Synopsis
Quantitative protein assays are needed in a wide range of biological studies. Traditional immunoassays are not available for a large number of proteins and development of new immunoassays requires a significant investment in time and money. The development of assays using peptide immunoaffinity enrichment coupled with targeted mass spectrometry has many advantages including versatility in design, ease of use, enhanced specificity, and good performance characteristics. This review presents recent developments in the characterization and implementation of immuno-SRM assays.
Keywords: quantitative proteomics, selected reaction monitoring, stable isotope dilution, SISCAPA, immuno-SRM, immuno-mass spectrometry
Introduction
Quantitative protein assays are a critical component in exploring the relationship between protein abundance and biological features such as phenotype, disease, or response to treatment. Traditionally, the sandwich immunoassay (e.g. Enzyme Linked Immunosorbent Assay, ELISA) has been the gold standard for protein quantification owing to the high sensitivity, high throughput, and low per sample cost (once an assay is developed and validated). However, the need for quantitative assays has far outpaced the rate of development. Modern strategies are capable of discovering hundreds to thousands of interesting genes and proteins that must be validated in follow-up experiments with a quantitative assay. Highly specific and sensitive assays are not available for quantifying the vast majority of human proteins, and de novo assay generation is associated with a high cost and long lead time. In addition, many existing assays suffer from poor specificity, a variety of interferences, lack of standardization or all of the above. Thus, the current situation is severely limiting to all areas of research, from basic biology to clinical chemistry. We desperately need alternative strategies for building assays to any protein (or modification) of interest without a prohibitive investment in time, money, and other resources.
One technology that holds particular promise in improving the situation is quantitative targeted mass spectrometry. Selected reaction monitoring (SRM) is a targeted mass spectrometry technique that has increased sensitivity compared to profiling modes of analysis while maintaining high specificity for the target analyte. This method is well established in clinical reference laboratories for accurate quantification of small molecules in plasma, such as metabolites that accumulate as a result of inborn errors of metabolism [1, 2]. SRM has been increasingly utilized in proteomics to measure the concentrations of target proteins in biological matrices [3-8]. To achieve quantitation of proteins, biological molecules are digested to component peptides using a proteolytic enzyme such as trypsin. One or more selected peptides whose sequences are unique to the target protein and are efficiently observed by the mass spectrometer (i.e. “proteotypic” peptides) are then measured as quantitative stoichiometric surrogates for the protein of interest. Quantitation is performed by measuring the surrogate peptide relative to a spiked stable isotope-labeled standard using conventional stable isotope dilution methods [9, 10]. The assays are specific, precise (%CV ≤ 20%), multiplex-able, and portable across laboratories and instrument platforms [11, 12]. They are also relatively inexpensive to develop, especially compared to other quantitative technologies.
Currently, a serious limitation to more widespread use of SRM-based assays is the limited sensitivity typically achieved in complex samples. For example, without enrichment, SRM is typically able to measure proteins present in the 100-1000 ng/mL concentration range in plasma [7], although many “biologically interesting” proteins are found several orders of magnitude below that range. An enrichment or fractionation step can enhance the sensitivity and extend detection to low abundance analytes. For example, previous studies have demonstrated the success of using abundant protein depletion with limited strong cation exchange fractionation [13] or glycopeptide enrichment [14] to analyze proteins in the low ng/mL range. However, this is unattractive for analysis of large numbers of samples due to the increase in cost and time associated with extra sample handling, as well as the potential impact of multiple sample handling steps on measurement variability and analyte recovery.
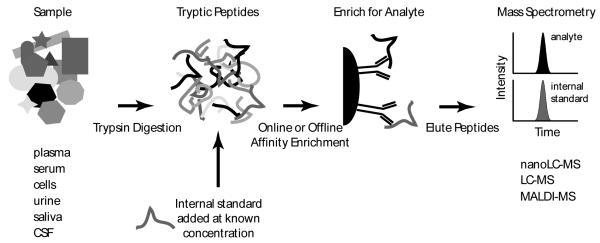
Another approach for improving sensitivity is to employ immunoaffinity techniques for selective enrichment of the analytes. Once captured, the enriched analyte is quantified using mass spectrometry. Several modes of implementation are possible [15-18] using antibodies for proteins or peptides. The design of the assay depends on several considerations including reagent availability, laboratory resources, requirements for throughput and sample handling, and the nature of the target (i.e. targeting specific forms or modifications can dictate how the assay is configured). One approach is to use anti-peptide antibodies to capture endogenous (i.e. light) peptides and a stable isotope-labeled (i.e. heavy) peptide internal standard (see Figure 1). This enrichment approach is referred to as SISCAPA (Stable Isotope Standards with Capture by Anti-Peptide Antibodies) [19] and when coupled with quantification using SRM targeted mass spectrometry can be generally referred to as an immuno-SRM assay. This chapter will review recent advancements in the area of peptide immuno-SRM assay development with a focus on technological aspects and recent applications.
Figure 1. Targeted enrichment and quantitation using an immuno-SRM assay.
The sample can be a variety of complex proteomic samples. To achieve quantitation of the targeted protein(s), these larger molecules are digested to component peptides using an enzyme such as trypsin. A stable isotope standard (SIS) is added to the sample at a known concentration for quantitative analysis. The selected peptides are then enriched using anti-peptide antibodies immobilized on a solid support. Following washing and elution from the anti-peptide antibody, the amount of surrogate peptide is measured relative to the stable isotope standard using targeted mass spectrometry.
Advantages of immuno-SRM assays
Coupling peptide immunoaffinity enrichment with mass spectrometry in an immuno-SRM assay has many advantages compared with traditional immunoassays and SRM assays lacking enrichment (see Table 1). First, compared with traditional sandwich immunoassays, immuno-SRM requires a single antibody with relaxed specificity requirements. The mass spectrometer acts like a second antibody with very high specificity. Restrictions typical to antibody development in traditional sandwich immunoassays are avoided because there are no requirements for a specific epitope and no constraints for finding multiple antibodies with independent epitope recognition. This substantially decreases the time involved in screening for working antibodies. Using a single antibody also simplifies the assay design and ultimately reduces the cost per sample. Another advantage to immuno-SRM assays is the standardization of synthetic peptide reagents. The use of stable isotope-labeled peptides as standards provides a reagent whose purity can be greatly controlled and highly characterized, the amount quantified using standard procedures (e.g. amino acid analysis), and the standard can be used in many laboratories with concordant results. Furthermore, enrichment of peptides avoids many pitfalls associated with traditional immunoassays. Interferences by endogenous auto-antibodies (recognizing the targeted protein) or anti-reagent antibodies (recognizing the capture antibody) are avoided because these endogenous proteins are enzymatically digested in the sample preparation [20-22]. Finally, the ability to readily multiplex a number of targets into a single assay is a tremendous advantage for immuno-SRM. Examples have been demonstrated for tens of peptides in a single assay [23, 24], but theoretically much higher limits are possible.
Table 1.
Summary of advantages and performance features of immuno-SRM assays based on peptide enrichment.
| Attribute | Description | Example(s) |
|---|---|---|
| Time | Assays configured in less than a year, multiplexing allows development in parallel |
[30] |
| Cost | Synthetic peptides and a single antibody per target |
[30] |
| Standards | Quality-controlled and quantifiable reagents | [11] |
| High specificity of mass spectrometer |
Relaxed requirements on antibody, reduces time to get working antibody |
|
| Avoids autoantibody and anti-reagent interference |
Autoantibodies and interfering proteins are digested in sample preparation, eliminating interactions with analytes and reagents |
[26] |
| Versatility in detecting protein modifications |
Modified peptides can be enriched and targeted for quantification |
[44, 51] |
| Large dynamic range |
Greater than 3 orders of magnitude | [27, 50] |
| Multiplexing | Currently tens of assays readily multiplexed | [24] |
| Sensitivity | Limits of detection on the order of ng/mL or better |
[23, 26, 27] |
| Precision | Less than 20% coefficient of variation | [24] |
The analytical performance of the assays is acceptable for a range of applications. Enrichment of the target peptide over 1000 fold is possible [19, 25]. This approach also provides reduced ion suppression enabling very sensitive detection of peptides enriched from complex samples. Demonstrations of detection limits in plasma have shown it is possible to analyze ng/mL protein concentrations with dynamic ranges over three orders of magnitude. Extending detection limits even lower is possible by increasing the amount of input sample [23]. The repeatability has been examined in several publications [19, 23, 25-27]. In most cases, the coefficient of variation for complete process replicates is below 20%. However, there are instances of individual proteins that show higher CVs, indicating some optimization in the digestion and capture protocols may be necessary for specific analytes. Overall, immuno-SRM assays show good figures of merit and a number of advantages over immunoassays and SRM alone.
Reagent and assay development
There are several steps involved in developing an immuno-SRM assay. These include target peptide selection, reagent generation, optimization, and configuration. Assay development begins with target peptide selection. Once a given protein or proteins are targeted, the general rules associated with picking peptides for SRM assays apply the same to immuno-SRM development [9]. These include selecting tryptic peptides that are unique to the protein of interest, respond well in mass spectrometry (relatively good ionization and production of several fragments suitable for SRM development), and do not contain known modifications (unless the modification is specifically being targeted). It is most common to select tryptic peptides in length 8-22 amino acids with moderate hydrophobicity (very hydrophilic and very hydrophobic peptides are less stable due to retention time variation in HPLC and loss to surfaces). Methionine residues (oxidation), N-terminal glutamine (cyclization), asparagine followed by glycine or proline (prone to deamidation), and dibasic termini (e.g. neighboring lysine or arginine residues such as KK, KR, RR, RK have the potential for variable digestion efficiency) are undesirable. For quantitation, stable isotope-labeled peptides are used as internal standards. It is important to ensure the labeled peptide is sufficiently separated in mass from the analyte peptide, ideally containing a mass shift that can be measured in the precursor and fragment ions. Thus, it is typical to incorporate heavy (e.g. 13C and 15N) labeled amino acids at the C-terminus of the peptide (i.e. K or R labeled) and design SRM transitions targeting y-ions.
There are a variety of antigenicity prediction algorithms available when developing antibodies to protein targets. These predictors are based principally on the location of the peptide sequence within the three-dimensional protein structure (i.e. the extent of surface exposure or location in beta-turns) [28]. However, these prediction algorithms have not yet found good utility in generating antibodies for the linear epitopes found in tryptic peptides used for immuno-SRM assays [29, 30]. As more assays are evaluated, there may be an improvement in the development of prediction algorithms.
The success rate in generating affinity reagents for use in immuno-SRM is relatively high. A large scale evaluation of rabbit polyclonal antibodies showed good success (>50%) for making individual anti-peptide antibodies with detection limits in the ng/mL to tens of ng/mL range [30]. By making antibodies to multiple peptides per protein, the success rate for generating an assay to a given protein of interest can be much higher (>90%). The antibody generation process can be multiplexed by injecting several peptides into a single animal, providing time and cost savings. The maximum number of multiplexed immunogens possible before the immune response to individual targets is diminished (i.e. immunodominance) has not been characterized. The time required to generate and characterize polyclonal antibodies makes it feasible for a laboratory to develop hundreds of assays per year, a considerable advantage compared to traditional immunoassays. Finally, polyclonal antibodies have a typical yield sufficient to generate assays for hundreds to thousands of samples, suitable for large biological experiments or biomarker verification studies.
While affinity-purified polyclonal antibodies offer an attractive and affordable source of affinity reagents, monoclonal antibodies are far more advantageous for many applications. Monoclonal antibodies provide a renewal affinity reagent providing the benefits of a standardized reagent and the capability of usage in an unlimited number of samples. Furthermore, monoclonals offer the potential to obtain higher binding affinities due to screening for the best performing clonal hybridomas. The use of a monoclonal antibody in an immuno-SRM assay response curve has been demonstrated to have good performance [31] and methods have been developed for determining the binding characteristics of anti-peptide monoclonal antibodies using a refined surface plasmon resonance technique [32]. These studies show the capabilities and promise of using monoclonals. Methods to increase the throughput of screening for high affinity antibodies using protocols near the final assay format have also been implemented on both electrospray [31] and MALDI (Matrix Assisted Laser Desorption Ionization) platforms [33]. Extending the approach to higher throughput makes the large-scale production of monoclonal reagents a distinct possibility.
In addition to analyte specific antibodies, group specific anti-peptide antibodies can achieve greater coverage of the proteome using a single affinity reagent. ‘Triple X Proteomics’ (TXP) antibodies [34] are designed for linear epitopes of 3-4 amino acids and the terminally charged group of peptides. The short sequence comprising the epitope can be chosen to overlap amongst many proteins; thus, the antibodies recognizing that epitope will enrich a whole class of peptides. Initial calculations show TXP antibodies can cover a large fraction of the human proteome with a fraction of the antibodies required for peptide-specific assays [35]. Demonstration of the approach showed success in enriching 38 signature peptides from cell lysates using two TXP antibodies [36]. Issues regarding the performance of these antibodies (i.e. analyte recovery, sensitivity, reproducibility) in comparison to other peptide specific approaches remain to be characterized, but the approach offers promise for efficient assay development to large numbers of proteins.
Once affinity reagents and synthetic peptides are obtained, they are assembled into the working assay. Configurations for peptide immunoaffinity enrichment are presented below. The reader is referred to several excellent reviews for more information on SRM assay configuration and optimization [9, 10].
Assay configuration
There are two predominant formats available for immunoaffinity enrichment of peptides. One format is enrichment using an affinity column, where antibodies are covalently coupled to chromatography media [19, 37-39]. The column format offers the advantage of an online analysis using established affinity chromatography techniques. The use of POROS (Applied Biosystems, Foster City, CA) nanocolumns was originally used in SISCAPA enrichment [19] and features high binding capacity, a relatively high concentration of antibodies allowing for rapid enrichment of target peptides, and the ability to prepare columns with a variety of functionalized groups. The column can also be regenerated and used to analyze many samples (e.g. hundreds) depending on the antibody and assay parameters. These features make online immunoaffinity enrichment attractive from a ruggedness/robustness standpoint. Disadvantages include the potential for sample carryover, inflexibility in changing the analytes, and difficulties passing a large sample volume over the column.
An alternative to immunoaffinity chromatography is offline enrichment using magnetic particles [25, 27]. Magnetic particles are available in a wide array of chemistries allowing for coupling to antibodies. Protein G coated particles offer the binding of antibodies in a preferred orientation, so these are commonly employed. Offline enrichment has the advantage that samples can be processed in parallel using larger volumes of relatively viscous samples or samples that might otherwise be problematic in chromatography systems. Magnetic particle processing has been automated in 96 well plates for the SISCAPA enrichment step with elution in the plates for analysis by mass spectrometry [23]. Alternatively, a novel bead trap device was developed to perform the bead handling steps inline with the nanoflow chromatography system [40]. This minimizes losses of peptides to containers between the elution and analysis steps. Offline enrichment has also been implemented by immobilizing antibodies in pipet tips [41].
In addition to electrospray mass spectrometry, MALDI has been used extensively in conjunction with peptide immunoaffinity enrichment. The largest advantage of MALDI compared to electrospray is the potential for detection of the enriched peptide without a chromatography step. This enables the analysis to be performed in a very short time (requiring minutes or less) enabling truly high throughput measurements. An additional advantage of MALDI is a greater tolerance to contaminating salts or detergents. The combination of immunoaffinity enrichment and MALDI detection was first configured with high throughput capabilities using the affinity reagent immobilized in pipet tips [41]. Although most commonly employed to analyze protein variants [42, 43], this approach is also capable of analyzing smaller peptide analytes [44]. A particular strength of this mass spectrometric immunoassay approach is the characterization of protein variants across a large number of samples. For targeting specific peptides using commercially available antibodies, assays have been configured with detection and quantification by MALDI covering two orders of magnitude [45]. Quantitative assays have also been configured for peptides from epidermal growth factor receptor (EGFR) [46], IglC - a protein used for detection of Francisella tularensis [47], and angiotensin [48]. The use of MALDI will likely continue to grow due to the tremendous potential for high throughput measurements.
Applications
There are several notable examples of immuno-SRM assays employed in the literature. The most prevalent use of the assays has been in the area of preclinical verification of biomarkers. For example, an assay for Fibulin-2 was developed to verify its discovery as a novel circulating marker in a mouse model of breast cancer [49]. The assay had a dynamic range of over 2 orders of magnitude and was employed to verify the over-abundance (30-fold) of Fibulin-2 in the plasma of mice with tumors. In this case, the assay was developed to increase the sensitivity for the target protein, which could not be quantitated by SRM alone. In another example, a multiplexed immuno-SRM assay was developed for Troponin I, an established marker of cardiac injury, and interleukin-33, an emerging cardiovascular biomarker [27]. The intent was to demonstrate the assay performance in a verification setting. The assays were easily multiplexed and showed sufficient precision, reproducibility, and sensitivity for utilization in verification studies. Finally, a highly multiplexed immuno-SRM assay was used to evaluate the potential for mass spectrometry-based technologies to impact a biomarker pipeline [24]. The 31-plex assay was used to provide initial biomarker verification in a mouse model of cancer by assaying 80 individual samples. The assay showed good reproducibility with a median limit of detection of 26 ng/mL (using 10 μL plasma).
The choice of surrogate peptide should be carefully considered and can provide additional possibilities in designing an immuno-SRM assay. An example of the versatility in building assays to surrogate peptides was illustrated in the development of an assay for pepsin and pepsinogen [50]. Peptide immunoaffinity enrichment was coupled with mass spectrometry for quantitating pepsin in saliva using an online column enrichment format. Notably for this assay, the C-terminal fragment peptide was chosen as the surrogate peptide to provide a combined measurement of pepsin and pepsinogen. The assay was capable of measuring pepsin concentrations in the low picomolar range with good reproducibility. Furthermore, the method was scalable to analyze larger volumes of initial material to customize the assay to expected sample concentrations. Another example highlighting the versatility in choosing peptides was the analysis of aberrant glycoforms of TIMP1 [51]. In this case, an immuno-SRM assay for TIMP1 was combined with lectin enrichment of the aberrantly glycosylated proteins in the serum of colon cancer patients. The assay was able to achieve detection limits below 1 ng/mL for the targeted isoforms using the combined enrichment approach.
Finally, there is the potential for immuno-SRM assays to be of great utility in the clinic. Thyroglobulin, a particularly challenging protein to assay [52], was quantified by immuno-SRM to provide an assay free of the interferences affecting traditional immunoassays [26]. A limit of detection of 2.6 ng/mL was achieved using the polyclonal antibody generated against a target peptide for thyroglobulin. Comparison to existing immunoassay showed good correlation and agreement, suggesting the assay was nearly good enough for clinical use. Improvements in the overall sensitivity, through higher affinity monoclonal antibodies or advancements in new mass spectrometers, should make the assay attractive for patient sample analysis.
Future areas of development
As presented above, the performance, multiplex-ability, ease of configuration, versatility, and specificity are considerable advantages to using immuno-SRM assays to meet the demand for quantitative protein assays. However, while the technology is promising, there remain several areas where the technique continues to see growth.
One area where optimization will be required is trypsin digestion. The imperfect nature of trypsin digestion, for which no current standards exist, is a source of error in using proteotypic peptides as stoichiometric surrogates of protein abundance. Incomplete tryptic digestion of parent proteins can result in underestimation of protein concentrations. One solution is to use stable isotope labeled proteins as standards in immunoaffinity enrichment coupled to quantitative mass spectrometry [53, 54]. A protein standard can be quantified independently (using amino acid analysis) and used as a reference for absolute protein quantification. This has the advantage of encompassing all analytical aspects of the assay, including trypsin digestion; however, this is also imperfect since post-translational modifications affecting trypsin digestion in the biospecimen under analysis may not be present in the recombinant protein standards. Perhaps more critical than the completion of trypsin digestion is the reproducibility of digestion, since precise relative quantitation could be achieved if the digestion were incomplete, yet reproducible. The efficiency of trypsin digestion is likely to vary considerably amongst proteins [55]; hence, for each analyte, it will be critical to take measures to minimize the effects of variable digestion and to detect it when it occurs.
A major cost associated with the development of any immuno-SRM assay is the generation of antibodies. One approach for reducing the cost is to multiplex the immunization process [30]. This approach also has the benefit of increasing the success rate for a given protein target. Other strategies for affinity reagent development must be developed to significantly reduce the cost. It remains to be seen whether less expensive, recombinant approaches to antibody production will be successful in making high affinity anti-peptide antibodies. In addition, the use of alternative reagents, like DNA aptamers [56], may provide a versatile tool for peptide enrichment.
Finally, the potential for multiplexing measurements using immuno-SRM assays has not been fully explored. We recently investigated the performance of multiplexing 50 peptides together into one assay [57] with good success. However, the mass spectrometer is capable of much higher multiplexing without compromising specificity. Thus future work will likely include the analysis of hundreds of analytes in a single assay, a tremendous advantage for saving the volume of precious samples and reducing the cost per analyte.
Summary
The ability to readily develop highly multiplexed, sensitive assays to any group of proteins would transform biological science. Such assays would enable large scale investigations into fundamental biology, improve our understanding of the relationships between molecular biology and disease, and provide a means to narrow the gap between biomarker discovery and personalized clinical medicine. Technologies utilized in such an endeavor must be versatile, time and cost effective. Immunoaffinity enrichment coupled with mass spectrometry has shown tremendous potential for meeting the demands of quantitative protein assays. The immuno-SRM approach provides a cost-effective alternative to traditional immunoassays with good performance capabilities and tremendous versatility.
Acknowledgments
This work was funded by the National Cancer Institutes Clinical Proteomic Technology Assessment for Cancer (CPTAC) Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author has nothing to disclose.
References
- 1.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6(11):1941–51. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 2.Chace DH, Kalas TA. A biochemical perspective on the use of tandem mass spectrometry for newborn screening and clinical testing. Clin Biochem. 2005;38(4):296–309. doi: 10.1016/j.clinbiochem.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Barr JR, Maggio VL, G. PD, Jr, et al. Isotope dilution--mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42(10):1676–82. [PubMed] [Google Scholar]
- 4.Gerber SA, Rush J, Stemman O, et al. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn E, Wu J, Karl J, et al. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4(4):1175–86. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 6.Barnidge DR, Goodmanson MK, Klee GG, et al. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3(3):644–52. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 7.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–88. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Agger SA, Marney LC, Hoofnagle AN. Simultaneous Quantification of Apolipoprotein A-I and Apolipoprotein B by Liquid Chromatography—Multiple Reaction Monitoring/Mass Spectrometry. Clin Chem. 2010 doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange V, Picotti P, Domon B, et al. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan S, Aebersold R, Chen R, et al. Mass spectrometry based targeted protein quantification: methods and applications. J Proteome Res. 2009;8(2):787–97. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addona TA, Abbatiello SE, Schilling B, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzyk MA, Smith D, Yang J, et al. MRM-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8(8):1860–77. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keshishian H, Addona T, Burgess M, et al. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–29. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79(15):5826–37. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 15.Ackermann BL, Berna MJ. Coupling immunoaffinity techniques with MS for quantitative analysis of low-abundance protein biomarkers. Expert Rev Proteomics. 2007;4(2):175–86. doi: 10.1586/14789450.4.2.175. [DOI] [PubMed] [Google Scholar]
- 16.Nedelkov D. Mass spectrometry-based immunoassays for the next phase of clinical applications. Expert Rev Proteomics. 2006;3(6):631–40. doi: 10.1586/14789450.3.6.631. [DOI] [PubMed] [Google Scholar]
- 17.Kiernan UA. Quantitation of target proteins and post-translational modifications in affinity-based proteomics approaches. Expert Rev Proteomics. 2007;4(3):421–8. doi: 10.1586/14789450.4.3.421. [DOI] [PubMed] [Google Scholar]
- 18.Parker CE, Pearson TW, Anderson NL, et al. Mass-spectrometry-based clinical proteomics--a review and prospective. Analyst. 2010;135(8):1830–8. doi: 10.1039/c0an00105h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson NL, Anderson NG, Haines LR, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3(2):235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347(1-2):3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate J, Ward G. Interferences in immunoassay. Clin Biochem Rev. 2004;25(2):105–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Ismail AA. Interference from endogenous antibodies in automated immunoassays: what laboratorians need to know. J Clin Pathol. 2009;62(8):673–8. doi: 10.1136/jcp.2008.055848. [DOI] [PubMed] [Google Scholar]
- 23.Whiteaker JR, Zhao L, Anderson L, et al. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9(1):184–96. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteaker JR, Lin C, Kennedy J, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011 doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteaker JR, Zhao L, Zhang HY, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362(1):44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoofnagle AN, Becker JO, Wener MH, et al. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54(11):1796–804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn E, Addona T, Keshishian H, et al. Developing Multiplexed Assays for Troponin I and Interleukin-33 in Plasma by Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry. Clin Chem. 2009;55(6):1108–17. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenbaum JA, Andersen PH, Blythe M, et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J Mol Recognit. 2007;20(2):75–82. doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 29.Blythe MJ, Flower DR. Benchmarking B cell epitope prediction: underperformance of existing methods. Protein Sci. 2005;14(1):246–8. doi: 10.1110/ps.041059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiteaker JR, Zhao L, Abbatiello SE, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoenherr RM, Zhao L, Whiteaker JR, et al. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring-mass spectrometry. J Immunol Methods. 2010;353(1-2):49–61. doi: 10.1016/j.jim.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pope ME, Soste MV, Eyford BA, et al. Anti-peptide antibody screening: selection of high affinity monoclonal reagents by a refined surface plasmon resonance technique. J Immunol Methods. 2009;341(1-2):86–96. doi: 10.1016/j.jim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Razavi M, Pope ME, Soste MV, et al. MALDI immunoscreening (MiSCREEN): a method for selection of anti-peptide monoclonal antibodies for use in immunoproteomics. J Immunol Methods. 2011;364(1-2):50–64. doi: 10.1016/j.jim.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poetz O, Hoeppe S, Templin MF, et al. Proteome wide screening using peptide affinity capture. Proteomics. 2009;9(6):1518–23. doi: 10.1002/pmic.200800842. [DOI] [PubMed] [Google Scholar]
- 35.Planatscher H, Supper J, Poetz O, et al. Optimal selection of epitopes for TXP-immunoaffinity mass spectrometry. Algorithms Mol Biol. 2010;5:28. doi: 10.1186/1748-7188-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoeppe S, Schreiber TD, Planatscher H, et al. Targeting peptide termini, a novel immunoaffinity approach to reduce complexity in mass spectrometric protein identification. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.002857. M110.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berna M, Schmalz C, Duffin K, et al. Online immunoaffinity liquid chromatography/tandem mass spectrometry determination of a type II collagen peptide biomarker in rat urine: Investigation of the impact of collision-induced dissociation fluctuation on peptide quantitation. Anal Biochem. 2006;356(2):235–43. doi: 10.1016/j.ab.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Li WW, Nemirovskiy O, Fountain S, et al. Clinical validation of an immunoaffinity LC-MS/MS assay for the quantification of a collagen type II neoepitope peptide: A biomarker of matrix metalloproteinase activity and osteoarthritis in human urine. Anal Biochem. 2007;369(1):41–53. doi: 10.1016/j.ab.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Nemirovskiy O, Li WW, Szekely-Klepser G. Design and validation of an immunoaffinity LC-MS/MS assay for the quantification of a collagen type II neoepitope peptide in human urine: application as a biomarker of osteoarthritis. Methods Mol Biol. 2010;641:253–70. doi: 10.1007/978-1-60761-711-2_15. [DOI] [PubMed] [Google Scholar]
- 40.Anderson NL, Jackson A, Smith D, et al. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics. 2009;8(5):995–1005. doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson RW, Krone JR, Bieber AL, et al. Mass spectrometric immunoassay. Anal Chem. 1995;67(7):1153–8. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 42.Oran PE, Jarvis JW, Borges CR, et al. Mass spectrometric immunoassay of intact insulin and related variants for population proteomics studies. Proteomics Clin Appl. 2011 doi: 10.1002/prca.201000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trenchevska O, Nedelkov D. Targeted quantitative mass spectrometric immunoassay for human protein variants. Proteome Sci. 2011;9(1):19. doi: 10.1186/1477-5956-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oran PE, Jarvis JW, Borges CR, et al. C-peptide microheterogeneity in type 2 diabetes populations. Proteomics Clin Appl. 2010;4(1):106–11. doi: 10.1002/prca.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren EN, Elms PJ, Parker CE, et al. Development of a protein chip: a MS-based method for quantitation of protein expression and modification levels using an immunoaffinity approach. Anal Chem. 2004;76(14):4082–92. doi: 10.1021/ac049880g. [DOI] [PubMed] [Google Scholar]
- 46.Jiang J, Parker CE, Hoadley KA, et al. Development of an immuno tandem mass spectrometry (iMALDI) assay for EGFR diagnosis. Proteomics Clin Appl. 2007;1(12):1651–9. doi: 10.1002/prca.200700009. [DOI] [PubMed] [Google Scholar]
- 47.Jiang J, Parker CE, Fuller JR, et al. An immunoaffinity tandem mass spectrometry (iMALDI) assay for detection of Francisella tularensis. Anal Chim Acta. 2007;605(1):70–9. doi: 10.1016/j.aca.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid JD, Holmes DT, Mason DR, et al. Towards the development of an immuno MALDI (iMALDI) mass spectrometry assay for the diagnosis of hypertension. J Am Soc Mass Spectrom. 2010;21(10):1680–6. doi: 10.1016/j.jasms.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 49.Whiteaker JR, Zhang H, Zhao L, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6(10):3962–75. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 50.Neubert H, Gale J, Muirhead D. Online high-flow peptide immunoaffinity enrichment and nanoflow LC-MS/MS: assay development for total salivary pepsin/pepsinogen. Clin Chem. 2010;56(9):1413–23. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 51.Ahn YH, Lee JY, Lee JY, et al. Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J Proteome Res. 2009;8(9):4216–24. doi: 10.1021/pr900269s. [DOI] [PubMed] [Google Scholar]
- 52.Hoofnagle AN, Wener MH. Serum thyroglobulin: A model of immunoassay imperfection. Clin Lab Int. 2006;12:12–4. [Google Scholar]
- 53.Kippen AD, Cerini F, Vadas L, et al. Development of an isotope dilution assay for precise determination of insulin, C-peptide, and proinsulin levels in non-diabetic and type II diabetic individuals with comparison to immunoassay. J Biol Chem. 1997;272(19):12513–22. doi: 10.1074/jbc.272.19.12513. [DOI] [PubMed] [Google Scholar]
- 54.Janecki DJ, Bemis KG, Tegeler TJ, et al. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal Biochem. 2007;369(1):18–26. doi: 10.1016/j.ab.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 55.Proc JL, Kuzyk MA, Hardie DB, et al. A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J Proteome Res. 2010;9(10):5422–37. doi: 10.1021/pr100656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Widen SG, Jamaluddin M, et al. Quantification of Activated NF-{kappa}B/RelA Complexes Using ssDNA Aptamer Affinity - Stable Isotope Dilution--Selected Reaction Monitoring--Mass Spectrometry. Mol Cell Proteomics. 2011;10(6) doi: 10.1074/mcp.M111.008771. M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whiteaker JR, Zhao L, Lin C, et al. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. doi: 10.1074/mcp.M111.015347. submitted for publication 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]