Abstract
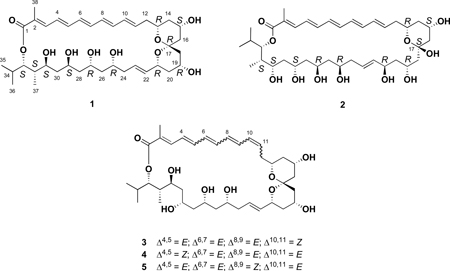
Two new polyene macrolides, marinisporolides A and B (1, 2), were isolated from the saline culture of the marine actinomycete, strain CNQ-140, identified as a member of the new marine genus “Marinispora.” The marinisporolides are 34 membered macrolides composed of a conjugated pentaene and several pairs of 1,3-dihydroxyl functionalities. Marinisporolide A (1) contains a bicyclic spiro-bis-tetrahydropyran ketal functionality, while marinisporolide B (2) is the corresponding hemiketal. The structures of these new compounds were assigned by combined spectral and chemical methods including extensive 2D NMR experiments and correlations of 13C NMR data with Kishi’s Universal NMR Database. Chemical modifications, including methanolysis, acetonide formation, and application of the modified Mosher method, provided the full stereostructures of these molecules. Three additional macrolides, marinisporolides C–E (3–5), which are olefin geometric isomers of marinisporolide A (1), were also isolated and their structures defined. Under room light, marinisporolides A and B readily photoisomerize to C–E indicating that they are most likely produced by photochemical conversion during the cultivation or isolation procedures. Although polyenes, marinisporolides A (1) and B (2) showed weak to no antifungal activity against Candida albicans.
Introduction
Since the discovery of actinomycin, cultured bacteria have been a prolific resource for drug discovery, with to date more than 13,000 bioactive metabolites described. More than 70% of these bioactive compounds have been isolated from cultured actinomycetes.1 The actinomycetes (Order Actinomycetales) are best known as soil bacteria, thus the majority of microbial drug discovery research has focused on terrestrial actinomycetes, and in particular strains of the chemically-prolific genus Streptomyces. Unfortunately, as the rate of known compound re-isolation from terrestrial actinomycetes has been increasing for more than a decade, the need to develop more efficient methods for the discovery of new chemical diversity has never been greater. Research on the metabolites of marine plants and animals began in the early 1970s, yielding to date large numbers of novel compounds that possess potent biological activities.2 Despite these productive results, marine discovery efforts were not equally extended to microorganisms in part due to the possibility that these bacteria are introduced into the marine environment from terrestrial sources such as wind blown spores or river runoff.3 It is now known that new actinomycete taxa occur in the ocean and that some display specific adaptations for life in the marine environment.4,5 These taxa include the chemically prolific genus Salinispora, members of which produce exciting new structural classes of secondary metabolites, such as the salinosporamides,6 sporolides,7 and cyanosporasides.8 A second new genus, for which the name “Marinispora" has been proposed, is also proving to be a productive source of new metabolites such as the marinomycins.9 These results provide clear evidence that targeting new marine actinomycete genera and species leads to the discovery of new chemotypes with significant biological activity and the potential to become leads for drug discovery.
To date, more than 20 “Marinispora” strains have been isolated from a diversity of marine habitats. The first chemical study of a cultured “Marinispora” strain (CNQ-140) led to the discovery of four novel macrodiolides, marinomycins A–D.9 Further chemical analysis of strain CNQ-140 has shown the presence of polyene macrolides that differ significantly from the marinomycins. Interestingly, the production of these macrolides has been observed in two additional “Marinispora” strains (CNP-027 and CNR-872) despite all three being isolated from diverse geographical locations (strain CNQ-140 from the Pacific Ocean near San Diego, CA, strain CNP-027 from the Sea of Cortès, Mèxico and strain CNR-872 from Palau). In this paper we report the structures of two new macrolides, marinisporolides A and B (1, 2), from culture extracts of Marinispora strain CNQ-140. The structures of these new macrolides, including their absolute stereostructures, were established by combined spectral and chemical methods. During isolation, marinisporolide A was observed to convert to complex olefin isomer mixtures, from which marinisporolides C–E (3–5) were subsequently isolated.
Results and Discussion
“Marinispora” strain CNQ-140 was isolated from marine sediment collected at a depth of 56 m offshore of La Jolla, California. The strain was cultured in 40 × 1 L volumes for six days and then extracted using Amberlite XAD-7 resin. The resin was filtered, eluted with acetone, the extraction solvent was concentrated under reduced pressure, and the residue was partitioned between ethyl acetate and water. The ethyl acetate-soluble fraction was subjected to a diversity of chromatographic purification steps to afford pure samples of marinisporolides A (1) and B (2), together with varying yields of three olefin geometrical isomers of 1, marinisporolides C (3), D (4) and E (5). Although solid data were obtained in the dark for 1 and 2, all of these compounds underwent facile photoisomerization to lead to complex olefin isomeric mixtures.
Marinisporolide A (1) was obtained as a yellow amorphous powder that analyzed for the molecular formula C38H58O10 by interpretation of HR-ESI-TOF MS (obsd [M+Na]+ at m/z 697.3922) and NMR data. The UV spectrum of 1 displayed absorption bands at 358 and 378 (sh) nm, indicating the presence of a conjugated polyene moiety.10 One and 2D 1H NMR spectral data (1H-1H COSY, TOCSY) illustrated signals attributable to a conjugated pentaene and a vinyl methyl group (Table 1). Interpretation of HMQC spectral data showed these proton signals correlated with 13C NMR signals at δ 138.9, 126.9, 140.4, 131.8, 136.5, 130.9, 134.6, 134.3, 131.4, and 12.5. Analysis of the 1H NMR spectrum of 1 showed the presence of six exchangeable protons (Table 1.), which illustrated that marinisporolide A (1) possessed at least 6 hydroxyl groups. Two dimensional NMR studies, emphasizing 1H-1H COSY and TOCSY experiments, allowed the conjugated pentaene to be connected to an aliphatic chain (C-12 – C-16) consisting of three sets of diastereotopic methylene protons and two oxymethine protons. In addition, analyses of 1H NMR, 1H-1H COSY and TOCSY spectral data for 1 showed the presence of seven additional oxymethine protons, six additional methylene protons, two nonconjugated trans olefinic protons, two aliphatic methine protons, and three secondary methyl groups. Interpretation of 13C NMR and HMQC spectral data for 1 allowed all protons to be assigned to their respective carbons. The oxymethine protons correlated with oxygenated carbons between in the region 62–80 ppm, while the methylene protons correlated with nine aliphatic methylene carbons at typical chemical shifts. The aliphatic methine protons showed correlations with carbon signals at δ 29.2 and 40.6, the aliphatic methyl group protons to carbon signals at δ 7.2, 18.7, and 19.5, and the two nonconjugated olefinic protons to olefinic carbon signals at δ 127.5 and 133.2. The 13C NMR spectrum of 1 also showed an oxygenated quaternary carbon signal at δ 98.7 (C-17), an olefinic carbon signal at δ 126.1, and an ester carbon signal at δ 167.8.
Table 1.
1H, 13C, and HMBC NMR Spectral Data for 1
| δ H mult (J, Hz)a | δ Cb | #H | HMBCa | |
|---|---|---|---|---|
| 1 | 167.8 | C | ||
| 2 | 126.1 | C | ||
| 3 | 7.09 d (11.5) | 138.9 | CH | C-1, 4, 5, 38 |
| 4 | 6.58 dd (14.0, 11.5) | 126.9 | CH | C-2, 6 |
| 5 | 6.70 dd (14.0, 10.5) | 140.4 | CH | C-3, 4, 6, 7 |
| 6 | 6.41 dd (14.5, 10.5) | 131.8 | CH | C-4, 8 |
| 7 | 6.54 dd (14.5, 10.5) | 136.5 | CH | C-5, 6, 9 |
| 8 | 6.28 dd (15.0, 10.5) | 130.9 | CH | C-6, 9, 10 |
| 9 | 6.40 dd (15.0, 11.0) | 134.6 | CH | C-7, 10 |
| 10 | 6.14 dd (14.5, 11.0) | 134.3 | CH | C-8, 9, 12 |
| 11 | 5.70 ddd (14.5, 9.0, 5.5) | 131.4 | CH | C-9, C-12 |
| 12a | 2.05 m | 38.9 | CH2 | C-10, 11, 13 |
| 12b | 2.42 br m | |||
| 13 | 3.42 br m | 67.4 | CH | |
| 14a | 1.03 md | 40.5g | CH2 | C-12, 13, 15, 16 |
| 14b | 1.81 mc | |||
| 15 | 3.72 br me | 63.7 | CH | |
| 16a / b | 1.11 md / 1.80 mc | 44.8 | CH2 | C-14, 15, 17, 18 |
| 17 | 98.7 | C | ||
| 18a | 0.90 m | 38.7 | CH2 | C-17, 19, 20 |
| 18b | 2.20 br dd (9.0, 3.0) | |||
| 19 | 3.54 br m | 62.2 | CH | |
| 20a / b | 1.06 md / 1.57 br d (10.5) | 42.3 | CH2 | C-19, 21, 22 |
| 21 | 4.20 br dd (10.5, 7.5) | 70.4 | CH | C-22, 23 |
| 22 | 5.31 dd (15.0, 7.5) | 133.2 | CH | C-23, 24 |
| 23 | 5.25 ddd (15.0, 8.0, 5.5) | 127.5 | CH | C-21, 22, 24 |
| 24a | 1.96 br ddd (14.0, 8.0, 6.0) | 41.3 | CH2 | C-22, 23, 25, 26 |
| 24b | 2.05 m | |||
| 25 | 3.44 br m | 68.0 | CH | |
| 26a / b | 1.12 md / 1.23 mf | 44.0h | CH2 | C-25, 27 |
| 27 | 3.71 br me | 65.8 | CH | |
| 28a / b | 1.06 mc / 1.23 mf | 44.2h | CH2 | C-29 |
| 29 | 3.63 br m | 69.0 | CH | |
| 30a / b | 1.34 m / 1.34 m | 39.5 | CH2 | C-31 |
| 31 | 3.46 br m | 73.8 | CH | |
| 32 | 1.82 mc | 40.6g | CH | C-37 |
| 33 | 4.83 dd (10.0, 1.0) | 79.9 | CH | C-1, 31, 32, 34, 35, 37 |
| 34 | 1.83 mc | 29.2 | CH | C-33, 36 |
| 35 | 0.81 d (6.0) | 19.5 | CH3 | C-33, 34, 36 |
| 36 | 0.88 d (6.0) | 18.7 | CH3 | C-33, 34, 35 |
| 37 | 0.87 d (7.5) | 7.2 | CH3 | C-31, 32 |
| 38 | 1.89 s | 12.5 | CH3 | C-1,2,3 |
| 15-OH | 4.70 br s | |||
| 19-OH | 4.57 br s | |||
| 25-OH | 4.48 br s | |||
| 27-OH | 4.34 br s | |||
| 29-OH | 4.70 br s | |||
| 31-OH | 4.53 br s |
500 MHz, DMSO-d6.
75 MHz, DMSO-d6.
Overlapping signals. Chemical shifts were assigned by interpretation of 1H-1H COSY data.
Interchangeable signals.
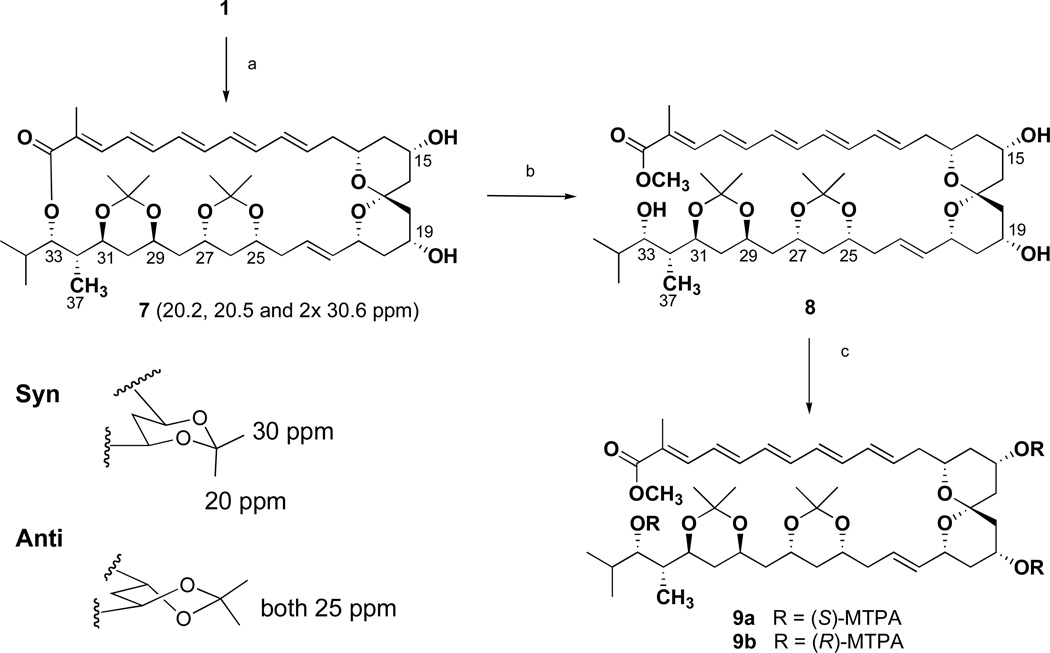
HMBC NMR correlations (Table 1) from the carbinol proton at δ 4.83 (H-33) and the olefinic proton at δ 7.09 (H-3) to the ester carbon at δ 167.8 allowed the ester and the pentaene chain to be connected to the C-33 position of the polyol chain. HMBC correlations from a vinyl methyl group (H3-38) to the C-2 and C-3 olefinic carbons and the C-1 ester carbon allowed the vinyl methyl group to be positioned at C-2. HMBC correlations also showed that the pentaene functionality from C-1 to C-16 was connected to the C-18 to C- 36 polyol chain through the C-17 ketal quaternary carbon. The structure of the C-13 – C-21 spiro-bis-tetrahydropyran ketal ring constellation was assigned by analysis of 2-D NMR data from ROESY and HMBC experiments. Combined NMR data indicated the presence of a spiro [5.5] ketal system. This was confirmed by comparison of the proton and carbon NMR data of 1 with that of the previously reported spirotetrahydropyran analogue of roflamycoin.11
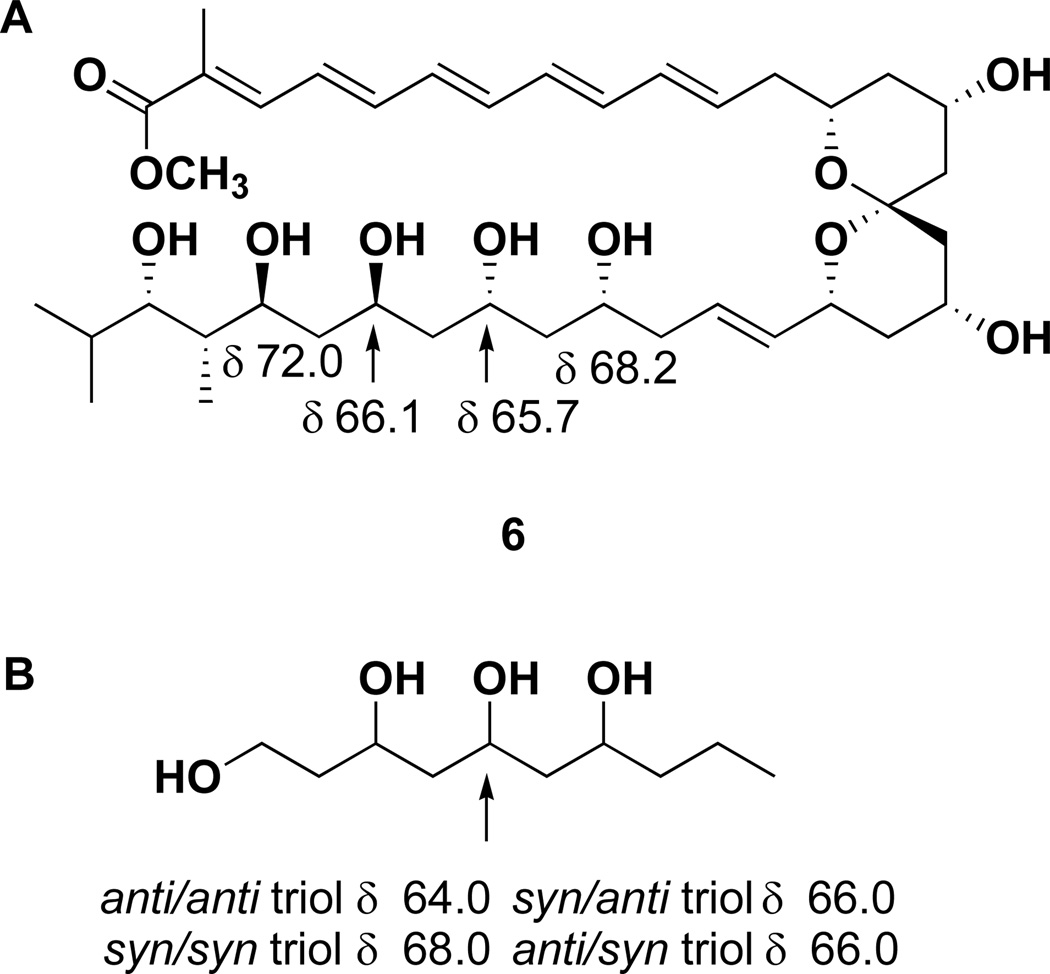
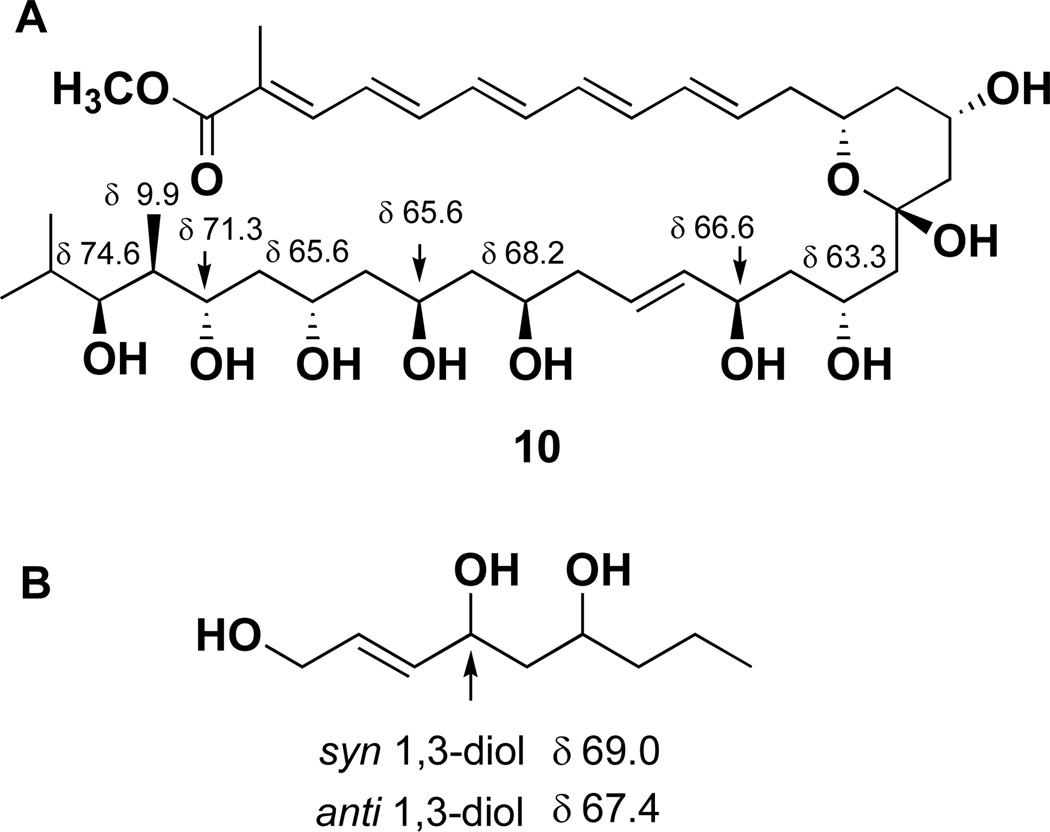
The geometries of the four double bonds (C-4 – C-11) in the pentaene chain were determined to be trans (E) on the basis of their characteristically large coupling constants (J ≥ 14.0 Hz) observed in the 1H NMR spectrum. The Δ2,3 trisubstituted olefin was assigned as E on the basis of a prominent ROESY correlation between the vinyl methyl protons (H3-38) and H-4. Next, the relative stereochemistry of marinisporolide A (1) was assigned by spectral analysis and chemical modification. The relative configurations of C-25, 27, 29, 31, 32, and 33 were initially suggested by comparison of NMR data from 1 with model compounds from Kishi’s Universal NMR Database12, and by conversion to their corresponding acetone ketals. Kobayashi et al.12 reported that the relative configuration of 1,3,5-triols can be predicted by comparison of 13C NMR chemical shifts of C-5 of the stereoisomers with the model compound decan-1,3,5,7-tetraol. Methanolysis of 1, by treatment with NaOMe and MeOH, yielded the methyl ester 6. The carbon chemical shifts of C-27 (δ 65.7) and C-29 (δ 66.1) in the HMQC spectrum of 6 (DMSO-d6), were very close to the C-5 value (δ 66.0) of the anti/syn triol or the syn/anti triol in the Kishi Database (Figure 1). This result suggested that the relative configurations of the hydroxyl groups at C-25, 27, 29, and 31 in 6 are syn/anti/syn or anti/syn/anti.
Figure 1.
Carbon chemical shift comparisons of the carbinol carbons in methyl ester 6 (A) with those of model compound (B).
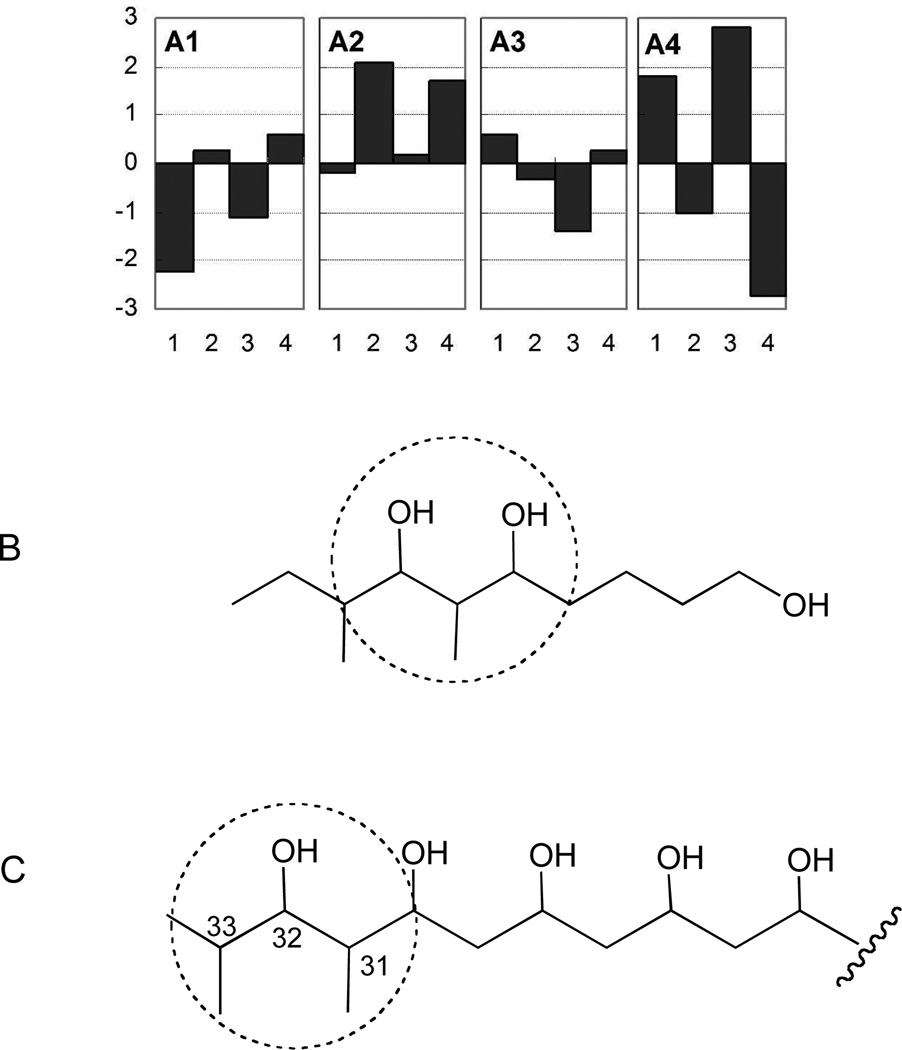
The relative configurations of the hydroxyl group at C-31, the methyl group (C-37) at C-32, and the hydroxyl group at C-33 of 6 were assigned as anti/syn on the basis of the carbon chemical shifts of C-31, C-32, C-33 and C-37 of 6, and the carbon chemical shifts of C-5, C-6, C-7, in comparison with the 6-methyl signal of each diastereomer of 6,8-dimethyl-deca-1,5,7-triol (Figure 2, Table S1).12a Treatment of 1 with 2,2-dimethoxypropane and pyridinium p-toluenesulfonate (PPTS) in methanol resulted in ketal formation at the C-25, C-27, C-29, and C-31 hydroxyl bearing carbons to afford the bis-acetonide 7 (Scheme 1). The carbon chemical shifts of the acetonide methyl groups were observed at δ 20.2, 20.5, 30.6 (× 2) in the HMQC spectrum of 7, indicating both six-membered 1,3-dioxane rings were in chair conformations. Rychnovsky’s method predicts that the carbon chemical shifts of methyl groups in a syn-acetonide (chair conformation) are at ca. δ 20 and ca. δ 30 (axial and equatorial methyl groups), while the anti 1,3-diol acetonide adopts the skew form to yield magnetically equivalent methyl groups observed at ca. δ 2513
Figure 2.
(A) Difference between the carbon NMR chemical shifts of C-31, 32, 33 and 37 of 6, and the values of each diastereomer of the model compound, with A1, A2, A3, and A4 representing α,α,β(hydroxyl group at C-1, methyl group at C-2, hydroxyl group at C-3)-, β,α,β-, β,α,α-, and α,α,α-diastereomers, respectively. In (A), the x- and y-axes represent carbon number and Δδ (Δδ = δModel compound - δ6). (B) The structure of the model compound. (C) The relevant polyol portion (C-31 – C-33) of 6.
Scheme 1.
Formation of acetonide 7 and conversion of 7 to ester 8, and conversion of 8 to the MTPA esters 9a,b.
Reagents and conditions: (a) MeOH, 2,2-dimethoxypyridine, PPTS, 0° C, 3h; (b) (i) 2.3% NaOMe/MeOH, RT, 1 h, (ii) Purification by C-18 RP HPLC using 45% MeCN/H2O; (c) (i) pyridine, DMAP, MTPA-Cl, RT, overnight, (ii) Purification to LC-MS single peak purity by silica normal phase chromatography (Sep-Pak, 1 g) using hexanes/EtOAc (5:1).
On the basis of the observed acetonide methyl chemical shifts, the hydroxyl groups at C-25 and C-27, and at C-29 and C-31 were assigned syn and syn configurations, respectively. The relative configurations of C-31, C-32, and C-33 of 1 were further confirmed to be anti/syn on the basis of NOE correlations between the H3-37 protons (methyl group at C-32), the α-equatorial proton at C-30 (H-30b; δ 1.45), and the α-axial proton at C-31 (H-31, δ 3.58) in the ROESY spectrum of 7 (Figure 3 and Figure S1). The relative configurations of the 1,3 diols were thus assigned as 25,27-syn, 27,29-anti, 29,31-syn, 31,32-anti, and 32,33-syn.
Figure 3.
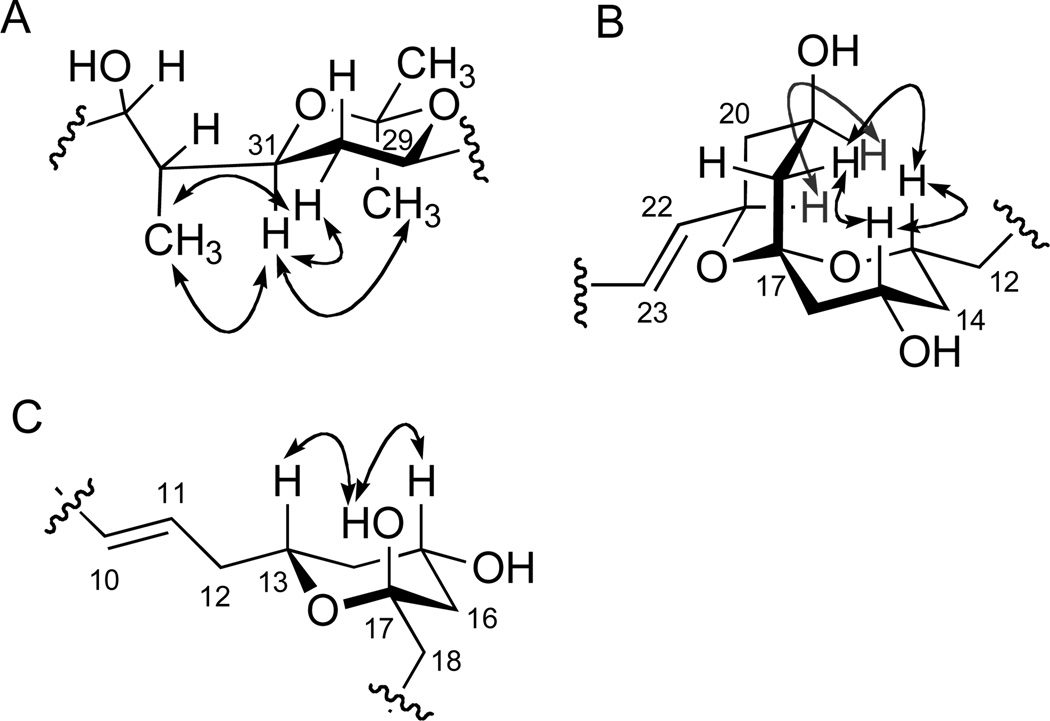
Key ROESY NMR correlation data (A) 1,3-diol acetonide at the C-29 and C-31 portion of compound 7. (B) The spiro-bis-tetrahydropyran ketal portion of compound 1. (C) The tetrahydropyran hemiketal portion of compound 2.
Assignment of the relative conformation of the spiro-bis-tetrahydropyran ketal portion of 1 was accomplished by comprehensive analysis of ROESY NMR correlation data (Figure 3). Briefly, the ROESY correlations between H-13 (δ 3.42), H-15 (δ 3.72), and equatorial proton at C-14 (H-14b, δ 1.81) indicated the 1,3 diaxial relation of H-13 and H-15. Similarly, the oxymethine protons at C-19 and C-21 were assigned axial configurations based on cross peaks in the ROESY spectrum between H-19 and H-21. In addition, one of the methylene protons at C-18 (H-18b) [δ 2.20 (1H, br dd, J = 9.0, 3.0 Hz)] was assigned in an equatorial configuration based on its small vicinal coupling constant observed in the 1H NMR spectrum, as well as a ROESY correlation with an adjacent axial methine proton at C-19. The ROESY correlations observed between H-13, H-15 and, H-18b suggested that C-18 was positioned in an axial position toward H-13 and H-15, as shown in Figure 3. On the basis of these overall analyses, the relative stereochemistry of the spiroketal portion of 1 was assigned with β-axial configurations of H-13, H-15 and C-18, and β-axial configurations of H-19 and H-21.
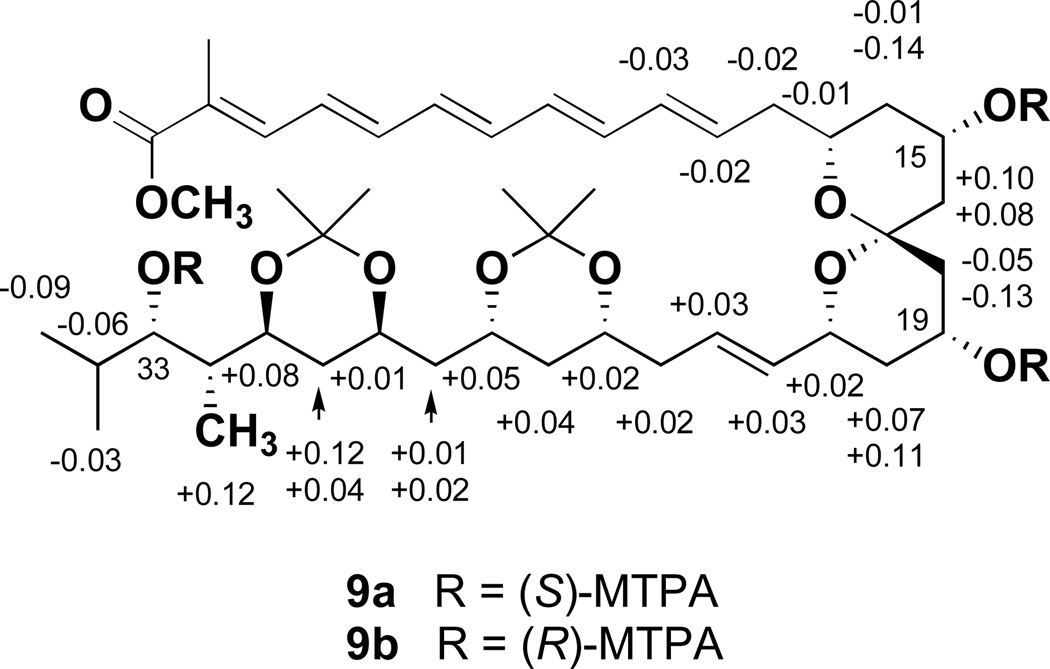
The absolute stereochemistry of marinisporolide A (1) was next determined by application of the modified Mosher ester NMR method14 using the methanolysis product 8 (Scheme 1). Treatment of 8 in pyridine with (R)-(−)-α-methoxy-α-(trifluoromethyl)phenylacetyl chloride (R-MTAP-Cl) and (S)-(+)-MTPA-Cl, in separate experiments, yielded the tris-S-Mosher ester 9a and tris-R-Mosher ester 9b, respectively. Analysis of the 1H NMR chemical shift differences (ΔδS-R) between 9a and 9b resulted in negative ΔδS-R values for H-11 – H-14 of the Mosher ester group at C-15, and positive values for H-16. Furthermore, negative ΔδS-R values for H2-18 of the C-19 Mosher ester were observed, while the H-20 to H-24 proton signals showed positive differences. These data allowed the absolute configurations at C-15 and C-19 to be assigned as S and R, respectively. In a similar fashion, NMR analysis of the H-24 to H-32 proton chemical shifts of 9a and 9b revealed the absolute configuration at C-33 to be S (Figure 4 and Figure S2).
Figure 4.
ΔδS-R values for the Mosher triesters 9a and 9b
Marinisporolide B (2) was isolated as a yellow amorphous powder that analyzed for the molecular formula C38H60O11 by HR-ESI-TOF MS data (obsd [M+Na]+ at m/z 715.4027) and analysis of NMR data (Table 2). The UV absorption spectrum of 2 was almost identical to that of marinisporolide A (1), which suggested the same chromophore as in 1. In a similar manner as with marinisporolide A, comprehensive analysis of the 2D NMR (1H-1H COSY, TOCSY, ROESY, HMQC, and HMBC experiments) data for 2, allowed the complete assignment of the proton and carbon signals, leading to the construction of a planar structure for 2. Unlike 1, the 1H NMR spectral data for 2 included HMBC correlations which illustrated signals for only eight exchangeable hydroxyl protons (Table 2). Analysis of 1H-1H COSY and TOCSY NMR data showed that only one of the eight hydroxyl groups [δ 5.56 (1H, br s)] did not correlate with an oxymethine proton. Further, the carbinol proton at C-13 did not couple to a hydroxyl proton signal, indicating these signals were part of an ether (hemiketal) linkage. An HMBC correlation (Table 2) from a hydroxyl proton at δ 5.56 to the quaternary C-17 (δ 97.3) indicated that C-17 is a hemiketal carbon. Analysis of key HMBC correlations then led to the construction of a tetrahydropyran hemiketal ring. The chemical shifts of the protons and carbons in the hemiketal ring were comparable to those from the previously reported constellation in roflamycoin.11
Table 2.
1H, 13C, and HMBC NMR Spectral Data for 2
| δ H mult (J, Hz)a | δ Cb | HMBC | ||
|---|---|---|---|---|
| 1 | 167.7 | C | ||
| 2 | 126.3 | C | ||
| 3 | 7.06 d (10.5) | 138.2 | CH | C-1, 2, 5, 38 |
| 4 | 6.59 dd (14.5, 10.5) | 127.0 | CH | C-2, 3, 5, 6 |
| 5 | 6.68 dd (14.5, 11.0) | 140.2 | CH | C-3, 4, 6, 7 |
| 6 | 6.42 dd (14.5, 11.0) | 131.5 | CH | C-4, 5, 7, 8 |
| 7 | 6.52 dd (14.5, 10.5) | 137.1 | CH | C-5, 6, 8, 9 |
| 8 | 6.26 dd (15.0, 10.5) | 130.4 | CH | C-6, 7, 9 10 |
| 9 | 6.34 dd (15.0, 10.5) | 135.5 | CH | C-7, 8, 10, 11 |
| 10 | 6.17 dd (15.0, 10.5) | 132.0 | CH | C-8, 9, 12 |
| 11 | 5.82 dt (15.0, 7.0) | 133.4 | CH | C-9, 12, 13 |
| 12a | 2.16 ddd (15.0, 7.5, 7.0) | 38.2 | CH2 | C-10, 11, 13, 14 |
| 12b | 2.33 br m | |||
| 13 | 3.90 br mc | 66.7 | CH | |
| 14a | 1.03 q (each 11.5) | 40.6h | CH2 | C-12, 13, 15, 16 |
| 14b | 1.76 md | |||
| 15 | 3.85 br mc | 62.8 | CH | |
| 16a /b | 1.12 me / 1.76 md | 44.7i | CH2 | C-14, 15, 17 |
| 17 | 97.3 | C | ||
| 18a | 1.53 m | 48.4 | CH2 | C-16, 17, 19 |
| 18b | 1.64 dd (13.5,8.0) | |||
| 19 | 4.05 br mf | 64.2 | CH | |
| 20a | 1.35 mg / 1.53 m | 44.8i | CH2 | C-19, 21, 22 |
| 20b | ||||
| 21 | 4.04 br mf | 68.5 | CH | |
| 22 | 5.38 dd (15.0, 6.0) | 136.0 | CH | C-21, 23, 24 |
| 23 | 5.49 dt (15.0, 6.5) | 125.4 | CH | C-21, 22, 24, 25 |
| 24a / b | 1.94 m / 1.99 m | 40.9j | CH2 | C-22, 23, 25, 26 |
| 25 | 3.57 br m | 69.6 | CH | |
| 26a / b | 1.22 m / 1.28 mg | 44.0 | CH2 | C-25, 27, 28 |
| 27 | 3.87 br mc | 66.0k | CH | |
| 28a / b | 1.13 m/ 1.13 me | 44.4i | CH2 | C-29 |
| 29 | 3.78 br m | 65.9k | CH | |
| 30a /b | 1.31 m / 1.31 mg | 41.6 | CH2 | C-28, 29, 31 |
| 31 | 3.30 m | 71.9 | CH | C-29, 33, 37 |
| 32 | 1.76 md | 40.8j | CH | C-30, 31, 37 |
| 33 | 4.87 d (10.0) | 79.1 | CH | C-1, 31, 32, 34, 35, 36, 37 |
| 34 | 1.85 m | 29.4 | CH | C-33, 35, 36 |
| 35 | 0.81 d (6.5) | 19.8 | CH3 | C-33, 34, 36 |
| 36 | 0.88 d (6.5) | 18.8 | CH3 | C-33, 34, 35 |
| 37 | 0.86 d (7.0) | 8.1 | CH3 | C-31, 32 |
| 38 | 1.89 s | 12.7 | CH3 | C-1, 2, 3 |
| 15-OH | 4.55 d (5.0) | C-14, 15, 16 | ||
| 17-OH | 5.56 br s | C-16, 17, 18 | ||
| 19-OH | 4.45 d (4.0) | C-18, 19, 20 | ||
| 21-OH | 4.47 d (4.5) | C-20, 21, 22 | ||
| 25-OH | 4.61 d (3.5) | C-24, 25, 26 | ||
| 27-OH | 4.50 d (4.0) | C-26, 27 | ||
| 29-OH | 4.40 d (5.0) | C-28, 29, 30, | ||
| 31-OH | 4.40 d (5.0) | 31, 32 |
500 MHz, DMSO-d6.
75 MHz, DMSO-d6.
Overlapping signals. Chemical shifts were assigned using 1H-1H COSY data.
Interchangeable signals.
The relative configuration of the tetrahydropyran hemiketal portion of 2 was assigned on the basis of ROESY NMR information (Figure 3 and Figure S3). Critical ROESY correlations between the H-13 oxymethine proton (δ 3.90), the H-15 oxymethine proton (δ 3.85), and the 17-OH hydroxyl proton indicated 1,3 diaxial relationships of H-13, H-15, and 17-OH on the same face of a six-membered chair conformation. Methanolysis of 2 with NaOMe in MeOH yielded the methyl ester 10. HMQC NMR data derived from 10 were utilized to analyze the relative configurations of the other oxymethine carbons (C-19, 21, 25, 27, 29, 31, 32, and 33) first by application of Kishi’s Universal NMR Database12, and then by comparison of the carbon chemical shifts of 10 with those from the methanolysis product 6. The carbon chemical shift of C-21 (δ 66.6, DMSO-d6) was very similar to the C-4 value (δ 67.4) of the anti-diol in the model compound, non-2-en-1,4,6-triol (Figure 5), indicating an anti 1,3-diol configuration at C-19 and C-21 in 10. The carbon chemical shifts of C-25, C-27, C-29, C-31, C-32 and C-33 in 10 (Table 2) were similar to those from 6 (Figure 1 and Figure 5), which suggested the same relative configuration at C-25–C-33 in 10. To confirm these assignments, 2 was treated with Dowex® X2-100 resin in MeOH for 10 min to generate marinisporolide A (1) as the major product. The 1H NMR (Figure S14) and CD spectra (Figure S59) of 1, derived from 2, were identical to spectral data from marinisporolide A (1). On the basis of these overall analyses, the absolute configuration of 2 was assigned as 13R, 15S, 17S, 19R, 21R, 25R, 27R, 29S, 31S, 32S, and 33S.
Figure 5.
(A) Carbon chemical shifts comparison of the carbinol carbons in methyl ester 10 with those of model compound (B).
Under the reaction conditions for methanolysis and acetone ketal formation, marinisporolides A (1) and B (2) degraded and were converted to complex mixtures that included geometric isomers. We examined several conditions, using HPLC analysis, for methanolysis and ketal formation in order to improve product yields. Treatment of 1 with 5% NaOMe in MeOH (anhyd.) for 1 hour at room temperature in the dark, cleanly provided the methanolysis product 6, albeit in low yield. Although acetonide formation was also problematic, bis-acetonide 7 was obtained in relatively good yield by reaction with excess pyridinium-p-toluenesulfonate and 2,2-dimethoxypropane for 3 hours at 0 °C in the dark.
Working with the marinisporolides A (1) and B (2) was also difficult due to their photochemical reactivity. When methanol solutions of 1 or 2 were exposed to ambient light in a capped clear vial, an equilibrium mixture of four main compounds was obtained within 2 hours. By HPLC peak area analysis, the equilibrium mixture from the photoconversion of 1 was 3:3:25:15 (marinisporolides E:D:C:A) (Figure S4). The compound ratio derived from 2 was 9:25:82:74 (unidentified isomers a:b:c:marinisporolide B) (Figure S5). It is known that the regioselectivity of cis-trans isomerization in polyenes is affected by the polarity of the solvent, the presence of electron donating and withdrawing substituents, and in particular the electron withdrawing group conjugated to the polyene is important.15 This suggested that steric effects and the conjugated carbonyl groups in structures 1 and 2 play a key role in limiting conversion to mainly 4 of 32 possible cis-trans isomers from 1 or 2. The photoisomerization products 3–5 were subsequently isolated from the culture extract and identified. Analysis of 2D NMR data allowed the structures of 3, 4, and 5 to be confidently assigned. However, in 3–5, and in several other cases, derivatives of these compounds were only analyzed with great difficulty. In many cases, analysis of the sample being evaluated was found to be composed of complex mixtures even when carefully treated in the dark. During attempts to record IR, UV, and some HRMS data, carefully light protected samples were found to photoisomerize. Comprehensive NMR analyses showed that the only differences between these isomers are that 3, 4, and 5 have Δ10,11 = Z, Δ4,5= Z, and Δ8,9= Z olefin configurations, respectively.
To examine the possible production of 3–5 under fermentation conditions, “Marinispora” strain CNQ-140 was cultured in the dark. This resulted in the production of primarily 1 and 2 (>95%). These observations strongly suggest that marinisporolides A (1) and B (2) appear to be true natural products, while isomers 3, 4 and 5 are artefacts subsequently produced by photochemical conversion of 1 in the fermentation broth or during isolation and purification procedures.
The [α]D values for marinisporolides A and B (1 and 2) were measured as +1.3 and +1.5, respectively, values which are approximately within the error range of the polarimeter used (+1 thru −1). In the beginning, this suggested that the marinisporolides could be racemic mixtures. However, the unambiguous results from the Mosher esterification cleanly ruled out the presence of racemic mixtures. Since there is a serious limitation in the comparison of small optical rotations, we examined the circular dichroism (CD) spectra of all four compounds isolated (Figure S59). The CD spectra of 1, and compound 1 derived from 2, showed identical positive peaks at 255 nm and broad complicated negative bands at 310 nm thru 400 nm. The CD spectrum of 2 also showed a positive peak at 255 nm and complex weak negative peaks between 310 and 400 nm, which correspond to the longest wavelength UV absorption band of 2. This weaker band appears at a longer wavelength than that of 1, suggesting that the bis-spiro-tetrahydropyran ketal rings of 1 and the tetrahydropyran hemiketal ring of 2, influence the strength of the optical chirality related to the polyene chain. The CD spectrum of 3 showed a broad positive band between 310 and 400 nm unlike those of 1 and 2. While these complex, long wavelength changes are difficult to interpret, the E to Z conversion of one double bond in the polyene chain clearly causes a major conformational change, which might be expected to modify spatial relationships between the polyene and spiro-tetrahydropyran moiety. Reversals in the optical properties of such structures have previously been reported in the geometric isomers of polyenes such as the marinomycins,9 clathrynamide, and hanliangicin.16
Marinisporolides A (1) and B (2) are composed of two structural units, an ester carbonyl conjugated pentaene and multiple 1,3-polyol functionalities. This structural composition can also be found in the dermostatins,17 mycoticins,18 roflamycoin,11 and roxaticin19. All of these macrolide lactones were isolated from terrestrial Streptomyces species and most possess antifungal activity particularly against Candida pathogens. Roflamycoin is one of only a few members possessing a tetrahydropyran hemiketal ring in this polyenone-polyol macrolides class, although this constellation is common in polyether macrolides and amphotericin B-like macrolides. The semisynthetic roflamycoin spiro-tetrahydropyran11 is the only previous example of a polyene spiro-bis- tetrahydropyran macrolide, while the marinisporolides are the first spiroketal-polyenone-polyol macrolides class reported from natural sources. Spiroketals are commonly found in milbemycin20 analogues and in polyether or polyol macrolide classes, such as spirastrellolides,21 cytovaricins,22 halistatin 1,23 and halichondrin B,24 inter alia.
As part of this project, we evaluated the cancer cell cytotoxicities, the antibacterial activities, and the antifungal activities of marinisporolides A–E (1–5) against the human colon carcinoma cell line HCT-116, and against the human pathogens methicillin-resistant Staphylococcus aureus (MRSA), and Candida albicans. Marinisporolide A (1) lacked significant cancer cell cytotoxity, and showed very weak to no antifungal activity against Candida albicans with an MIC value of 22 µM. None of the other marinisporolide isomers (B–E, 2–5) showed any significant bioactivity against these targets.
In summary, the marinisporolides possess unusual substitution patterns of polyol groups compared to the known polyene-polyol macrolides, and show interesting photoreactivity, and chiroptical properties. The discovery of these new compounds continues to indicate that new actinomycetes of the genus “Marinispora” will be a significant resource structurally interesting molecules.
Experimental Section
Isolation of CNQ-140 Strain, Cultivation, Extraction
“Marinispora” strain CNQ-140 was isolated on medium A1+C (10 g of starch, 4 g of peptone, 2 g of yeast extract, 1 g of calcium carbonate, 18 g of agar, 1 L of seawater) from a marine sediment sample collected at a depth of 56 m 1 mile Northwest of the Scripps Institution of Oceanography Pier (La Jolla, California).25 The 16S rRNA gene sequence for this strain has been deposited with GenBank under accession number BANKIT 1128592. This strain was cultured in 40 replicate 2.8 L Fernbach flask each containing 1 L of fermentation medium A1 (10 g of starch, 4 g of peptone, 2 g of yeast extract, 18 g of agar, 1 L of seawater) while shaking at 230 rpm for 6 days. At the end of the fermentation period, 20 g/L Amberlite XAD-7 adsorbent resin was added to each flask and shaking was continued for 2 additional hours at a reduced speed. The resin was then collected by filtration through cheesecloth, washed with deionized water, and eluted twice with acetone. The acetone solution was concentrated under reduced pressure and the resulting residue was partitioned between ethyl acetate and water to afford, after solvent removal under vacuum, approximately 6.8 g of ethyl acetate extract.
Isolation and Purification of Marinisporolides A–E (1–5)
The EtOAc extract (6.8 g) was adsorbed on Celite (7 g) and subjected to C-18 (50 g) reversed-phase column chromatography eluting with a step gradient of acetonitrile (MeCN)-H2O solvent mixtures (increasing the acetonitrile by 10% per 500 mL from 30% to 100% acetonitrile). The 50% MeCN/H2O fraction (900 mg) showed conspicuous LC-MS peaks that were suggestive of polyene macrolides because of their characteristic UV spectra (λmax ca. 360 nm) and relatively large molecular ions ([M+Na]+ m/z 715 or m/z 697). This fraction was then further fractionated by C-18 reversed-phase HPLC with a methanol (MeOH)-H2O gradient solvent system (50% MeOH/H2O for 10 min, then 50–100% MeOH/H2O for 30 min; 20 × 250 mm waters C-18 column, 10 mL/min flow rate) to obtain subfractions containing the marinisporolides [retention time (Rt): fraction 50-1: 25–35 min; fraction 50-2: 35–38 min; fraction 50-3: 38–42 min]. Fraction 50-2 was further purified by repeated C-8 reversed-phase HPLC (Phenomenex Luna C-8(2) 10 × 250 mm column; 2 mL/min flow rate) with MeCN-H2O (43:67) to obtain marinisporolide A (1) (16 mg/40L, Rt 32 min). Marinisporolide B (2) (42 mg/40L, Rt 25 min) was obtained from fraction 50-3 by repeated C-8 RP HPLC with MeCN-H2O (43:67). The geometric isomers of marinisporolide A (1) were found in fraction 50-1, and were isolated by repeated C-18 reversed phase HPLC (Varian Dynamax C-18 10 × 250 mm column; 2 mL/min flow rate) with MeCN-H2O (46:54) to afford marinisporolides C (3) (4 mg, Rt 21.5 min), D (4) (2 mg, Rt 21 min), and E (5) (1 mg, Rt 19 min).
Marinisporolide A (1)
Yellow amorphous powder, [α]D20 +1.3 (c 0.15, MeOH). UV (MeOH) λmax: 262 nm (ε= 14,000), 358 nm (ε= 43,300), 378 nm sh (ε= 34,200). IR (film) νmax: 3380, 2930, 1690, 1560, 1240, 1100 cm−1. HR-ESI-TOF MS: m/z 697.3922 [M+Na]+, C38H58O10Na requires m/z 697.3922. See Table 1 for 1H and 13C NMR data.
Marinisporolide B (2)
Yellow amorphous powder, [α]D20 +1.5 (c 0.33, MeOH). UV (MeOH) λmax: 259 (ε= 6,100), 363 (ε= 47,200), 378 sh (ε= 43,700) nm. IR (film) νmax: 3400, 2930, 1690, 1560, 1250, 1100 cm−1. HR-ESI-TOF MS: m/z 715.4027 [M+Na]+, C38H60O11Na requires m/z 715.4028. See Table 2 for 1H and 13C NMR data.
Marinisporolide C (3)
Yellow amorphous powder. 1H NMR (500 MHz, DMSO-d6): δ 0.80 (3H, d, J = 6.5 Hz, H3-35), 0.84 (1H, m, H2-18a), 0.87 (3H, d, J = 6.5 Hz, H3-36), 0.91 (3H, d, J = 7.0 Hz, H3-37), 1.00 (1H, ddd, J = 11.5, 11.5, 11.5 Hz, H2-20a), 1.07 (1H, ddd, J = 11.0, 11.0, 11.0 Hz, H2-14a), 1.16 (1H, m, H2-16a), 1.16 (1H, m, H2-26a), 1.35 (2H, m, H2-26b, H2-28a), 1.39 (1H, m, H2-28b), 1.42 (1H, m, H2-30a), 1.53 (1H, br m, H2-30b), 1.57 (1H, br d, J = 11.5 Hz, H2-20b), 1.72 (1H, dq, J = 7.0, 7.0 Hz, H-32), 1.80 (2H, m, H2-14b, H2-16b), 1.82 (1H, m, H2-24a), 1.86 (1H, m, H-34), 1.89 (3H, s, H3-38), 1.90 (1H, m, H2-24b), 2.09 (1H, br ddd, J = 6.0, 6.0, 6.0 Hz, H2-12a), 2.22 (1H, br dd, J = 13.0, 3.0 Hz, H2-18b), 2.45a (1H, m, H2-12b), 3.14 (1H, br m, H-31), 3.30a (1H, m, H-13), 3.48 (1H, br m, H-25), 3.50 (1H, br m, H-19), 3.71 (1H, br m, H-15), 3.80 (1H, br m, H-27), 3.82 (1H, br m, H-29), 4.15 (1H, br ddd, J = 11.5, 7.5, 2.0 Hz, H-21), 4.40 (1H, d, J = 5.0 Hz, 25-OH), 4.43 (1H, d, J = 6.0 Hz, 31-OH), 4.50 (1H, br d, J = 5.0 Hz, 19-OH), 4.53 (1H, br d, J = 4.0 Hz, 27-OH), 4.56 (1H, br d, J = 4.5 Hz, 29-OH), 4.93 (1H, d, J = 9.5 Hz, H-33), 4.73 (1H, br d, J = 5.0 Hz, 15-OH), 5.26 (1H, dd, J = 15.5, 7.5 Hz, H-22), 5.34 (1H, ddd, J = 15.5, 7.5, 6.0 Hz, H-23), 5.56 (1H, br ddd, J = 11.5, 11.0, 6.0 Hz, H-11), 6.21 (1H, dd, J = 11.5, 11.5 Hz, H-10), 6.34 (1H, dd, J =14.5, 11.0 Hz, H-8), 6.45 (1H, dd, J = 14.5, 10.0 Hz, H-6), 6.62 (1H, dd, J = 15.0, 9.5 Hz, H-4), 6.62 (1H, dd, J = 14.5, 11.0 Hz, H-7), 6.66 (1H, dd, J = 15.0, 10.0 Hz, H-5), 6.72 (1H, dd, J = 14.5, 11.5 Hz, H-9), 7.05 (1H, d, J = 9.5 Hz, H-3),a overlapping signals with solvent peaks. 13C NMR (75 MHz, DMSO-d6): δ 8.9 (C-37), 13.0 (C-38), 18.9 (C-36), 19.9 (C-35), 29.5 (C-34), 34.1 (C-12), 38.3b (C-18), 40.5b (C-24, C-32), 40.8b (C-30), 41.5b (C-14, C-20), 42.1 (C-28), 44.9 (C-26), 45.5 (C-16), 61.8 (C-19), 63.8 (C-15), 65.6 (C-27), 66.7 (C-29), 68.1 (C-25), 69.3 (C-13), 69.9 (C-21), 70.7 (C-31), 78.3 (C-33), 98.8 (C-17), 126.3 (C-2), 127.2 (C-4), 128.4 (C-23), 130.3 (C-11), 131.0 (C-10), 131.7 (C-8), 131.7 (C-6), 132.1 (C-9), 132.8 (C-22), 137.5 (C-3, C-5), 140.1 (C-7), 167.4 (C-1),b chemical shifts were assigned using gHMQC spectral data because of overlapping with solvent signals. 1H NMR (500 MHz, pyridine-d5): δ 0.87 (3H, d, J = 6.5 Hz, H3-35), 0.98 (3H, d, J = 6.5 Hz, H3-36), 1.24 (3H, d, J = 7.0 Hz, H3-37), 1.51 (1H, m, H2-26a), 1.56 (1H, m, H2-18a), 1.64 (1H, m, H2-14a), 1.68(1H, m, H2-20a), 1.80 (1H, m, H2-26b), 1.92 (1H, m, H2-28a), 1.95 (1H, m, H2-16a), 1.98 (1H, m, H-34), 2.02 (1H, m, H2-28b), 2.04 (3H, s, H3-38), 2.13 (1H, m, H2-12a), 2.14 (1H, m, H-32), 2.16 (1H, m, H2-30a), 2.18 (3H, m, H2-14b, H2-24), 2.20 (1H, m, H2-20b), 2.26 (1H, m, H2-30b), 2.48 (1H, m, H2-16b), 2.76 (1H, m, H2-12b), 2.80 (1H, m, H2-18b), 3.50 (1H, t, J = 10.5 Hz, H-13), 3.94 (1H, br t, J = 7.0 Hz, H-31), 4.10 (1H, br m, H-25), 4.29 (1H, br m, H-19), 4.31 (1H, br m, H-15), 4.66 (2H, br m, H-27, H-29), 4.80 (1H, br ddd, J =11.5, 7.5, 2.0 Hz, H-21), 5.57 (1H, mc, H-33), 5.58 (1H, mc, H-11), 5.84 (2H, br mc, H-22, H-23), 5.87 (1H, br s, 27-OH), 5.99 (2H, br s, 29-OH, 31-OH), 6.02 (1H, dd, J = 14.5, 11.0 Hz, H-7), 6.23 (1H, br d, J =5.0 Hz, 19-OH), 6.29 (1H, dd, J = 11.5, 11.5 Hz, H-10), 6.35 (1H, dd, J = 14.5, 11.0 Hz, H-8), 6.51 (1H, br s, 15-OH), 6.41 (1H, dd, J = 14.5, 10.5 Hz, H-6), 6.62 (2H, mc, H-4, H-5), 6.88 (1H, dd, J = 14.5, 11.5 Hz, H-9), 6.91 (1H, br s, 25-OH), 7.50 (1H, d, J = 9.5 Hz, H-3),c indistinguishable multiplicities due to signal overlap. 13C NMR (125 MHz, pyridine-d5): δ 9.1 (C-37), 13.3 (C-38), 19.1 (C-35), 20.2 (C-36), 30.5 (C-34), 35.1 (C-12), 39.5 (C-18), 40.8 (C-30), 42.2 (C-32), 42.6 (C-24), 42.8 (C-14, C-28), 43.1 (C-20), 45.4 (C-26), 46.6 (C-16), 63.1 (C-19), 65.2 (C-15), 69.2 (C-27), 69.7 (C-29), 70.0 (C-13), 70.4 (C-25), 71.6 (C-21), 72.7 (C-31), 79.4 (C-33), 100.2 (C-17), 127.6 (C-2), 128.1 (C-4), 129.8 (C-23), 130.7 (C-11), 131.9 (C-10), 132.2 (C-8), 132.3 (C-6), 133.1 (C-9), 134.6 (C-22), 137.8 (C-7), 138.2 (C-3), 140.3 (C-5), 168.3 (C-1). UV (MeOH) λmax: 263 (ε =7,700), 358 (ε = 42,900), 378 sh (ε = 35,600) nm. IR (film) νmax 3350, 2940, 1670, 1230, 1120 cm−1. HR-ESI-TOF MS: m/z 697.3918 [M+Na]+, C38H58O10Na requires m/z 697.3928.
Marinisporolide D (4)
Yellow amorphous powder. 1H NMR (500 MHz, pyridine-d5): δ 0.91 (3H, d, J = 7.0 Hz, H3-35), 1.00 (3H, d, J = 7.0 Hz, H3-36), 1.27 (3H, d, J = 7.0 Hz, H3-37), 1.57 (1H, ddd, J = 11.0, 11.0, 11.0, H2-14a), 1.60 (1H, dd, J = 13.5, 12.0 Hz, H2-18a), 1.71 (1H, m, H2-20a), 1.72 (1H, m, H2-26a), 1.82 (1H, m, H2-26b), 1.94 (2H, m, H2-28), 2.00 (1H, m, H2-16a), 2.03 (3H, s, H3-38), 2.06 (1H, m, H-34), 2.07 (1H, m, H2-30a), 2.13 (1H, m, H2-14b), 2.15 (2H, m, H2-30b, H-32), 2.26 (1H, m, H2-12a), 2.28 (1H, m, H2-24a), 2.32 (1H, m, H2-20b), 2.36 (1H, m, H2-12b), 2.44 (1H, m, H2-24b), 2.52 (1H, br dd, J = 12.5, 4.0 Hz, H2-16b), 2.87 (1H, br dd, J = 13.5, 3.0 Hz, H2-18b), 3.62 (1H, br. t, J = 10.5 Hz, H-13), 4.02 (1H, br m, H-25), 4.08 (1H, br m, H-31), 4.32 (1H, br m, H-15), 4.37 (1H, br m, H-19), 4.58 (1H, br m, H-27), 4.74 (1H, br m, H-29), 4.96 (1H, br m, H-21), 5.65 (1H, dd, J = 9.0, 1.0 Hz, H-33), 5.76 (1H, dd, J = 15.5, 6.0 Hz, H-22), 5.82 (1H, ddd, J = 14.5, 9.0, 5.0 Hz, H-11), 5.93 (1H, dt, J = 15.5, 8.0 Hz, H-23), 6.09 (1H, br d, J = 3.5 Hz, 31-OH), 6.12 (1H, br d, J = 3.5 Hz, 27-OH), 6.17 (1H, dd, J = 14.5, 10.0 Hz, H-10), 6.23 (1H, br d, J = 3.5 Hz, 29-OH), 6.26 (1H, dd, J = 12.0, 11.0 Hz, H-4), 6.32 (1H, br d, J = 3.5 Hz, 25-OH), 6.32 (1H, dd, J = 14.0, 11.5 Hz, H-8), 6.34–6.37 a (3H, m, H-5, H-7, H-9), 6.40 (1H, br d, J = 5.0 Hz, 19-OH), 6.49 (1H, br d, J =5.0 Hz, 15-OH), 6.84 (1H, dd, J = 14.0, 11.5 Hz, H-6), 7.99 (1H, br d, J = 12.0 Hz, H-3),.a undistinguishable due to signal overlap. 13C NMR (125 MHz, pyridine-d5): δ 10.5 (C-37), 13.0 (C-38), 19.4 (C-35), 19.8 (C-36), 31.0 (C-34), 39.7 (C-18), 40.5 (C-12), 41.9 (C-32), 42.2 (C-24), 42.4 (C-14), 42.9 (C-30), 43.0 (C-20), 43.5 (C-26), 44.7 (C-28), 46.7 (C-16), 63.5 (C-19), 65.0 (C-15), 68.9 (C-29), 69.0 (C-27), 69.6 (C-13), 71.1 (C-21), 71.2 (C-25), 73.3 (C-31), 78.3 (C-33), 100.2 (C-17), 123.9b (C-4), 127.3 (H-6), 128.2 (C-23), 128.5 (C-2), 131.3 (C-8), 132.4 (C-3), 133.2 (C-11), 133.8 (C-10), 133.9 (C-22), 135.0b (× 2) and 137.1 (C-5, 7, and 9), 168.5 (C-1), bchemical shifts were assigned using gHSQC spectral data because of solvent signal overlap. UV (MeCN/H2O) λmax (rel. int.): 262 (26), 360 (100), 376 sh (91) nm. IR (film) νmax: 3320, 2940, 1670, 1250, 1100 cm−1. ESI MS: m/z 697 [M+Na]+.
Marinisporolide E (5)
Yellow amorphous powder. 1H NMR (500 MHz, pyridine-d5): δ 0.88 (3H, d, J = 7.0 Hz, H3-35), 1.00 (3H, d, J = 7.0 Hz, H3-36), 1.30 (3H, d, J = 7.0 Hz, H3-37), 1.58 (1H, ddd, J = 11.5, 11.5, 11.5 Hz, H2-14a), 1.59 (1H, m, H2-18a), 1.62 (1H, m, H2-26a), 1.68 (1H, ddd, J = 11.5, 11.5, 11.5, H2-20a), 1.87 (1H, ddd, J = 13.5, 9.0, 9.0 Hz, H2-26b), 1.96 (1H, m, H2-16a), 1.98 (1H, m, H2-28a), 2.01 (1H, m, H2-28b), 2.01 (3H, s, H3-38), 2.03 (1H, m, H-34), 2.11 (1H, m, H2-30a), 2.12 (1H, m, H-32), 2.16 (1H, m, H2-14b), 2.24 (1H, m, H2-24a), 2.25 (1H, m, H2-20b), 2.27 (1H, m, H2-30b), 2.34 (1H, m, H2-24b), 2.35 (1H, m, H2-12a), 2.38 (1H, m, H2-12b), 2.54 (1H, dd, J = 11.5, 4.0 Hz, H2-16b), 2.85 (1H, br dd, J = 12.5, 3.0 Hz, H2-18b), 3.55 (1H, br t, J = 10.5 Hz, H-13), 3.97 (1H, br m, H-31), 4.05 (1H, br m, H-25), 4.28 (1H, br m, H-19), 4.33 (1H, br m, H-15), 4.59 (1H, br m, H-27), 4.78 (1H, br m, H-29), 4.92 (1H, m, H-21)a, 5.65 (1H, d, J = 10.0 Hz, H-33), 5.75 (1H, dd, J = 15.5, 6.0 Hz, H-22)b, 5.76 (1H, ddd, J = 15.0, 9.0, 5.5 Hz, H-11)b, 5.98 (1H, ddd, J = 15.5, 8.0, 7.0 Hz, H-23), 6.06 (1H, dd, J = 11.5, 11.5 Hz, H-8)b, 6.08 (1H, br d, J = 4.0 Hz, 31-OH), 6.13 (27-OH)a, 6.14 (1H, dd, J = 11.5, 11.5 Hz, H-9) b, 6.19 (1H, br d, J = 4.0 Hz, 29-OH), 6.34 (2H, br m, 19-OH, 25-OH)a, 6.36 (1H, dd, J = 14.5, 10.5 Hz, H-6)b, 6.46 (1H, br d, J = 3.0 Hz, 15-OH), 6.56 (1H, dd, J = 14.5, 11.5 Hz, H-4), 6.70 (1H, dd, J = 14.5, 10.5 Hz, H-5), 6.90 (1H, dd, J = 15.0, 11.5 Hz, H-10), 6.95 (1H, dd, J = 14.5, 11.5 Hz, H-7), 7.56 (H-3)a; a indistinguishable multiplicities due to peak overlap. bPeak multiplicities and coupling constants were assigned by interpretation of homo 2D J-resolved 1H NMR data. 13C NMRc (125 MHz, pyridine-d5): δ 9.4 (C-37), 12.9 (C-38), 18.9 (C-35), 19.8 (C-36), 30.5 (C-34), 39.2 (C-18), 40.0 (C-12), 42.1 (C-32), 42.2 (C-24), 42.6 (C-14), 42.8 (C-20, C-30), 44.4 (C-26), 44.5 (C-28), 46.3 (C-16), 62.9 (C-19), 64.7 (C-15), 68.3 (C-27), 68.6 (C-29), 69.0 (C-13), 70.4 (C-25), 70.7 (C-21), 72.3 (C-31), 78.5 (C-33), 100.0 (C-17), 127.4 (C-2), 127.8 (C-4), 127.8 (C-8), 128.2 (C-23), 129.8 (C-10), 131.9 (C-9), 132.2 (C-7), 132.5 (C-6), 133.5 (C-11), 133.7 (C-22), 138.3 (C-3), 140.4 (C-5), 168.5 (C-1); c 13C NMR chemical shifts were assigned from HMQC and HMBC spectra. UV (MeCN/H2O) λmax (rel. int.): 262 (18), 360 (100), 376 sh (87) nm. ESI MS: m/z 697 [M+Na]+.
Methanolysis of 1 To Yield Ester 6
Marinisporolide A (1, 5 mg) was dissolved in 5% NaOMe in MeOH and stirred for 1 hour at RT. The reaction mixture was neutralized with 1N aqueous HCl, the aqueous phase was extracted with EtOAc, and the residue, after solvent removal, was purified by C18 HPLC using MeCN-H2O (45:55) to yield compound 6 (1.4 mg): 1H NMR (500 MHz, DMSO-d6): δ 0.74 (3H, d, J = 7.0 Hz, H3-35), 0.77 (3H, d, J = 7.0 Hz, H3-37), 0.88 (3H, d, J = 7.0 Hz, H3-36), 0.93 (1H, m, H2-18a), 0.96 (1H, m, H2-14a), 1.09 (1H, ddd, J = 11.5, 11.5, 11.5 Hz, H2-20a), 1.17 (1H, m, H2-16a), 1.38 (1H, m, H2-26a), 1.40 (2H, m, H2-28), 1.41 (1H, m, H2-30a), 1.43 (1H, m, H2-26b), 1.51 (3H, m, H2-30b, H-32, H-34), 1.80 (1H, m, H2-20b), 1.81 (2H, m, H2-14b, H2-16b), 1.90 (3H, s, H3-38), 2.05 (1H, m, H2-24a), 2.07 (1H, m, H2-24b), 2.28 (2H, br m, H2-12), 2.30 (1H, m, H2-18b), 3.29 (1H, m, H-33), 3.40 (1H, br m, H-13), 3.59 (1H, br m, H-31), 3.62 (2H, br m, H-19, H-25), 3.67 (3H, s, OCH3), 3.70 (1H, br m, H-15), 3.83 (1H, br m, H-27), 3.85 (1H, br m, H-29), 4.14 (1H, d, J = 4.5 Hz, 33-OH), 4.30 (1H, br m, H-21), 4.50 (1H, br d, J = 4.0 Hz, 27-OH), 4.58 (2H, br m, 25-OH, 29-OH)a, 4.62 (1H, br d, J = 4.5 Hz, 19-OH), 4.72 (2H, br m, 15-OH, 31-OH), 5.40 (1H, dd, J = 15.0, 6.0 Hz, H-22), 5.60 (1H, dt, J = 15.0, 6.5 Hz, H-23), 5.83 (1H, dt, J = 14.5, 7.5 Hz, H-11), 6.20 (1H, dd, J = 14.5, 11.0 Hz, H-10), 6.28 (1H, dd, J = 15.0, 10.5 Hz, H-8), 6.41 (1H, dd, J = 15.0, 11.0 Hz, H-9), 6.44 (1H, dd, J = 14.5, 10.5 Hz, H-6), 6.50 (1H, dd, J = 14.5, 10.5 Hz, H-7), 6.62 (1H, dd, J = 14.0, 11.5 Hz, H-4), 6.72 (1H, dd, J = 14.0, 10.5 Hz, H-5), 7.18 (1H, d, J = 11.5 Hz, H-3); a indistinguishable multiplicities due to peak overlap. 13C NMR (75 MHz, DMSO-d6): δ 9.5 (C-37), 12.4 (C-38), 18.9 (C-35), 19.3 (C-36), 30.4 (C-34), 38.7 (C-12, C-18), 39.2 (C-32), 39.8 (C-24), 40.8 (C-14), 41.2a (C-20), 41.3a (C-28, C-30), 44.5 (C-26), 45.1 (C-16), 51.5 (OCH3), 61.9 (C-19), 65.7 (C-15, C-27), 66.1 (C-29), 68.2 (C-25), 69.2 (C-21), 69.6 (C-13), 72.0 (C-31), 74.7 (C-33), 99.1 (C-17), 123.6 (C-4), 126.6 (C-2), 127.6 (C-23), 130.9 (C-8), 131.7 (C-6), 132.1 (C-10), 132.4 (C-11), 132.9 (C-22), 134.9 (C-9), 135.4 (C-7), 138.3 (C-3), 139.9 (C-5), 167.0 (C-1), asignals interchangeable. UV (MeCN/H2O) λmax (rel. int.): 260 (6), 342 sh (68), 360 (100), 373 (97) nm. ESI MS m/z 729 [M+Na]+.
Bis-acetonide 7
Marinisporolide A (1, 5 mg) was dissolved in 2,2-dimethoxypropane (2 mL) and methanol (1 mL), and pyridinium-p-toluenesulfonate (5 mg) was added. The reaction was allowed to stir for 3 h in ice bath, then quenched with 5% aqueous NaHCO3, and the aqueous phase was extracted twice with CH2Cl2. The CH2Cl2 solvent was removed under reduced pressure, and the residue was purified by C-18 RP HPLC (85:15 MeCN-H2O) to provided compound 7 (2 mg). 1H NMR (500 MHz, CD3CN): δ 0.89 (3H, d, J = 6.0 Hz, H3-35), 0.90 (3H, d, J = 6.0 Hz, H3-36), 0.90 (1H, m, H2-26a), 1.94 (1H, m, H-34), 0.97 (3H, d, J = 7.0 Hz, H3-37), 1.05 (1H, ddd, J = 11.5, 11.5, 11.5 Hz, H2-30a), 1.11 (2H, m, H2-14a, H2-16a), 1.13 (3H, s, acetonide ax. CH3), 1.19 (3H, s, acetonide eq. CH3), 1.20 (3H, s, acetonide eq. CH3), 1.25 (1H, m, H2-28a), 1.26 (1H, m, H2-20a), 1.31 (3H, s, acetonide ax. CH3), 1.34 (1H, m, H2-26b), 1.38 (1H, m, H2-28b), 1.42 (1H, m, H2-18a), 1.43 (1H, m, H2-30b), 1.79 (1H, qd, J = 7.0, 2.0 Hz, H-32), 1.87 (1H, m, H2-16b), 1.88 (3H, s, H3-38), 1.94 (2H, m, H2-18b, H2-20b), 2.06 (2H, m H2-24), 2.17 (1H, m, H2-12a), 2.20 (1H, m, H2-12b, H2-14b), 2.75 (1H, br d, J = 4.5 Hz, 15-OH), 2.94 (1H, br d, J = 6.0 Hz, 19-OH), 3.58 (1H, br ddd, J = 11.5, 7.0, 2.5 Hz, H-31), 3.73 (2H, br m, H-13, H-15), 3.76 (1H, br m, H-29), 3.82 (1H, br dddd, J = 11.0, 5.5, 5.5, 2.5 Hz, H-25), 3.88 (1H, br m, H-19, H-27), 3.98 (1H, br dt, J = 11.0, 3.0 Hz, H-21), 4.79 (1H, dd, J = 10.5, 2.0 Hz, H-33), 5.54 (2H, m, H-22, H-23)a, 6.02 (1H, ddd, J = 15.0, 10.0, 5.5 Hz, H-11), 6.18 (1H, dd, J = 15.0, 11.0 Hz, H-10), 6.26 (1H, dd, J = 14.5, 10.5 Hz, H-8), 6.38 (1H, dd, J = 14.5, 11.0 Hz, H-9), 6.39 (1H, dd, J = 14.5, 10.5 Hz, H-6), 6.48 (1H, dd, J = 14.5, 10.5 Hz, H-7), 6.56 (1H, dd, J = 14.5, 11.0 Hz, H-4), 6.67 (1H, dd, J = 14.5, 10.5 Hz, H-5), 7.21 (1H, d, J = 11.0 Hz, H-3), aunresolved multiplicities due to signal overlap. 13C NMR (75 MHz, CD3CN): δ 8.4 (C-37), 13.1 (C-38), 19.2 (C-36), 19.7 (acetonide (1) ax. CH3), 21.1 (C-34), 20.2 (acetonide ax. CH3), 20.5 (C-35), 30.6 (acetonide eq. CH3 ×2), 35.7 (C-30), 37.6 (C-26), 39.9 (C-24), 40.0 (C-12)b, 40.1 (C-20)b, 40.9 (C-32), 41.6 (C-14), 42.1 (C-16), 45.3 (C-18), 44.0 (C-28), 64.2 (C-15), 65.0 (C-27), 66.0 (C-19), 66.7 (C-29), 69.7 (C-25), 71.4 (C-21), 72.1 (C-31), 72.4 (C-13), 80.4 (C-33), 99.2 (acetonide, ketal), 99.4 (acetonide, ketal), 100.5 (C-17), 126.8 (C-23), 127.3 (C-2), 127.5 (C-4), 131.4 (C-8), 131.9 (C-6), 132.3 (C-10), 134.1 (C-22), 136.1 (C-11), 137.4 (C-9), 139.1 (C-7), 140.6 (C-3), 142.3 (C-5), 169.0 (C-1), binterchangeable signals. UV (MeCN/H2O) λmax (rel. int.): 258 (16), 338 sh (68), 360 (100), 375 (93) nm. ESI MS: m/z 777 [M+Na]+.
Methanolysis of 7 To Yield Ester 8
Bis-acetonide 7 (2 mg) was treated with 2.5% NaOMe in methanol for 1 h and the reaction mixture was processed as described in the methanolysis of 1. Compound 8 (1.4 mg) was purified by C-18 RP HPLC (9:1 MeCN-H2O). 1H NMR (500 MHz, CD3CN) : δ 0.76 (3H, d, J = 7.0 Hz, H3-35), 0.80 (3H, d, J = 7.0 Hz, H3-37), 0.93 (3H, d, J = 7.0 Hz, H3-36), 1.01 (1H, ddd, J = 12.0, 12.0, 12.0 Hz, H2-30a), 1.06 (2H, m, H2-14a, H2-16a), 1.15 (1H, m, H2-20a), 1.16 (1H, m, H2-28a), 1.26 (3H, s, acetonide CH3), 1.27 (3H, s, acetonide CH3), 1.27 (1H, m, H2-18a), 1.38 (6H, s, acetonide CH3 ×2), 1.45 (3H, m, H2-26, H2-30b), 1.58 (2H, m, H-32, H-34), 1.87 (1H, m, H2-16b), 1.89 (1H, m, H2-20b), 1.91 (1H, m, H2-18b), 1.93 (overlapping with solvent signal, H3-38), 2.12 (2H, m, H2-24), 2.24 (1H, m, H2-12a), 2.28 (2H, m, H2-12b, H2-14b), 1.46 (1H, m, H2-28b), 3.87 (1H, br m, H-31), 2.74 (1H, d, J = 5.5 Hz, 15-OH), 2.89 (1H, d, J = 5.5 Hz, 19-OH), 2.66 (1H, d, J = 5.0 Hz, 33-OH), 3.37 (1H, br ddd, J = 8.5, 5.0, 2.0 Hz, H-33), 3.69 (3H, s, OCH3), 3.72 (1H, br m, H-15), 3.73 (1H, br m, H-13), 3.88 (1H, br m, H-19), 3.95 (2H, br m, H-21, H-29), 4.02 (2H, br m, H-25, H-27), 5.31 (1H, dd, J = 15.5, 5.5 Hz, H-22), 5.63 (1H, dt, J = 15.5, 6.5 Hz, H-23), 5.81 (1H, dt, J = 15.0, 7.5 Hz, H-11), 6.19 (1H, dd, J = 15.0, 10.5 Hz, H-10), 6.27 (1H, dd, J = 15.0, 10.5 Hz, H-8), 6.36 (1H, dd, J = 15.0, 10.5 Hz, H-9), 6.40 (1H, dd, J = 15.0, 10.0 Hz, H-6), 6.46 (1H, dd, J = 15.0, 10.5 Hz, H-7), 6.58 (1H, dd, J = 14.5, 11.0 Hz, H-4), 6.65 (1H, dd, J = 14.5, 10.0 Hz, H-5), 7.18 (1H, d, J = 11.0 Hz, H-3). UV (MeCN/H2O) λmax (rel. int.): 258 (7), 338 sh (69), 358 (100), 372 (95) nm. ESI MS: m/z 809 [M+Na]+.
Mosher esters 9a and 9b
Compound 8 (0.7 mg) was dissolved in 500 µL of pyridine, and dimethylaminopyridine (1 mg) and (R)-MTPACl (15 µL) were then added in sequence. The reaction mixture was stirred for 24 hours at RT and two drop of water was then added. The solution was concentrated under reduced pressure and the residue was subjected to silica gel normal phase Sep-Pak (1 g) using ethyl acetate-methanol (5:1) to afford 0.5 mg of the tri-(S)-MTPA ester 9a (see supporting information for spectral data; Figure S2, S50, S51, and S52). In a same fashion, a solution of 8 (0.7 mg) µL of pyridine was treated with dimethylaminopyridine (1 mg) and (S)-MTPACl (15 µL) to afford, after the isolation as above, 0.5 mg of the (R)-MTPA ester 9b (see supporting information for spectral data; Figure S2, S53, S54, and S55).
Methanolysis of 2 To Yield Ester 10
Marinisporolide B (1, 5 mg) was treated with 5% NaOMe in MeOH for 1 hour at room temperature in the same manner as in the methanolysis of 1. The reaction mixture, after neutralizing and solvent removal under vacuum, was purified by C18 HPLC using MeCN-H2O (45:55) to yield compound 10 (0.8 mg): 1H NMR (500 MHz, DMSO-d6): δ 0.74 (3H, d, J =7.0 Hz, H3-35), 0.77 (3H, d, J =7.0 Hz, H3-37), 0.88 (3H, d, J = 7.0 Hz, H3-36), 1.06 (1H, m, H2-14a), 1.10 (1H, m, H2-16a), 1.35 (2H, m, H2-20), 1.38 (1H, m, H2-26a), 1.40 (2H, m, H2-28), 1.47 (1H, m, H2-18a), 1.42 (1H, m, H2-26b), 1.44 (2H, m, H2-30), 1.52 (2H, m, H-32, H-34), 1.58 (1H, m, H2-18b), 1.76 (1H, m, H2-14b), 1.85 (1H, m, H2-16b), 1.90 (3H, s, H3-38), 2.02 (1H, m, H2-24a), 2.07 (1H, m, H2-24b), 2.17 (1H, m, H2-12a), 2.23 (1H, m, H2-12b), 3.28 (1H, br m, H-33), 3.59 (1H, m, H-31), 3.63 (1H, br m, H-25), 3.67 (3H, s, OCH3), 3.83 (3H, br m, H-13, H-27, H-29), 3.83 (1H, br m, H-15), 4.08 (1H, br m, H-21), 4.12 (1H, br m, H-19), 4.18 (1H, br s, 33-OH), 4.47 (1H, br m, 15-OH)a, 4.49 (1H, br m 19-OH)a, 4.51 (1H, br m 25-OH)a, 4.53 (1H, br m 21-OH)a, 4.57 (2H, br m, 27-OH, 29-OH)a, 4.74 (1H, br s, 31-OH), 5.42 (1H, dd, J = 15.5, 6.0 Hz, H-22), 5.55 (1H, dt, J = 15.5, 6.5 Hz, H-23), 5.83 (1H, dt, J = 14.5, 7.5 Hz, H-11), 6.14 (1H, dd, J = 14.5, 10.5 Hz, H-10), 6.26 (1H, dd, J = 15.0, 10.0 Hz, H-8), 6.37 (1H, dd, J = 15.0, 10.5 Hz, H-9), 6.43 (1H, dd, J = 14.5, 10.0 Hz, H-6), 6.49 (1H, dd, J = 14.5, 10.0 Hz, H-7), 6.61 (1H, dd, J = 14.5, 11.0 Hz, H-4), 6.73 (1H, dd, J = 14.5, 10.0 Hz, H-5), 7.17 (1H, d, J = 11.0 Hz, H-3); a the multiplicity of these signals could not be resolved and because of signal overlap, the chemical shifts were assigned using 1H-1H-COSY spectral data. UV (MeCN/H2O) λmax (rel. int.): 260 (10), 340sh (71), 360 (100), 373 (95) nm. ESI MS: m/z 747 [M+Na]+. See Figure S58 for the gHMQC spectrum.
Photoisomerization of Marinisporolide A (1) and B (2)
In separate experiments, a solution of marinisporolide A (1, 0.01 mg/mL) or B (2, 0.02 mg/mL) in methanol was exposed to strong room light in a capped clear pyrex vial. After irradiation, each solution was analyzed by C18 reversed-phase HPLC, which showed the presence of four major peaks. The time course of the reaction was monitored by reversed-phase HPLC, which showed the formation of an equilibrium mixture of mainly four or five peaks within 2 hours (Figures S4 and S5).
Supplementary Material
Acknowledgment
This research is a result of financial support from the National Cancer Institute, under grant CA44848, and from the California Sea Grant College Program of the U.S. Department of Commerce's National Oceanic and Atmospheric Administration under NOAA Grant # NA04OAR4170038, project # R/MP-96, through the California Sea Grant College Program; and in part by the California State Resources Agency. The views expressed herein do not necessarily reflect the views of any of those organizations.
Footnotes
Supporting Information Available: General experimental procedures and spectral data of 1–10; HRMS, UV, 1H, 13C, and 2D NMR spectra for 1–3; UV and ESI-MS data derived from HPLC-DAD-MSD for 1b and 4–10; 1H NMR spectrum for 1b; 1H and 13C NMR (or gHMQC) spectra for 4, 5, 6, 7, 10; 1H NMR and 1H-1H COSY spectra for 8, 9a, and 9b; the 13C NMR data for model compounds in Figure 2; Illustrations of ROESY correlations for 1, 2, 3, 4 and 7: HPLC chromatogram of products derived from photoisomerization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bédy J. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2. Faulkner JD. Nat. Prod. Rep. 2000;17:1–6. doi: 10.1039/a909113k. Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2008;25:35–94. doi: 10.1039/b701534h. and references therein.
- 3.Goodfellow M, Haynes JA. Biological, Biochemical, and Biomedical Aspects of Actinomycetes. New York: Academic Press; 1984. pp. 453–472. [Google Scholar]
- 4.Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Appl. Environ. Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Environ. Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 6.(a) Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]; (b) Macherla VR, Mitchell SS, Manam RR, Reed KA, Chao T-H, Nicholson B, Deyanat-Yazdi G, Mai B, Jensen PR, Fenical W, Neuteboom STC, Lam KS, Palladino MA, Potts BCM. J. Med. Chem. 2005;48:3684–3687. doi: 10.1021/jm048995+. [DOI] [PubMed] [Google Scholar]; (c) Williams PG, Buchanan GO, Feling RH, Kauffman CA, Jensen PR, Fenical W. J. Org. Chem. 2005;70:6196–6203. doi: 10.1021/jo050511+. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Org. Lett. 2005;7:2731–2734. doi: 10.1021/ol050901i. [DOI] [PubMed] [Google Scholar]
- 8.Oh D-C, Williams PG, Kauffman CA, Jensen PR, Fenical W. Org. Lett. 2006;8:1021–1024. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 9.Kwon HC, Kauffman CA, Jensen PR, Fenical W. J. Am. Chem. Soc. 2006;128:1622–1632. doi: 10.1021/ja0558948. [DOI] [PubMed] [Google Scholar]
- 10.Bruno TJ, Svoronos PDN. Handbook of basic tables for chemical analysis. Florida: CRC press; 2000. p. 222. [Google Scholar]
- 11.Rychnovsky SD, Griesgraber G. J. Am. Chem. Soc. 1994;116:2623–2624. [Google Scholar]
- 12.(a) Kobayashi Y, Tan C-H, Kishi Y. J. Am. Chem. Soc. 2001;123:2076–2078. doi: 10.1021/ja004154q. [DOI] [PubMed] [Google Scholar]; (b) Kobayashi Y, Czechtizky W, Kishi Y. Org. Lett. 2003;5:93–96. doi: 10.1021/ol0272895. [DOI] [PubMed] [Google Scholar]
- 13.Rychnovsky SD, Rogers BN, Richardson TI. Acc. Chem. Res. 1998;31:9–17. [Google Scholar]
- 14.Ohtani I, Kusumi T, Kashman Y, Kakisawa H. J. Am. Chem. Soc. 1991;113:4092–4096. [Google Scholar]
- 15.Sonoda Y, Morii H, Sakuragi M, Suzuki Y. Chem. Lett. 1998:349–350. [Google Scholar]
- 16.(a) Ojika M, Itou Y, Sakagami Y. Biosci. Biotechnol. Biochem. 2003;67:1568–1573. doi: 10.1271/bbb.67.1568. [DOI] [PubMed] [Google Scholar]; (b) Kundim BA, Itou Y, Sakagami Y, Fudou R, Iizuka T, Yamanaka S, Ojika M. J. Antibiot. 2003;56:630–638. doi: 10.7164/antibiotics.56.630. [DOI] [PubMed] [Google Scholar]
- 17.(a) Pandey RC, Rinehart KL, Jr, Millington DS. Hindustan Antibiot. Bull. 1980;22:47–61. Chemical Abstract 95:42296. [PubMed] [Google Scholar]; (b) Sinz CJ, Rychnovsky SD. Tetrahedron. 2002;58:6561–6576. [Google Scholar]
- 18.Wasserman HH, Van Verth JE, McCaustland DJ, Borowitz IJ, Kamber B. J. Am. Chem. Soc. 1967;89:1535–1536. doi: 10.1021/ja00982a052. [DOI] [PubMed] [Google Scholar]
- 19.Maehr H, Yang R, Hong LN, Liu CM, Hatada MH, Todaro LJ. J. Org. Chem. 1989;54:3816–3819. [Google Scholar]
- 20.(a) Takiguchi Y, Mishima H, Okuda M, Terao M, Aoki A, Fukuda R. J. Antibiot. 1980;33:1120–1127. doi: 10.7164/antibiotics.33.1120. [DOI] [PubMed] [Google Scholar]; (b) Mishima H, Ide J, Muramatsu S, Ono M. J. Antibiot. 1983;36:980–990. doi: 10.7164/antibiotics.36.980. [DOI] [PubMed] [Google Scholar]
- 21.Williams DE, Roberge M, Van Soest R, Andersen RJ. J. Am. Chem. Soc. 2003;125:5296–5297. doi: 10.1021/ja0348602. [DOI] [PubMed] [Google Scholar]; (b) Williams DE, Lapawa M, Feng X, Tarling T, Roberge M, Andersen RJ. Org. Lett. 2004;6:2607–2610. doi: 10.1021/ol0490983. [DOI] [PubMed] [Google Scholar]
- 22.Kihara T, Kusakabe H, Nakamura G, Sakurai T, Isono K. J. Antibiot. 1981;34:1073–1074. doi: 10.7164/antibiotics.34.1073. [DOI] [PubMed] [Google Scholar]
- 23.Pettit GR, Tan R, Gao F, Williams MD, Doubek DL, Boyd MR, Schmidt JM, Chapuis JC, Hamel E. J. Org. Chem. 1993;58:2538–2543. [Google Scholar]
- 24.Hirata Y, Uemura D. Pure Appl. Chem. 1986;58:701–710. [Google Scholar]
- 25.The isolation of this strain and details of its taxonomy have been described, see Prieto-Davó A, Fenical W, Jensen PR. Aquat. Microbial Ecol. 2008;52:1–11.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.