Abstract
Querying MS/MS spectra against a database containing only proteotypic peptides reduces data analysis time due to reduction of database size. Despite the speed advantage, this search strategy is challenged by issues of statistical significance and coverage. The former requires separating systematically significant identifications from less confident identifications, while the latter arises when the underlying peptide is not present, due to single amino acid polymorphisms (SAPs) or post-translational modifications (PTMs), in the proteotypic peptide libraries searched. To address both issues simultaneously, we have extended RAId’s knowledge database to include proteotypic information, utilized RAId’s statistical strategy to assign statistical significance to proteotypic peptides, and modified RAId’s programs to allow for consideration of proteotypic information during database searches. The extended database alleviates the coverage problem since all annotated modifications, even those occurred within proteotypic peptides, may be considered. Taking into account the likelihoods of observation, the statistical strategy of RAId provides accurate E-value assignments regardless whether a candidate peptide is proteotypic or not. The advantage of including proteotypic information is evidenced by its superior retrieval performance when compared to regular database searches.
Keywords: peptide identification, statistical significance, E-value, proteotypic peptides, knowledge integration
1. INTRODUCTION
In mass spectrometry (MS) based proteomics experiments, only a small subset of database peptides can be consistently observed or identified [1–3]. These consistently observable peptides, also termed proteotypic peptides [1, 3], apparently play an important role in protein identification. One should note that other definition of proteotypic peptide also exists. For example, proteotypic peptide is sometimes defined, in addition to being consistently observed, as that uniquely identifies a specific protein or protein isoform [2]. Through this manuscript, we adopt the definition of proteotypic peptide given in Craig, Cortens and Beavis [1]. That is, a proteotypic peptide in this manuscript represents one that can be consistently detected but not necessarily one that uniquely identifies a specific protein or protein isoform.
The fact that proteotypic peptides do not necessarily belong to the most abundant proteins also makes them important for protein quantitation [3]. Recent research efforts involving proteotypic peptides include, but are not limited to, construction of peoteotypic peptide libraries for protein identifications [1, 2, 4], cataloguing of proteotypic peptides in failing/nonfailing human heart [5], and de novo proteotypic peptide prediction [6] that may aid designs/choices of synthetic peptides for absolute peptide quantitation [7].
Querying MS/MS spectra against a database containing only proteotypic peptides can significantly reduce the data analysis time [1]. This speed advantage comes from a reduction of the effective database size, which also makes methods searching spectral libraries [8–12] fast. Interestingly, it is likely that proteotypic peptides are the ones ending up present in spectral libraries because a consensus spectrum in any spectral library is usually constructed by averaging over sufficient number of good quality spectra of the same peptide. Another advantage of searching only proteotypic peptides is that by concentrating on the pool of most observable ones, one may reduce the likelihood of false identifications. This is an implicit use of our proteomics knowledge that not every possible peptide appears with equal probability.
Despite the advantage of speed and focusing on the most observable set by searching a database consisting of only proteotypic peptides, there are potential issues need to be addressed. For example, it is possible that such a database restriction may introduce false negatives, especially when, for a given query spectrum, the best candidate from the peptide libraries does not seem a very confident identification. Further, even when there exists library peptide that scores very well, it is still possible that another database peptide, not present in the library, can score even better. The central question arises from these concerns is: how does one assign the statistical significance to candidate peptides from searching a proteotypic peptide library? Because peptides are not observed with equal probability [1], how does one include such an experimental fact into statistical analysis without risking the introduction of false negatives? Furthermore, is it possible to draw a line to separate significant identifications from less confident identifications?
In addition to the problem of statistical significance assignment, another important question arises upon considering personalized/targeted proteomics studies. It is foreseeable that in experiments involving complex protein mixtures, not every proteotypic peptide can be observed. That is, in a complex mixture it is quite likely that only one out of a few proteotypic peptides of a given protein can be observed. One then asks, what if for an individual this proteotypic peptide segment happens to contain a single amino acid polymorphism (SAP) or a post-translational modification (PTM) and this specific variant is not present in the standard proteotypic peptide libraries.
In this paper, we propose a possible solution to simultaneously address the aforementioned issues such as statistical significance assignment, incorporation of the fact that proteotypic peptides are more observable than others, and problems related to the presence of SAPs and PTMs within proteotypic peptides. The basic idea is to incorporate proteotypic peptide information to the knowledge-enhanced databases of RAId DbS as well as to utilize the statistical framework of RAId_DbS [13] and RAId_aPS [14] to assign statistical significance to proteotypic peptides. The remainder of this paper is organized as follows. In the Methods section, we will briefly review the statistical strategies of RAId_DbS and RAId_aPS, describe how the knowledge-enhanced databases of RAId can incorporate proteotypic peptide information. In the Results and Discussion section we present the results obtained from integrating proteotypic peptide information in RAId’s database, assess the utility of RAId’s enhanced database, evaluate statistical significance accuracy of RAId’s statistical strategy, and we comment on retrieval improvement when incorporating proteotypic information. Strengths and limitations of this approach, some technical remarks, and a short description of future directions will be provided in the Conclusion section.
Materials
The proteotypic peptide spectra used for this study were downloaded from the following spectral libraries: (a) National Institute of Standards and Technology (NIST) at http://www.peptideatlas.org/speclib/#NIST, (b) Institute for Systems Biology (ISB) at http://www.peptideatlas.org/speclib/, and (c) the Global Proteome Machine Organization (GPM) at ftp://ftp.thegpm.org/projects/xhunter/libs/. From the above spectral libraries, we have downloaded spectra files for the twenty organisms listed in Table 1. Note that not every organism used in our study has corresponding spectral files on all three libraries. Throughout this manuscript we use Homo sapiens spectra files to demonstrate how we include proteotypic peptide information in RAId’s database. To augment RAId’s knowledge databases, we processed, without performing database searches, the downloaded spectral library files using RAId_aPS (version 08.18.2009; available from ftp://ftp.ncbi.nlm.nih.gov/pub/RAId/Software/RAId/).
Table 1.
List of the downloaded files that were used to construct enhanced knowledge database of RAId.
| Organism | GPM | NIST | ISB |
|---|---|---|---|
| Homo sapiens | human_cmp_20.mgf | NIST_human_IT_v3.0_2009_02_04_7AA.sptxt | ISB_Hs_plasma_20070706_Q0.sptxt |
| Homo sapiens | NIST_human-7-22-2008-qtof.sptxt | ISB_Hs_plasma_20070706_Q1.sptxt | |
| Homo sapiens | ISB_Hs_plasma_20070706_Q2.sptxt | ||
| Homo sapiens | ISB_human_consensus_Q0.sptxt | ||
| Aspergillus fumigatus | afumigatus_cmp_20.mgf | ||
| Arabidopsis | ath1_cmp_20.mgf | ||
| Bos taurus | cow_cmp_20.mgf | NIST_bsa_IT_v3.0_2009-05-06.msp.gz | |
| Caenorhabditis elegans | worm_cmp_20.mgf | ISB_Ce-phospho_20080313.gz | |
| Canis familiaris | dog_cmp_20.mgf | ||
| Cavia porcellus | cavia_cmp_20.mgf | ||
| Danio rerio | fish_cmp_20.mgf | ||
| Drosophila melanogaster | NIST_drosophila_IT_v3.0_2008-07-14.msp.gz | ISB_Dm-phospho_20080313.gz | |
| Equus caballus | horse_cmp_20.mgf | ||
| Escherichia coli | NIST_ecoli_IT_v3.0_2009-05-21.msp.gz | ||
| Felis catus | cat_cmp_20.mgf | ||
| Gallus gallus | chicken_cmp_20.mgf | NIST_chicken_IT_v3.0_2009-04-15.msp.gz | |
| Macaca mulatta | rhesus_cmp_20.mgf | ||
| Mus musculus | mouse_cmp_20.mgf | NIST_mouse_IT_v3.0_2009-04-29.msp.gz | |
| Oryctolagus cuniculus | rabbit_cmp_20.mgf | ||
| Pan troglodytes | chimp_cmp_20.mgf | ||
| Rattus norvegicus | rat_cmp_20.mgf | NIST_rat_IT_v3.0_2009-05-21.msp.gz | |
| Saccharomyces cerevisiae | yeast_cmp_20.mgf | NIST_yeast_IT_v3.0_2009-05-04.msp.gz | ISB_Sc-phospho_20080313.gz |
| Saccharomyces cerevisiae | NIST_yeast_QTOF_v3.0_2009-05-06.msp.gz | ||
| Schizosaccharomyces pombe | spombe_cmp_20.mgf |
For evaluation of statistical significance accuracy and retrieval performance, two spectral library data sets and two experimentally generated spectra sets were used. The two spectral library data sets, ISB_Human_Plasma_Q1 (containing 23, 841 spectra) and ISB_Human_Plasma_Q0 (containing 29, 109 spectra), are downloaded from http://www.peptideatlas.org/speclib/. The two experimentally generated spectra sets, PRIDE_Exp_mzData_Ac_8421.xml (containing 15, 916 spectra) and PRIDE_Exp_mzData_Ac_8422.xml (containing 15, 561 spectra) are downloaded from the PRoteomics IDEntifications (PRIDE) database (http:www.ebi.ac.ukprideppp2_links.do).
All database searches performed for this study are done using RAId_DbS (version 08.18.2009). RAId_aPS and RAId_DbS have been implemented to handle as input different spectral file formats such as PKL, DTA, MGF, pepXML, and others as well as to handle the file format used by different spectral libraries, all of which helped the data processing pipeline. All the computational analysis for this study was done in the Biowulf computer cluster at the National Institutes of Health.
Methods
In the introduction, we have described the need of (i) assigning accurate statistical significance to proteotypic peptides and (ii) treating the possibility of false negatives. In the first subsection, we will briefly review the statistical strategy used by the software package RAId (including both RAId_DbS and RAId_aPS) and show that this strategy may be applied to simultaneously mitigate both issues mentioned above. In the second subsection, we will describe how the knowledge-enhanced databases of RAId_DbS can accommodate SAPs and PTMs within proteotypic peptides and thus may help advances towards personalized medicine and targeted/individualized proteomics. We have also noticed that in some spectral libraries there exist documented amino acid modifications from pull-down experiments [15, 16] and also from in vitro amino acid modifications (such oxidation and deamidation) that may be integrated to our knowledge database to potentially improve search sensitivity of RAId.
Assigning statistical significance to proteotypic peptides
The expected number of false positive hits increases with the number of independent entries compared. In terms of mathematical expression, this statement is essentially
| (1) |
where σ indicates the query spectrum, Nd is the total number of qualified peptides (whose molecular masses difference to the parent ion mass are within the mass error tolerance) in a database, P(S∣σ) represents the P-value associated with the score cutoff S when using spectrum σ as query, and E(S∣σ) is the E-value associated with the score cutoff S when using spectrum σ as query. For a given spectrum and a score threshold, the P-value associated with that score threshold is defined to be the probability of finding a false hit that have score better than or equal to that threshold. On the other hand, the E-value associated with that score threshold is defined to be the expected number of false hits that have score better than or equal to that threshold. In simple terms, the E-value associated with a candidate peptide may be viewed as the number of false hits anticipated, from querying a spectrum, before calling the peptide at hand a true hit.
The statistical inference thus depends on how P-value is obtained. A simple method to obtain P-value is via score histogram of peptides considered. The area underneath the score histogram is normalized to be one, and the partial area with score above a given score threshold yields the P-value associated with that score cutoff. Unfortunately, when considering only database peptides, the best P-value obtained this way is 1/Nd, and when multiplied by the factor Nd, limiting the best E-value to be 1 and preventing meaningful statistical inference. This problem can be overcome by considering the score histogram of all possible peptides, be they in the databases or not, within the appropriate mass range [14, 17]. One then assumes that the score distribution thus obtained can be used to assign P-values for peptides of all categories, proteotypic or not. Since peptides within any category is a subset of all possible peptides considered, this assumption is quite reasonable and is employed by RAId_aPS [14]. There also exists other way to obtain a score distribution function by properly extrapolating/interpolating score histogram of peptides considered. By extending the central limit theorem to take into account the skewness of the m/z peak intensity distribution per spectrum, the score distribution function for P-value assignment used by RAId_DbS can be theoretically derived [13] to reach the regime of arbitrarily small P-value. In principle, one may use a similar approach to derive score distribution functions associated with proteotypic and nonproteotypic peptides. However, extrapolating the score histogram of a small number of proteotypic peptides to form a score distribution function can be error prone. For this reason, RAId_DbS [13] use the same score distribution function for both proteotypic and nonproteotypic peptides.
Founded on the idea above, the statistical strategy of RAId_DbS and RAId_aPS is to further categorize peptides into different sets so that the total number of qualified peptides becomes set-dependent. For example, it is known that when using a digesting enzyme such as trypsin, it is more likely to observe the tryptic peptides with correct cleavages than peptides with incorrect cleavages. Therefore, one would like to consider peptides with correct cleavages at both terminus as more probable to be observed than peptides with incorrect N-terminal cleavages. We therefore have two different counters set up, one only counts qualified peptides with canonical cleavages while the second one counts the number of all qualified peptides (with correct or incorrect N-terminal cleavages). The basic idea is to give each candidate peptide the best benefit of the doubt. When assigning the statistical significance to a candidate peptide with correct cleavages on both terminus, we will set Nd to be the number recorded in the first counter, while for a candidate peptide with an incorrect N-terminal cleavage, we will set Nd to be the number recorded in the second counter. Assuming that we have for a query spectrum two candidate peptides of identical scores (and thus identical P-values) and that one candidate has canonical cleavages while the other has an incorrect N-terminal cleavage, upon employing formula (1) to assign statistical significance, the one with canonical cleavages will be assigned a smaller (or more significant) E-value than the one with an incorrect N-terminal cleavage due to the fact that the former is multiplied by a smaller Nd. Looking at this strategy from a different view point, we are effectively assigning a smaller prior probability of being false positives to peptides with canonical cleavages than to those with incorrect N-terminal cleavages. As described below, this idea can be easily extended to accommodate the fact that proteotypic peptides are more likely to be observed than others.
Let’s introduce another counter that counts the number of proteotypic peptides within the set of qualified peptides. Evidently, since the set of proteotypic peptides is a subset of qualified peptides, the proteotypic counter will accumulate a smaller number than the qualified-peptide (qp) counter. For non-proteotypic peptide hit, the corresponding Nd used for computing E-value is the number from the qp counter, which sums the total number of proteotypic and non-proteotypic peptides. Consequently, it is equivalent to say that we are assigning a smaller prior probability of being false positives to proteotypic peptides than to non-proteotypic peptides. What we have implemented is a bit more complicated than just described. In fact, for a given molecular weight range, the counters for proteotypic and non-proteotypic groups are each further extended to consider the cases of whether the N-terminal cleavage is canonical and the number of miscleavage sites present in the peptides. To be more precise, we define below the notation for various counters and provide some examples.
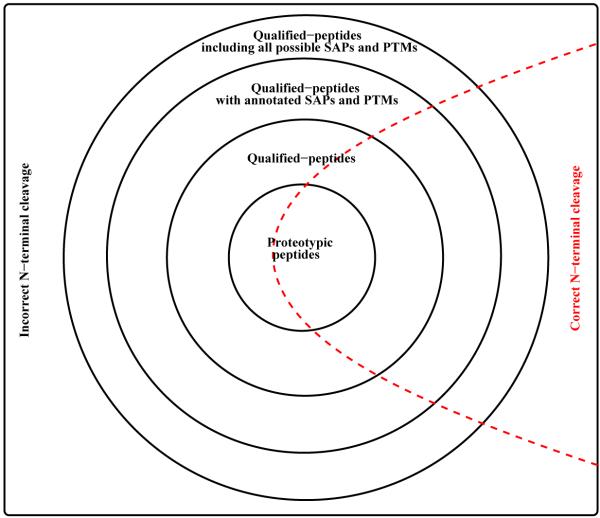
The notion Nd(k; fp∣fN·fC·) denotes the counter needed for peptides with k miscleavages and with proteotypic flag fp, N-terminal cleavage flag fN, and C-terminal cleavage flag fC. Proteotypic (qp) counter is indicated by fp → p (fp → n), while correct (incorrect) N-terminal cleavage is indicated by fN → c (fN → i). Since we always demand correct C-terminal cleavage, in our study, we always have fC → c. Therefore the qualified-peptide (qp) counter with incorrect N-terminal cleavage needed for peptides with 3 miscleavages is denoted by Nd(3; n∣i.c.). The counter for k miscleavage sites records the total number of peptides containing j ≤ k miscleavages. As an example, for k = 2 and with canonical cleavages on both terminus, the proteotypic counter Nd(2; p∣c.c.) sums the total number of proteotypic peptides with zero, one, and two miscleavage sites conditioned upon canonical cleavages are on both terminus. The qp counter Nd(k; n∣i.c.), needed for non-proteotypic peptides with k miscleavages and with incorrect N-terminal cleavage, sums the total number of non-proteotypic and proteotypic peptides with k or less miscleavages regardless of whether the N-terminal cleavage being correct or not. Therefore, the Bonferroni correction factor Nd is always smaller for proteotypic peptides, i.e., each of them has a smaller prior probability to be a false positive than that associated with a non-proteotypic peptide. Figure 1 illustrates the relationship between different type of counters.
Figure 1.
A schematic illustration of different counters. The venn diagram above shows how different groups of qualified peptides are related. Note that a peptide can be counted towards many different counters. Proteotypic peptide set is included in the qualified-peptide set, which is included in the qualified-peptide with annotated SAPs and PTMs set, and so on. The red dashed line is used to illustrate schematically that each candidate peptide set shown in this figure can be further divided into groups with correct and incorrect N-terminal cleavages.
The key step therefore is to set up a few new counters to accommodate the fact that one would like to assign a smaller prior probability of being false positives to proteotypic peptides, provided that P-values can be obtained accurately. However, obtaining accurate P-values is in general a nontrivial task. In our earlier publications, we have shown that one may choose to use either the score distribution of database peptides [13] or use the score distribution of all possible peptides [14, 17] to obtain accurate P-values. We have thus incorporated the proteotypic peptide search feature in both RAId_DbS [13] and RAId_aPS [14].
Integrating proteotypic information into RAId’s knowledge-enhanced database
In general, what is proteotypic in a particular protocol, on a given instrument, might not be proteotypic in another setting. Therefore, in principle, one may have protocol-specific library of proteotypic peptides and lumping these protocol-specific proteotypic peptides together may seem strange. Nevertheless, aggregating proteotypic peptides from different protocols does have certain advantages. First, it provides a maximal proteotypic library that is likely to saturate (as argued in [1]) as more and more spectral data are generated. This maximal proteotypic library is still expected to be much smaller than the organismal database, and thus still distinguish proteotypic peptides from the others. Second, this aggregation increases the number in proteotypic peptide counter and thus prevents E-value from becoming too small (significant). Therefore, within our framework, we do not assume the existence of generic proteotypic peptides across different protocols, but rather, take the union of proteotypic peptides under all existing protocols.
The task of integrating proteotypic information is achieved as follows. Within the knowledge-enhanced databases constructed for RAId_DbS [18] (also used by RAId_aPS), by specifying the beginning and terminating points we mark proteotypic peptides extracted from either a given spectral library or from a prescribed list. Peptides marked this way when scored are counted towards the newly devised proteotypic counter(s). Formula (1) is then applied with Nd = Nd(k; p∣c.c.)(Nd = Nd(k; p∣i.c.)) when assigning E-values to proteotypic peptides containing k miscleavages and with correct(incorrect) N-terminal cleavages.
In order to integrate proteotypic information into RAId’s enhanced databases, we processed spectral libraries files for the twenty organisms downloaded from three spectral libraries (GPM, ISB, NIST) to decide which peptides to mark proteotypic. Each of the downloaded library spectra has an “consensus peptide” associated with it. Using RAId_aPS’s reassigning E-value mode and using multiple scoring functions (XCorr [19], Hyperscore [20], K-score [21], and RAId score [13]), we rescored each consensus peptide using its associated library spectrum to obtain a combined E-value. This process is spectrum-centric. That is, we treat each spectrum as an independent entity even if some spectra were assigned the same “consensus peptide”. Because different scoring functions have different noise filters,[14] it seems reasonable to use the combined E-value as a quality control measure to ensure that only confidently identified peptides within the spectral libraries were labelled as proteotypic.
By simultaneously scoring all possible peptides (typically of order 1025 or more peptides for a given molecular mass larger than 2, 000 Da.), RAId_aPS[14] is able to obtain score distributions for the selected scoring functions and use these distributions to obtain respectively the corresponding P-values. By transforming each of these P-values into a database P-value and by applying Fisher’s formula [22, 23] to combine the database P-values, one can obtain a combined P-value and hence a combined E-value. Since details of this procedure is given in [23], we only provide the recipe to make the paper self-contained and will not delve into further elaboration.
Given a spectrum σ, the E-value associated with score S is obtained using eq. (1). Assumes that the occurrence of a high-scoring random hit is a rare event and thus can be modeled by a Poisson process with expected number of occurrence E(S∣σ), one may then define another P-value, which is called the database P-value, via
| (2) |
The database P-value Pdb(S) represents the probability of seeing at least one hit in a given random database with quality score larger than or equal to S. Fisher’s formula combines L independent P-values , p1; p2, … , pL. With , the combined P-value is given by
| (3) |
When L = 2, Fisher’s formula yields Pcomb(p1p2) = p1p2 [1 – ln(p1p2)]. Once Pcomb is obtained, we may invert the formula in eq. (2) to get a combined E-value Ecomb via
| (4) |
In carrying out the procedure above, it is necessary to include the Bonferroni factor Nd, see eq. (1). Since the database search parameters (such as mass error tolerance used) associated with each consensus peptide is not always available, we set Nd = 10, 000 when obtaining the combined E-value via eq. (1). This is because a regular database search with post-translational modifications option turned off yields approximately 10, 000 candidate peptides (see Figure 2) when the parent ion molecular weight is greater than 800 Da and with a mass error tolerance of ±3 Da. A peptide is labelled proteotypic if it has a combined E-value less than 10−3. This cutoff means that one would expect one out of 1, 000 labelled proteotypic peptides to be a false identification.
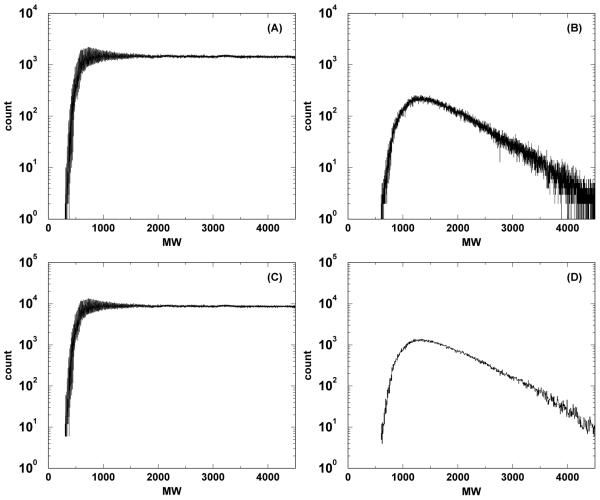
Figure 2.
Number of qualified peptides present in RAId’s Homo sapiens database as a function of precursor ion mass. Panel A(C) displays the result when counting all qualified peptides allowing up to two miscleavages when the precursor ion mass error tolerance is set to ±0.5 Da (±3 Da). Results when including only proteotypic peptides are shown correspondingly in panels B and D for mass error tolerance ±0.5 Da and ±3 Da.
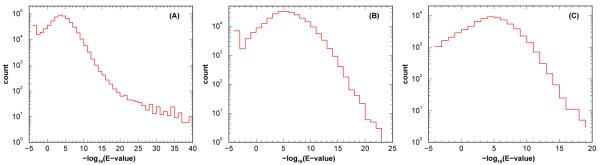
Figure 3 displays the combined E-value distributions for the consensus peptides from different spectral libraries. As one may see, all three distributions of the combined E-value reach their maxima at the range 10−4 to 10−5, along with long tails of smaller E-values. This indicates that our choosing 10−3 as the E-value cutoff allows one to view the majority of “consensus peptides” in spectral libraries as peoteotypic.
Figure 3.
Distribution of the combined E-values obtained from rescoring each of the consensus peptides associated with spectra in various Homo sapiens spectral libraries using RAId_aPS’s four scoring functions: XCorr, Hyperscore, Kscore, and RAId. Panels A, B, and C display results obtained respectively from the GPM, NIST, and ISB spectral libraries, which contain respectively 614,753, 260,491 and 69,616 spectra.
After applying the cutoff 10−3 in E-value, there are in total 734, 509 spectra from all three spectral libraries passed this threshold. Many of those spectra do share the same consensus peptides due to the fact that the raw spectra used by different libraries may overlap and a peptide may be annotated with different charge states. To avoid redundancy, duplicate consensus peptides (with combined E-value less than 10−3) within spectral libraries were removed to create a set of nonredundant peptides, which are to be marked proteotypic in RAId’s databases. Table 2 summarizes the results from analyzing Homo sapiens spectral library. After removing duplicate peptides, we obtained 271, 557 nonredundant proteotypic peptides. These proteotypic peptides were mapped to their corresponding proteins with the proteotypic label and length information inserted after each peptide’s C-terminal, and information associated with these proteotypic peptides was also integrated to RAId’s enhanced Homo sapiens database.
Table 2.
Number of analyzed Homo sapiens peptides from various spectral libraries. A consensus peptide may appear more than once in a given spectral library. The second column lists the total number of consensus peptides (with redundancy) within each library. The third column shows the total number of consensus peptides (with redundancy) that have combined E-values less than 10−3 assigned by RAId_aPS. The non-redundant version of this column is recorded in the fourth column. Due to the inter-library redundancy, the total number, 271,557, of nonredundant consensus peptides is smaller than the sum from that of each library. These 271,557 nonredundant consensus peptides were labeled proteotypic in RAId’s Homo sapiens database.
| spectral library |
no. of pept. |
no. of pept. with E ≤ 10−3 |
no. of nonredundant pept. with E ≤ 10−3 |
|---|---|---|---|
| GPM | 614,753 | 456,674 | 186,906 |
| NIST | 260,491 | 223,094 | 163,290 |
| ISB | 69,616 | 54,741 | 19,526 |
| Total | 944,860 | 734,509 | 271,557 |
Results and Discussion
To enable the use of proteotypic information in spectral data analysis, we have modified RAId program to accommodate new options. Basically, the baseline option L− means not to include proteotypic information, reducing to the original RAId_DbS [13] search. Instead of scoring only proteotypic peptides, our proteotypic on (L+) option scores all qualified peptides in the database but with proteotypic peptides counted towards both the proteotypic counter and the regular counter while the non-proteotypic peptides are only counted towards the regular counter. The motivation is given below using human proteome as an example.
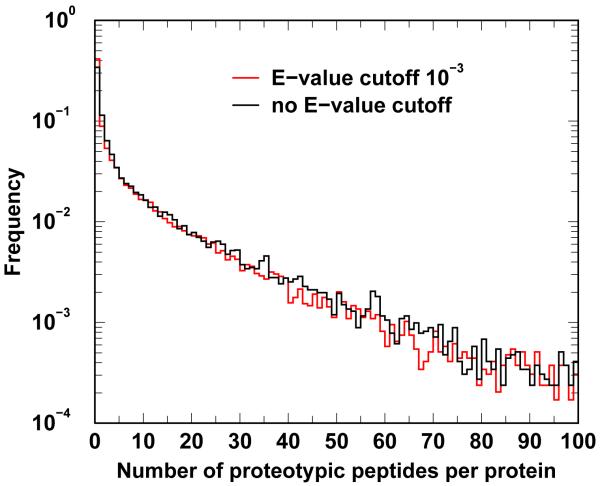
The human protein coverages by proteotypic peptides are shown in Figure 4. Without E-value cutoff, we find that about 35% of human proteins do not contain any proteotypic peptide. With combined E-value less than 10−3 required, the percentage of human protein void of proteotypic peptides increases to 40%. This indicates that if one were to search a database of proteotypic peptides only, there will be no chance to identify these (35 to 40%) of proteotypic-peptide-deficient human proteins. That is why among all RAId options, scoring only proteotypic peptide is not one of them.
Figure 4.
Normalized protein counts. The ordinate represents percentage of Homo sapiens proteins containing a certain number, specified by the abscissa, of proteotypic peptides. The E-value cutoff here refers to the quality control E-value in proteotypic annotation as described in the Methods section.
For the remainder of this section, we report in the first subsection the overall status of RAId’s enhance databases constructed including proteotypic information. In the second subsection, we evaluate the accuracy of reported E-values by RAId program through querying RAId’s reversed human protein database with both library spectra and real experimental spectra. In the third subsection, we assess the retrieval gain when proteotypic information is included in the spectral data analysis.
Knowledge-enhanced databases with proteotypic information
Figure 5 describes in detail the structure of RAId’s enhanced database and how proteotypic peptides, PTM’s and SAP’s are labelled. As shown in the figure, it only costs a few additional bytes per proteotypic peptides. Since the number of proteotypic peptides are of order 104 ~ 105, the total increase in the database size due to inclusion of preteotypic peptides per organism is in general much smaller than the original organismal database. During the proteotypic mapping, PTMs and SAPs were all considered to gain maximum matches. PTMs that are in proteotypic peptides but not yet present in RAId’s databases are also added to RAId’s databases.
Figure 5.
Protein NP_001484 is used as an example to demonstrate the structure of our sequence file, part of the enhanced database. A “[” character is always inserted after the last amino acid of each protein to serve as a separator. Annotated SAPs (red color), PTMs (black color) and proteotypic information (green color) are included in a pair of angular brackets. Annotated SAPs and PTMs associated with an amino acid are included following that amino acid. SAPs are further enclosed by a pair of curly brackets, while PTMs are further enclosed by a pair of round brackets. Similarly, the length of a proteotypic peptide is indicated, after its C-terminal residue, within a pair of vertical bars. Amino acid followed by two zeros indicates an annotated SAP. Every annotated PTM has a two-digit positive integer that is used to distinguish different modifications. When there is more than one length enclosed by a pair of vertical bars, it means that there are multiple annotated proteotypic peptides sharing the same C-terminal.
It is worth pointing out that in vitro amino acid modifications such as oxidations and deamidations may occur during a proteomics experiment. Some occurrences of such were included in some spectral libraries. While extracting peptides from a spectral library, we record their associated in vitro and in vivo modifications, and integrate such information into our knowledge-enhanced database. Incorporation of these modifications may in principle increase detection sensitivity of searches.
RAId’s enhanced database (Figure 5) includes the protein sequence file and a definition file (Table 3), where the information sources related to SAPs, PTMs, diseases, and proteotypic peptides are documented. The definition file records for each proteotypic peptide its spectral indices associated with spectral libraries. The caption of Table 3 explains the structure and the content of RAId’s definition file. For each proteotypic peptide hit resulting from a database search, the peptide’s full information, including spectral indices, is reported along with the peptide. Thus users can easily investigate from which library spectrum/spectra did the reported proteotypic peptide arise.
Table 3.
Two sequences are used to demonstrate the structure of our processed information file. The text line after the “>” symbol contains accession numbers associated with the sequence. The other rows each contains six entries separated by tabs. The first column indicates the residue position. The second column indicates the modified residue(s) that can occur at the position specified in the first column. The third column, labeled by either SAP or PTM, indicates the modification type. The fifth column contains the accession number of the source of modification, this may be a protein sequence or mRNA. When the source of modification is obtained from proteotypic consensus peptide within a spectral library, the fifth column shows the proteotypic peptide instead. The fourth column explains the nature of the modification; a lower case letter indicates residue content in the source sequence, the upper case letter indicates the modified residue in the variant sequence. The notation, v → I, indicates the source sequence with amino acid V can change into I, ie, a SAP. The notation, gT C → A, is a short hand for codon change from gtc to atc, ie, a SNP that changes the coded amino acid from V to I as well. The sixth column contains additional information for the fourth column. It may include disease information, database entry index, or spectral indices when the modification is associated with a proteotypic peptide. As an example, the post-translational modification (M06) at position 59 of the second sequence is observed in the following proteotypic peptides: PDETM06VIGNYR, CFIEEIPDETM06VIGNYR, FHIGETEKKC-FIEEIPDETM06VIGNYR and CFIEEIPDETM06VIGNYR. The first three are obtained from spectra of GPM (human_cmp_20) spectral library with spectral indices 134010, 442918, 442918, 442920, and 589710, while the fourth one came from the NIST (#NIST_human_IT_v3.0) spectral library with spectral indices 25094 and 25102.
|
>NP_775259,NM_173167,Q8IWX7 | |||||
| 60 | I00 | SAP | gTC→A | ∣ NM 173167 ∣ | dbSNP:16970659 |
| 60 | I00 | SAP | v→I | ∣ Q8IWX7 ∣ | dbSNP:16970659, FTId=VAR 027506 |
| 199 | V00 | SAP | GcA→T | ∣ NM 173167 ∣ | dbSNP:35749208 |
| 377 | R00 | SAP | AaG→G | ∣ NM 173167 ∣ | dbSNP:41389545 |
| 496 | H00 | SAP | d→H | ∣ Q8IWX7 ∣ | (breast cancer). FTId=VAR 035870 |
| ⋥ | ⋥ | ⋥ | ⋥ | ⋥ | ⋥ |
| >NP_059980,NM_017510 | |||||
| 55 | Q00 | SAP | CcG→A | ∣ NM 017510 ∣ | #dbSNP:11545866 |
| 59 | M06 | PTM | m Hydroxylated | PDETM06VIGNYR | #human_cmp_20:134010 |
| 59 | M06 | PTM | m Hydroxylated | CFIEEIPDETM06VIGNYR | #human_cmp_20:442918,442919,442920 |
| 59 | M06 | PTM | m Hydroxylated | FHIGETEKKCFIEEIPDETM06VIGNYR | #human_cmp_20:589710 |
| 59 | M06 | PTM | m Hydroxylated | CFIEEIPDETM06VIGNYR | #NIST_human_IT_v3.0:25094,25102 |
| 77 | Q07 | PTM | q Deamidated | EEYQ07PATPGLGMFVEVKDPEDK | #human_cmp_20:524138 |
| 78 | Q00 | SAP | CcG→ A | ∣ NM 017510 ∣ | #dbSNP:11545867 |
| 85 | M06 | PTM | m Hydroxylated | EEYQPATPGLGM06FVEVK | #human_cmp_20:393423,393424,393425 |
| 85 | M06 | PTM | m Hydroxylated | EEYQPATPGLGM06FVEVK | #NIST_human_IT_v3.0:50099 |
| ⋥ | ⋥ | ⋥ | ⋥ | ⋥ | ⋥ |
Although, we have analyzed the spectral library files for twenty organisms, the proteotypic information was integrated into knowledge-enhanced databases only for the fourteen organisms listed in Table 4. This is because for the remaining six organisms, we need to work on incorporating SAPs, PTMs, and disease information first. We also plan to construct enhanced databases for other organisms in the future as data becomes available.
Table 4.
The header abbreviations in this table are explained as follows. The second column, headed by DB_name, documents the abbreviated database name for searches using standalone version of RAId_DbS. The column headed by “Protein” indicates the final number of protein clusters in the processed organismal databases. The columns headed by NP, NM, and SP summarize the break down of the total number of accession numbers included respectively from protein products, transcript products, and SwissProt protein entries. The columns headed by PPs, SAPs and PTMs indicate respectively the total number of annotated proteotypic peptides (PP), single amino acid polymorphisms (SAPs) and post-translational modifications (PTMs) included. The last column shows the database size in bytes.
| Organism | DB_name | Protein | NP | NM | SP | PPs | SAPs | PTMs | DB_size (byte) |
|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | hsa | 29284 | 35059 | 35031 | 15030 | 271557 | 116073 | 84406 | 16,265,018 |
| Arabidopsis thaliana | artha | 29651 | 31740 | 31711 | 5527 | 48707 | 5207 | 11977 | 12,318,213 |
| Bos taurus | botau | 23796 | 26504 | 26491 | 3979 | 102130 | 3295 | 15810 | 11,188,490 |
| Canis familiaris | cafam | 31705 | 33834 | 33821 | 528 | 65224 | 2766 | 4196 | 18,458,474 |
| Danio rerio | darer | 31192 | 36150 | 36137 | 1552 | 37113 | 7358 | 3841 | 14,477,794 |
| Drosophila melanogaster | drmel | 17232 | 20207 | 20207 | 2568 | 69104 | 5611 | 9290 | 9,796,785 |
| Equus caballus | eqcab | 17300 | 17637 | 17624 | 171 | 101657 | 485 | 1045 | 9,404,150 |
| Gallus gallus | gagal | 18154 | 18724 | 18681 | 1455 | 52576 | 1109 | 6522 | 8,728,501 |
| Macaca mulatta | mamul | 32547 | 38141 | 38128 | 207 | 139505 | 1370 | 1262 | 14,498,187 |
| Mus musculus | mumus | 28506 | 35503 | 35451 | 12170 | 179017 | 27614 | 61684 | 14,363,491 |
| Oryctolagus cuniculus | orsat | 26636 | 26784 | 26777 | 1205 | 66014 | 1291 | 2182 | 10,679,924 |
| Pan troglodytes | patro | 41464 | 52130 | 52117 | 482 | 166691 | 3721 | 3734 | 20,217,986 |
| Rattus norvegicus | ranor | 28914 | 39425 | 39389 | 5569 | 1181710 | 9297 | 33240 | 15,879,569 |
| Saccharomyces cerevisiae | sacer | 5699 | 5880 | 0 | 5807 | 95456 | 5507 | 13220 | 2,927,330 |
Evaluation of statistical significance accuracy
Even though RAId program was shown to report reasonably accurate E-values [13, 14], on should still test the accuracy of significance assignment by RAId’s statistical strategy (1) when proteotypic information is included. The four data sets mentioned in the Materials section are used for this purpose. As described in the Methods section, the Bonferroni corection factor Nd associated with a peptide is determined by whether the peptide is proteotypic, whether it has correct N-terminal cleavage, and the number of miscleavages within it.
For each of the four data sets, we search RAId’s reverse Homo sapiens database using respectively the L− (baseline) and the L+ (proteotypic) options. Every peptide hit in the reverse database is considered a false positive. When the search is performed with L−, only qp counters, such as Nd(k; n∣c.c.) and Nd(k; n∣i.c.), are deployed. For searches performed with L+ option, the proteotypic counters such as Nd(k; p∣c.c.) and Nd(k; p∣i.c.) are set up in addition to qp counters.
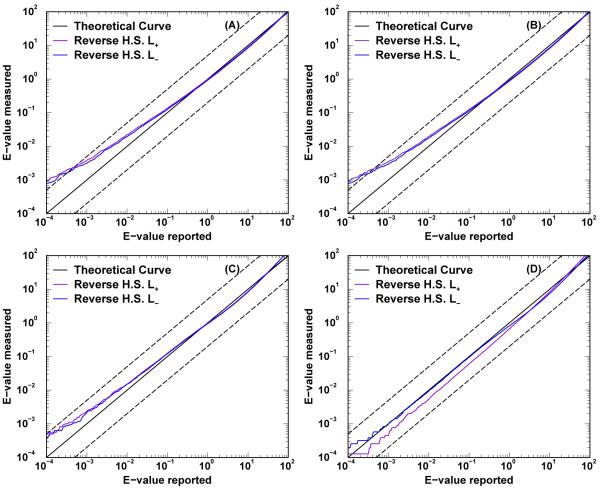
The E-value accuracy is tested by plotting the average number of false positives whose reported E-values by RAId_DbS are smaller than prescribed cutoffs. Figure 6 displays the E-value accuracy using the four datasets mentioned in the Materials section. All the E-value curves trace reasonably well the theoretical line x = y over six orders of magnitude with more observable deviations near small E-value region. When the query spectra used are obtained experimentally, panels (C) and (D) of Figure 6, the reported E-values seem to agree better with textbook definition than when using library spectra as queries, panels (A) and (B) of Figure 6. Investigation of this observed difference in accuracy of significance assignment between different query types is beyond the scope of the current manuscript. The important point, however, is that although the statistical significance assignment is not most accurate (when queried with library spectra), it is still good enough for our proteotypic curation. That is, when used as a quality control, the significance assignment obtained can still exclude library spectra with low confidence consensus peptides. In most applications where the query spectra are experimentally obtained, as indicated by panels (C) and (D) of Figure 6, RAId program can report accurate E-values even when proteotypic information is used.
Figure 6.
E-value accuracy assessment. The averaged cumulative number of false positives is plotted against the theoretical E-value. In addition to a theoretical line, each plot contains two curves: searching the reverse Homo sapiens (H.S.) database with L+ and L− options. Panels A and B display the results respectively from data sets ISB_Human_Plasma_Q1 (containing 23, 841 spectra) and ISB_Human_Plasma_Q0 (containing 29, 109 spectra). Both sets are part of the ISB library spectra that entered our proteotypic curation. Panels C and D display the results respectively from data sets PRIDE_Exp_mzData_Ac_8421.xml containing 15, 916 spectra) and PRIDE_Exp_mzData_Ac_8422.xml containing 15, 561 spectra). Both sets are experimentally acquired spectra downloaded from PRIDE database.
Retrieval performance evaluation
The utility of RAId’s proteotypic-information-included enhanced database is assessed in two steps. First, we examine what is the optimal retrieval gain achievable by using subsets of library spectra as queries. We then use two experimentally acquired data sets to evaluate what might be the typical retrieval gain. Throughout this section, the retrieval is shown as a function of the false discovery rate (FDR). When the true positives are know a priori, computing FDR is straightforward. When the true positives are unknown, the FDR may be estimated via target-decoy methods [24, 25]. To focus on the effect of including proteotypic information, we always and only compare retrieval results from running with L− (baseline) option and with L+ (proteotypic) option.
Two human plasma data sets, ISB_Human_Plasma_Q0 and ISB_Human_Plasma_Q1, both downloaded from ISB spectral library, are used for the first task. In these two data sets, each library spectrum is assigned a consensus peptide. We use the consensus peptides to form the true positive (TP) set. Knowing the true positive set, we don’t need a decoy database for this task. Any peptide hit outside the true positive set is considered a false positive (FP). The FDR in this case can be readily computed
| (5) |
where Ec is the specified E-value cutoff, tFP(E ≤ Ec) is the total number of FPs with E ≤ Ec, and T(E Ec) is the total number of candidate peptides, with E ≤ Ec, out of all query spectra.
For the second task, two experimentally generated spectra sets, PRIDE_Exp_mzData_Ac_8421.xml and PRIDE_Exp_mzData_Ac_8422.xml, are used. Since the TPs are unknown except that the samples are from Homo sapiens, we estimate the FDR by using a target(forward)-decoy(reverse) approach. The FDR is estimated by
| (6) |
where Ec is the specified E-value cutoff, D(E ≤ Ec) is the total number of hits with E ≤ Ec in decoy (reverse) database, and T(E ≤ Ec) is the total number of hits with E ≤ Ec in the target (forward) database. In this context, the retrieval is measured by number of peptides identified within the target database versus the FDR. When a peptide is identified with E ≤ Ec in k different query spectra, it will contribute k counts in the retrieval measurement.
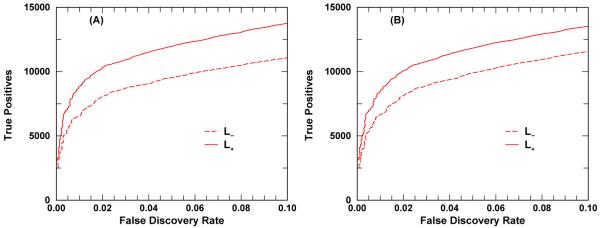
Each panel of Figure 7 displays the cumulative number of TPs versus FDR for a given query dataset under both L− and L+ options. Both panels exhibit the same trend: the retrieval effectiveness is better when proteotypic information is considered during data analysis. At FDR equals 5%, for the data set ISB Human_Plasma_Q0 (ISB_Human_Plasma_Q1), the L− option retrieved 9, 882 (9, 552) TPs while the L+ option retrieved 11, 783 (11, 895) TPs corresponding to a retrieval gain of approximately 19% ( 24%). However, the large increase of TPs retrieved by L+ option over L− option should not be regarded as typical gains by turning on the proteotypic option, but should be viewed as some kind of upper bounds instead. This is because a good fraction of the spectra used for this test have their consensus peptides marked proteotypic in our enhanced Homo sapiens database. The expected amount of gain will be tested in the second task along with some control tests.
Figure 7.
Enhanced database utility test. The cumulative number of true positives identified is plotted against the FDR, see eq. (5). Panel A (B) displays the results from ISB_Human_Plasma_Q1 (ISB_Human_Plasma_Q0) spectral library data set containing a total of 23, 841 (29, 109) spectra. Since these two spectral data sets were also included for proteotypic annotation, the performance gain indicates some sort of upper bounds rather than typical.
Upon compiling the search results of the first task, we have noticed the presence of significant false positives for each data set under both options. Many of those significant false positives come from library spectra whose consensus peptides received large (poor) combined E-values. For each data set and each search option, we have assembled the significant false positives, their E-values, their corresponding spectra’s consensus peptides, and consensus peptides’ statistical significance assigned by RAId aPS [14] into a supplementary excel file. It seems quite likely that the consensus peptides listed in these four supplementary excel files were incorrectly assigned to their corresponding library spectra since database peptides that can much better match the spectra exist. If this is true and if those significant false positives are actually true positives, then a small retrieval improvement might be reached compared to what is shown in Figure 7. This also illustrate the importance of our quality control mentioned in the Methods section.
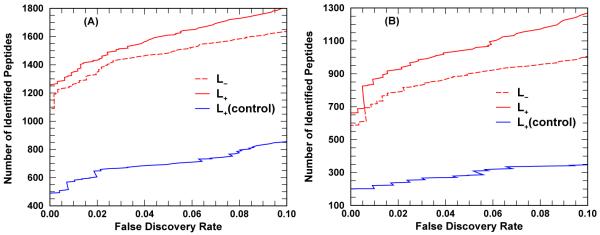
Figure 8 illustrates what typical retrieval gain may be when proteotypic information is included. Panel A (B) of Figure 8 shows retrieval results of both L− and L+ options using the PRIDE_Exp_mzData_Ac_8421.xml (PRIDE_Exp_mzData_Ac_8422.xml) data set. As expected, the retrieval gain is not as much as shown in Figure 7. At FDR equals 5%, for the data set 1; 486 (900) TPs while the L+ option retrieved 1; 610 (1; 050) TPs corresponding to a retrieval gain of approximately 8% ( 16%).
Figure 8.
RAId_DbS database search results for (A) PRIDE_Exp_mzData_Ac_8421.xml (15,916 spectra), and (B) PRIDE_Exp_mzData_Ac_8422.xml (15,561 spectra). The cumulative number of identified peptides within the target database is plotted against the FDR, see eq. (6). The blue (control) curves result from searches using a target database with fake proteotypic annotations: proteotypic peptides are randomly assigned. The blue curves indicate much damage in retrieval can be introduced if false proteotypic information is introduced.
Evidently, the effective database size of proteotypic peptides is smaller in L+ option than in L− option. A natural question arises. Is the retrieval gain solely due to the reduction of effective database or is it due to inclusion of correct proteotypic information? To address this question, we constructed a control target database that differs from RAId’s enhanced Homo sapiens database only in the proteotypic annotations. In the control target database, the proteotypic peptides, although randomly selected, are chosen to have the same molecular mass distribution as that of the proteotypic peptides in RAId’s enhanced Homo sapiens database. A control decoy database is generated from reversing the protein sequences in the control target database. Care was taken to ensure the knowledge annotations are also properly kept. The retrieval curves using the control target and decoy databases show much worse performance than the case when using RAId’s enhanced Homo sapiens database as target and its reverse as decoy for both L− and L+ options. The worse performance resulting from using the control target and decoy databases indicates the importance to have high quality annotation since low confidence annotation may result in poor performance. Not only indicating the importance to have a quality filtering when incorporating proteotypic information into databases, this also indicates that blindly shrink the effective database size will not boost the retrieval performance.
Conclusion
We have constructed for fourteen organisms enhanced databases containing proteotypic peptide information, and we intend to construct databases for more organisms in the future as data becomes available. It is worth mentioning that RAId’s database structure can in principle accommodate extra layers of information. [18] This means that proteotypic peptide information may have additional flags associated respectively with spectral counts (number of experimental spectra used to construct the library spectrum), tissue types, diseases, and experimental conditions. These additional layer of information can provide users with specific information that can be useful for clinical studies and allow for more flexibility. For example, the spectral count flag, once implemented, allows the users to dynamically set the threshold of calling a peptide proteotypic.
A problem with the current RAId’s enhanced database is that it does not yet contain correlation information between/within PTMs and SAPs. However, this does not emerge from the design of RAId’s database, but has to do with the lack available correlation information. Once available from the literature, the correlation information can be incorporated to RAId’s database [18].
In summary, we have demonstrated that proteotypic peptide information available in spectral libraries can be processed and incorporated into organism specific databases. We have shown how the proteotypic peptide information can be used by database search tools during database search to improve peptide identification. We have also shown that, when using experimentally acquired spectra as queries, the statistical strategy used by RAId_DbS produces E-values obeying the text book definition of E-value which can be employed to assign statistical significance to proteotipic peptides during database searches.
The enhanced organismal databases including proteotypic information can be downloaded from ftp://ftp.ncbi.nlm.nih.gov/pub/RAId/Software/RAId/. Binaries of modified RAId program for Linux, Windows, and Mac OS X are available from the same page.
Supplementary Material
Acknowledgement
We thank the administrative group of the NIH Biowulf clusters, where all the computational tasks were carried out. This work was supported by the Intramural Research Program of the National Library of Medicine of the National Institutes of Health/DHHS. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data For each of the two spectral library data sets, an Excel file listing significant false positives is provided for both the L− and L+ options.
References
- [1].Craig R, Cortens JP, Beavis RC. Rapid Commun. Mass Spectrom. 2005;19:1844–1850. doi: 10.1002/rcm.1992. [DOI] [PubMed] [Google Scholar]
- [2].Kuster B, Schirle M, Mallick P, Aebersold R. Nat. Rev. Mol. Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- [3].Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- [4].Yen CY, Russell S, Mendoza AM, Meyer-Arendt K, Sun S, Cios KJ, Ahn NG, Resing KA. Anal. Chem. 2006;78:1071–1084. doi: 10.1021/ac051127f. [DOI] [PubMed] [Google Scholar]
- [5].Kline KG, Frewen B, Bristow MR, Maccoss MJ, Wu CC. J. Proteome Res. 2008;7:5055–5061. doi: 10.1021/pr800239e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Webb-Robertson BJ, Cannon WR, Oehmen CS, Shah AR, Gurumoorthi V, Lipton MS, Waters KM. Bioinformatics. 2008;24:1503–1509. doi: 10.1093/bioinformatics/btn218. [DOI] [PubMed] [Google Scholar]
- [7].Kirkpatrick DS, Gerber SA, Gygi SP. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- [8].Yates JR, Morgan SF, Gatlin CL, Griffin PR, Eng JK. Anal. Chem. 1998;70:3557–3565. doi: 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- [9].Craig R, Cortens JC, Fenyo D, Beavis RC. J. Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- [10].Deutsch EW, Lam H, Aebersold R. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Anal. Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- [12].Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- [13].Alves G, Ogurtsov AY, Yu Y-K. Biology Direct. 2007;2:25. doi: 10.1186/1745-6150-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alves G, Ogurtsov AY, Yu Y-K. 2010. arXiv.org:0806.2685.
- [15].Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, Panse C, Lee HK, Schlapbach R, Aebersold R. Mol. Syst. Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. Nat. Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Alves G, Yu YK. Physica A. 2008;387:6538–6544. doi: 10.1016/j.physa.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Alves G, Ogurtsov AY, Yu YK. BMC Genomics. 2008;9:505. doi: 10.1186/1471-2164-9-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eng JK, McCormack AL, Yates JR., III J. Amer. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [20].Fenyo D, Beavis RC. Anal. Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- [21].MacLean B, Eng JK, Beavis RC, McIntosh M. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- [22].Fisher RA. Statistical Methods for Research Workers. 2nd ed Hafner; New York, NY: 1958. [Google Scholar]
- [23].Alves G, Wu WW, Wang G, Shen RF, Yu YK. J. Proteome Res. 2008;7:3102–3113. doi: 10.1021/pr700798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Elias JE, Gygi SP. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [25].Navarro P, Vazquez J. J. Proteome Res. 2009;8:1792–1796. doi: 10.1021/pr800362h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.