p (Lo et al. 2007). While LMP1 plays a key role in the EBV-induced transformation of B cells in culture by activating a number of progrowth cellular pathways, LMP1 overexpression can be toxic, so this result may imply that viral miRNAs are acting to fine-tune the level of LMP1 expression.
The final human γ-herpesvirus that I discuss here is KSHV, which expresses 12 pre-miRNAs in latently infected PEL cells (Umbach and Cullen 2010). A substantial number of potential mRNA targets for KSHV miRNAs have been reported (Grundhoff and Sullivan 2011), and I here restrict myself to discussing a small number of cellular mRNA targets that are of interest from the perspective of viral transformation.
As noted above, the KSHV miRNA miR-K11 functions as an ortholog of the cellular oncomir miR-155 and shares a very large number of mRNA targets with miR-155 (Gottwein et al. 2007; Skalsky et al. 2007). Presumably, by analogy to the key role played by induced cellular miR-155 in B-cell transformation by EBV and by viral miR-M4 in T-cell transformation by MDV (Linnstaedt et al. 2010; Zhao et al. 2011), this viral miRNA also plays a key role in B-cell transformation by KSHV. However, as KSHV will not replicate in animals or transform cells in culture, this hypothesis is difficult to experimentally address.
Other interesting cellular mRNA targets for KSHV miRNA include the transcripts encoding p21, a downstream effector of the p53 pathway that induces cell cycle arrest (Gottwein and Cullen 2010). Endogenous levels of the viral miRNA miR-K1 were found to effectively block p21 expression upon activation of p53 and prevent the resultant inhibition of cell growth. This result has obvious implications for the transforming potential of KSHV. Three KSHV miRNAs—miR-K5, miR-K9, and miR-K10—have been reported to down-regulate the cellular factor BCLAF1, a protein that has proapoptotic activity (Ziegelbauer et al. 2009). Ziegelbauer et al. (2009) also reported that BCLAF1 inhibits lytic reactivation of KSHV, implying that these miRNAs may predispose latently KSHV-infected cells to reactivation. Unfortunately, the role of KSHV miRNAs in either promoting or inhibiting lytic reactivation in culture remains unresolved, with published reports supporting both possible activities (Grundhoff and Sullivan 2011). Other reported targets down-regulated by KSHV miRNAs include MAF, which plays a role in regulating the differentiation of lymphatic endothelial cells, a known target for KSHV infection (Hansen et al. 2010), as well as TWEAKR, a cellular proapoptotic protein (Abend et al. 2010), and thrombospondin 1 (THBS1), which is thought to function as a tumor suppressor by inhibiting tumor angiogenesis (Samols et al. 2007). These results suggest that KSHV, like EBV, uses viral miRNAs to promote the growth and viability of latently infected cells, at least in part by inhibiting innate immune responses. Preliminary data obtained using the novel photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) technique (Hafner et al. 2010) to globally recover and sequence mRNA target sites occupied by RISCs programmed by virally encoded miRNAs in fact suggest that the miRNAs encoded by EBV and KSHV may repress a highly overlapping set of cellular mRNAs, despite the lack of sequence homology between these viral miRNAs (E Gottwein and BR Cullen, unpubl.).
Conclusions
While efforts to identify virally encoded miRNAs are approaching completion, our understanding of how viruses interact with cellular miRNAs and how viral miRNAs promote viral replication and pathogenesis remains quite rudimentary. This will likely change quite rapidly, as efforts to phenotypically characterize viral miRNA mutants increase and novel, highly powerful CLIP techniques are used to globally identify all of the mRNA targets that are bound by the cellular or viral miRNAs expressed in infected cells. Of course, as this will likely identify >100 mRNA targets for each miRNA, we will still be far from reaching a full understanding of how miRNAs function, especially if their phenotypes require the coordinate down-regulation of several mRNAs simultaneously and/or the combined action of several viral and/or cellular miRNAs acting in concert. However, it is already clear that a full appreciation of how viruses subvert the cellular biosynthetic machinery while avoiding elimination by host immune responses is impossible, especially in the case of herpesviruses, if the actions of viral miRNAs are not taken into account.
Acknowledgments
Research described in this review was supported by National Institutes of Health grants R01-AI067968, R21-AI088327, R01-DA030086, and R56-AI083644.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.17352611.
References
- Abend JR, Uldrick T, Ziegelbauer JM 2010. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol 84: 12139–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol 79: 9556–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res 36: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22: 607–619 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B 2003. Human immunodeficiency virus type 1 escape from RNA interference. J Virol 77: 11531–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 37: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol 81: 13761–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, Marcinowski L, Dölken L, Pfeffer S 2010. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol 80: 2234–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2: e23 doi: 10.1371.journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK 2008. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 82: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawood R, Chen HH, Carroll F, Bazan-Peregrino M, van Rooijen N, Seymour LW 2009. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog 5: e1000440 doi: 10.1371/journal.ppat.1000440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328: 1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 205: 2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HL, Mischo HE, Steitz JA 2004. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol Cell Biol 24: 4522–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA 2009. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol 83: 2357–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM 2009. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2004. Transcription and processing of human microRNA precursors. Mol Cell 16: 861–865 [DOI] [PubMed] [Google Scholar]
- Cullen BR 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol 7: 563–567 [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Dölken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol 81: 13771–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, et al. 2010a. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog 6: e1001150 doi: 10.1371/journal.ppat.1001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M, et al. 2010b. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human γ-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7: 324–334 [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR, Delecluse HJ 2011. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog 7: e1001294 doi: 10.1371/journal.ppat.1001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M 2008. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-κB pathway. Nucleic Acids Res 36: 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol 84: 5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi J-A, Braich R, Manoharan M, Soutschek J, Ohler U, et al. 2007. A viral microRNA functions as an ortholog of cellular miR-155. Nature 450: 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol 79: 12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 3: e163 doi: 10.1371/journal.ppat.0030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog 6: e1000967 doi: 10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS 2011. Virus-encoded microRNAs. Virology 411: 325–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, et al. 2010. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev 24: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al. 2011. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9: 58–69 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Jopling CL, Schutz S, Sarnow P 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol 84: 4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Greiner S, Russell SJ 2008. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med 14: 1278–1283 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol 12: 513–519 [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560 [DOI] [PubMed] [Google Scholar]
- Lin J, Cullen BR 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 81: 12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Sullivan CS 2011. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci 108: 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS 2010. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA 16: 1540–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol 84: 11670–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci 104: 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol 78: 12868–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Marquitz AR, Mathur A, Nam CS, Raab-Traub N 2011. The Epstein-Barr virus BART microRNAs target the pro-apoptotic protein Bim. Virology 412: 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna DJ, McDade SS, Patel D, McCance DJ 2010. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol 84: 10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, et al. 2010. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res 38: 6234–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melar-New M, Laimins LA 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol 84: 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighty KK, Laimins LA 2011. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J Virol 85: 8863–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, et al. 2008. Sequence conservation and differential expression of Marek's disease virus microRNAs. J Virol 82: 12213–12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci 105: 5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5: 376–385 [DOI] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, et al. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR 2004. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P 2008. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 36: 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734–736 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Plaisance-Bonstaff K, Renne R 2011. Viral miRNAs. Methods Mol Biol 721: 43–66 [DOI] [PubMed] [Google Scholar]
- Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B 2009. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology 386: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW 2010. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol 84: 10344–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Steitz JA 2010. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J Virol 84: 5148–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouha H, Thurner C, Mandl CW 2010. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res 38: 8328–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 3: e65 doi: 10.1371/journal.ppat.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi VR, Steel LF 2011. A re-examination of global suppression of RNA interference by HIV-1. PLoS ONE 6: e17246 doi: 10.1371/journal.pone.0017246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS 2008. Evolutionarily conserved function of a viral microRNA. J Virol 82: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog 6: e1001063 doi: 10.1371/journal/ppat.1001063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR 2010. Viruses, microRNAs, and host interactions. Annu Rev Microbiol 64: 123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 81: 12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D 2009. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology 387: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci 105: 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Patel A, Krause PR 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol 83: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Margolis TP, Krause PR 2011. Herpes simplex virus-2 miR-H6 is a novel LAT-associated microRNA, but reduction of its expression does not influence viral latency establishment or recurrence phenotype. J Virol 85: 4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol 84: 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR 2009. Analysis of human α-herpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83: 10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Strelow LI, Wong SW, Cullen BR 2010. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology 405: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR 2010. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci 107: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res 33: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res 68: 1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Kieff E 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol 81: 9967–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC 2011. Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J Virol 82: 4007–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK 2008. MicroRNA-155 is an Epstein-Barr Virus induced gene that modulates Epstein Barr virus regulated gene expression pathways. J Virol 82: 5295–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR 2004. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 32: 4776–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, Kgosana L, Wang X, Nair V 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol 83: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V 2011. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog 7: e1001305 doi: 10.1371/journal.ppat.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]