CUP is an eIF4E-binding protein (4E-BP) that is recruited to maternal mRNAs during embryonic development. Izaurralde and colleagues delineate a novel role for the noncanonical eIF4E-binding motif of the CUP protein whereby CUP not only represses translation, but also prevents decapping and subsequent degradation of deadenylated mRNA targets.
Keywords: translational repression, decapping, deadenylation
Abstract
CUP is an eIF4E-binding protein (4E-BP) that represses the expression of specific maternal mRNAs prior to their posterior localization. Here, we show that CUP employs multiple mechanisms to repress the expression of target mRNAs. In addition to inducing translational repression, CUP maintains mRNA targets in a repressed state by promoting their deadenylation and protects deadenylated mRNAs from further degradation. Translational repression and deadenylation are independent of eIF4E binding and require both the middle and C-terminal regions of CUP, which collectively we termed the effector domain. This domain associates with the deadenylase complex CAF1–CCR4–NOT and decapping activators. Accordingly, in isolation, the effector domain is a potent trigger of mRNA degradation and promotes deadenylation, decapping and decay. However, in the context of the full-length CUP protein, the decapping and decay mediated by the effector domain are inhibited, and target mRNAs are maintained in a deadenylated, repressed form. Remarkably, an N-terminal regulatory domain containing a noncanonical eIF4E-binding motif is required to protect CUP-associated mRNAs from decapping and further degradation, suggesting that this domain counteracts the activity of the effector domain. Our findings indicate that the mode of action of CUP is more complex than previously thought and provide mechanistic insight into the regulation of mRNA expression by 4E-BPs.
The asymmetric distribution of cellular components plays a critical role in cell polarization, the specification of cell fates during differentiation and embryonic patterning (for review, see Czaplinski and Singer 2006). One powerful mechanism that generates asymmetry is the spatial and temporal regulation of translation of selected mRNAs, which are maintained in a silent, repressed state until their translation is activated at the location and time at which the encoded protein is needed (for review, see Richter and Sonenberg 2005; Wilhelm and Smibert 2005).
The translational repression of localized mRNAs is mediated by multiple proteins, which are recruited in a sequence-specific manner and inhibit translation by diverse mechanisms (for review, see Richter and Sonenberg 2005; Wilhelm and Smibert 2005). One common mechanism for translation inhibition is interference with the assembly of the eukaryotic initiation factor eIF4F, which consist of the cap-binding protein eIF4E, the scaffolding protein eIF4G, and the RNA helicase eIF4A. The eIF4E subunit binds the mRNA 5′ cap structure and interacts with eIF4G, which in turn promotes translation initiation by recruiting the 43S preinitiation complex (composed of the 40S ribosomal subunit, eIF2-GTP-Met-tRNAMeti, eIF3, eIF1, eIF1A, and probably eIF5) (Jackson et al. 2010). A diverse group of translational repressors inhibits translation initiation by binding to eIF4E and disrupting its interaction with eIF4G (Haghighat et al. 1995; Mader et al. 1995). These eIF4E-binding proteins (4E-BPs) share a common eIF4E-binding motif with eIF4G (4E-BM: YxxxxLφ, where x is any amino acid and φ is a hydrophobic amino acid), and therefore compete with eIF4G for binding to the same surface on eIF4E (Haghighat et al. 1995; Mader et al. 1995; Altmann et al. 1997; Marcotrigiano et al. 1999).
The Drosophila melanogaster protein CUP is a member of the 4E-BP protein family that specifically associates with oskar and nanos mRNA through interactions with the RNA-binding proteins Bruno and Smaug, respectively, and represses the expression of these mRNAs prior to their posterior localization (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004; for review, see Piccioni et al. 2005). Similar to all members of the 4E-BP protein family, CUP contains an eIF4E-binding motif (4E-BM1) and competes with eIF4G for eIF4E binding (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004). In contrast to other 4E-BPs, however, CUP contains a second, noncanonical motif (4E-BM2) that contributes to, but is not essential for, eIF4E binding (Nelson et al. 2004). Although this second binding motif likely binds a different surface on eIF4E, it also blocks eIF4E–eIF4G interaction in vitro (Nelson et al. 2004). These observations have led to the model that CUP inhibits the expression of mRNA targets by disrupting eIF4E–eIF4G interaction, thereby inhibiting translation initiation. However, the contribution of the noncanonical 4E-binding motif to CUP-mediated translational repression is unknown.
The two eIF4E-binding motifs in CUP are located within the N-terminal domain, which is followed by a middle region (Mid) and a glutamine-rich C-terminal region (Q-rich). The Mid and Q-rich regions have been implicated in mediating binding to the Nanos protein and Bruno, respectively, as well as to additional protein partners, including Miranda and Staufen (Verrotti and Wharton 2000; Nakamura et al. 2004; Piccioni et al. 2009). Whether the Mid and Q-rich regions play a direct role in the repression of CUP-associated mRNAs, in addition to mediating protein–protein interactions, has not been addressed.
The regulation of oskar and nanos mRNA expression involves not only translation inhibition, but also changes in the poly(A) tail length (by deadenylation and polyadenylation), which affect both translation efficiency and mRNA stability (Chang et al. 1999; Juge et al. 2002; Castagnetti and Ephrussi 2003; Benoit et al. 2005; Semotok et al. 2005; Chekulaeva et al. 2006; Jeske et al. 2006, 2011; Zaessinger et al. 2006). Currently, it is not known whether CUP plays a role in poly(A) tail regulation.
In this study, we investigate the mechanism of CUP-mediated mRNA repression and the role of the noncanonical eIF4E-binding motif. Additionally, we elucidate the contribution of the Mid and Q-rich regions to this repression. We show that, in Drosophila Schneider 2 cells (S2 cells), CUP binding to an mRNA reporter represses protein production and promotes mRNA deadenylation. Although deadenylated mRNAs are normally rapidly decapped and degraded in S2 cells, mRNAs that are associated with CUP are protected from decapping and further degradation. Remarkably, this protection depends on the N-terminal domain and the noncanonical 4E-BM2 motif. Contrary to our expectation, the canonical 4E-BM1 motif is dispensable for CUP-mediated translational repression and mRNA deadenylation. Instead, these activities reside in the Mid and Q-rich regions of the protein, which collectively we termed the effector domain. Alone, the effector domain has potent mRNA degradation activity. However, when the N-terminal domain and the noncanonical 4E-BM2 motif are present, the degradative activity of the effector domain is inhibited so that the target mRNA is deadenylated but not degraded further. These results indicate that the N-terminal domain counteracts the activity of the effector domain. Taken together, our results uncover an unprecedented role for the N-terminal domain and the noncanonical eIF4E-binding motif in preventing the decapping and subsequent degradation of deadenylated mRNA targets and reveal novel and essential functions for the Mid and Q-rich regions of CUP in post-transcriptional mRNA regulation.
Results
CUP promotes deadenylation of target mRNAs
To obtain insight into the mechanism by which CUP inhibits the expression of target mRNAs, we used a tethering assay in D. melanogaster S2 cells. This assay allowed us to study CUP-intrinsic activities (i.e., those independent of Bruno and Smaug). The expressed CUP protein included two tags: a peptide derived from the bacteriophage λ N protein (λN tag) (Gehring et al. 2005) to enable tethering to a firefly luciferase (F-Luc) reporter, and an HA tag (hemagglutinin) to allow detection of the expressed protein by Western blot. The F-Luc reporter contains five Box B hairpins (5BoxB) inserted in the 3′ untranslated region (UTR); these bind the λN tag with high affinity and thus recruit CUP to the F-Luc-5BoxB mRNA.
We transfected S2 cells with the F-Luc-5BoxB reporter, a plasmid expressing either λN-HA-CUP or the λN-HA tag alone, and a plasmid encoding Renilla luciferase as a transfection control (R-Luc). We observed that λN-HA-CUP repressed the expression of the F-Luc-5BoxB reporter relative to the λN-HA tag alone (Fig. 1A). The repression was specific: The expression of an F-Luc reporter lacking the Box B hairpins was unaffected by λN-HA-CUP (Supplemental Fig. S1A–D), indicating that CUP must bind to the mRNA in order to repress its expression.
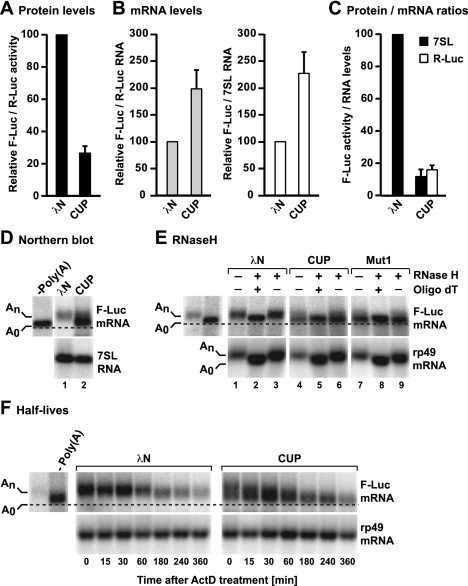
Figure 1.
CUP represses translation and promotes the deadenylation of bound mRNAs. (A–D) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-5BoxB reporter, another expressing the Renilla luciferase (R-Luc) as a transfection control, and a third expressing λN-HA or λN-HA-CUP, as indicated. Additionally, all transfection mixtures contained a plasmid expressing a short uncapped and unadenylated RNA derived from the 7SL RNA. (A) Firefly luciferase activity was normalized to Renilla luciferase and set to 100 in cells expressing λN-HA. (B) F-Luc-5BoxB mRNA levels were normalized to those of the Renilla mRNA or 7SL RNA and set to 100 in cells expressing λN-HA. Similar values were obtained independently of the normalization control. Mean values ± standard deviations from three independent experiments are shown. (C) The normalized F-luc activity values were divided by the corresponding normalized mRNA levels for each condition. These ratios were set to 100 in cells expressing λN-HA. (D) Northern blot analysis of RNA samples corresponding to those shown in B. F-Luc-5Box mRNA lacking a poly(A) tail (A0) was loaded as reference. The dashed line indicates the position of the deadenylated F-Luc-5BoxB mRNA. (E) RNA samples isolated from tethering assays (e.g., D) were treated with RNase H in the absence or presence of oligo(dT) and analyzed by Northern blot; rp49 mRNA served as a positive control for the RNase H treatment. (F) S2 cells were transfected as described in A. Three days after transfection, cells were treated with actinomycin D (5 μg/mL) and harvested at the indicated time points. F-Luc-5BoxB mRNA lacking a poly(A) tail was loaded as reference, and rp49 mRNA served as a loading control.
To determine whether CUP inhibits the expression of the F-Luc-5BoxB reporter by repressing translation directly or indirectly (i.e., by reducing mRNA levels), we analyzed the steady-state levels of the F-Luc-5BoxB mRNA by Northern blotting. Unexpectedly, in cells expressing λN-HA-CUP, the abundance of the F-Luc-5BoxB mRNA increased ∼2.2-fold relative to the R-Luc mRNA transfection control or a 7SL RNA derivative that lacks a poly(A) tail (Fig. 1B), resulting in a net sixfold reduction in F-Luc activity (Fig. 1C). Furthermore, in cells expressing λN-HA-CUP, most of the accumulated F-Luc-5BoxB transcripts migrated as a diffuse band, and a fraction comigrated with the corresponding transcript lacking the poly(A) tail, which was loaded as a reference (Fig. 1D). These results suggest that CUP promotes the deadenylation of its target mRNAs. An F-Luc mRNA reporter lacking the Box B hairpins was not deadenylated when coexpressed with λN-HA-CUP (Supplemental Fig. S1C).
We performed two independent experiments to provide more direct evidence that CUP promotes the deadenylation of bound mRNAs. First, we used an oligo(dT) targeted ribonuclease H (RNase H) cleavage assay to confirm that the F-Luc-5BoxB transcripts that accumulated in λN-HA-CUP-expressing cells were indeed deadenylated (Fig. 1E). In control cells (i.e., cells expressing λN-HA), both the F-Luc-5BoxB reporter and an endogenous rp49 mRNA (encoding ribosomal protein L32) migrated faster after cleavage with oligo(dT)-directed RNase H had removed the poly(A) tail (Fig. 1E, lane 1 or 3 vs. 2). In contrast, in cells expressing λN-HA-CUP, the F-Luc-5BoxB mRNA migrated as a diffuse band, and a large fraction of this mRNA was already deadenylated (Fig. 1E, lanes 4,6). Consistent with this interpretation, RNase H treatment in the presence of oligo(dT) resulted in the accumulation of only the fast-migrating band and the disappearance of the longer polyadenylated forms (Fig. 1E, lane 4 or 6 vs. 5).
In the second experiment, we exposed transfected cells to actinomycin D to inhibit transcription and then analyzed the levels of F-Luc-5BoxB mRNA over time (compared with the long-lived rp49 mRNA, which has a half-life >8 h). In cells expressing λN-HA, the half-life of F-Luc-5BoxB mRNA was ∼160 min, whereas its half-life in cells expressing CUP was 240 min, consistent with the observed twofold increase in mRNA abundance. Moreover, the F-Luc-5BoxB transcripts were rapidly deadenylated in the presence of CUP (Fig. 1F).
Together, these results indicate that CUP promotes deadenylation of bound mRNAs. Notably, although deadenylated mRNAs are normally rapidly degraded in S2 cells (Behm-Ansmant et al. 2006; Haas et al. 2010), the F-Luc-5BoxB mRNA accumulated in a deadenylated, repressed form in cells expressing λN-HA-CUP, suggesting that CUP-bound mRNAs are protected from decapping and subsequent degradation.
A noncanonical eIF4E-binding motif in CUP protects deadenylated mRNA targets from degradation
Sequence alignments and secondary structure predictions indicate that the CUP protein contains an N-terminal domain (N-term, amino acids 1–417), a middle domain (Mid, amino acids 417–770), and a C-terminal region that is rich in glutamine residues (Q-rich, 21% glutamine) (Fig. 2A). The N-terminal domain contains a canonical eIF4E-binding motif (4E-BM1) and a second motif (4E-BM2) that has also been implicated in mediating binding to eIF4E (Fig. 2A; Nelson et al. 2004). The Mid domain also exhibits limited similarity to the eIF4E transporter protein (4E-T) (Dostie et al. 2000).
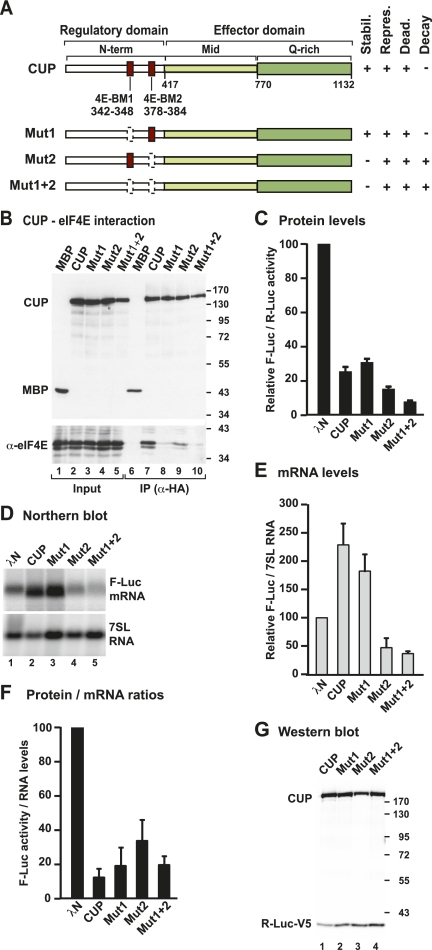
Figure 2.
CUP represses mRNA expression independently of eIF4E binding. (A) Domain organization of CUP protein. CUP contains an N-terminal regulatory domain (N-term) containing two eIF4E-binding motifs (4E-BM1 and 4E-BM2), a middle region (Mid), and a glutamine-rich C-terminal region (Q-rich). The Mid and Q-rich regions define the effector domain. The bottom panel shows the CUP point mutants used in this study and a summary of their activities. (stabil.) mRNA stabilization; (repres.) repression of protein production; (dead.) deadenylation; (decay) degradation of the mRNA body. (B) Interactions between HA-CUP wild type or mutants and endogenous eIF4E. Proteins were immunoprecipitated from cell lysates using a monoclonal anti-HA antibody. An HA-tagged version of maltose-binding protein (MBP) served as negative control. Inputs (1%) and immunoprecipitates (10%) were analyzed by Western blotting using anti-HA and anti-eIF4E antibodies. (C–F) The activity of CUP mutants relative to wild type was tested in tethering assays using the F-Luc-5BoxB reporter and analyzed as described in Figure 1. The mean values ± standard deviations from three independent experiments are shown. (G) Wild-type CUP and mutants were expressed at comparable levels. R-Luc-V5 served as a transfection control.
To investigate whether the interaction of CUP with eIF4E is required for the translational repression and deadenylation of target mRNAs, we generated CUP mutants carrying alanine substitutions in 4E-BM1 (Y342A and L347A; Mut1), 4E-BM2 (L379A and L383A; Mut2), or both (Mut1+2) (Fig. 2A). These substitutions were previously shown to abolish or reduce eIF4E binding, respectively (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004). We first confirmed that these mutations inhibited the CUP–eIF4E interaction using coimmunoprecipitation assays in S2 cells. HA-tagged CUP coimmunoprecipitated endogenous eIF4E, as well as a GFP-tagged eIF4E that was coexpressed (Fig. 2B, lane 7; Supplemental Fig. S1E). The substitution of two conserved amino acids in 4E-BM1 (Mut1) strongly reduced CUP interaction with eIF4E, as reported previously (Fig. 2B, lane 8; Supplemental Fig. S1E; Nakamura et al. 2004; Nelson et al. 2004). Substitutions of residues at similar positions in 4E-BM2 (Mut2) reduced but did not abolish eIF4E binding (Fig. 2B, lane 9; Supplemental Fig. S1E). Mutations in 4E-BM2 did not exacerbate the effect of the mutations in 4E-BM1 (Fig. 2B, lane 10, mutant Mut1+2; Supplemental Fig. S1E). Our results are consistent with those reported by Nelson et al. (2004), showing that 4E-BM1 provides a high-affinity eIF4E-binding site and that 4E-BM2 contributes to, but is not essential for, CUP binding to eIF4E.
We next tested whether the mutant CUP proteins that were impaired in eIF4E binding could promote the translational repression and deadenylation of the F-Luc-5BoxB reporter. We observed that all three mutants (Mut1, Mut2, and Mut1+2) repressed the expression of the F-Luc-5BoxB reporter (Fig. 2C). However, when mRNA levels were analyzed by Northern blotting, only Mut1, which is strongly impaired in eIF4E binding, behaved as wild-type CUP: It promoted reporter mRNA deadenylation but stabilized the mRNA in the deadenylated form (Fig. 2D,E). The F-Luc-5BoxB transcripts associated with CUP Mut1 were indeed deadenylated, as shown using an oligo(dT) targeted ribonuclease H (RNase H) cleavage assay (Fig. 1E, lanes 7–9). In contrast, Mut2 or Mut1+2 reduced the abundance of the F-Luc-5BoxB mRNA (Fig. 2D,E). Although for these two mutants the reduction of mRNA levels contributes to the reduction in protein production, a net decrease in F-Luc activity was nevertheless observed (Fig. 2F). All mutants were expressed at comparable levels (Fig. 2G) and did not affect the expression of an F-Luc reporter lacking the BoxB hairpins (Supplemental Fig. S1D).
Two important conclusions can be drawn from these observations. First, the noncanonical eIF4E-binding motif (4E-BM2) is required to protect the deadenylated mRNA reporter from degradation. Indeed, reporter mRNA was degraded when bound to a CUP mutant carrying amino acid substitutions in 4E-BM2 (Fig. 2D [lanes 4,5], E). Second, a CUP mutant carrying amino acid substitutions in the canonical eIF4E-binding motif (4E-BM1) represses reporter mRNA expression as efficiently as wild-type CUP, even though this mutant is strongly impaired in eIF4E binding (Fig. 2B,C,F). Thus, according to this assay, the canonical eIF4E-binding motif is dispensable for CUP to repress the expression and promote deadenylation of the mRNA reporter.
Our finding that 4E-BM1, and thus eIF4E binding, is dispensable for CUP-mediated repression contrasts with previous studies suggesting that CUP inhibits translation by binding to eIF4E and competing with eIF4G (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004). It was therefore important to investigate whether tethered CUP was able to displace eIF4G from the mRNA target. We observed that eIF4G association with the F-Luc-5BoxB mRNA was reduced when the reporter was bound to CUP (Supplemental Fig. S2A–C). These results suggest that eIF4G displacement takes place and may contribute to, but is not necessary for, CUP-mediated translational repression.
CUP Mid and Q-rich regions are required for mRNA repression and deadenylation
To more precisely map the domains in CUP that are required to repress translation and promote target mRNA deadenylation, we generated a series of deletion mutants lacking the N-term, Mid, or Q-rich regions individually or in combination (Fig. 3A). We observed that an N-terminal fragment of CUP (N-term) that contains the two eIF4E-binding motifs but lacks the Mid and Q-rich regions did not repress the expression of the F-Luc-5BoxB reporter and failed to promote its deadenylation (Fig. 3B–D; Supplemental Fig. S2D–F). However, this protein fragment retained the ability to bind to eIF4E (Fig. 3E, lane 11) and stabilized the mRNA reporter, leading to an increase in both protein expression and mRNA levels (Fig. 3B–D, lane 3; Supplemental Fig. S2D–F). In contrast, full-length CUP causes a similar increase in reporter mRNA levels but represses protein expression and promotes mRNA deadenylation (Fig. 3B–D). As for full-length CUP, the ability of the N-term fragment to stabilize the mRNA reporter depends on the presence of the 4E-BM2; when this motif was mutated, the N-terminal fragment no longer stabilized the mRNA (Supplemental Fig. S2D–F).
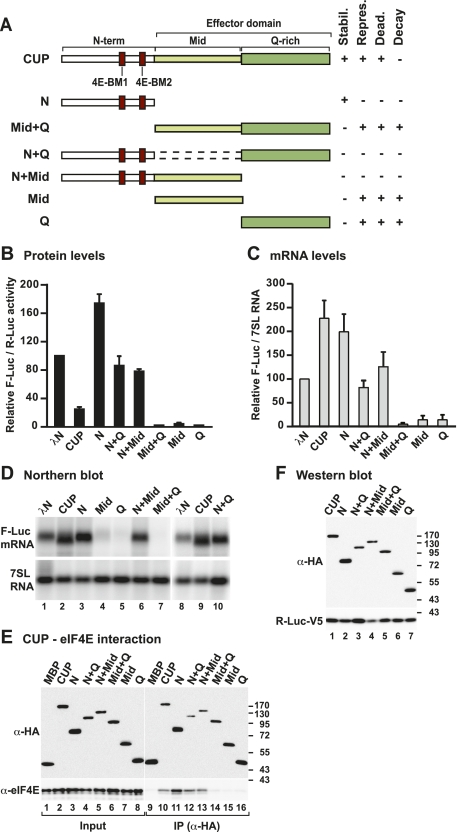
Figure 3.
The Mid and Q-rich regions define the CUP effector domain. (A) Schematic representation of CUP deletion mutants used in this study and a summary of their activities, as described in Figure 2A. (B–D) The activities of CUP deletion mutants relative to wild type were tested using the F-Luc-5BoxB reporter as described in Figure 1. (E) The interaction between HA-tagged wild-type or mutant CUP protein and endogenous eIF4E was analyzed by coimmunoprecipitation as described in Figure 2B. (F) Wild-type and mutant CUP proteins were expressed at comparable levels. R-Luc-V5 served as a transfection control.
Notably, deleting either the Mid or Q-rich regions (N+Q or N+Mid) also abrogated the translational repression and deadenylation of the F-Luc-5BoxB reporter (Fig. 3–D, lanes 6,10, respectively), although these deletion mutants were expressed at comparable levels and still interacted with eIF4E (Fig. 3E,F). These findings provide further evidence that CUP binding to eIF4E is not sufficient to promote the translational repression and deadenylation of CUP-associated mRNAs. A second important conclusion from these results is that the Mid and Q-rich regions are both required to repress translation and promote deadenylation, as deletion of either of these regions inhibits CUP function. Therefore, we conclude that these regions act together to repress and deadenylate mRNA targets and collectively define the effector domain of CUP.
The CUP effector domain is a potent trigger of mRNA degradation
As stated above, the CUP Mid and Q-rich regions (the effector domain) are both required for CUP-mediated mRNA repression and deadenylation. We were therefore interested to determine whether these regions were sufficient for CUP function. Remarkably, binding of the CUP effector domain to the F-Luc-5BoxB reporter triggered efficient mRNA degradation (Mid+Q) (Fig. 3B–D). Further analyses showed that either the Mid or the Q-rich region alone was sufficient to efficiently degrade the mRNA reporter (Figs. 3B–D). All three protein fragments were expressed at comparable levels (Fig. 3F) and none affected the expression of an F-Luc reporter lacking BoxB elements (Supplemental Fig. S1D). Thus, the CUP effector domain elicits degradation of its associated mRNAs.
Given the strong degradative activity of the effector domain, it is remarkable that this activity is counteracted by the N-terminal region in the context of full-length CUP so that bound mRNAs are deadenylated but not degraded further. Similarly, although the isolated Mid or Q-rich regions are able to trigger efficient target mRNA degradation, fusion of either of these regions to the N-terminal domain (e.g., CUP N+Q or N+Mid) inhibits this activity. We conclude that the CUP N-terminal domain prevents the effector domain from degrading the target mRNA after deadenylation. The inhibitory effect of the N-terminal domain requires the 4E-BM2 motif; however, additional N-terminal sequences also contribute to the inhibition because CUP Mut2 and Mut1+2 are less active in promoting mRNA degradation than the isolated effector domain or the Mid and Q-rich regions (Figs. 2E vs. 3C).
CUP represses the expression of a reporter containing the oskar mRNA 3′ UTR
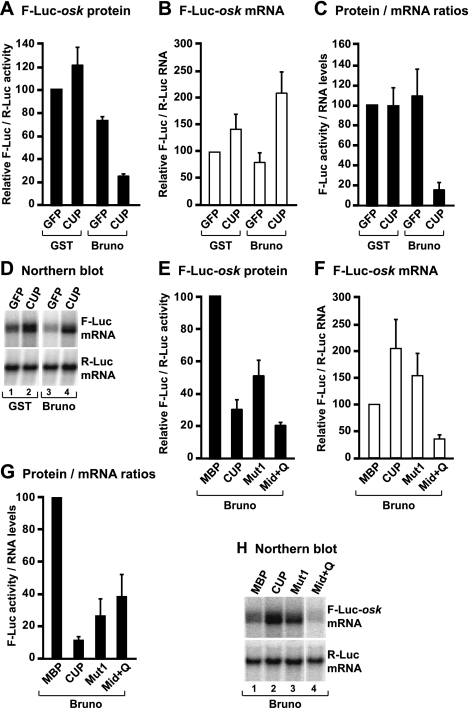
CUP is recruited to natural mRNA targets (e.g., oskar and nanos mRNAs) via interactions with RNA-binding proteins (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; for review, see Piccioni et al. 2005). One such protein is Bruno, which recruits CUP to multiple Bruno response elements (BREs) present in the oskar mRNA 3′ UTR (Kim-Ha et al. 1995; Castagnetti and Ephrussi 2003; Nakamura et al. 2004). To validate the observations obtained using the tethering assay, we generated an F-Luc reporter containing the oskar 3′ UTR. We observed that, individually, neither CUP nor Bruno repressed this reporter (Fig. 4A–C). However, in the presence of both CUP and Bruno, the oskar reporter was repressed, although mRNA levels increased (Fig. 4A–C). Furthermore, the transcripts accumulated in a deadenylated form, as observed in the tethering assay (Fig. 4D). Importantly, the effects of wild-type CUP and the CUP mutants on this natural 3′ UTR were similar to those observed in the tethering assay. In particular, a CUP mutant carrying substitutions in the canonical eIF4E-binding motif (Mut1) repressed the expression of the F-Luc-oskar mRNA (Fig. 4E–G), albeit less efficiently than wild-type CUP, indicating that eIF4E binding may contribute to, but is not necessary for, CUP to repress a natural target. As observed in the tethering assay, both CUP and Mut1 caused an increase in reporter mRNA levels (Fig. 4F,H). Moreover, the effector domain, which confers binding to Bruno, strongly reduced reporter mRNA abundance, as shown in tethering assays (Fig. 4F,H). In summary, in isolation, the effector domain of CUP causes strong mRNA degradation independent of whether it is directly tethered to the mRNA or recruited via interactions with Bruno. In the context of full-length CUP, however, the degradative activity of the effector domain is suppressed by the N-terminal regulatory domain.
Figure 4.
CUP represses an mRNA reporter containing the oskar 3′ UTR. (A–D) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-oskar reporter, another expressing the Renilla luciferase (R-Luc) as a transfection control, and a third expressing GFP or CUP. Additionally, transfection mixtures contained a plasmid expressing GST or Bruno as indicated. (A) Firefly luciferase activity was normalized to Renilla luciferase and set to 100 in control cells (i.e., expressing GFP and GST). (B) F-Luc-oskar mRNA levels were normalized to those of the Renilla mRNA and set to 100 in control cells. Mean values ± standard deviations from three independent experiments are shown. (C) F-Luc activity was normalized to the mRNA levels. (D) Northern blot analysis of RNA samples corresponding to those shown in B. (E–H) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-oskar reporter, another expressing the Renilla luciferase (R-Luc) as a transfection control, and a third expressing MBP or CUP. Additionally, all transfection mixtures contained a plasmid expressing Bruno. Luciferase activity and mRNA levels were analyzed as described in A–D.
The CUP effector domain interacts with components of the deadenylase and decapping complexes
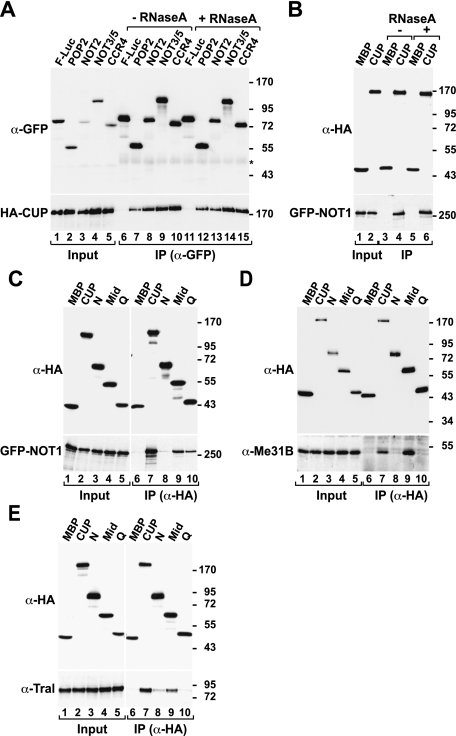
It has been reported that CUP interacts with decapping activators, including Me31B, DCP1, and Trailer hitch (Tral) (Nakamura et al. 2004; Wilhelm et al. 2005; Tritschler et al. 2008). The finding that CUP triggers deadenylation of bound mRNA raised the possibility that it may also interact with components of the deadenylase complex. Supporting this hypothesis is the observation that CUP copurifies with the CAF1–CCR4–NOT complex isolated from D. melanogaster embryonic extracts (Temme et al. 2010). We performed coimmunoprecipitation assays to determine whether these interactions also occur in S2 cells. CUP coimmunoprecipitated with all core components of the D. melanogaster CAF1–CCR4–NOT deadenylase complex (POP2, CCR4, NOT2, NOT3/5, and NOT1) (Fig. 5A–C; Temme et al. 2004, 2010). These interactions were observed in the presence of RNase A, suggesting that they are not mediated by RNA (Fig. 5A,B). As a control, and consistent with previous studies (Nakamura et al. 2004; Wilhelm et al. 2005; Tritschler et al. 2008), CUP also coimmunoprecipitated endogenous Me31B and Tral in RNase A-treated extracts (Fig. 5D,E). In contrast, wild-type CUP or mutants (Mut1, Mut2, and Mut1+2) failed to coimmunoprecipitate EDC4, DCP2, and XRN1 (Supplemental Fig. S3A–D), suggesting large ribonucleoprotein complexes such as P-bodies are not coimmunoprecipitated under the conditions used here.
Figure 5.
The effector domain of CUP confers binding to deadenylase and decapping complexes. (A,B) Interaction between HA-CUP and GFP-tagged subunits of the CAF1–CCR4–NOT complex. Cell lysates were immunoprecipitated using a polyclonal anti-GFP antibody (A) or a monoclonal anti-HA antibody (B). When indicated, cell lysates were treated with RNase A before immunoprecipitation. F-Luc-GFP and HA-MBP served as negative controls. (C–E) Interactions between wild-type CUP or the indicated CUP fragments with binding partners. (C) Interaction of CUP and GFP-NOT1. (D,E) Interactions of HA-CUP with endogenous Me31B or Tral.
We next defined the domains of the CUP protein required for its interaction with deadenylation and decapping factors. We observed that the CUP Mid region was capable of binding NOT1, Me31B, and Tral (Fig. 5C–E), although binding to NOT1 and Tral was reduced compared with full-length CUP (Fig. 5C,E). NOT1 also interacted with the CUP Q-rich region (Fig. 5C, lane 10). Thus, CUP interacts with subunits of the deadenylase and decapping complexes through the effector domain, providing one plausible mechanism for how this domain could mediate efficient target degradation.
The CUP effector domain elicits deadenylation-dependent decapping
Given that the effector domain interacts with components of both the deadenylase and decapping complexes (Fig. 5; Nakamura et al. 2004; Wilhelm et al. 2005; Tritschler et al. 2008), we hypothesized that this domain directs bound mRNAs to the 5′-to-3′ mRNA decay pathway. In this pathway, mRNAs are first deadenylated and then decapped by DCP2, which requires additional cofactors for full activity and/or stability (Franks and Lykke-Andersen 2008). In metazoans, these cofactors include DCP1, EDC3, EDC4 (also known as Ge-1), Pat, and the DEAD-box protein RCK/Me31B. Once decapped, mRNAs are ultimately degraded by the major cytoplasmic 5′-to-3′ exonuclease XRN1 (Franks and Lykke-Andersen 2008).
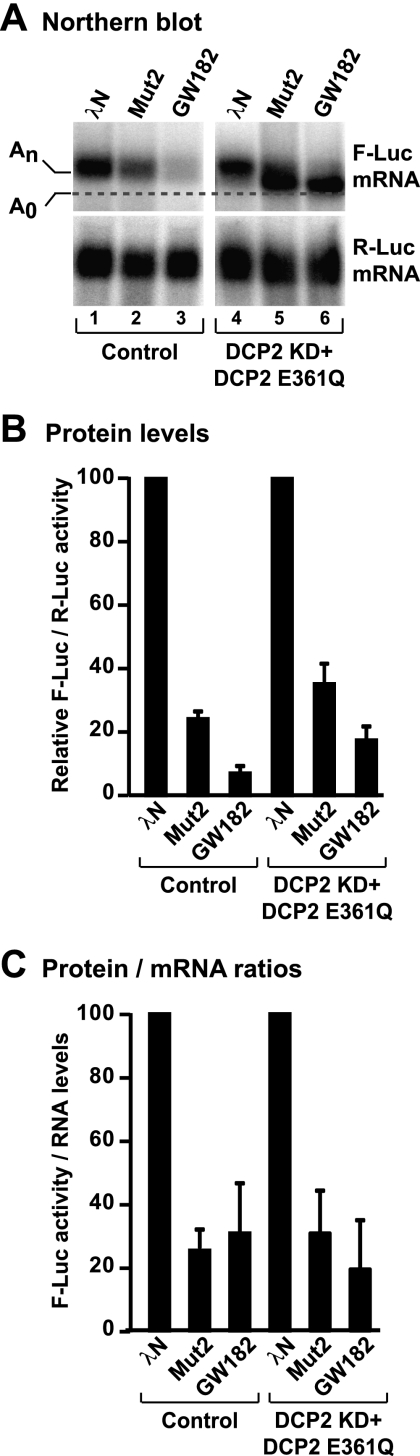
To determine whether the effector domain promotes deadenylation-dependent decapping, we performed the reporter tethering assay in cells in which decapping was inhibited. For this assay, we used CUP Mut2 because this mutant differs from wild-type CUP by only two amino acids in 4E-BM2, but in contrast to wild-type CUP, triggers mRNA degradation. We inhibited decapping by overexpressing a catalytically inactive DCP2 mutant (E361Q) in cells depleted of endogenous DCP2. In these cells, degradation caused by tethered CUP Mut2 was inhibited, leading to the accumulation of deadenylated transcripts (Fig. 6A, lane 2 vs. 5). Similar results were obtained with tethered GW182 (Fig. 6A, lane 3 vs. 6), which triggers deadenylation-dependent decapping and thus served as a positive control (Behm-Ansmant et al. 2006). For both proteins, we observed no net increase in F-Luc activity (Fig. 6B,C), most likely because the accumulated transcripts are deadenylated and thus are not efficiently translated. The accumulation of deadenylated transcripts upon decapping inhibition indicates that CUP Mut2 promotes deadenylation that is normally followed by decapping.
Figure 6.
CUP Mut2 promotes deadenylation-dependent decapping. (A–C) A tethering assay with CUP Mut2 or GW182 was performed in control cells (treated with GST dsRNA and expressing GFP) and in cells depleted of DCP2 (DCP2 knockdown; KD) and expressing a catalytically inactive GFP-DCP2 mutant (E361Q). Samples were analyzed as described in Figure 1. The dashed line in A indicates the position of the deadenylated F-Luc-5BoxB mRNA.
It is important to note that the translational repression and deadenylation mediated by wild-type CUP is not affected in cells in which decapping factors such as Me31B or Tral were depleted, consistent with the conclusion that mRNAs associated with CUP are deadenylated but not decapped (Supplemental Fig. S4A–C).
Collectively, our results indicate that a CUP mutant carrying two amino acid substitutions in 4E-BM2 promotes deadenylation that is followed by decapping and degradation of the mRNA body. In the context of wild-type CUP, however, target mRNAs are deadenylated but not degraded, reinforcing the conclusion that 4E-BM2 interferes with decapping and subsequent mRNA degradation.
CUP-mediated mRNA deadenylation requires the CAF1–CCR4–NOT complex
Having established that CUP elicits deadenylation and associates with the CAF1–CCR4–NOT complex, we sought to determine whether this complex is involved in deadenylating mRNAs associated with CUP. We also explored whether mRNA translational repression and deadenylation were mediated directly by tethered CUP or indirectly through the recruitment of Bruno or Smaug. Indeed, both proteins interact with CUP, and Smaug is also known to interact with the CAF1–CCR4–NOT complex; thus, Smaug could have acted as a bridging factor in our assays (Nelson et al. 2004; Semotok et al. 2005; Zaessinger et al. 2006; Tadros et al. 2007).
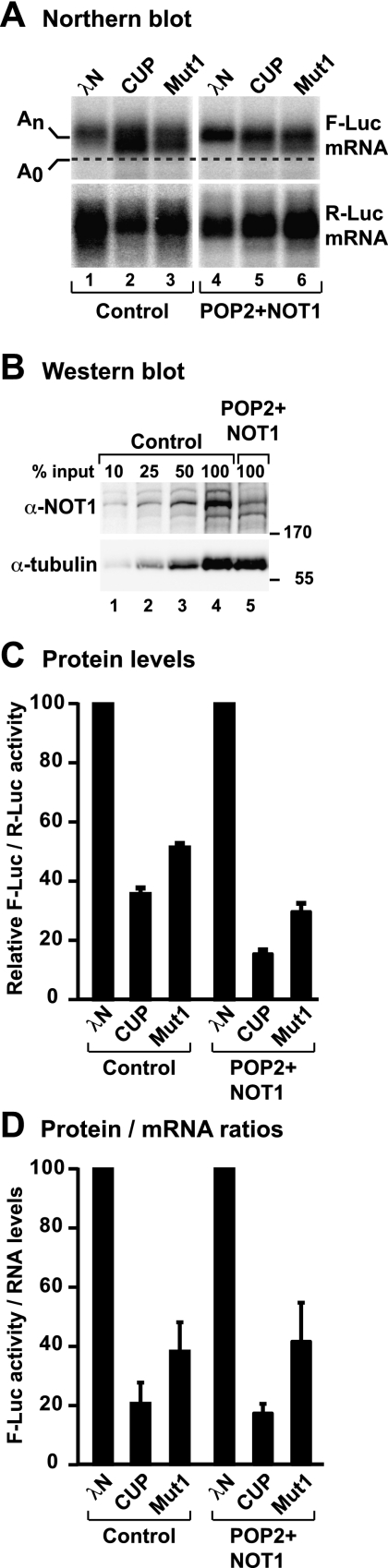
First, we established that neither Bruno nor Smaug depletion had an effect on CUP-mediated repression or deadenylation of the F-Luc-5BoxB mRNA, indicating that CUP activity is independent of Bruno and Smaug in tethering assays (Supplemental Fig. S5A–F). Next, we performed the tethering assay in cells depleted of two components of the CAF1–CCR4–NOT complex (NOT1 and the D. melanogaster CAF1 ortholog POP2) that have been shown to be required for deadenylation in S2 cells (Temme et al. 2004, 2010; Behm-Ansmant et al. 2006). Codepletion of POP2 and NOT1 partially suppressed CUP-mediated deadenylation, reducing the pool of fast-migrating (i.e., deadenylated) transcripts (Fig. 7A, lane 2 vs. 5). These results indicate that CUP requires the CAF1–CCR4–NOT complex to deadenylate bound mRNAs. Similar results were obtained for CUP Mut1 (Fig. 7A, lane 3 vs. 6), which behaves similarly to wild-type CUP in all assays. In POP2- and NOT1-depleted cells, NOT1 levels were reduced below 50% of wild-type levels (Fig. 7B). Importantly, the restoration of the mRNA poly(A) tail was not accompanied by a corresponding increase in firefly luciferase activity, indicating that the F-Luc-5BoxB mRNA was still translationally repressed (Fig. 7C,D). On the basis of these results, we conclude that deadenylation mediated by tethered CUP is independent of Bruno and Smaug and involves the CAF1–CCR4–NOT complex.
Figure 7.
The CAF1–CCR4–NOT complex deadenylates mRNAs associated with CUP. (A,B) S2 cells were treated with the indicated dsRNAs on days 0 and 4. Control cells were treated with an unrelated dsRNA targeting Neomycin. On day 6, cells were cotransfected with a mixture of three plasmids: one expressing the F-Luc-5BoxB mRNA, another expressing Renilla luciferase (R-Luc), and a third expressing the indicated λN-HA-tagged proteins. (A) Northern blot analysis of representative RNA samples. (B) Western blot analysis of control and NOT1-depleted cells. α-Tubulin served as a loading control. Dilutions of control cell lysates were loaded in lanes 1–4 to estimate the efficacy of the depletion. (C) Firefly luciferase activity was normalized to that of Renilla luciferase. For each condition, the normalized values of F-Luc activity were set to 100 in cells expressing the λN-HA tag. Mean values ± standard deviations from three independent experiments are shown. (D) The normalized F-luc activity values were divided by the corresponding normalized mRNA levels for each condition. These ratios were set to 100 in cells expressing λN-HA.
CUP can promote translational repression in the absence of deadenylation
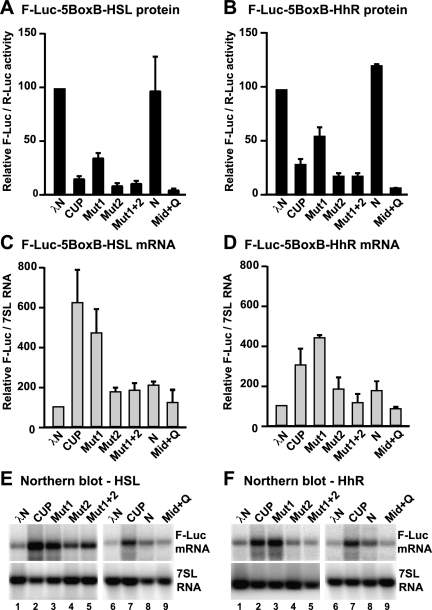
The above observations suggest that CUP represses translation even when deadenylation is partially blocked. We therefore analyzed whether CUP could repress the expression of mRNAs lacking a poly(A) tail. In these experiments, we use F-Luc-5BoxB reporters in which the cleavage and polyadenylation signal had been replaced by a histone H4 3′-terminal stem–loop or a self-cleaving hammerhead ribozyme (HhR) (Eulalio et al. 2009). Remarkably, these reporters were repressed by CUP as well as by the CUP mutants Mut1, Mut2, and Mut1+2 (Fig. 8A,B), although Mut1 was impaired relative to wild-type CUP. Furthermore, tethered CUP or Mut1 stabilized the reporters lacking a poly(A) tail, resulting in fourfold to sixfold increase in mRNA levels (Fig. 8C–F). Again, this stabilization required 4E-BM2 and was not as prominent when this motif was mutated (CUP Mut2 or Mut1+2) (Fig. 8C–F).
Figure 8.
Deadenylation is not required for CUP-mediated mRNA repression. (A–F) S2 cells were transfected with F-Luc5BoxB reporters in which the cleavage and polyadenylation signal had been substituted with either a histone H4 3′-terminal stem–loop (HSL; A,C,E) or a self-cleaving hammerhead ribozyme (HhR; B,D,F). Plasmids expressing Renilla luciferase (R-Luc) or 7SL RNA served as transfection controls. Firefly luciferase activity and mRNA levels were analyzed as described in Figure 1. A Northern blot analysis of representative RNA samples is shown below the corresponding graphs.
In contrast to the results observed with the polyadenylated reporter, however, the tethering of CUP Mut2 or Mut1+2 to nonadenylated reporters did not cause mRNA decay (Fig. 8C–F). Nevertheless, CUP Mut2 and Mut1+2 efficiently repressed the expression of these reporters (Fig. 8A,B), suggesting that translational repression can occur in the absence of deadenylation or mRNA destabilization.
To further define the regions in CUP that are required for translational repression in the absence of deadenylation, we tethered either the CUP N-terminal or the effector domains to the nonadenylated reporters. We observed that the N-terminal domain slightly increased reporter expression and mRNA abundance (Fig. 8A–D). In contrast, tethering the effector domain strongly repressed reporter expression in the absence of mRNA destabilization (Fig. 8A–F).
The analysis of the nonadenylated reporters yielded three important observations. First, the canonical 4E-BM1 may contribute to, but is not essential for, CUP-mediated translational repression. Second, the noncanonical 4E-BM2 is required for the stabilization of mRNAs associated with CUP. This stabilization occurs even for mRNAs that do not undergo deadenylation, supporting the idea that 4E-BM2 exerts its stabilizing effect by preventing decapping. Third, CUP can promote translational repression independently of deadenylation and eIF4E binding (e.g., Mut1+2); this activity resides in the effector domain.
Discussion
CUP is an eIF4E-binding protein that is recruited to target mRNAs through interactions with RNA-binding proteins and is thought to repress the translation of these mRNAs by blocking the eIF4E–eIF4G interaction (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004; for review, see Piccioni et al. 2005). In this study, we show that CUP not only represses translation, but also promotes mRNA deadenylation. However, binding to eIF4E is not required for CUP-mediated translational repression and deadenylation of target mRNAs. Instead, the Mid and Q-rich regions, which we collectively termed the CUP effector domain, are essential for these functions. The isolated effector domain is a potent trigger of mRNA degradation and promotes deadenylation-dependent decapping and subsequent decay of the mRNA. In the context of full-length CUP, however, the N-terminal domain counteracts the activity of the effector domain, allowing deadenylation to occur but inhibiting mRNA decapping and subsequent degradation. The protective function of the N-terminal domain requires the noncanonical 4E-BM2 motif, which contributes to, but is not essential for, eIF4E-binding. Taken together, our results show that CUP uses multiple mechanisms to regulate protein output of its targets and further reveal unexpected functions for the noncanonical eIF4E-binding motif (4E-BM2) and the Mid and Q-rich regions in this regulation.
The CUP effector domain
In this study, we show that CUP consists of two functional domains: a regulatory N-terminal domain and a C-terminal effector domain. The effector domain consists of the Mid and Q-rich regions and elicits mRNA degradation. Although the isolated Mid and Q-rich regions are also able to trigger mRNA degradation (Fig. 3C,D), in the context of full-length CUP, these regions are both required for translational repression and mRNA deadenylation; a protein containing the N-terminal region but lacking either the Mid or Q-rich region does not repress translation or promote deadenylation (Fig. 3B). Thus, the Mid and Q-rich regions act together as a single effector domain. This domain serves as a binding platform for deadenylating and decapping factors. Further work will be required to determine whether the effector domain interacts with these factors directly, and whether these interactions occur simultaneously or consecutively.
One important observation from our studies is that the effector domain can repress translation in the absence of deadenylation, suggesting that deadenylation is required not to establish repression, but rather to sustain the repressed state. This observation is consistent with previous studies showing that deadenylation contributes to, but is not essential for, the repression of nanos and oskar mRNAs (Lie and Macdonald 1999; Chekulaeva et al. 2006; Jeske et al. 2006, 2011).
A second important finding is that CUP can promote deadenylation independent of Smaug. This finding provides one explanation for the observation that the repression of oskar mRNA involves deadenylation (Tadros et al. 2007), even though Smaug is not involved in the repression of this mRNA. Our results suggest that CUP could be responsible for the observed deadenylation of unlocalized oskar mRNA, maintaining it in a repressed state. In agreement with this possibility, the deadenylation of oskar mRNA is catalyzed by CCR4 (Benoit et al. 2005). Furthermore, the translation activation of oskar mRNA at the posterior pole requires polyadenylation mediated by Orb, the Drosophila homolog of cytoplasmic polyadenylation element-binding protein (CPEB) and Drosophila poly(A) polymerase (PAP) (Chang et al. 1999; Juge et al. 2002; Castagnetti and Ephrussi 2003; Benoit et al. 2005), reinforcing the hypothesis that oskar mRNA repression involves deadenylation.
The canonical eIF4E-binding motif
The model that CUP inhibits translation by competing with eIF4G for binding to eIF4E is supported in vivo by the observation that a CUP mutant (CUPΔ212) that lacks N-terminal residues, including the canonical eIF4E-binding motif, fails to repress the expression of unlocalized oskar mRNA in Drosophila oocytes (Nakamura et al. 2004). Thus, our observation that the 4E-BM1 motif is not required for CUP-mediated repression is contrary to the current model for CUP function. However, the results obtained with CUPΔ212 mutant (which is thought to initiate translation at Met348) are difficult to interpret because this mutant lacks a large portion of the N-terminal region of the protein, the function of which remains unknown. Moreover, it is important to note that direct evidence that CUP binding to eIF4E is required for translational repression is lacking; for example, it has not been directly shown that point mutations in the canonical 4E-BM1 relieve CUP-mediated translational repression (for review, see Piccioni et al. 2005).
Furthermore, in agreement with our findings, several lines of evidence indicate that the mRNA 5′ cap structure, and thus eIF4E binding, is not essential for CUP-mediated translational repression (Lie and Macdonald 1999; Nakamura et al. 2004; Chekulaeva et al. 2006; Jeske et al. 2011). First, the Δ212 mutation does not impair CUP function during oogenesis and only affects oskar mRNA expression in the embryo, whereas other CUP alleles have strong effects on oocyte maturation and ovary development, leading to female sterility (Schüpbach and Wieschaus 1991; Keyes and Spradling 1997; Verrotti and Wharton 2000; Nakamura et al. 2004). Second, oskar mRNA was repressed in Drosophila embryo extracts isolated from flies carrying the Δ212 mutation (Chekulaeva et al. 2006), although it is not clear whether this repression was mediated by the N-terminal-truncated form of CUP. Third, the repression of oskar and nanos mRNA reporters was independent of the presence of a 5′ cap structure in cell-free extracts (Chekulaeva et al. 2006; Jeske et al. 2011). Fourth, mRNA reporters containing a nanos 3′ UTR, the translation of which was initiated in a cap-independent manner (by the cricket paralysis virus IRES), were repressed in vitro (Jeske et al. 2011). Finally, our results show that the 4E-BM1 motif is dispensable for the repression of a reporter containing the oskar 3′ UTR to which CUP was recruited through interactions with Bruno, indicating that binding to eIF4E may contribute to, but is neither necessary nor sufficient for, CUP repression of its targets.
Thus, it is possible that the 4E-BM1 motif has other functions. For example, through its binding to eIF4E, this motif may stabilize eIF4E binding to localized mRNAs so that translation can resume immediately after CUP-mediated repression is relieved. Consistent with this interpretation, the posterior localization of eIF4E has been shown to depend on CUP (Wilhelm et al. 2003; Zappavigna et al. 2004). Thus, the effects of the Δ212 mutation may be partially related to a defect in eIF4E localization and/or on eIF4E phosphorylation (CUP also controls the eIF4E phosphorylation status in the ovary) (Wilhelm et al. 2003; Nakamura et al. 2004; Zappavigna et al. 2004). A role for the canonical 4E-BM1 motif in recruiting eIF4E to mRNA targets is also supported by the observation that CUP can recruit eIF4E to mRNAs that lack an m7G cap structure (Chekulaeva et al. 2006; Jeske et al. 2011). Thus, the 4E-BM1 motif may enable eIF4E to piggyback on transported mRNAs.
Role of the regulatory domain and the noncanonical eIF4E-binding motif
The noncanonical 4E-BM2 motif also contributes to eIF4E binding, although to a lesser extent than the canonical 4E-BM1 (Fig. 2B; Nelson et al. 2004). This observation raised the question of whether this motif plays a role in translational repression. A major finding in the present study is that this motif is required to inhibit the potent decapping and degradative activity of the CUP effector domain in tethering assays. However, additional sequences within the N-terminal regulatory domain also contribute to protect CUP-associated mRNAs from degradation, as the isolated effector domain exhibits a stronger degradative activity than CUP Mut2 and Mut1+2.
It is unclear how the regulatory domain and the 4E-BM2 motif can counteract the activity of the effector domain. One possible mechanism could be that the regulatory domain and 4E-BM2 block decapping indirectly by making the cap structure less accessible to DCP2. This protection could be achieved by increasing eIF4E affinity for the cap structure or, alternatively, competing with unknown proteins that facilitate eIF4E dissociation. However, the interaction of 4E-BM2 with eIF4E is weak (Fig. 2B; Nelson et al. 2004), suggesting that the inhibitory activity of 4E-BM2 may be independent of eIF4E binding. Another possible mechanism could be that the regulatory domain and/or 4E-BM2 directly antagonizes DCP2 recruitment and/or activation at mRNAs associated with CUP. This possibility is consistent with the observation that when the regulatory domain is deleted or 4E-BM2 is mutated, CUP promotes the decapping of deadenylated targets, indicating that DCP2 is recruited to and activated at CUP-associated mRNAs in the absence of the regulatory domain or the 4E-BM2 motif. It is also possible that the regulatory domain or 4E-BM2 directly interferes with the activity of the effector domain. However, we consider this possibility unlikely because the regulatory domain of CUP is sufficient to stabilize bound mRNAs (Fig. 3C). Finally, neither wild-type CUP nor a 4E-BM2 mutant (Mut2) interacted with DCP2, EDC4, or XRN1, indicating that 4E-BM2 does not inhibit decapping by directly preventing the interaction between CUP and decapping enzymes.
Regardless of the specific mechanism, the observation that the regulatory domain and the 4E-BM2 motif inhibit decapping of associated mRNAs suggest that their activity could be subject to regulation, possibly via post-translational modifications or binding partners, so that mRNAs associated with CUP could be either fully degraded (e.g., if unlocalized) or stored in a repressed deadenylated form.
More generally, the finding that the regulatory domain and the 4E-BM2 motif can block mRNA decapping in cis is both striking and unprecedented. This result opens up the exciting possibility that similar domains and motifs are present in other proteins, where they could specify alternative fates for bound mRNAs: complete degradation or storage in a deadenylated, repressed form for translation at a later time point. Therefore, our study has important implications for the understanding of translational regulation by 4EBPs. This topic warrants further investigation, as the repression of 4EBP targets may also be achieved through diverse, potentially 4E-independent mechanisms.
Materials and methods
DNA constructs
Luciferase reporters and plasmids for expression of GFP-, V5-, or HA-tagged cDNAs encoding full-length DCP2, DCP2 catalytic mutant (E361Q), GW182, MBP, and GST were described previously (Behm-Ansmant et al. 2006; Tritschler et al. 2008; Haas et al. 2010). An F-luc-oskar reporter was generated by inserting the oskar 3′ UTR into the NheI and SalI sites of plasmid pAc5.1-F-Luc. A plasmid for the expression of XRN1-V5 was obtained by inserting the XRN1 ORF into the Not1 and Xba1 sites of the pAc5.1A vector. The GFP-NOT1 construct was obtained by inserting the corresponding cDNA into the pAc5.1B-EGFP vector using EcoRI and XbaI restriction sites. A cDNA fragment corresponding to the Bruno ORF was cloned into the EcoRI and XhoI sites of pAc5.1B-EGFP. Plasmids for the expression of additional subunits of the deadenylase complex were described previously (Haas et al. 2010). Plasmids for the expression of HA- or GFP-tagged wild-type or mutant CUP were obtained by inserting the corresponding cDNAs into the EcoRV and NotI sites of pAc5.1B-λN-HA and pAc5.1B-EGFP vectors. Mutant D. melanogaster CUP constructs were generated by site-directed mutagenesis using the QuickChange Mutagenesis kit from Stratagene. CUP mutants Mut1, Mut2, and Mut1+2 carry alanine substitutions of residues Y342 and L347 (Mut1); L379 and L383 (Mut2); or Y342, L347, L379, and L383 (Mut1+2). For expression of the 7SL RNA, a fragment of 1200 base pairs (bp), including the 7SL enhancer, promoter, and terminator sequences, was amplified from genomic DNA and cloned in vector pAc5.1A.
Functional assays, dsRNAi, and RT–PCR analysis
For the λN-tethering assay, 2 × 106 cells were cotransfected with the following plasmids: 0.1 μg of reporter plasmid (F-Luc-5BoxB or F-Luc), 0.4 μg of pAc5.1C-R-Luc-V5, 0.5 μg of 7SL reporter, and 0.2 μg of plasmid expressing λN-HA or λN-HA-CUP protein fusions. For the assay using F-Luc-oskar, 2 × 106 cells were cotransfected with the following plasmids: 0.1 μg of reporter plasmid (F-Luc-oskar), 0.4 μg of pAc5.1C-R-Luc-V5, 0.5 μg of 7SL reporter, 0.2 μg of plasmid expressing MBP or CUP protein fusions, and 0.1 μg of plasmid expressing GST or GFP-Bruno. RNAi, Northern blotting, RNase H cleavage assays, and RT–PCR analysis were performed as described (Behm-Ansmant et al. 2006). Detailed procedures are provided in the Supplemental Material.
Coimmunoprecipitation assays and Western blotting
For coimmunoprecipitation assays, S2 cells (10 × 106 to 12 × 106 cells) were collected 3 d after transfection. Immunoprecipitations were performed as described by Tritschler et al. (2008). HA-tagged and GFP-tagged proteins were detected using an HRP-conjugated monoclonal anti-HA antibody (1:5000; Roche 3F10) and an anti-GFP antibody (1:2000; Roche 11814460001), respectively. V5-tagged proteins were detected using anti-V5 antibodies (1:5000; QED Bioscience, Inc.). Endogenous proteins were detected with rat or rabbit polyclonal antibodies eIF4E (1:3000), Tral (1:1000), EDC4 (1:500), NOT1 (1:1000), XRN1 (1:2000), and Me31B (1:2000). All Western blots were developed with the ECL Western blotting detection system (GE Healthcare) according to the manufacturer's recommendations. The association of the F-Luc-5BoxB mRNA with eIF4G was analyzed by coimmunoprecipitation followed by quantitative RT–PCR as described by Zekri et al. (2009).
Acknowledgments
We are grateful to A. Eulalio for generating some of the CUP expression plasmids, T. Nishihara for providing a plasmid for expression of a 7SL derivative, J.E. Braun for providing the F-Luc-oskar reporter, and E. Wahle for the gift of the anti-NOT1 antibody. This study was supported by the Max Planck Society, and by grants from the Deutsche Forschungsgemeinschaft (DFG; FOR855 and the Gottfried Wilhelm Leibniz Program awarded to E.I.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17136311.
References
- Altmann M, Schmitz N, Berset C, Trachsel H 1997. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J 16: 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M 2005. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell 9: 511–522 [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A 2003. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 130: 835–843 [DOI] [PubMed] [Google Scholar]
- Chang JS, Tan L, Schedl P 1999. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol 215: 91–106 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Singer RH 2006. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci 31: 687–693 [DOI] [PubMed] [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J 19: 3142–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E 2009. Deadenylation is a widespread effect of miRNA regulation. RNA 15: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE 2005. Exon–junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E 2010. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14: 5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E 2006. Rapid ATP-dependent deadenylation of nanos mRNA in a cell-free system from Drosophila embryos. J Biol Chem 281: 25124–25133 [DOI] [PubMed] [Google Scholar]
- Jeske M, Moritz B, Anders A, Wahle E 2011. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J 30: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge F, Zaessinger S, Temme C, Wahle E, Simonelig M 2002. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J 21: 6603–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes LN, Spradling AC 1997. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development 124: 1419–1431 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM 1995. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412 [DOI] [PubMed] [Google Scholar]
- Lie YS, Macdonald PM 1999. Translational regulation of oskar mRNA occurs independent of the cap and poly(A) tail in Drosophila ovarian extracts. Development 126: 4989–4996 [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol 15: 4990–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA 2004. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 23: 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccioni F, Zappavigna V, Verrotti AC 2005. A cup full of functions. RNA Biol 2: 125–128 [DOI] [PubMed] [Google Scholar]
- Piccioni F, Ottone C, Brescia P, Pisa V, Siciliano G, Galasso A, Gigliotti S, Graziani F, Verrotti AC 2009. The translational repressor Cup associates with the adaptor protein Miranda and the mRNA carrier Staufen at multiple time-points during Drosophila oogenesis. Gene 428: 47–52 [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA 2005. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 15: 284–294 [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155 [DOI] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E 2010. Subunits of the Drosophila CCR4–NOT complex and their roles in mRNA deadenylation. RNA 16: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V 2008. A similar mode of interaction enables Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol 28: 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrotti AC, Wharton RP 2000. Nanos interacts with cup in the female germline of Drosophila. Development 127: 5225–5232 [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Smibert CA 2005. Mechanisms of translational regulation in Drosophila. Biol Cell 97: 235–252 [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ 2003. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 163: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685 [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M 2006. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133: 4573–4583 [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC 2004. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci 101: 14800–14805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstädt S, Izaurralde E 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]