Abstract
Salient features of the first meiotic division are independent segregation of chromosomes and homologous recombination (HR). In non-sexually reproducing, homozygous species studied to date HR is absent. In this study, we constructed the first linkage maps of homozygous, bivalent-forming Oenothera species and provide evidence that HR was exclusively confined to the chromosome ends of all linkage groups in our population. Co-segregation of complementary DNA-based markers with the major group of AFLP markers indicates that HR has only a minor role in generating genetic diversity of this taxon despite its efficient adaptation capability. Uneven chromosome condensation during meiosis in Oenothera may account for restriction of HR. The use of plants with ancient chromosomal arm arrangement demonstrates that limitation of HR occurred before and independent from species hybridizations and reciprocal translocations of chromosome arms—a phenomenon, which is widespread in the genus. We propose that consecutive loss of HR favored the evolution of reciprocal translocations, beneficial superlinkage groups and ultimately permanent translocation heterozygosity.
Keywords: Oenothera, homologous recombination, reciprocal translocation, permanent translocation heterozygosity, AFLP mapping
Introduction
Why sexual reproduction should be virtually ubiquitous in eukaryotes is one of the most intriguing puzzles in evolutionary biology (Otto and Lenormand, 2002; de Visser and Elena, 2007). It is generally thought that the long-term advantage of sexual recombination lies in the coupling of beneficial mutations and the elimination of deleterious mutations thereby, preventing degeneration of chromosomes. Thus, homologous recombination (HR) should promote genetic variance in fitness and facilitate an efficient adaptation to changing environments. With rare exceptions and in contrast to an asexual life style members of sexually reproducing populations need to produce females and males, whereas only the first is capable of giving birth to the progeny. In competition with same-gender individuals both have to find each other to ensure fertilization. Therefore, evolutionary driving forces responsible for the origin and maintenance of sex and HR are still enigmatic particularly with regard to the disadvantage of this ‘twofold' cost of producing males (Smith, 1978; West et al., 1999; Rice and Chippindale 2001; Butlin, 2002; Rice, 2002; Nielsen, 2006; Otto and Gerstein, 2006).
In no bivalent forming, sexually reproducing species studied to date HR is absent in both sexes (Hadany and Comeron 2008). However, the frequency of HR can vary among chromosomes and chromosomal regions, between sexes, and in species that differ in life history or ecology (Butlin, 2005; Agrawal, 2006; Wilfert et al., 2007). HR seems central to meiosis because it ensures both interchromosomal recombination between homologous pairs, rather than sister chromatids, and accurate segregation of the entire chromosome set. Thus, HR could also be regarded as an incidental consequence of crossing over required for stabilizing pairing and for avoiding aneuploidy (Cavalier-Smith, 2002; Wijnker and de Jong, 2008). The genus Oenothera (evening primrose) provides a model system for investigating these issues. It represents a well-known group of flowering plants, for which a rich source of taxonomic, biosystematic and genetic information is available (Cleland, 1972; Harte, 1994; Dietrich et al., 1997; Levin, 2002; Mráček et al., 2006; Golczyk et al., 2008; Rauwolf et al. 2008; Greiner et al., 2008a; Johnson et al., 2010). Furthermore, Oenothera is an interesting genus in which many species are essentially sexual clones (Rauwolf et al., 2008; Johnson et al., 2010). They reproduce sexually via seeds but free segregation of chromosomes and HR is restricted in permanent translocation heterozygote (PTH) species (Cleland, 1972; Holsinger and Ellstrand, 1984; Harte, 1994). In PTH species chromosome arms 1–14 instead of chromosomes I–VII are counted because reciprocal translocations of all chromosome arms encompass the entire chromosome complement. Therefore, the genotype designation in Oenothera is based on the disposition of chromosome arms of two haploid chromosome sets, so called Renner complexes, for example, O. biennis strain suaveolens Grado consists of the two Renner complexes Galbicans and Gflavens with the chromosome arm arrangement 1·12 3·6 5·7 9·4 10·11 13·8 2·14 and 1·4 3·2 5·6 7·10 9·8 11·12 13·14, respectively (Cleland, 1972; Harte, 1994; Rauwolf et al., 2008). In consequence, in the two Renner complexes of PTH species no chromosome is fully homologous to any other and no bivalents are formed. Instead, all chromosomes are arranged end to end in a ring during diakinesis and the parental chromosomes are oriented in an alternate order (Cleland, 1936, 1972; Burnham, 1962; Golczyk et al., 2005 and 2008). Self-incompatibility, selective fertilization, gamtetophytic and/or zygotic lethal systems often lead to sex linkage of two different chromosome complexes—typically one passes through the pollen and the other through the egg cell, which maintains heterozygote advantages (Cleland, 1972; Rauwolf et al., 2008). Not only in Oenothera, but also in about a dozen other plant species—mostly in Onagaceae species—reciprocal translocation of chromosome arms is known (Holsinger and Ellstrand, 1984; Levin, 2002; Golczyk et al., 2005).
Oenothera is a classical model for suppression of HR in PTH species. Mechanistically, two contradictory models for this phenomenon are found. As originally proposed by Darlington, (1931) break points supporting reciprocal translocations of chromosomes arms in Oenothera can be elsewhere in a chromosome than the centromere. As a result, interchanged chromosome regions are homologous for their terminal segment but not for their proximal part. In consequence, meiotic paring and crossing over in PTH species is restricted to the homologous, terminal, (sub)telomeric regions, and suppressed in the non-homologous proximal part. It has therefore long been assumed that HR is repressed in ring-forming PTH species but not necessarily in homozygous ones, not involved in interchanges. This concept is widely accepted throughout the literature (refer Stebbins, 1950; Stebbins, 1971; Levin, 2002; Ranganath, 2008; Johnson et al., 2009a, 2010; Johnson, 2011). Cleland, (1972) introduced another view on suppression of HR in Oenothera. He assumed that the breaking point of chromosomes involved in arm interchanges occurs within the heterochromatic centromeres. This should still allow recombination in ring-formers—interchanged chromosomes arms are still fully homologues and could potentially pair over their entire length. According to this hypothesis the observed suppression of HR in PTH is mediated by uneven condensation of chromosomes and limitation of chiasmata formation to the telomeric regions. The situation for bivalent formers remained unclear so far because available genetic data are insufficient to allow definitive conclusions and studies using lines with untranslocated chromosomes arms in original species are missing (Cleland, 1972; Harte, 1994).
To test these both models, we generated the first genetic maps of Oenothera species using amplified fragment length polymorphism (AFLP) and derived complementary DNA markers (Bensch and Akesson, 2005, Meudt and Clarke, 2007). We show for the first time that even in bivalent-forming, homozygous Oenothera species, HR is absent on the majority of all chromosomes in both sexes and restricted to the very distal chromosomal regions. The material used in this study displays the so-called ‘Johansen' arm arrangement of chromosomes (1·2 3·4 5·6 7·10 9·8 11·12 13·14) and is considered to be the most ancient arrangement of the entire population of the subgenus Oenothera. It is widely distributed in North America, present in different open pollinating species, and free of lethal factors (Cleland, 1972). Furthermore, bivalent-forming chromosomes in partial complex heterozygous species were generally not involved in interchanges of chromosomes and often represent unaltered, original chromosomes (Cleland, 1972). As we used homozygous species with this ancient chromosomal arrangement they likely harbor unaltered ‘original' chromosomes never involved in interchanges (Cleland, 1972), we suggest that restriction of HR occurred before evolution of reciprocal translocation and independent of hybridization events. We propose that restriction of HR in homozygous species favored the evolution of reciprocal translocations and ultimately permanent translocation heterozygosity by rearranging single chromosomes to superlinkage groups allowing co-segregation of beneficial mutations.
Materials and methods
Plant material
Details of genetic constitution and corresponding references for Oenothera strains used in this work, as well as field growth and crossing experiments were described elsewhere (Rauwolf et al., 2008). An overview about strains used in this work is given in the Table 1 . We first chose to genotype the F2 generation obtained from an interspecific cross of the inbred and homozygous parental plants O. grandiflora strain grandiflora Tuscaloosa IIItusca (female genotype: BB or htuscaloosa·htuscaloosa) and O. elata subsp. hookeri strain johansen Standard IIIlamS (male genotype: A1A1 or hjohansen Standard·hjohansen Standard). The two diploid lines share the same ancient chromosome arrangement (1·2 3·4 5·6 7·10 9·8 11·12 13·14) and carry plastome type III, which ensures regular segregation of bivalent-forming chromosomes and fully fertile plastome-genome constitutions in the progeny, respectively (Rauwolf et al., 2008). To refine our data we secondly selected two homozygous subspecies poor of the A genotype with identical chromosome arrangement (1·4 3·2 5·9 7·10 6·8 11·12 13·14), both originating from the same habitat in Mexico (Steiner, 1955). O. elata subsp. elata strain elata Cholula (A2A2) served as the maternal parent and strain elata Puebla (A3A3) as the pollen donor.
Table 1. Oenothera strains used in this work.
| Species or hybrid | Straina | Basic genotypeb | Renner complexc | Segmental arrangementd | Diakinesis | Relevant reference |
|---|---|---|---|---|---|---|
| Oe. elata subsp. elata | elata Cholula | A2A2-I | hcholula | 1·4 3·2 5·9 7·10 6·8 11·12 13·14 | 7 pairs | Steiner, 1955 |
| hchoulua | 1·4 3·2 5·9 7·10 6·8 11·12 13·14 | |||||
| Oe. elata subsp. elata | elata Puebla | A3A3-I | hpuebla | 1·4 3·2 5·9 7·10 6·8 11·12 13·14 | 7 pairs | Steiner, 1955 |
| hpuebla | 1·4 3·2 5·9 7·10 6·8 11·12 13·14 | |||||
| Oe. grandiflora | grandiflora Tuscaloosa | BB-III | htuscaloosa | 1·2 3·4 5·6 7·10 9·8 11·12 13·14 | 7 pairs | Steiner and Stubbe, 1984 |
| htuscaloosa | 1·2 3·4 5·6 7·10 9·8 11·12 13·14 | |||||
| Oe. elata subsp. hookeri x Oe. glaziovianae | johansen Standard IIIlamS | A1A1-III | hjohansen Standard | 1·2 3·4 5·6 7·10 9·8 11·12 13·14 | 7 pairs | Rauwolf et al., 2008 |
| hjohansen Standard | 1·2 3·4 5·6 7·10 9·8 11·12 13·14 |
For detailed taxonomy of the section Oenothera see Dietrich et al., 1997.
The first name of the strains refers to the species, which were originally described in the literature, the second one to the collection site and/or the genetic name of a strain.
Latin capital letters refer to the basic nuclear genotype and roman numbers to the basic plastome genotype. For details see Cleland (1972) and Harte (1994).
The subscript ‘h' refers to haplo- or haploid-complex. This designation is used throughout the literature for lethal factor-free Renner complexes.
Segmental arrangemts follow the Cleland system.
The strain is a hybrid of the nuclear genome of Oe. elata subsp. hookeri strain johansen Standard (Cleland, 1935) (Syn: hookeri Johansen, Johansen, johansen) and the chloroplast genomes of Oe. glazioviana strain rr-lamarckiana Sweden (Heribert-Nilsson, 1912). For details see Rauwolf et al., 2008.
AFLP analysis
Isolation of genomic DNA, design of AFLP reactions and DNA fragment detection were performed essentially as reported recently (Rauwolf et al., 2008). The primer combinations yielding the highest number of polymorphic bands for genotyping are listed in (Supplementary Tables S3-S8). AFLP markers were designated as described (Peters et al., 2001). Co-dominant expression sequence tag-derived markers were designated as described (Mráček et al., 2006; Rauwolf et al., 2008).
Statistical analysis
LOD scores and genetic linkage maps using the Kosambi function were calculated using the JoinMap program (Kyazma BV, Wageningen, The Netherlands). GeneScan analysis and Genotyper software (Applied Biosystems Inc, Lincoln, NE, USA) were used to capture genotyping and segregation data for each polymorphic marker. Polymorphic bands were selected with 0.5-bp tolerances. The data were stored in the Access database (Microsoft, Redmond, WA, USA).
Cytological analysis
The configuration of meiotic chromosomes was determined from inflorescences of appropriate age essentially as described (Golczyk et al., 2008; Rauwolf et al., 2008). To depict meiotic configurations in a single focal plane, frames were captured, stacked and combined using Combine ZM software (http://www.hadleyweb.pwp.blueyonder.co.uk).
Results
Reduction of HR in homozygous Oenothera species
The frequency of HR between markers can be utilised to establish genetic maps. This approach has been used in this study to construct the first molecular linkage maps of the diploid Oenothera strains O. elata subsp. hookeri strain johansen Standard IIIlamS (A1 genome) and O. grandiflora strain grandiflora Tuscaloosa IIItusca (B genome).
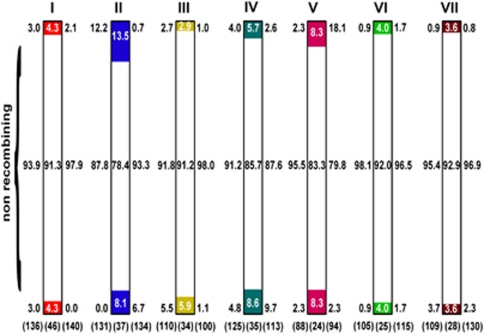
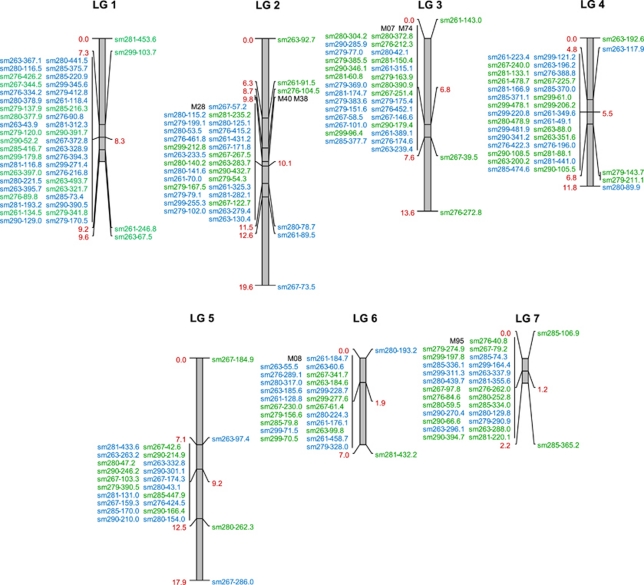
Of 10 245 AFLP fragments obtained using 120 primer combinations, 39.8% displayed polymorphism between the parental lines, which is within the range observed in other interspecific mapping populations (Supplementary Table S1). First, maps with sex-specific markers were generated based on 80 meioses and 1582 polymorphic markers. These fell into seven linkage groups congruent with the seven chromosomes of Oenothera genomes (Table 2 ) (Supplementary Table S2). Surprisingly, most markers in each linkage group (93.3% on average) strictly co-segregated (Table 2) (Supplementary Figure S1). The population size was therefore increased to 244 plants, and 222 markers were used to obtain reliable estimates of recombination frequencies. Segregation distortion in several linkage groups (Table 2) indicated some inbreeding depression and/or epistatic interactions (Grant-Downton and Dickinson, 2004). But, remarkably, among 3,416 meiotic chromosomes examined, an average of 88.5% of the markers did not recombine even once and were again grouped in single clusters (Figure 1, Table 2). Furthermore, 93.2% of additionally integrated, expression sequence tag-derived, co-dominant markers also failed to recombine and clustered in the same groups, allowing determination of the cluster genotypes and merging of the two maps (Figure 2) (Supplementary Figure S1). The major marker groups were flanked by only a few, very distally localized, rarely recombining markers in all chromosomes. The small fraction of recombining markers and their surprisingly low recombination frequency (0.4–7.8%) severely limit the genetic resolution of the map (Figures 1 and 2).
Table 2. Numbers of recombining and non-recombining molecular markers and segregation ratios of all linkage groups calculated from F2 plants of hjohansen Standard/htuscaloosa (A1B) hybrids.
| Linkage groups |
Number of A1 markers based on 80 meioses |
Number of B markers based on 80 meioses |
Number of integrated markers based on 488 meioses |
Segregation ratios of single linkage groups |
|||
|---|---|---|---|---|---|---|---|
| Clustered | Recombining | Clustered | Recombining | Clustered | Recombining | A1A1:A1B:BB | |
| 1 | 124 | 8 | 137 | 3 | 42 | 4 | 0.4:2.1:1.5 |
| 2 | 115 | 16 | 125 | 9 | 29 | 8 | 0.8:2:1.2 |
| 3 | 101 | 9 | 98 | 2 | 31 | 3 | 1.2:2.0:0.8 |
| 4 | 114 | 11 | 99 | 14 | 30 | 5 | 0.9:2.1:1 |
| 5 | 84 | 4 | 75 | 19 | 20 | 4 | 1.1:2:0.9 |
| 6 | 103 | 2 | 111 | 4 | 23 | 2 | 0.6 1.8:1.6 |
| 7a | 104 | 5 | 126 | 4 | 26 | 2 | 0.8:2.2:1 |
| Sum | 745 | 55 | 771 | 55 | 201 | 28 | |
| Percentage | 93.1 | 6.9 | 93.3 | 6.7 | 87.8 | 12.2 | |
Chromsome 9·8, see Supplementary Figure S1 and Rauwolf et al., 2008.
Figure 1.
Schematic view of recombining regions of all seven Oenothera chromosomes. Non-recombining (white) and recombining (color) chromosomal regions are located in large proximal and much shorter, far distal regions, respectively. The lengths of the regions correspond to the relative numbers of markers per chromosome. The relative numbers of markers specific for hjohansen Standard (A1) and htuscaloosa (B), indicated to the left and right of chromosomes, respectively, are based on the analysis of 80 meioses. The relative numbers of markers merged in the integrated map are based on 488 meioses and indicated within the ideograms. The total numbers of markers used in this study are indicated in parentheses. Because of the low genetic resolution of the maps the sizes of the chromosomes were normalized
Figure 2.
Integrated AFLP map of Oenothera species. The molecular maps of hjohansen Standard and htuscaloosa are based on the analysis of 488 meioses and 229 molecular markers. Co-dominant markers are marked in black, AFLP markers of hjohansen Standard and htuscaloosa are marked in green and blue, respectively. The size of the chromosomes in centimorgan (red) corresponds to the genetic resolution.
Thus, on the basis of co-segregation of expression sequence tag-derived markers with the major groups of AFLP markers and the cytological data (see below), we suppose that the recombining, short terminal chromosomal regions are gene-poor and that HR has only a minor role in generating genetic diversity even in homozygous species. Moreover, some terminal AFLP markers that do show recombination tended to form small subclusters comprising up to seven loci (Supplementary Figure S1), reflecting the presence of preferential sites of recombination and ensuring exchanges of presumably co-evolved alleles located on defined chromosomal segments.
Reduction of HR is independent of sequence divergence
To exclude the possibility that the degree of sequence divergence between pairing homologous chromosomes of the basic A1 and B genomes is responsible for lack of recombination, a second F2 linkage analysis was performed using an intraspecific cross. For this purpose we chose to genotype the F2 generation of the inbred and homozygous parental plants with the genotypes A2A2 and A3A3 (see Materials and methods section). Overall, 10.9% of the bands detected were polymorphic that is four times less than the value for the interspecific cross and reflects the close relationship between the two strains. Again, only a small portion of 6.5% of investigated markers recombined, based on the analysis of 560 meiotic chromosomes (Supplementary Table S1).
Unusual chromosome behavior during meiosis
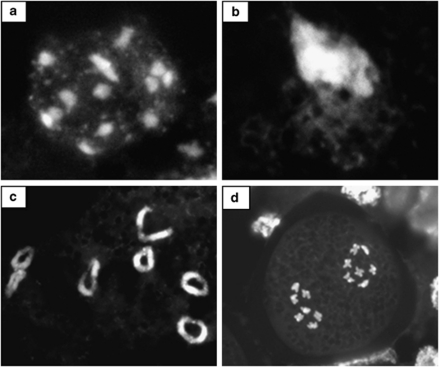
Cytological studies were performed to elucidate whether reduction of recombination is accompanied by alterations in chromosome behavior during meiosis (Figures 3a and d). The data of both A1 and B parental species (data not shown) and their F1 hybrids consistently confirmed pairing of seven homologs at diakinesis and regular meiotic segregation at late anaphase I (Figures 3c and d). During the remaining stages the chromosome behavior was quite unusual. Briefly, in contrast to the usual pattern, all unpaired 14 chromocenters, which represent large pericentromeric segments (Figure 3a) cluster in early meiotic prophase and remain clustered until late pachytene within a highly polarized Rabl configuration (Figure 3b). The observed exceptional uneven chromosome condensation in homozygous A1B hybrids seems to be characteristic for all Oenothera species studied so far (Wisniewska, 1935; Kurabayashi et al., 1962; Cleland, 1972; Golczyk et al., 2008). The chromosomes consist of large and highly condensed centromere-proximal segments and display relaxed distal regions during prophase I (Figure 3b). The decondensed distal chromosome portions are becoming smaller as the meiosis progresses (Figures 3b and c). The less condensed distal chromosome segments in the seven bivalents are terminally attached indicating crossing over events (Figure 3c). Only at very late diakinesis all chromosomes finally become evenly compacted throughout their entire length, as we described recently for other Oenothera species (Golczyk et al., 2008). This structural peculiarity may account for the lack of HR over almost the entire lengths of all chromosomes in the genus.
Figure 3.
Cytology of pre-meiotic and meiotic phases in hjohansen Standard htuscaloosa (A1B) hybrids. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) to visualize DNA. (a) Pre-meiotic nuclei with 14 chromocenters. (b) Rabl configuration at zygotene, showing a strongly stained condensed pericentromeric pole and decondensed distal chromosome segments. (c) Early diakinesis. Less condensed distal chromosome segments in the seven bivalents are terminally attached. (d) Late anaphase I. Chromosomes have segregated regularly.
Discussion
Collectively, our data uncover the striking fact that HR is repressed over about 90% length of all chromosomes. The observed terminal attachments of chromosomes may be necessary to ensure regular segregation of chromosomes and indicate that crossing over and presumably HR frequently occurs but is exclusively restricted to the very distal, gene-poor, chromosomal regions in homozygous, bivalent-forming Oenothera species. Distal recombination events possibly escaped detection analyzing 80 and 488 meiosis with 1,582 and 222 polymorphic markers, respectively, either because of their low frequency or because of their terminal position. At least in ring-forming PTH species it remains possible that the number of recombination at chromosome ends is somewhat underestimated due to a presumed higher degree of homology presumably pointing to a limited number of polymorphisms in this region (Cleland, 1972; Rauwolf et al., 2008). However, the relatively high degree of genetic diversity detected between the parental species O. elata subsp. hooerki and O. grandiflora and predating their divergence about 500,000 years ago (Levy and Levin, 1975; Greiner et al., 2008b) argues against an underestimation of HR at chromosome ends in our mapping population.
This is the first case in which sex and HR have been found to be uncoupled over a length of ∼90% of all chromosomes in a bivalent-forming, homozygous and sexually reproducing eukaryotic organism, allowing all chromosomes in both sexes to be maintained essentially unchanged over generations. The only recombination events observed at the chromosome ends suggest that HR is spatially regulated. As most complementary DNA markers used in this study also cluster in the co-segregated group of most markers, HR is insufficient to provide significant genetic diversity. Nevertheless, the successful cosmopolitan subgenus Oenothera demonstrated a remarkably efficient adaptation capability and occupation of new environments in its evolutionary history (Dietrich et al., 1997).
So far, the limited number of available phenotypic markers and the appearance of reciprocal translocations, the occurrence of balanced lethals, as well as megaspore, embryo sac and pollen tube competition between distinct chromosomal complexes have previously prevented precise determination of overall rates of recombination in Oenothera species (Cleland, 1972; Harte, 1994; Rauwolf et al., 2008). Suppression of HR in ring-forming Oenothera species was long thought to be a consequence of reciprocally translocated chromosome arms (Darlington, 1931; Levin, 2002; Ranganath, 2008; Johnson, 2010). Our data clearly disprove this longstanding hypothesis. Furthermore, we could demonstrate that HR is also suppressed in bivalent-forming Oenothera species.
Ecological and evolutionary conclusions of recent papers are based on the assumption that HR is normal in homozygous Oenothera species as compared with PTH species (Johnson et al., 2009a, 2010). According to our data only free segregation of chromosomes but not HR may account for ecological and reproductive differences between homozygous and complex-heterozygous species.
It was already speculated by Cleland, (1972) that a poorly known kinetics of uneven chromatin condensation during meiosis may be responsible for restriction of HR in Oenothera. Perhaps, only the less condensed terminal regions at the defined meiotic period might be available for recombination and presumably provide chromosome-specific anchors for pairing in Oenothera. Chromatin conformation is a key factor limiting HR in plants (Kirik et al., 2006; Goodstadt and Ponting, 2010). Therefore, uneven condensation is likely to be a major impediment to meiotic recombination not only in complex-heterozygous PTH species, but also in homozygous Oenothera. However, we cannot rule out the possibility, that also the supposed ancestral ‘Johansen' arrangement in A1 and B genomes results from a so far unknown complex karyotype evolution, which may have a general influence on HR in the whole genus.
Nevertheless, the data presented here strongly suggest that restriction of HR is not a by-product of the structural consequences of reciprocal translocations, but may have arisen in homozygous, bivalent-forming Oenothera species already before its occurrence. This view is substantiated by the fact that most PTH Oenothera species derived independently from bivalent-forming ancestors and that the hjohansen Standard and htuscaloosa genomes used in this study possess ancestral chromosome arrangements (Cleland, 1972; Dietrich et al., 1997; Levin, 2002).
These findings shed new light on the development of alternative genetic strategies in adaptive evolution. Apparently, a sequence of three distinct modes of coupling the genetic material has conferred coherent evolutionary advantages on Oenothera. On the basis of our data we propose that progressive restriction of HR at proximal regions was the first step fixing genetic material to defined chromosomal segments and eventually to entire linkage groups, which were still able to segregate in a Mendelian manner. These original species may have existed as normal open-pollinating diploid species, formed seven bivalents during meiosis and were characterized by median centromers (Cleland, 1972). Because of restriction of pairing and crossing over to the distal segments of the chromosomes tight co-evolution of linked alleles was established. The genetically fixed gene combinations in translocated segments acquired particular selective value favoring stability of translocations (refer Stebbins, 1950). Subsequently, consecutive reciprocal translocations initially stabilized a few, and ultimately all, chromosomes in a single linkage group. This prevented free segregation of individual chromosomes and, together with general species interfertility, partially compensated for the reduction of HR, while retaining the advantages of heterosis. This is particularly relevant when species switched from outbreeding to autogamy (Johnson et al., 2009b). Finally, hybridization of species, in combination with the acquisition of gametophytic and sporophytic lethals, generated a balanced genetic system as a refinement of a consistent evolutionary scenario. Stable heterozygosity of the nuclear genotype encompassing all chromosomes was ultimately accomplished.
Oenothera is a highly successful genus, which originated in the Americas, reached Europe after 1500 A.D. and has since invaded all continents (Harte, 1994; Dietrich et al., 1997). This demonstrates that it is possible to establish an intermediate position between sexual and asexual modes of life, in which HR is decoupled from sexual reproduction. Furthermore, asexual lineages are known from most major plant and animal groups and their persistence over geological time proves that they retain sufficient capacity to adapt (Welch and Meselson, 2000; Ozias-Akins and van Dijk, 2007). Originally, when sporophytic lethals still did not accumulate in partial and/or permanent complex heterozygotes, the introduction of one or more beneficial foreign chromosomes into a stable genetic system by cross-pollination can lead to a steady improvement of the chromosome set, and potentially represents an effective form of adaptation.
The acquisition of new gene combinations by repeated successful inter- and intraspecific hybridization over generations with regard to individual chromosomes or—more advanced—to entire chromosome sets also contributes to ecological divergence and provides an escape from mutational load.
Furthermore, the PTH state provides the benefit to the species of maintaining seed set as sexual ‘clones' with a greater probability of spreading over a larger area versus more typical methods of asexual propagation.
Conclusions
Our data question the long-standing assumption that HR is primarily repressed due to reciprocal translocation of chromosome arms in ring-forming Oenothera species. The factors responsible for evolution of reciprocal translocations are still enigmatic. A pre-requisite seemed to be a similar size of chromosomes with heterochromatic centromeres representing potential break points. Translocations at centromeres would also avoid partial deletions in resulting hybrids und maintain the chromosomal morphology and fertility (Cleland, 1972). In the resulting hybrids of often-unrelated complexes heterosis effects and accumulation of deleterious mutations may have favored complex-heterozygous selection. Consecutive repression of HR in Oenothera seemed to be restricted in an early evolutionary stage in bivalent formers and is likely to be a pre-requisite for the evolution of reciprocal translocations and superlinkage groups, because it ensured perpetuation of their advantages. Together with interspecific hybridization, these factors provide an alternative strategy for recombining the genetic material to facilitate stacking of beneficial mutations. Our results show that Oenothera provides an ideal model for examining hypotheses regarding the evolutionary advantages of sex, as well as of speciation.
Acknowledgments
We thank E Gerick and U Wissnet for technical help with plant crosses and propagation. D Leister is acknowledged for his support. This work was made possible by grants from the Hanns-Seidel-Stiftung (supported by the Bundesministerium für Bildung und Forschung) to S Greiner, and from the German Science Foundation to RG Herrmann and J Meurer.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Agrawal AF. Evolution of sex: Why do organisms shuffle their genotypes. Curr Biol. 2006;16:R696–R704. doi: 10.1016/j.cub.2006.07.063. [DOI] [PubMed] [Google Scholar]
- Bensch S, Akesson M. Ten years of AFLP in ecology and evolution: why so few animals. Mol Ecol. 2005;14:2899–2914. doi: 10.1111/j.1365-294X.2005.02655.x. [DOI] [PubMed] [Google Scholar]
- Burnham CR. Discussions in Cytogenetics. Burgess Publishing, Minneapolis, Minnesota; 1962. [Google Scholar]
- Butlin R. The costs and benefits of sex: New insights from old asexual lineages. Nat Rev Genet. 2002;3:311–317. doi: 10.1038/nrg749. [DOI] [PubMed] [Google Scholar]
- Butlin RK. Recombination and speciation. Mol Ecol. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity. 2002;88:125–141. doi: 10.1038/sj.hdy.6800034. [DOI] [PubMed] [Google Scholar]
- Cleland RE. Cyto-taxonomic studies on certain Oenotheras from California. Proc Am Phil Soc. 1935;75:339–429. [Google Scholar]
- Cleland RE. Some aspects of the cyto-genetics of Oenothera. Bot Rev. 1936;2:316–348. [Google Scholar]
- Cleland RE. Oenothera—cytogenetics and evolution. Academic Press Inc., London, United Kingdom; 1972. [Google Scholar]
- Darlington CD. The cytological theory of inheritance in Oenothera. J Genet. 1931;24:405–474. [Google Scholar]
- de Visser JAGM, Elena SF. The evolution of sex: Empirical insights into the roles of epistasis and drift. Nat Rev Genet. 2007;8:139–149. doi: 10.1038/nrg1985. [DOI] [PubMed] [Google Scholar]
- Dietrich W, Wagner WL, Raven PH. Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae) The American Society of Plant Taxonomists, Laramie, Wyoming; 1997. [Google Scholar]
- Golczyk H, Hasterok R, Joachimiak AJ. FISH-aimed karyotyping and characterization of Renner complexes in permanent heterozygote Rhoeo spathacea. Genome. 2005;48:145–153. doi: 10.1139/g04-093. [DOI] [PubMed] [Google Scholar]
- Golczyk H, Musiał K, Rauwolf U, Meurer J, Herrmann RG, Greiner S. Meiotic events in Oenothera—a non-standard pattern of chromosome behaviour. Genome. 2008;51:952–958. doi: 10.1139/G08-081. [DOI] [PubMed] [Google Scholar]
- Goodstadt L, Ponting CP. Is the control of recombination conserved among eukaryotes. Heredity. 2010;106:710–711. doi: 10.1038/hdy.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton RT, Dickinson HG. Plants, pairing and phenotypes—two's company. Trends Genet. 2004;20:188–195. doi: 10.1016/j.tig.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Greiner S, Wang X, Herrmann RG, Rauwolf U, Mayer K, Haberer G, et al. The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol Biol Evol. 2008a;25:2019–2030. doi: 10.1093/molbev/msn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 2008b;36:2366–2378. doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L, Comeron JM. Why are sex and recombination so common. Ann N Y Acad Sci. 2008;1133:26–43. doi: 10.1196/annals.1438.011. [DOI] [PubMed] [Google Scholar]
- Harte C. Oenothera—Contributions of a Plant to Biology. Springer-Verlag, Heidelberg, Germany; 1994. [Google Scholar]
- Heribert-Nilsson N. Die Variabilität der Oenothera Lamarckiana und das Problem der Mutation. Z indnkt Abstamm- u Vererb-Lehre. 1912;9:89–231. [Google Scholar]
- Holsinger KE, Ellstrand NC. The evolution and ecology of permanent translocation heterozygotes. Am Nat. 1984;124:48–71. [Google Scholar]
- Johnson MT. The contribution of evening primrose (Oenothera biennis) to a modern synthesis of evolutionary ecology. Popul Ecol. 2011;53:9–21. [Google Scholar]
- Johnson MT, Smith SD, Rausher MD. Plant sex and the evolution of plant defenses against herbivores. Proc Natl Acad Sci. 2009a;106:18079–18084. doi: 10.1073/pnas.0904695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MT, Smith SD, Rausher MD. Effects of plant sex on range distributions and allocation to reproduction. New Phytol. 2010;186:769–779. doi: 10.1111/j.1469-8137.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- Johnson MT, Vellend M, Stinchcombe JR. Evolution in plant populations as a driver of ecological changes in arthropod communities. Philos Trans R Soc Lond B Biol Sci. 2009b;364:1593–1605. doi: 10.1098/rstb.2008.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006;18:2431–2442. doi: 10.1105/tpc.106.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi M, Lewis H, Raven PH. A comparative study of mitosis in the Onagraceae. Amer J Bot. 1962;49:1003–1026. [Google Scholar]
- Levin DA. The Role of Chromosomal Change in Plant Evolution. Oxford University Press, New York; 2002. [Google Scholar]
- Levy M, Levin DA. Genic heterozygosity and variation in permanent translocation heterozygotes of the Oenothera biennis complex. Genetics. 1975;79:493–512. doi: 10.1093/genetics/79.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Mráček J, Greiner S, Cho WK, Rauwolf U, Braun M, Umate P, et al. Construction, database integration, and application of an Oenothera EST library. Genomics. 2006;88:372–380. doi: 10.1016/j.ygeno.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Evolution: Why Sex. Science. 2006;311:960–961. doi: 10.1126/science.1124663. [DOI] [PubMed] [Google Scholar]
- Otto SP, Gerstein AC. Why have sex? The population genetics of sex and recombination. Biochem Soc Trans. 2006;34:519–522. doi: 10.1042/BST0340519. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, van Dijk PJ. Mendelian genetics of apomixis in plants. Annu Rev Genet. 2007;41:509–537. doi: 10.1146/annurev.genet.40.110405.090511. [DOI] [PubMed] [Google Scholar]
- Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, et al. A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol. 2001;127:1579–1589. [PMC free article] [PubMed] [Google Scholar]
- Ranganath RM. Meiotic chromosome pairing and recombination take refuge in the telomeres. Nat Rev Genet. 2008;9:318. doi: 10.1038/nrg2224-c1. [DOI] [PubMed] [Google Scholar]
- Rauwolf U, Golczyk H, Meurer J, Herrmann RG, Greiner S. Molecular marker systems for Oenothera genetics. Genetics. 2008;180:1289–1306. doi: 10.1534/genetics.108.091249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Experimental tests of the adaptive significance of sexual recombination. Nat Rev Genet. 2002;3:241–251. doi: 10.1038/nrg760. [DOI] [PubMed] [Google Scholar]
- Rice WR, Chippindale AK. Sexual recombination and the power of natural selection. Science. 2001;294:555–559. doi: 10.1126/science.1061380. [DOI] [PubMed] [Google Scholar]
- Smith JM. The Evolution of Sex. Cambridge University Press, New York; 1978. [Google Scholar]
- Stebbins GL. Variation and Evolution in Plants. Columbia Universtiy Press, New York.; 1950. [Google Scholar]
- Stebbins GL. Chromosomal Evolution In Higher Plants. Addison-Wesley Publishing Company, Reading, Menlo Park, London, Don Mills; 1971. [Google Scholar]
- Steiner EE. A cytogenetic study of certain races of Oenotheraelata. Bull Torrey Bot Club. 1955;82:292–297. [Google Scholar]
- Steiner EE, Stubbe W. A contribution to the population biology of Oenothera grandiflora L'Her. Am J Bot. 1984;71:1293–1301. [Google Scholar]
- Welch DM, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- West SA, Lively CM, Read AF. A pluralist approach to sex and recombination. J Evol Biol. 1999;12:1003–1012. [Google Scholar]
- Wijnker E, de Jong H. Managing meiotic recombination in plant breeding. Trends Plant Sci. 2008;13:640–646. doi: 10.1016/j.tplants.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity. 2007;98:189–197. doi: 10.1038/sj.hdy.6800950. [DOI] [PubMed] [Google Scholar]
- Wisniewska E. Badania cytologiczne nad Oenothera Hookeri de Vries. Acta Soc Bot Pol. 1935;12:113–164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.