Rinn and colleagues present an integrative approach to define a reference catalog of >8000 human lincRNAs. This annotation catalog provides a comprehensive mapping and analysis of human lincRNAs to date, and is important for characterizing long noncoding RNAs into different classes based on various properties.
Keywords: long noncoding RNAs, RNA sequencing, lincRNAs
Abstract
Large intergenic noncoding RNAs (lincRNAs) are emerging as key regulators of diverse cellular processes. Determining the function of individual lincRNAs remains a challenge. Recent advances in RNA sequencing (RNA-seq) and computational methods allow for an unprecedented analysis of such transcripts. Here, we present an integrative approach to define a reference catalog of >8000 human lincRNAs. Our catalog unifies previously existing annotation sources with transcripts we assembled from RNA-seq data collected from ∼4 billion RNA-seq reads across 24 tissues and cell types. We characterize each lincRNA by a panorama of >30 properties, including sequence, structural, transcriptional, and orthology features. We found that lincRNA expression is strikingly tissue-specific compared with coding genes, and that lincRNAs are typically coexpressed with their neighboring genes, albeit to an extent similar to that of pairs of neighboring protein-coding genes. We distinguish an additional subset of transcripts that have high evolutionary conservation but may include short ORFs and may serve as either lincRNAs or small peptides. Our integrated, comprehensive, yet conservative reference catalog of human lincRNAs reveals the global properties of lincRNAs and will facilitate experimental studies and further functional classification of these genes.
A few dozen long noncoding RNAs (lncRNA) are known to play important regulatory roles in diverse processes, such as X inactivation (XIST) (Zhao et al. 2008), imprinting (H19 and KCNQ1OT1) (Leighton et al. 1995; Pandey et al. 2008), and development (HOTAIR and COLDAIR) (Rinn et al. 2007; Heo and Sung 2011). Recent genomic studies have shown that a substantial portion of the mammalian genome may be transcribed (Carninci et al. 2005), suggesting the presence of many more noncoding transcripts and spurring efforts to catalog them (Carninci et al. 2005; Harrow et al. 2006) using data collected with tiling microarrays (Bertone et al. 2004; Kapranov et al. 2007), shotgun sequencing of expressed sequence tags (ESTs) and cloned cDNA (Carninci et al. 2005; Birney et al. 2007), and maps of histone modification patterns (Guttman et al. 2009). In particular, recent studies have focused on large intergenic noncoding RNAs (lincRNAs) (Ponjavic et al. 2007; Guttman et al. 2009; Khalil et al. 2009; Orom et al. 2010), which do not overlap annotated protein-coding regions, as this facilitates experimental manipulation and computational analysis.
Recent work has suggested various functions and molecular mechanisms for lincRNAs (Mercer et al. 2009; Ponting et al. 2009), including the regulation of epigenetic marks and gene expression (Rinn et al. 2007; Nagano et al. 2008; Pandey et al. 2008; Zhao et al. 2008, 2010; Khalil et al. 2009; Koziol and Rinn 2010). Other studies have inferred and tested the functional role of lincRNAs in processes such as pluripotency and p53 response pathways by associating the expression of lincRNAs with those of protein-coding genes (Guttman et al. 2009; Huarte et al. 2010; Loewer et al. 2010; Hung et al. 2011). More globally, a recent comprehensive screen identified dozens of lincRNAs required to maintain pluripotency and suggested that these lincRNAs work in trans (Guttman et al. 2011). Another class of “enhancer RNAs” may either be by-products of transcription (De Santa et al. 2010; Kim et al. 2010) or serve to activate gene expression in cis (Orom et al. 2010; Wang et al. 2011). Despite these intriguing studies of individual lincRNAs, generalizing these findings to thousands of lincRNAs remains a substantial challenge. Collectively, lincRNAs are likely to reflect different families with distinct roles.
A first requirement toward functional categorization is a systematic catalog of lincRNA transcripts and their expression across tissues. In practice, however, researchers studying human lincRNAs are faced with an excessive set of noncoding transcripts of varying or unknown reliability that may not be well defined (Khalil et al. 2009) and have little or no expression data (Harrow et al. 2006), or with very small sets of experimentally validated ones (Amaral et al. 2010). Transcripts in current annotations of the human transcriptome from the GENCODE/HAVANA (Harrow et al. 2006) or the University of California at Santa Cruz (UCSC) Genome Browser (Hsu et al. 2006) are valuable resources, but it is hard to evaluate their biological characteristics in the absence of expression levels and further processing.
Recent advances in RNA sequencing (RNA-seq) (Mortazavi et al. 2008) and computational methods for transcriptome reconstruction (Guttman et al. 2010; Trapnell et al. 2010; Garber et al. 2011) now provide an opportunity to comprehensively annotate and characterize lincRNA transcripts. Indeed, an initial application of this approach in three mouse cell types characterized the gene structure of more than a thousand mouse lincRNAs, most of which were not previously identified (Guttman et al. 2010).
Here, we present an integrative approach to define a reference set of lincRNAs that unifies existing annotation sources with transcripts reconstructed from >4 billion RNA-seq reads collected across 24 human tissues and cell types. We developed a conservative, broadly applicable pipeline to identify transcripts that are sufficiently expressed and have a negligible potential to encode proteins. We identified 8195 putative lincRNAs, of which 4662 (57%) form a “stringent” set. We characterized each lincRNA in the catalog by a panorama of structural, sequence, and expression features as an initial step toward fine categorization.
We used these features to test some of the proposed roles and characteristics of lincRNAs in a global and systematic way. For example, we found that lincRNAs—at all expression levels—are expressed in a highly tissue-specific manner—much more so than protein-coding genes. We observed no significant enrichment of correlated coexpression between lincRNAs and their neighboring genes beyond that expected for any two neighboring protein-coding genes. We identified expressed orthologous transcripts in another vertebrate species for 993 (12%) human lincRNAs. An additional set of 2305 other transcripts with high evolutionary conservation but ambiguous coding potential may function as noncoding RNAs or as small peptides. Finally, we highlight 414 lincRNAs that reside within intergenic regions previously associated with specific diseases/traits by genome wide association studies (GWAS) as candidates for future disease-focused studies. Our reference catalog will facilitate future experimental and computational studies to uncover lincRNA functions.
Results
A computational approach for comprehensive annotation of lincRNAs
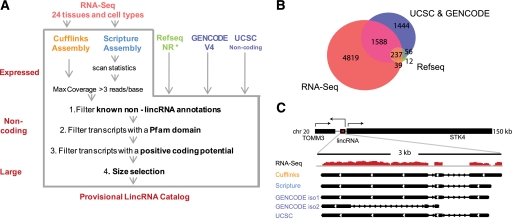
To comprehensively identify human lincRNAs, we developed a computational approach that integrates RNA-seq data with available annotation resources (Fig. 1A) and consists of four key steps (see the Materials and Methods): (1) transcriptome reconstruction of each sample from RNA-seq data using two transcript assemblers: Cufflinks (Trapnell et al. 2010), and Scripture (Guttman et al. 2010); (2) compilation of all noncoding and unclassified transcripts previously annotated; (3) integration of RNA-seq reconstructions with all annotation resources, using Cuffcompare (Trapnell et al. 2010) to determine a unique set of isoforms for each transcript locus; and (4) processing of the collected transcripts to identify lincRNAs, defined as transcripts that are reliably expressed, large, multiexonic, noncoding, and intergenic.
Figure 1.
lincRNA catalog generation. (A) An integrative computational pipeline to map, reconstruct, and determine the coding potential of lincRNAs based on known annotations and computational methods, and its application to human lincRNAs. The pipeline takes as input RNA-seq data (top, red) and existing annotation sources (top) (RefSeq NR, Gencode, and UCSC annotation for humans). RNA-seq data are assembled by two assemblers: Cufflinks (gold) and Scripture (blue). Transcripts from all inputs are filtered by known annotations, presence of a Pfam domain, and positive coding potential. Transcripts annotated by RefSeq NR (*) were not filtered by the Pfam domain scan and the coding potential score. Finally, only multiexonic transcripts >200 base pairs (bp) are retained. (B) The number of lincRNA loci identified and their overlap with other annotation sources. The Venn diagram shows the overlap between transcripts from RNA-seq assembly (red), GENCODE and UCSC (purple), and RefSeq (green). (C) A representative example of a noncoding transcript that was reconstructed by Cufflinks and Scripture and was also curated in GENCODE and UCSC. (Top) The human genomic locus of the human lincRNAs (red) and its protein-coding neighbors. (Black, arrowhead) Direction of transcription. (Bottom) Magnified view of the lincRNA locus showing the coverage of RNA-seq reads from the testes (red) and the transcripts identified by each source (black). (iso) Isoform.
There are two main challenges in applying this integrative approach to annotate lincRNA gene loci: (1) distinguishing lowly expressed lincRNAs (Guttman et al. 2010) from the tens of thousands of lowly expressed, single-exon, unreliable fragments assembled from RNA-seq; and (2) distinguishing novel transcripts encoding proteins or short peptides from bona fide noncoding ones. To address the first challenge, we removed unreliable lowly expressed transcripts using a learned read coverage threshold (Supplemental Material) and focus only on multiexonic transcripts. To address the second challenge, we evaluated the coding potential of each of the remaining putative lincRNAs using two methods. First, we removed any putative ORFs that are evolutionarily constrained to preserve synonymous amino acid content, as reflected by a positive phylogenetic codon substitution frequency (PhyloCSF) metric (Lin et al. 2011) calculated for each locus across 29 mammals (Supplemental Material). Second, we scanned each transcript in all three reading frames to exclude transcripts that encode any of the 31,912 protein domains cataloged in the protein family database Pfam (Finn et al. 2010).
An annotated human lincRNA catalog
To generate a human lincRNA catalog, we applied our pipeline to polyadenylated RNA-seq data collected from 24 human tissues and cell lines. These included both single- and paired-end reads that are 50 or 75 bases long, sequenced on Illumina platforms (∼4 billion reads total; ∼175 million reads per sample on average) (Materials and Methods). We integrated those with annotations from RefSeq (Pruitt et al. 2002), the UCSC Genome Browser (Hsu et al. 2006), and GENCODE (version 4) (Harrow et al. 2006) that were processed through our pipeline. We eliminated all annotated non-lincRNA transcripts (e.g., annotated protein-coding genes, microRNAs, tRNAs, and pseudogenes).
The initial catalog consists of a provisional set of 8195 intergenic transcripts (Fig. 1B). Although many of the previously annotated transcripts are also captured by the ones assembled from the sequencing data (1864 lincRNAs identified by both) (Fig. 1B,C), most (4819) novel lincRNAs were only identified using RNA-seq. Based on the three samples for which we had two biological replicates (brain, testes, and lung fibroblasts), the reconstructed transcripts are highly reproducible: 70%–80% of assembled transcripts in the lower coverage replicate are also assembled in the higher coverage replicate (Supplemental Table 1; Supplemental Material).
Despite the high correspondence between protein-coding transcripts reconstructed by Cufflinks and Scripture (∼85% of coding genes) (Supplemental Material; Supplemental Fig. 1A), there were larger differences between the noncoding transcripts assembled by the two methods, due to the differences in how each assembler reconstructs low-abundance transcripts (∼46% of the putative lincRNAs were identified by only one source) (Supplemental Fig. 1B). This is comparable with previously observed discrepancies in reconstruction of lowly expressed protein-coding genes (Garber et al. 2011) and is handled below.
We annotated each putative lincRNA in the provisional catalog with a comprehensive “profile” listing dozens of traits, such as its chromatin state, maximal expression level, proximity to coding genes, and evolutionary conservation (Materials and Methods, Supplemental Data Sets 1, 2). Below, we use these features to define particular criteria by which we focus our analysis. Future users may leverage the annotated catalog through criteria of their choosing.
A stringent set of 4662 human lincRNAs
We defined a stringent lincRNA set that includes those loci for which at least one lincRNA isoform was reconstructed in at least two different tissues or by two assemblers in the same tissue (Supplemental Material). This leverages the unique benefits of each assembler, while in principle removing transcripts with insufficient coverage. The stringent set includes 4662 lincRNA loci (14,353 transcripts), 2798 of which (∼60%) were not identified by RefSeq, UCSC, and GENCODE. We focused on the characteristics of this stringent set.
lincRNAs are alternatively spliced and preferentially proximal to developmental regulators
We characterized the basic features of lincRNAs, comparing them with protein-coding genes when appropriate. First, the size of lincRNAs is smaller than that of protein-coding transcripts, and they have fewer exons (on average, 2.9 exons and a transcript length of ∼1 kb for lincRNAs vs. 10.7 exons and ∼2.9 kb for protein-coding transcripts) (Supplemental Fig. 2A,B). Notably, we may underestimate the length and exon number of lincRNAs, since their lower abundance may result in incomplete assembly. Second, lincRNAs are alternatively spliced (on average, ∼2.3 isoforms per lincRNA locus) (Supplemental Fig. 2C). Third, lincRNA loci are located from a few bases to >3 Mb from a protein-coding locus, with 28% within 10 kb of their coding neighbor (median = ∼40 kb) (Supplemental Fig. 2D). Finally, protein-coding genes proximal (≤10 kb) to lincRNAs are enriched for those associated with development and transcriptional regulation (e.g., GATA2, GZF1, and NEUROG2 all have lincRNA neighbors) (Supplemental Fig. 3), consistent with previous reports (Guttman et al. 2009; Ponjavic et al. 2009).
Many lincRNAs are characterized by K4–K36 domains
We next explored the chromatin features of lincRNA loci as reflected in chromatin state maps from the nine ENCODE cell lines and other cells (Khalil et al. 2009; Ernst et al. 2011). We examined each locus for the presence of a “K4–K36 domain,” a chromatin signature of actively transcribed genes that we previously used to identify lincRNAs (Guttman et al. 2009). This domain consists of histone 3 Lys 4 trimethylation (H3K4me3) at the promoter followed by histone 3 Lys 36 trimethylation (H3K36me3) along the transcribed region. Despite the lack of paired matched samples of histone modifications and RNA-seq, 24% of the lincRNAs in our catalog have previously defined chromatin K4–K36 domains and ∼40% have such domains when using less stringent criteria (with the remaining exhibiting partial signatures) (Supplemental Fig. 4; Supplemental Material).
lincRNA genes are no more likely to overlap enhancers than protein-coding genes
Recent studies reported short transcripts derived from enhancer elements, termed eRNAs, that are most likely not polyadenylated (Kim et al. 2010). While this suggests that eRNAs and lincRNAs come from different classes, it is possible that longer polyadenylated transcripts may also be derived from enhancer elements and hence be related to eRNAs. To test this possibility, we examined the overlap between lincRNAs' exons and two recent annotations of human enhancers based on genome-wide chromatin state maps. Twenty-seven percent of our lincRNAs and 44% of coding genes overlap 111,362 genomic regions previously suggested to function as enhancers (Ernst et al. 2011) in nine ENCODE cell lines (each overlap, P < 0.001, permutation test) (Supplemental Material). When considering a more stringent subset of regions that are more likely to function only as enhancers (Supplemental Material), ∼10% and 14% of lincRNAs and coding genes, respectively, overlap such regions (both P < 0.001). Both lincRNAs and protein-coding genes have even lower overlap (both <3%, P < 0.001) with an enhancer set from human embryonic stem (ES) cells (Rada-Iglesias et al. 2010), possibly due to the lack of biological correspondence between the cell types and the tissue-specific nature of both lincRNAs and enhancers. Notably, this low overlap persists even when comparing more closely matched samples. Thus, only 15% of lincRNAs defined in mouse ES cells (Guttman et al. 2010) overlap enhancers defined in mouse ES cells (Zentner et al. 2011) (Supplemental Methods), and <1% of lincRNA defined in mouse neuronal progenitor cells (Guttman et al. 2010) overlap enhancer elements that express eRNAs in mouse cortical neurons (Supplemental Material; Kim et al. 2010). Taken together, these data suggest that lincRNAs and eRNAs represent different subtypes of lncRNAs.
lincRNAs are expressed in a more tissue-specific manner than protein-coding genes
The maximal expression levels of lincRNAs are lower than those of protein-coding genes across the 24 samples (Fig. 2A), with a ∼10-fold lower median maximal expression level (expression estimated with Cufflinks) (Fig. 2B; Materials and Methods; Trapnell et al. 2010). Importantly, lincRNAs identified by RefSeq annotations were similarly lowly expressed relative to coding genes (∼10 fold lower) (Supplemental Fig. 5). These lower expression levels are consistent with previous reports (Ravasi et al. 2006; Guttman et al. 2010), suggesting a general property of lincRNAs.
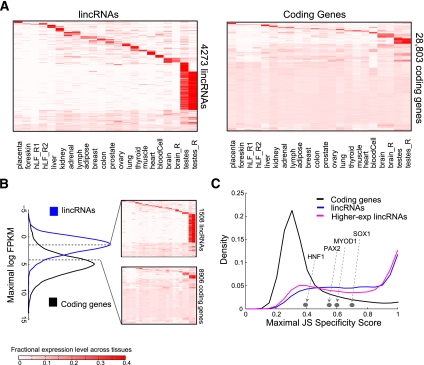
Figure 2.
Tissue specificity of lincRNAs and coding genes. (A) Abundance of 4273 lincRNA (rows, left panel) and 28,803 protein-coding genes (rows, right panel) across tissues (columns). Rows and columns are ordered based on a k-means clustering of lincRNAs and protein-coding genes. Color intensity represents the fractional density across the row of log-normalized FPKM counts as estimated by Cufflinks (saturating <4% of the top normalized expression values) (Supplemental Methods). (B) lincRNAs are more lowly expressed than protein-coding genes. Maximal expression abundance (log2-normalized FPKM counts as estimated by Cufflinks) of each lincRNA (left panel, blue) and coding (left panel, black) transcript across tissues. The right panel shows the expression levels of 1508 lincRNAs (top right) and 8906 coding genes (bottom right) that have a maximal expression level within the range bounded by the dashed segments in the left panel ([1.6–4.3] log2 FPKM) (see Supplemental Material). Heat maps are clustered and visualized as in A. (C) Tissue-specific expression. Shown are distributions of maximal tissue specificity scores calculated for each transcript across the tissues from the data in A for coding genes (black), lincRNAs (blue), and the 1508 highly expressed lincRNAs (pink; as in B). Examples of the tissue specificity score of coding genes with known tissue-specific patterns are marked by gray dots.
The vast majority of lincRNAs exhibit tissue-specific expression patterns—more so than protein-coding genes—based on unsupervised clustering of expression profiles (Fig. 2A). We further calculated a tissue specificity score for each transcript using an entropy-based metric that relies on Jensen-Shannon (JS) divergence (Materials and Methods). This specificity metric (ranging from 0 to 1) quantifies the similarity between a transcript's expression pattern across tissues and another predefined pattern that represents the extreme case in which a transcript is expressed only in one tissue. Thus, a perfect tissue-specific pattern will be scored as JS = 1.
Based on this measure, the majority of lincRNAs (78%) are tissue-specific, relative to only ∼19% of coding genes (P < 10−300, Fisher exact test) (Fig. 2C; Supplemental Fig. 6). These differences are not the result of the low expression levels of lincRNAs and hold true for lincRNAs and protein-coding genes expressed at similar levels (Fig. 2B,C; Supplemental Fig. 6). This was particularly true for the 35% of more highly expressed lincRNAs (and comparably expressed protein-coding genes, each with a maximal expression level of 3–20 FPKM [fragments per kilobase of exons per million fragments mapped]). Thus, lincRNAs exhibit more tissue specificity than protein-coding genes at different expression ranges.
Approximately a third of our lincRNAs are specific to testes. Very few (<2%) of those overlap with a previously defined set of testes-specific small piRNAs (∼30 nucleotides long) (Girard et al. 2006). Thus, testes-specific lincRNAs may define a new class of RNAs in this organ. Testes-specific lincRNAs do not bias the global transcriptional characteristics above: lincRNAs that are not testes-specific are also lowly expressed and tissue-specific (presenting a qualitatively similar distribution with only moderately reduced tissue specificity scores) (Supplemental Figs. 5, 6A).
Finally, we predicted putative functions for our lincRNAs based on the known functions of protein-coding genes with similar expression patterns. We clustered lincRNAs and protein-coding genes using k-means clustering with the tissue specificity distance measure (Supplemental Material) and annotated each cluster with enriched functions of the protein-coding gene members. Clusters of tissue-specific lincRNAs and protein-coding genes are enriched for processes specific to that tissue or its differentiation (e.g., a liver-specific cluster is enriched with functional terms such as cholesterol and lipid transport and homeostasis) (Supplemental Fig. 7; Supplemental Data Sets 2, 3).
lincRNAs are coexpressed with neighboring coding genes at levels similar to those expected for any pair of chromosomal neighbors
The enrichment of specific gene functions in protein-coding genes neighboring lincRNAs and the presence of some pairs of neighboring lincRNA:protein-coding genes within expression clusters raise the hypothesis that such organization may be important for the regulatory function of lincRNAs. In particular, recent studies suggested that some lincRNAs may act in cis and affect the gene expression of their chromosomal neighborhood (Ponjavic et al. 2009; Orom et al. 2010).
One expectation from this hypothesis is that the expression of lincRNAs and their neighboring gene loci would be correlated across our samples. To test this hypothesis, we focused on the expression patterns of 1361 (28%) of our stringent lincRNAs that are located within 10 kb from a coding gene. Indeed, these lincRNAs and their coding neighbors were more correlated to each other than random gene pairs (P < 5 × 10−46, Kolomogorv-Smirnov [KS] test; P < 10−307, Student's t-test, effect size = 0.86) (Fig. 3A; Supplemental Material).
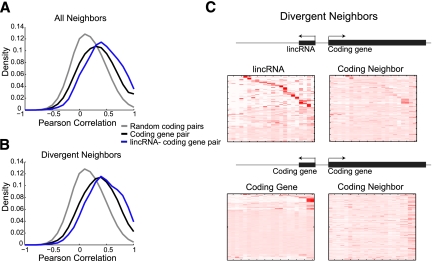
Figure 3.
Chromosomal domains of gene expression. (A) Correlation of expression patterns between pairs of neighboring genes. Shown are distributions of Pearson correlation coefficients in expression levels across the tissues in Figure 2A between either 6524 pairs of coding gene neighbors (black), 497 pairs of lincRNAs and their neighboring coding gene (blue), or 10,000 random pairs of protein-coding genes (gray; null model) (*). (B) Shown are distributions of Pearson correlation coefficients calculated as in A, but only for 223 pairs of divergently transcribed pairs of lincRNA and protein-coding gene (blue) or 1575 pairs of divergently transcribed protein-coding genes (*). (C) Expression patterns of pairs of divergently expressed genes. Shown are expression patterns (presented as in Fig. 2A) for pairs of divergently transcribed lincRNA (rows, top left) and protein-coding genes (rows, top right), or pairs of divergently transcribed protein-coding genes (rows, bottom left and right panels) (*). (*) Only lincRNAs that have spliced read support when maximally expressed and that are not testes-specific are presented (refer to Supplemental Material, “Estimating expression abundance,” for further details).
We must interpret this result with care, since the coexpression between a lincRNA and its protein-coding neighbor may result from either (1) a true cis effect of lincRNAs on its neighbor or (2) proximal transcriptional activity in the surrounding open chromatin (Ebisuya et al. 2008), since coexpression of chromosomal protein-coding gene neighbors was previously shown across eukaryotes (Cohen et al. 2000; Hurst et al. 2004). Supporting the second possibility, pairs of neighboring protein-coding genes were also more correlated to each other than random pairs (P < 3.4 × 10−159, KS test) (Fig. 3A). Furthermore, the correlation between lincRNA:protein-coding gene neighbors was only modestly higher than between protein-coding gene:protein-coding gene neighbors of a similar distance (effect size = 0.23, P < 4.3 × 10−7, KS test; P < 6.9 × 10−7, Student's t-test) (Fig. 3A).
To further distinguish between these two possibilities, we focused on those protein-coding genes that had a lincRNA neighbor on one side and a coding neighbor on the other side, and used a paired test to compare the correlation between each protein-coding gene and its lincRNA neighbor with that between the same protein-coding gene and its protein-coding gene neighbor. This paired comparison showed a weak opposite trend, where pairs of coding gene neighbors are slightly more correlated to each other than neighboring lincRNA:protein-coding gene pairs (P < 0.001 paired Student's t-test; effect size = 0.23), thus favoring option 2, an effect of gene proximity.
Taken together, this analysis suggests that, overall, lincRNAs are not more correlated to their protein-coding gene neighbors than expected for a pair of neighboring protein-coding gene loci. Yet, the ultimate test of cis- or trans-regulatory mechanisms for lincRNAs requires experimental gain-of-function or loss-of-function data.
Divergently transcribed lincRNAs
Unstable, likely noncoding, transcripts can also be derived from divergent (bidirectional) transcription in both yeast and mammals (Core et al. 2008; Preker et al. 2008; Seila et al. 2008). These may be either by-products of chromatin remodeling and recruitment of the transcription machinery to the neighboring gene's promoter or functional transcripts (Kanhere et al. 2010). Due to limited read length and computational methods, previous studies did not determine whether these transcripts are spliced. Interestingly, several functionally studied lincRNAs, including Tug1 (Young et al. 2005), HOTAIR (Rinn et al. 2007), and HOTTIP (Wang et al. 2011), are divergent transcripts. We therefore hypothesized that other divergently transcribed transcripts may be spliced and polyadenylated lincRNAs.
Indeed, 588 (∼13%) of our stringent lincRNAs are spliced transcripts divergently transcribed within 10 kb of a coding gene promoter, with a majority (∼65%) that initiate within 1 kb of a coding gene's annotated transcription start site (Supplemental Fig. 8). Furthermore, ∼35% of the 588 pairs share a H3K4me3 domain (a hallmark of active promoters), based on the ENCODE chromatin state maps (Supplemental Material), although we cannot definitively determine whether these divergently encoded pairs are also divergently transcribed from the same promoter. These divergent coding gene neighbors are enriched for developmental and metabolic processes (Supplemental Fig. 3B). Focusing on the 68% that are spliced in the tissue where they are maximally transcribed (Supplemental Material, “Estimating expression abundance”), there is only a slightly higher correlation between divergent lincRNAs and neighboring coding genes than for divergent coding gene pairs (effect size = 0.27, P < 0.008 KS test; P < 0.009, Student's t-test) (Fig. 3B). Furthermore, while ∼49% of the divergently transcribed lincRNAs are tissue-specific, for approximately half of those, the neighboring gene is ubiquitously expressed (Fig. 3C). Thus, although there are clearly bidirectionally transcribed, spliced lincRNAs in our catalog, we found no clear additional distinguishing features for this set.
Expressed syntenic orthologs of human lincRNAs in mammals and vertebrates
We and others have previously reported evidence for purifying selection at different sets of mammalian lincRNAs (Ponjavic et al. 2007; Guttman et al. 2009; Orom et al. 2010). A recent study has also identified expressed orthologs of a few highly conserved and brain-expressed mouse lncRNAs in species as distant as opossums and chickens (Chodroff et al. 2010). However, the number of human lincRNAs that have an orthologous, actively expressed, transcript in other species remains unknown.
To identify human lincRNAs with orthologous expressed transcripts in other species (supported by experimental evidence), we surveyed a catalog of mammalian and nonmammalian vertebrate transcripts that were syntenicaly mapped to the human genome by TransMap (Zhu et al. 2007), a cross-species mRNA alignment method. TransMap maps all known transcripts (e.g., full-length cDNAs and others in RefSeq or UCSC) and ESTs across vertebrate species using syntenic BLASTZ alignments (Schwartz et al. 2003) that use conserved gene order (synteny). Since EST coverage varies between species (Supplemental Table 2), TransMap can only provide a lower-bound estimate of orthologous transcripts.
Of the 8195 lincRNAs, 993 are syntenically paired with an orthologous transcript (Fig. 4A–D), comprising a trans-mapped lincRNA set (∼135 expected by random permutations) (Materials and Methods; Supplemental Material). Seven-hundred-two of the trans-mapped lincRNAs are in the stringent lincRNA set (∼15% of stringent lincRNAs). The majority (53%) of the trans-mapped lincRNAs was not previously annotated in the human transcriptome (GENCODE, RefSeq, or UCSC) (Supplemental Fig. 9A). Trans-mapped lincRNAs have tissue specificity and low expression, comparable with that of all other lincRNAs (Supplemental Figs. 6A, 9B,C). Fifty-nine percent of the trans-mapped lincRNAs were mapped to annotated transcripts that had evidence beyond ESTs. Supporting our noncoding classification scheme, only 18% of the 641 lincRNAs with trans-mapped orthologous transcripts in mice were classified as coding in mice and only ∼11% have a positive PhyloCSF score (Materials and Methods; Supplemental Fig. 9A). Trans-mapped lincRNAs have orthologs in species from mice to fish, with closer species that have more transcriptome data showing more orthologs than distant ones (Fig. 4D).
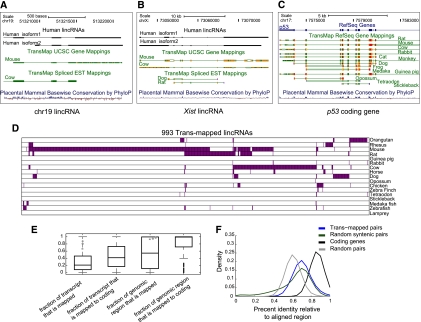
Figure 4.
Orthologous transcripts of human lincRNAs in mammals and other vertebrates. (A) A human lincRNA with syntenic trans-map mappings to mice and cows. Shown are UCSC browser (Kent et al. 2002) tracks showing two isoforms of the human lincRNA (black, top tracks), the mouse and cow transcripts (green, middle tracks) that were trans-mapped to their human locus, and the base-wise conservation calculated by PhyloP at this locus (red–blue, bottom track). (B) Syntenic trans-mapping to XIST. Tracks presented as in A. (C) Syntenic trans-mapping to p53. (D) Species distribution of 993 human lincRNAs with trans-mapped orthologs (columns) and the species in which the trans-mapped transcripts were found (rows, purple). (E) Characteristics of trans-mapping to human lincRNAs. Box plots of the fraction of the human lincRNA transcript that is aligned to an ortholog (first and second boxes) and the fraction of the lincRNA genomic locus covered by the syntenic mapping of the ortholog (third and fourth boxes) for all trans-mapped lincRNAs (first and third boxes) or only for those lincRNAs that were mapped to mouse coding transcripts (second and fourth boxes). The gray square, star, and circle represent XIST, HOTAIR, and the lincRNA shown in A, respectively. (F) Distribution of the percentage of identical bases across the FSA (Bradley et al. 2009) pairwise alignments between human and mouse trans-mapped transcript pairs. (Blue) lincRNAs and their mouse orthologs; (black) human coding genes and their mouse orthologs; (green) randomly selected 1-kb human and mouse syntenic blocks; (gray) random pairing of human lincRNAs and mouse transcripts (from the set marked in blue). All statistics presented in this figure were calculated at the locus level (i.e., each lincRNA loci was accounted for once, rather than accounting for all of its isoforms).
Orthologous lincRNAs exhibit modest sequence homology
We evaluated the degree of sequence similarity between the trans-mapped transcripts. We measured the portion of each lincRNA transcript's length that is aligned to the orthologous transcript. The majority of trans-mapped lincRNAs are only moderately spanned by an orthologous mapped transcript (a median of 21% and 56% of their transcript or genomic locus, respectively, aligned) (Fig. 4E). In loci where lincRNAs are trans-mapped to mouse coding transcripts, a larger portion of the human locus but a smaller portion of the mouse transcript aligns between the species (Fig. 4E; Supplemental Fig. 10A,B). This may be due to either cryptic small peptides in the human transcript or the evolution of a noncoding transcript from a coding one. The available data are insufficient to distinguish between these hypotheses, which can be tested as paired cross-species RNA-seq samples are collected.
We next compared the fraction of identical bases aligned between the lincRNAs and their orthologs with that of random sequence pairs, randomly selected syntenic blocks, or orthologous coding genes. Trans-mapped lincRNAs and their orthologous transcripts show sequence identity similar to that of randomly selected syntenic blocks, which is lower than pairs of orthologous protein-coding genes and higher than for random pairs of genomic regions of similar size (Fig. 4F; Supplemental Fig. 10C,D; Materials and Methods). With only 34% of the human genome syntenically mapped to the mouse genome (Kent et al. 2003), the resemblance of trans-mapped lincRNAs to random syntenic blocks still implies evolutionary constraint to preserve sequence elements.
Novel transcripts with potential coding capacity
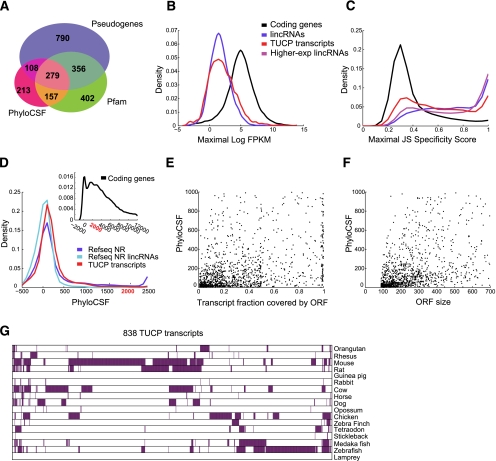
While our stringent lincRNA classification strategy focused on noncoding transcripts, we also characterized 2305 transcripts that were excluded by our coding potential criteria (a Pfam domain, a positive PhyloCSF score, or previously annotated as pseudogenes) and termed them the transcripts of uncertain coding potential (TUCP) set (Supplemental Material). These may include lincRNAs as well as other transcripts. The majority (1533; ∼66%) was previously annotated as pseudogenes that, due to our focus on multiexonic transcripts, are probably not retrotransposed, spliced mRNAs that were integrated back to the genome (Fig. 5A). Similar to the stringent set, TUCP transcripts are expressed at lower and more tissue-specific patterns than protein-coding genes (Fig. 5B,C).
Figure 5.
Novel transcripts with potential coding capacity. (A) Characteristics of TUCP transcripts. Shown is a Venn diagram of the 2305 TUCP set transcripts annotated as pseudogenes (purple), containing a Pfam domain (green), having a PhyloCSF score higher than the pipelines set criteria (pink), or combinations thereof. (B) Expression levels of TUCP transcripts. Shown are distributions of maximal expression abundance (log-normalized FPKM counts as estimated by Cufflinks) in TUCP (red), stringent set lincRNA (blue), and coding (black) transcripts. (C) Tissue specificity of TUCP transcripts. Shown are distributions of maximal tissue specificity scores calculated for each transcript in the TUCP set (red), stringent lincRNA set (blue), coding (black), and higher-expressed lincRNAs (magenta) (transcripts defined as in Fig. 2C). (D) PhyloCSF scores of TUCP transcripts. Shown is the distribution of PhyloCSF scores of the TUCP transcripts (red), all noncoding genes in RefSeq (blue), or the subset of RefSeq classified as lincRNA by our pipeline (light blue). (Inset) The corresponding distribution for protein-coding genes that spans a much wider range of positive scores. (E,F) Putative ORFs in TUCP transcripts. Shown are scatter plots of the fraction of each transcript spanned by an ORF (E; X-axis) or of the ORF size (F, in nucleotides; X-axis) versus the PhyloCSF score of that ORF (Y-axis), for the 1404 TUCP transcripts that had a PhyloCSF score >0. (G) Orthologs for TUCP transcripts. Shown are 838 TUCP transcripts (columns) with trans-mapped orthologs and the species in which the trans-mapped transcripts were found (rows, purple).
The coding potential of most of these transcripts was very low compared with known coding genes, and only 32% (757) exceeded our PhyloCSF score criteria (Fig. 5A,D; Materials and Methods). The evolutionarily constrained ORFs in these transcripts are mostly short (51% are <70 amino acids long) and cover a small portion of the transcript (53% cover <25%) (Fig. 5E,F). Thus, some of these transcripts may encode small functional peptides (Kondo et al. 2010), whereas others may function as noncoding RNA.
TUCP transcripts are under stronger purifying selection than stringent lincRNAs. First, the exonic sequence in TUCP transcripts is more highly conserved than that of stringent lincRNAs (P < 10−116, effect size = 0.77) (Supplemental Fig. 11; Supplemental Material), even when excluding pseudogenes (Supplemental Fig. 11). Second, a larger fraction of them has a trans-mapped syntenic ortholog (∼36% [838], or ∼34% when excluding pseudogenes, compared with ∼15% [702] of stringent lincRNAs), and the syntenic alignments cover a slightly larger portion of the transcript (Supplemental Fig. 12). Third, 74% of the trans-mapped transcripts have an ortholog in a species more distant than mice (vs. 37% of the trans-mapped lincRNAs; ∼67% when excluding TUCP pseudogenes) (Fig. 5G).
lincRNAs in disease-associated regions
Although GWAS have identified thousands of common genetic variants related to specific traits or disease phenotypes, many of these variants (∼43%) (Hindorff et al. 2009) lie in intergenic regions and hence remain largely unexplained. We identified 414 lincRNAs from our comprehensive catalog (215 of the stringent set) that are located within 1146 disease- or trait-associated regions from the published GWAS catalog (Hindorff et al. 2009) that do not contain annotated coding genes (Supplemental Material; Supplemental Data Set 2). Notably, 30 and 81 of those lincRNAs overlap a common variant that was associated with a disease phenotype within their exon or their intron, respectively (both tag and imputed SNPs). Another 76 intergenic disease/trait regions overlap 84 TUCP transcripts (Supplemental Data Set 6).
The 215 stringent lincRNAs in these regions are typically expressed in a tissue-specific manner, which in a few cases directly corresponds to the tissue relevant to the associated disease (Supplemental Table 3). For example, a lincRNA positioned ∼3 kb downstream from a thyroid cancer-associated SNP in chromosome 14q13.3 (rs944289, odds ratio [OR] = 1.37; P = 2.0 × 10−9) (Gudmundsson et al. 2009) is strongly expressed specifically in the thyroid (∼5.4 log2 FPKM). The “tag SNP” and the proximal lincRNA are within a 249-kb linkage disequilibrium (LD) region that does not contain any known genes. rs944289 is ∼3.5 kb upstream of the transcription start site of the thyroid-specific lincRNA. rs944289[T] is predicted to be part of a binding motif for C/EBP-α (Supplemental Material; Supplemental Fig. 13) and may affect the lincRNA's expression. The LD region is ∼250 kb upstream of the gene NKX2-1 (TTF1), a transcription factor with a prominent role in thyroid development and a previously suggested candidate gene for this SNP association. The lincRNA may be an additional candidate, playing a role in thyroid-specific processes (possibly in coordination with the neighboring NKX2-1) and in thyroid cancer.
Discussion
We generated a reference catalog of 8195 human lincRNAs based on integrating RNA-seq data from 24 tissues and cell types with publicly available transcript annotations. Fifty-eight percent of the transcripts in our catalog are novel and are now identified for the first time using RNA-seq. We annotated each lincRNA with a broad range of structural, expression, and evolutionary features, shedding new light on their global properties and testing or generalizing previous hypotheses.
lincRNAs are remarkably tissue-specific compared with protein-coding genes. This possibility was previously raised (Mercer et al. 2009; Ponting et al. 2009) based on differential expression patterns in specific biological systems and has several implications. First, researchers studying a particular system may benefit from RNA-seq profiling followed by de novo assembly in that system. Second, it is consistent with the hypothesis that some lincRNAs interact with chromatin modulators and provide their target specificity. Third, it may indicate that lincRNAs could serve as specific fine-tuners. Fourth, the low level of lincRNA expression in a complex tissue such as the brain may in fact be a by-product of their expression in only a few specific cells. Future targeted perturbations of tissue-specific lincRNAs defined in our study may elucidate their role in tissue-specific processes.
Could many lincRNAs act as enhancer elements, promoting the transcription of their neighboring coding genes? Recent studies have demonstrated that several lincRNAs have enhancer-like functions (Orom et al. 2010; Wang et al. 2011). While our coexpression analysis is consistent with this notion, it is insufficient to suggest a global trend in which lincRNAs act as enhancers of their neighbors, since neighboring coding genes exhibit similar coexpression patterns. Further systematic perturbation studies in individual systems (as in Orom et al. 2010) may help assess the scope of this function. Notably, a very recent study that systematically perturbed 150 lincRNAs expressed in mouse ES cells suggested that lincRNAs primarily affect gene expression in trans (Guttman et al. 2011). Collectively, this suggests that some lincRNAs can work in cis, while others work in trans.
Nine-hundred-ninety-three lincRNAs have an orthologous transcript expressed from a syntenic region in another species, ∼50% of which were identified for the first time in this study. These lincRNAs had only moderate sequence identity and alignment to their orthologs. This moderate conservation may indicate the importance of transcription from a specific genomic location, the reduced selective pressure on the primary sequence of noncoding RNAs (Brown et al. 1992; Zhao et al. 2008), or the rapid evolution of new functions. It may also be due to alignment to orthologous ESTs that are incomplete transcripts. Our analysis was limited by available transcript data in other species, and will be enhanced as more transcriptomes are sequenced in other organisms.
TUCP intergenic transcripts did not pass our stringent classification criteria as lincRNA due to evidence of possible protein-coding potential. These transcripts have expression levels and tissue specificity similar to the stringent lincRNA set, but a significantly higher level of sequence conservation. Many could encode small peptides, similar to those that function in Drosophila melanogaster embryogenesis (Kondo et al. 2010). Another 1533 TUCP transcripts are classified as pseudogenes, and may represent pseudogenes that have evolved to function as noncoding regulatory agents. Ribosome profiling (Ingolia et al. 2009) and mass spectrometry of small peptides will help to resolve which of the TUCP transcripts are more likely to be coding.
Substantial progress has been recently made toward the essential goal of annotating long noncoding RNA loci. Our study presents an integrative yet conservative computational approach to mapping lincRNA transcripts that can be used for mapping new transcripts in other species. This is critical to overcome major barriers for future experiments (e.g., cloning, expression profiling, gain of function, and loss of function), as well as for the interpretation of genetic association studies. Indeed, 414 lincRNAs in our catalog stand out as located within intergenic regions associated with common disease. Future work will be necessary to identify RNA sequence domains that relate to function (Zhao et al. 2008; Kanhere et al. 2010), and to further classify lincRNAs into families. Our panorama of lincRNA properties will greatly advance these goals.
Materials and methods
RNA-seq data sets
We used two data sets of RNA-seq for transcriptome reconstruction. The first includes polyadenylated RNA samples from 16 tissues that were sequenced using Illumina HiSeq 2000 as part of the Human Body Map 2 project (235 million reads per sample on average) (Supplemental Table 4). The second data set included eight additional tissues and cell lines, each sequenced by the Illumina Genome Analyzer II (GAII) (54 million reads per sample on average) (Supplemental Table 4). The Human Body Map 2 data are accessible from ArrayExpress (accession no. E-MTAB-513; http://www.ebi.ac.uk/arrayexpress/browse.html?keywords=E-MTAB-513&expandefo=on).
The eight additional tissues and cell lines are available at Gene Expression Omnibus (GEO) (accession no. GSE30554) (see the Supplemental Material).
Publicly available annotations
All known annotations that were used for the analysis of this study are specified in Supplemental Table 5.
lincRNA classification pipeline
Once the transcriptome of each tissue sample was assembled (Supplemental Material), we further processed the assemblies and used Cuffcompare (Trapnell et al. 2010) to eliminate intron and polymerase run-on fragments surrounding all transcripts annotated by GENCODE 4. We then used Cuffcompare to generate a unique set of assembled isoforms from all processed tissue assemblies. Next, we ran the unique transcript set through the following filters: (1) size selection, (2) minimal read coverage threshold, (3) filter of known non-lincRNAs annotations, (4) positive coding potential threshold, (5) known protein domain filter, and (6) intergenic classification (see the Supplemental Material).
To derive a unique set of lincRNAs that includes previous annotations, we used Cuffcompare to integrate the RNA-seq-derived lincRNAs with the predetermined set of lincRNAs previously annotated by RefSeq, UCSC, or GENCODE 4. The publicly available lincRNA sets were derived by running specific steps of our lincRNA classification pipeline on the transcripts annotated in the public data sets (Fig. 1A; see Supplemental Table 5 for specific details).
lincRNAs catalog and annotation
The complete lincRNA catalog (including the TUCP transcripts) as well as all RNA-seq alignments and transcriptome reconstructions are available at http://www.broadinstitute.org/genome_bio/human_lincrnas. Specific descriptions of all characterization fields are provided on the site. The catalog is also provided as Supplemental Data Sets 1–6.
Estimating expression abundance and normalization
We estimated the expression abundance of all lincRNAs and protein-coding genes by running Cufflinks in its expression abundance estimation mode across our 24 samples (Trapnell et al. 2010). We used the complete noncoding transcripts catalog and all coding transcripts annotated in UCSC for a comprehensive representation of transcripts along the genome while performing abundance estimation. FPKM calls were log2-normalized (after addition of ε = 0.05). The HeLa and liver samples from the eight-sample set were eliminated from further expression analysis due to low coverage and a lower expression range in comparison with other samples.
Tissue specificity score
To evaluate the tissue specificity of a transcript, we relied on Trapnell et al. (2010) and devised an entropy-based measure that quantifies the similarity between a transcript's expression pattern and another predefined pattern that represent an extreme case in which a transcript is expressed in only one tissue. This specificity measure relies on the JS divergence. The JS divergence of two discrete probability distributions, p1,p2, is defined to be
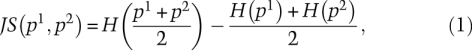 |
where H is the entropy of a discrete probability distribution:
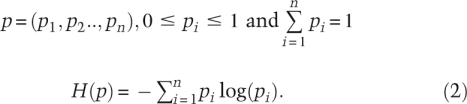 |
Relying on the theorem that the square root of the JS divergence is a metric (Fuglede and Topsoe 2004), we define the distance between two tissue expression patterns, 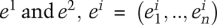 , as
, as
The tissue specificity of a transcript's expression pattern, e, across n tissues with respect to tissue t can then be defined as
where et is a predefined expression pattern that represents the extreme case in which a transcript is expressed in only one tissue. Formally, 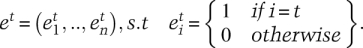
Finally, we define the tissue specificity score of a transcript as the maximal tissue specificity score across all n tissues of the transcripts expression pattern e:
Refer to the Supplemental Material for further details on the normalization of expression vector for tissue specificity calculation.
Identification of trans-mapped syntenic orthologs of human lincRNAs
We downloaded all available TransMap mappings of expressed transcripts to the human genome (NCBI39/Hg19) from the UCSC Genome Browser (http://genome.ucsc.edu; Zhu et al. 2007). The TransMap methodology maps all annotated transcripts of one species to the other by using the syntenic BLASTZ alignments between two species (Schwartz et al. 2003). First, it aligns all mRNA sequences of species a to its own genome. Then, it uses the syntenic alignment between species a and b to project the mRNA sequence of a to the genome of b and finally refines this mapping. We crossed all UCSC, RefSeq, mRNA, and EST transcripts trans-mapped to humans with our lincRNA set and included every lincRNA that had an exon overlap with a trans-mapped transcript in the trans-mapped lincRNA set. We used the UCSC classification of coding and noncoding transcripts applied to human and mouse transcripts known to UCSC (and downloaded from the UCSC Genome Browser) (Hsu et al. 2006).
Refined alignment of human lincRNAs and their mouse orthologs
To assess the alignment quality of the trans-mapped lincRNAs and their syntenic orthologs, we realigned the transcript sequence of all human lincRNAs and their mouse orthologs using the fast statistical alignment algorithm with default parameters (see the Supplemental Material; Bradley et al. 2009).
Acknowledgments
We gratefully thank Manuel Garber, Mitchell Guttman, Or Zuk, and Noam Shoresh for insightful discussions and code. C.T. is a Damon Runyon Cancer Foundation Fellow. L.G. is an NSF Post-doctoral Fellow. M.K. is a Human Frontier Science Program Fellow. A.R. is supported by an NIH Pioneer Award, the Burroughs Wellcome Fund Career Award at the Scientific Interface, and HHMI. J.L.R. is a Damon Runyon-Rachleff, Searle, Smith Family Scholar, and is supported by Director's New Innovator Awards (1DP2OD00667-01). This work was funded in part by a Center of Excellence in Genome Science from the NHGRI (A.R. and J.L.R.). A.R. and J.L.R. are investigators of the Merkin Foundation for Stem Cell Research at the Broad Institute. The body map data were kindly provided by the Gene Expression Applications research group at Illumina.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.17446611.
References
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2010. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 doi: 10.1093/nar/gkq1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L 2009. Fast statistical alignment. PLoS Comput Biol 5: e1000392 doi: 10.1371/journal.pcbi.1000392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP 2010. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol 11: R72 doi: 10.1186/gb-2010-11-7-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26: 183–186 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 doi: 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E 2008. Ripples from neighbouring transcription. Nat Cell Biol 10: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 doi: 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglede B, Topsoe F 2004. Jensen-Shannon divergence and Hilbert space embedding. In Proceedings of the IEEE International Symposium on Information Theory, p. 31 doi: 10.1109/ISIT.2004.1365067 [Google Scholar]
- Garber M, Grabherr MG, Guttman M, Trapnell C 2011. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8: 469–477 [DOI] [PubMed] [Google Scholar]
- Girard Al, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, et al. 2009. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41: 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Yang X, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. 2006. GENCODE: producing a reference annotation for ENCODE. Genome Biol 7: S4 doi: 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D 2006. The UCSC known genes. Bioinformatics 22: 1036–1046 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. 2011. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D 2003. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci 100: 11484–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL 2010. RNA traffic control of chromatin complexes. Curr Opin Genet Dev 20: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375: 34–39 [DOI] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 doi: 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. 2010. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP 2009. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5: e1000617 doi: 10.1371/journal.pgen.1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen Sr, Christensen MS, Mapendano CK, Schierup MH, Jensen TH 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Pruitt K, Tatusova T, Maglott D 2002. The reference sequence (RefSeq) project. In The NCBI handbook (ed. McEntyre J, Ostell J), chapter 18 National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/books/NBK21091 [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL 2005. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 15: 501–512 [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D 2007. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput Biol 3: e247 doi: 10.1371/journal.pcbi.0030247 [DOI] [PMC free article] [PubMed] [Google Scholar]