Abstract
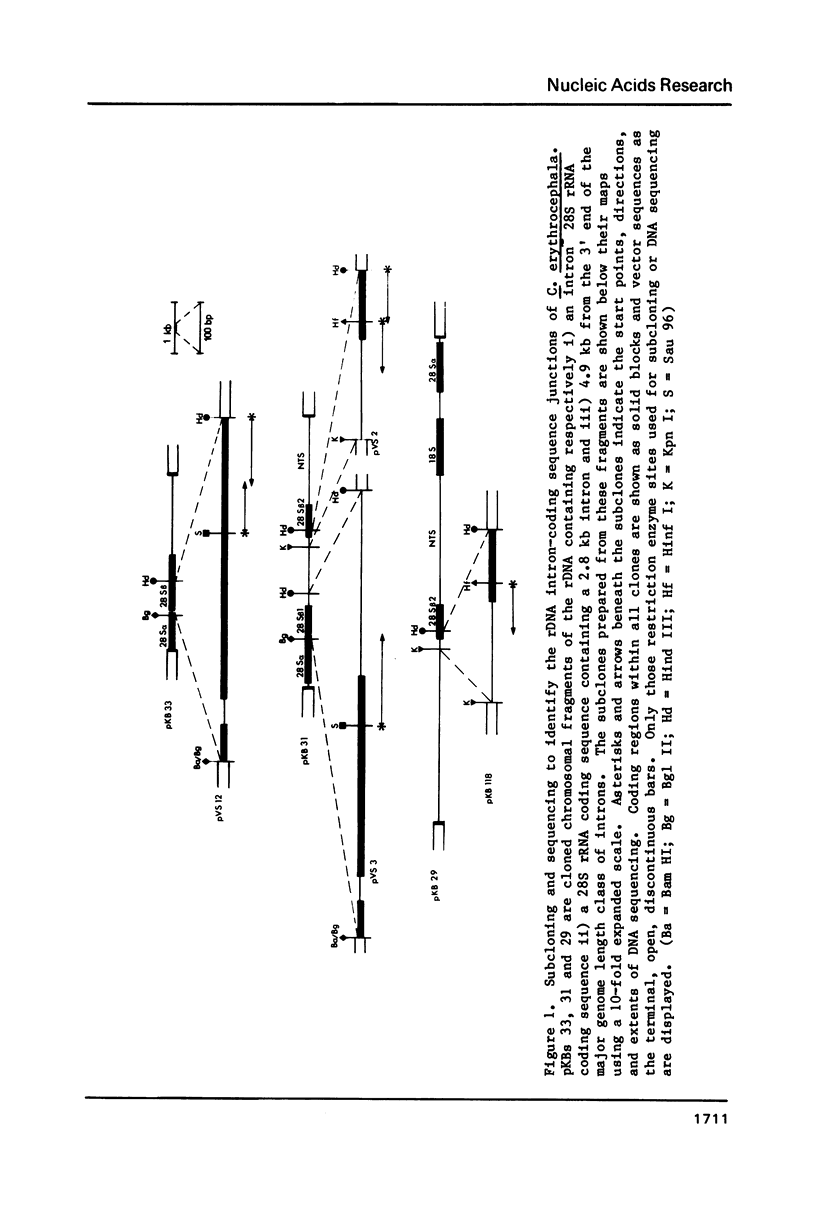
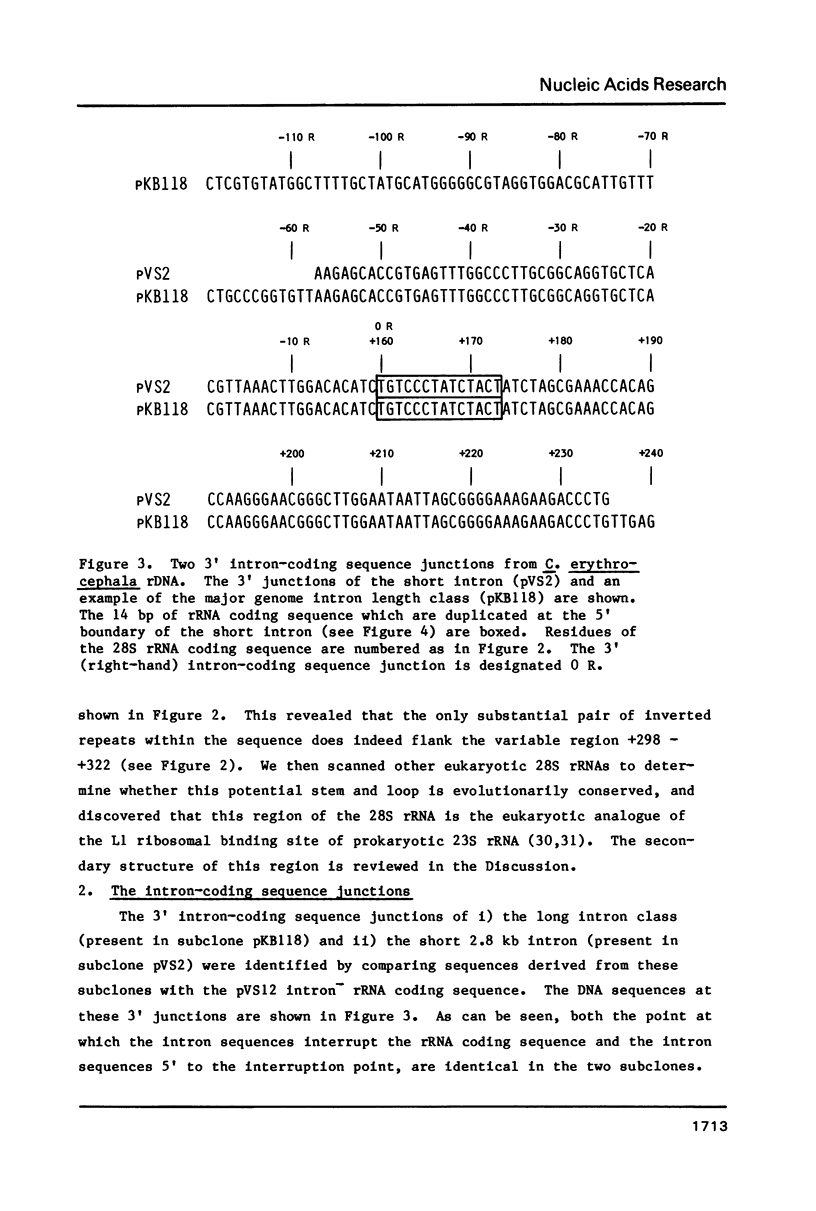
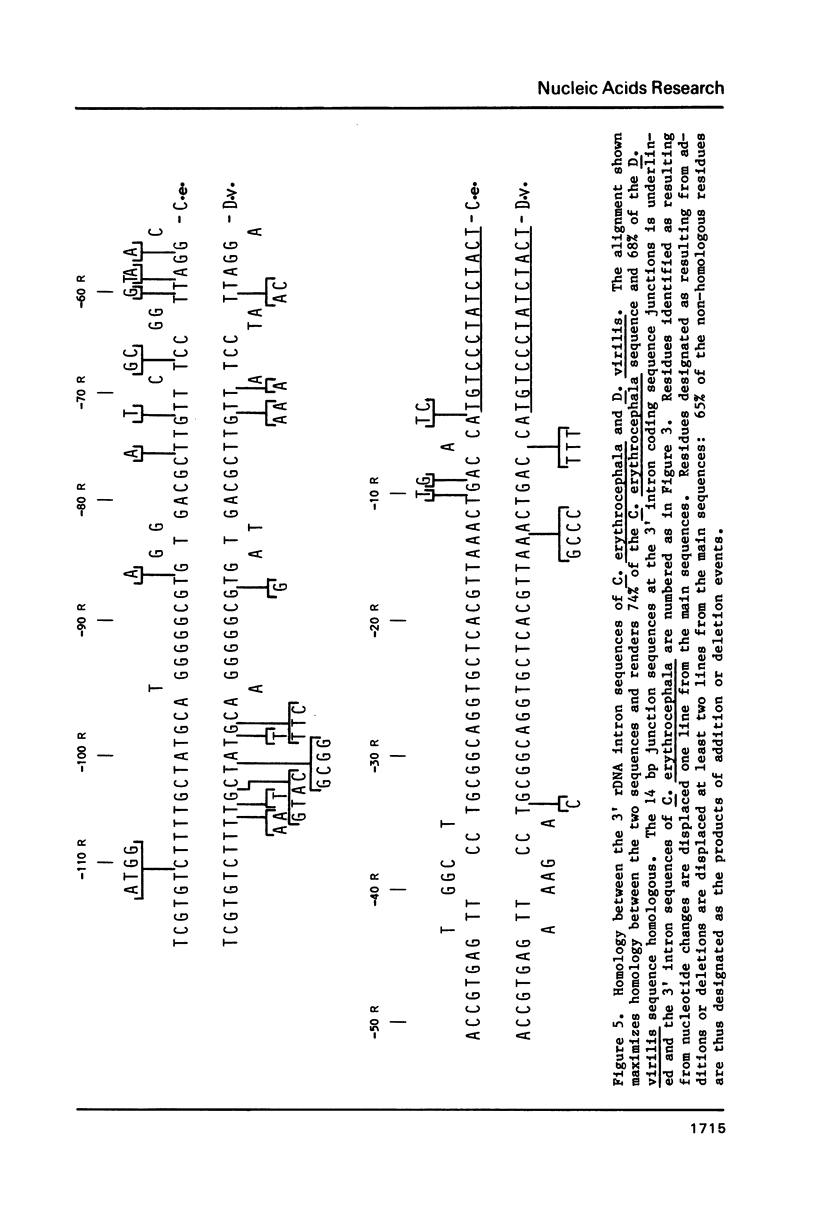
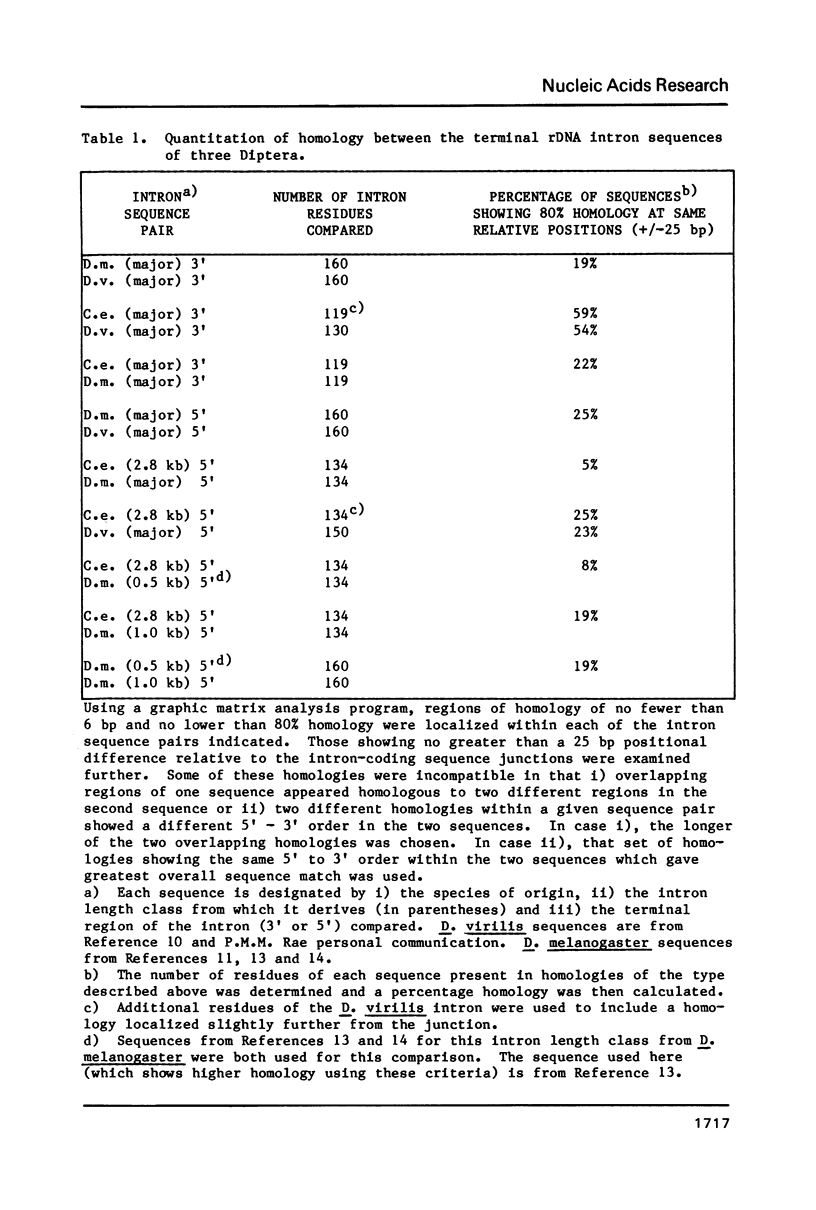
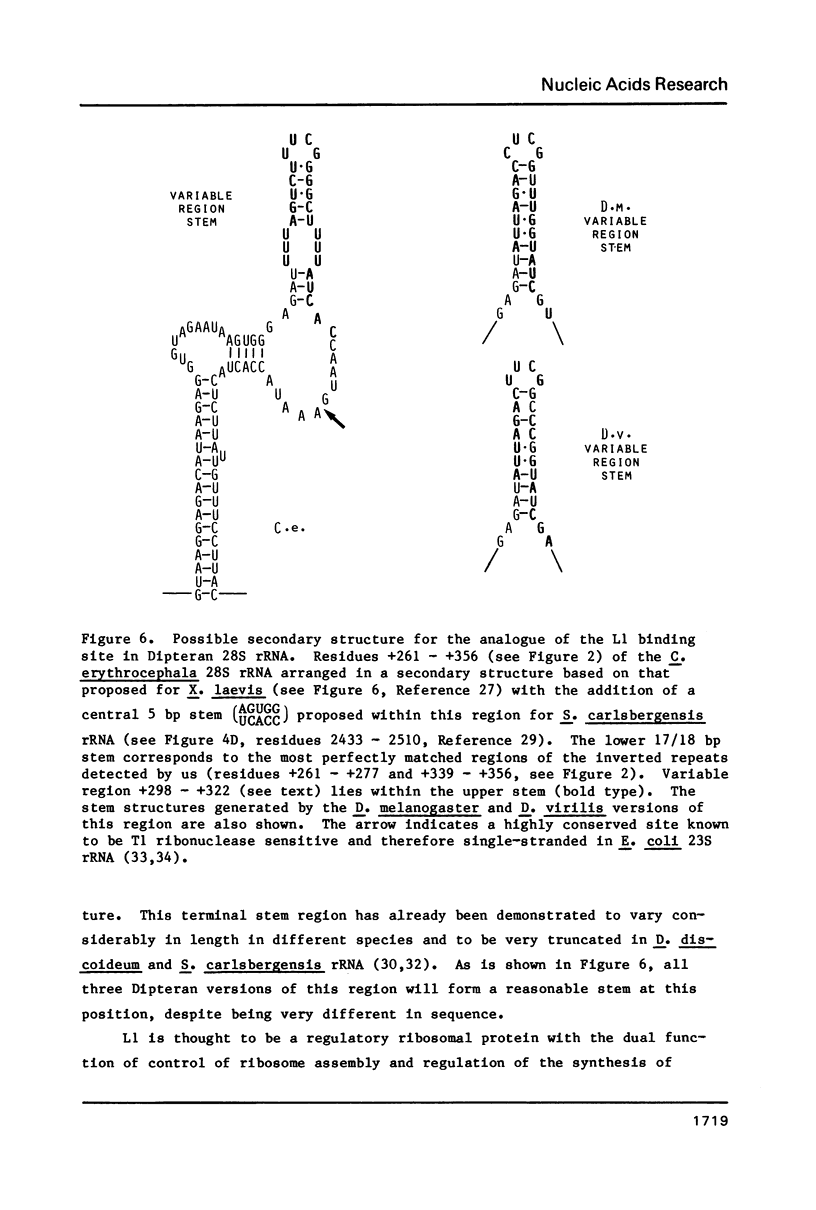
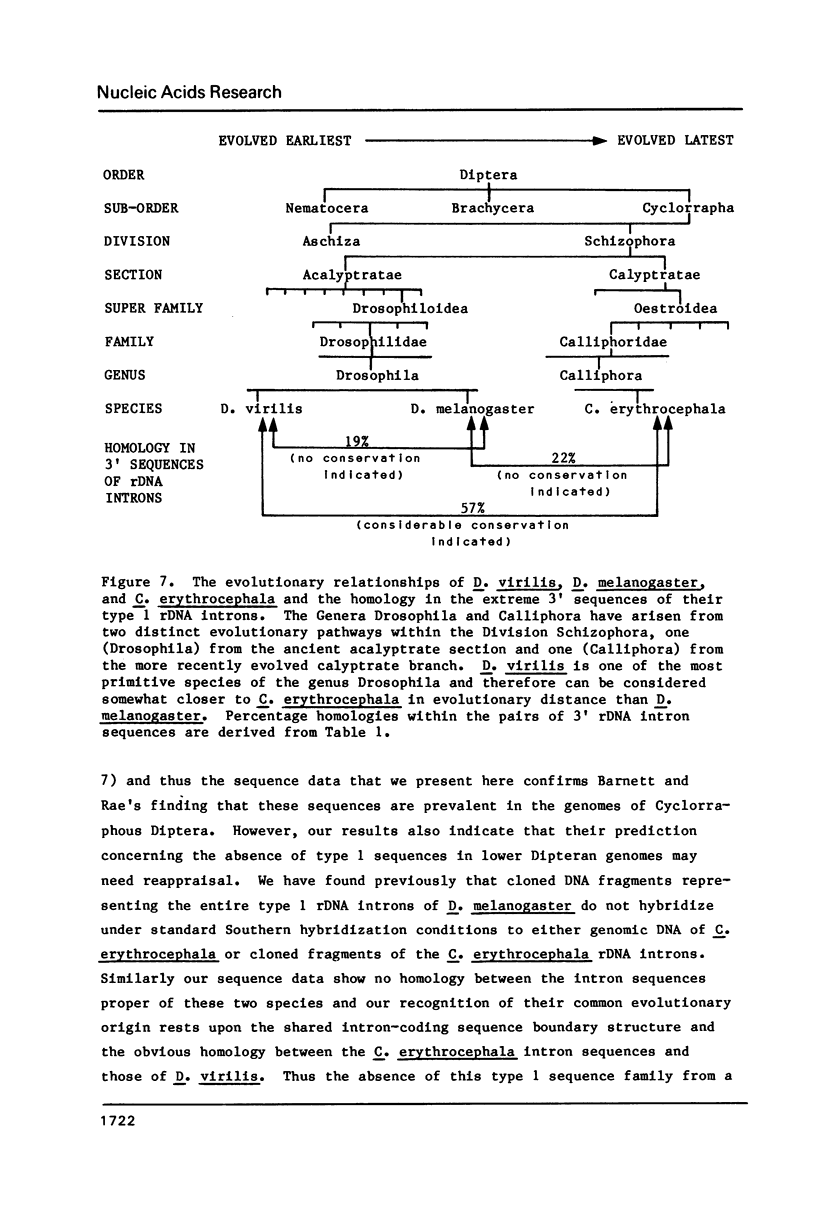
We have sequenced the available cloned examples of the intron-coding sequence junctions for the rDNA of the higher Dipteran, Calliphora erythrocephala. The introns interrupt the rDNA at the same position as the type 1 intron family detected in Drosophila melanogaster and D. virilis (10,11). A duplication of 14 base pairs of the 28S rRNA coding sequence surrounds a short version of the major genomic length class of introns. This same duplication is associated with boundaries of the type 1 introns in D. virilis and D. melanogaster (10, 13,14). We have detected considerable homology between the 3' intron sequences of C. erythrocephala and D. virilis. The rRNA coding sequences flanking the introns are extremely homologous in C. erythrocephala, D. melanogaster and D. virilis, with only one small region of significant divergence. This corresponds to a variable stem region previously identified in eukaryotic 28S rRNA at a site analogous to the L1 ribosomal protein binding site of prokaryotic 23S rRNA (27).
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett T., Rae P. M. A 9.6 kb intervening sequence in D. virilis rDNA, and sequence homology in rDNA interruptions of diverse species of Drosophila and other diptera. Cell. 1979 Apr;16(4):763–775. doi: 10.1016/0092-8674(79)90092-8. [DOI] [PubMed] [Google Scholar]
- Beckingham K. A plasmid cloning vector for Kpnl-cleaved DNA. Plasmid. 1980 Nov;4(3):354–356. doi: 10.1016/0147-619x(80)90074-8. [DOI] [PubMed] [Google Scholar]
- Beckingham K. The ribosomal DNA of Calliphora erythrocephala. The cistron classes of total genomic DNA. J Mol Biol. 1981 Jun 25;149(2):141–169. doi: 10.1016/0022-2836(81)90296-5. [DOI] [PubMed] [Google Scholar]
- Beckingham K., White R. The ribosomal DNA of Calliphora erythrocephala; and analysis of hybrid plasmids containing ribosomal DNA. J Mol Biol. 1980 Mar 15;137(4):349–373. doi: 10.1016/0022-2836(80)90162-x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Rebbert M. L. Nucleotide sequences at the boundaries between gene and insertion regions in the rDNA of Drosophilia melanogaster. Nucleic Acids Res. 1981 Oct 10;9(19):5011–5020. doi: 10.1093/nar/9.19.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. K., Fouts D. L., Manning J. E. Sequence arrangement of the rRNA genes of the dipteran Sarcophaga bullata. Nucleic Acids Res. 1981 Jun 11;9(11):2563–2576. doi: 10.1093/nar/9.11.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T., Brown D. M. Base-specific reactions useful for DNA sequencing: methylene blue--sensitized photooxidation of guanine and osmium tetraoxide modification of thymine. Nucleic Acids Res. 1978 Feb;5(2):615–622. doi: 10.1093/nar/5.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., White R. L., Finnegan D. J., Hogness D. S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell. 1975 Jun;5(2):149–157. doi: 10.1016/0092-8674(75)90023-9. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan N. C., Gall J. G. The intervening sequence of the ribosomal RNA gene is highly conserved between two Tetrahymena species. Nucleic Acids Res. 1982 May 11;10(9):2809–2822. doi: 10.1093/nar/10.9.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Munjaal R. P., Chandra T., Woo S. L., Dedman J. R., Means A. R. A cloned calmodulin structural gene probe is complementary to DNA sequences from diverse species. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2330–2334. doi: 10.1073/pnas.78.4.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M. Coding region deletions associated with the major form of rDNA interruption in Drosophila. Nucleic Acids Res. 1981 Oct 10;9(19):4997–5010. doi: 10.1093/nar/9.19.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Kohorn B. D., Wade R. P. The 10 kb Drosophila virilis 28S rDNA intervening sequence is flanked by a direct repeat of 14 base pairs of coding sequence. Nucleic Acids Res. 1980 Aug 25;8(16):3491–3504. doi: 10.1093/nar/8.16.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M. Unequal crossing-over accounts for the organization of Drosophila virilis rDNA insertions and the integrity of flanking 28S gene. Nature. 1982 Apr 8;296(5857):579–581. doi: 10.1038/296579a0. [DOI] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Glätzer K. H., Kunz W. Characterization of cloned ribosomal DNA from Drosophila hydei. Nucleic Acids Res. 1980 Oct 24;8(20):4593–4611. doi: 10.1093/nar/8.20.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Matsumoto L., Gerbi S. A. Two distinct intervening sequences in different ribosomal DNA repeat units of Sciara coprophila. Nucleic Acids Res. 1981 Aug 11;9(15):3747–3764. doi: 10.1093/nar/9.15.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H., Glover D. M. Duplicated rDNA sequences of variable lengths flanking the short type I insertions in the rDNA of Drosophila melanogaster. Nucleic Acids Res. 1981 Nov 11;9(21):5521–5532. doi: 10.1093/nar/9.21.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H., Miller J. R., Woods L. C., Glover D. M. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981 Apr 30;290(5809):749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Tracy S. Improved rapid methodology for the isolation of nucleic acids from agarose gels. Prep Biochem. 1981;11(3):251–268. doi: 10.1080/00327488108061767. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Tartof K. D. X and Y chromosomal ribosomal DNA of Drosophila: comparison of spacers and insertions. Cell. 1978 Jun;14(2):269–278. doi: 10.1016/0092-8674(78)90113-7. [DOI] [PubMed] [Google Scholar]
- Zaha A., Leoncini O., Hollenberg C. P., Lara F. J. Cloning and characterization of the ribosomal RNA genes of Rhynchosciara americana. Chromosoma. 1982;87(1):103–116. doi: 10.1007/BF00333512. [DOI] [PubMed] [Google Scholar]


