Abstract
The transition from self-incompatibility to self-compatibility is a common transition in angiosperms often reported in populations at the edge of species range limits. Geographically distinct populations of wild tomato species (Solanum section Lycopersicon (Solanaceae)) have been described as polymorphic for mating system with both self-incompatible and self-compatible populations. Using controlled pollinations and sequencing of the S-RNase mating system gene, we test the compatibility status of a population of S. peruvianum located near its southern range limit. Pollinations among plants of known genotypes revealed strong self-incompatibility; fruit set following compatible pollinations was significantly higher than following incompatible pollinations for all tested individuals. Sequencing of the S-RNase gene in parents and progeny arrays was also as predicted under self-incompatibility. Molecular variation at the S-RNase locus revealed a diverse set of alleles, and heterozygosity in over 500 genotyped individuals. We used controlled crosses to test the specificity of sequences recovered in this study; in all cases, results were consistent with a unique allelic specificity for each tested sequence, including two alleles sharing 92% amino-acid similarity. Site-specific patterns of selection at the S-RNase gene indicate positive selection in regions of the gene associated with allelic specificity determination and purifying selection in previously characterized conserved regions. Further, there is broad convergence between the present and previous studies in specific amino-acid positions inferred to be evolving under positive selection.
Keywords: molecular evolution, positive selection, self-incompatibility, Solanaceae, S-RNase gene, tomato
Introduction
The majority of flowering plants are simultaneous hermaphrodites, yet numerous strategies to avoid the detrimental effects of self-fertilization and inbreeding depression have evolved among angiosperm species. These include mechanical or temporal separation of male and female gender function, separation of sexes onto different individuals, and genetically encoded self-incompatibility (SI) systems. Genetically controlled SI systems are also widespread in angiosperms, having evolved convergently upwards of 20 times and in many lineages (Weller et al., 1995; Allen and Hiscock, 2008); classically, SI systems fall into one of two categories: either gametophytic or sporophytic SI, though a large number of lineages possess late acting SI (Seavey and Bawa, 1986).
In gametophytic systems, the incompatibility phenotype is determined by the haploid pollen grain (that is, the male gametophyte). Several types of gametophytic SI (GSI) exist and can be differentiated on the basis of molecular mechanisms involved in recognition and rejection of self (and closely related) pollen. The best characterized GSI system is S-RNase based GSI, which was initially described in Solanaceae, and is also present in additional plant families. In fact, S-RNase based GSI is the inferred ancestral condition of all asterids and rosids, making it ancestral in ∼75% of all eudicots (Igic and Kohn, 2001; Steinbachs and Holsinger, 2002; Vieira et al., 2008).
Two tightly linked genes at the S-locus control the recognition reaction in S-RNase based GSI. The female determinant is an S-RNase, which is expressed in styles of mature flowers. The male determinant, which has been identified in Solanaceae as the S-locus F-box, is expressed in pollen grains. Following pollination, if the specificity of the pollen matches with either of the two specificities expressed in the style of a maternal plant, pollen tubes abort and fertilization fails. Individual plants functioning as maternal parents are thus unable to accept pollen from themselves or pollen from other plants bearing a matching specificity in the pollen grain. As a result, only gametes with different S-locus specificities can form offspring and individuals are obligately heterozygous at the S-locus.
Seventy years ago, Wright, 1939 first described the importance of negative frequency-dependent selection in maintaining extensive allelic diversity of SI alleles in natural populations. In particular, he noted that plants with relatively rare alleles would enjoy a mating advantage, and that such negative frequency dependence would lead to the maintenance of many alleles at equal frequencies in populations at equilibrium. The efficacy of negative frequency dependence in maintaining variation at this locus extends beyond allelic diversity in present day populations, and also affects the preservation of allelic variation over evolutionary timescales. Ioerger et al., 1990 were the first to document the presence of trans-specific or trans-generic polymorphism, in which allelic polymorphism predates the divergence of distantly related species.
The tomato plant family (Solanaceae) has served as a model system for the study of GSI in angiosperms from both a mechanistic (reviewed in McClure, 2004) and a population genetic perspective (Igic et al., 2004; Kohn, 2008). In particular, the development of reverse-transcriptase PCR methods for amplifying S-alleles using RNA extracted from styles (Richman et al., 1995) has resulted in a number of population surveys of S-RNase variation in several species and genera (Lycium, Savage and Miller, 2006, Miller et al., 2008, 2010; Nicotiana, Roldán et al., 2010; Petunia, Wang et al., 2001; Physalis, Richman et al. 1996, Lu, 2002; Solanum, Richman et al., 1995, Igic et al., 2007, Mena-Ali and Stephenson, 2007; Witheringia, Stone and Pierce, 2005). Data from studies of SI taxa generally meet expectations; that is, populations contain numerous alleles and allelic lineages persist over millions of years. Such data can also be used to examine the pattern and strength of selection at mating system genes, and previous studies have documented a characteristic signature of sequence evolution at the S-RNase gene (Savage and Miller, 2006; Igic et al., 2007; Miller et al., 2008). In particular, those regions of the S-RNase gene associated with allelic specificity evolve under diversifying (positive) selection, whereas purifying selection predominates in regions associated with protein folding and stability (Ida et al., 2001; Vieira et al., 2010). Two recent studies illustrate convergence in positively selected amino-acid (AA) positions for different data sets in Solanaceae (Takebayashi et al., 2003; Vieira et al., 2007) or between closely related species (Savage and Miller, 2006). Such convergence across genera and species in Solanaceae implies that specific amino-acid positions may be important in specificity determination. Connected to this, comparisons of positively selected amino-acid positions among closely related alleles, coupled with empirical evidence that these alleles represent distinct specificities are needed.
Although Solanaceae have served as a model system for the study of incompatibility, it is also the case that SI has been lost independently many times and in many species. Indeed, the evolution of self-compatibility (SC) is considered one of the most common transitions in angiosperm evolutionary biology (Stebbins, 1974), and many examples of multiple transitions within plant lineages have been documented (Kohn et al., 1996; Schoen et al., 1997; Goodwillie, 1999; Igic et al., 2004, 2006). The evolutionary transition from outcrossing to selfing at species range limits (or on oceanic islands) is a recurrent pattern among unrelated lineages of plants (Busch, 2005; Crawford et al., 2008 and references below). Both SI and SC populations have been described for several species of wild tomatoes (Solanum section Lycopersicon), and the presence of SC is often associated with populations located at species range limits (Rick, 1986). Rick and Tanksley, 1981 surveyed populations of S. pennellii along a north-south axis in Peru and Chile; the majority of populations were SI, but their survey revealed several SC populations from the southernmost range limit. Likewise, toward both their northern and southern limits, populations of S. habrochaites are SC, whereas in the interior of the range, populations remain SI (Rick and Chetelat, 1991; Rick et al., 1979). Peralta and Spooner, 2001 and Peralta et al., 2008 also document polymorphism in compatibility status in wild tomato species, including S. pennellii, S. habrochaites and S. peruvianum. Most recently, Igic et al. (2008) reviewed the loss of SI in Solanum section Lycopersicon and identified intraspecific polymorphism in compatibility status in five species including new records for S. arcanum and S. chilense. As pointed out by Igic et al., (2006, 2008), polymorphism at the S-RNase locus is expected to decline relatively rapidly following the loss of SI, as S-alleles are no longer maintained by frequency-dependent selection. Thus, examination of the SI status and S-RNase diversity in species of Solanum section Lycopersicon known to harbor SI/SC polymorphism and in populations located within species range peripheries are needed.
In this study, we investigate the S-RNase mating system locus in greenhouse-raised plants of Solanum peruvianum collected from a population near the southern edge of the species range. In particular, we genotype 30 parental plants and ∼500 offspring to determine whether individuals are heterozygous (expected in SI populations), as well as to determine the extent of allelic diversity in our sample. We also analyze the pattern of molecular evolution at the S-RNase locus and compare this with the pattern identified in SI populations in previous studies of Solanaceae. Second, we use controlled pollinations among plants to ascertain the compatibility status of this peripheral population. Third, we genotype offspring from a subset of these crosses to confirm that the S-RNase sequences isolated represent allelic specificities, and use these crosses to assess whether highly similar S-RNase sequences represent distinct alleles.
Methods
Study species and tissue collection
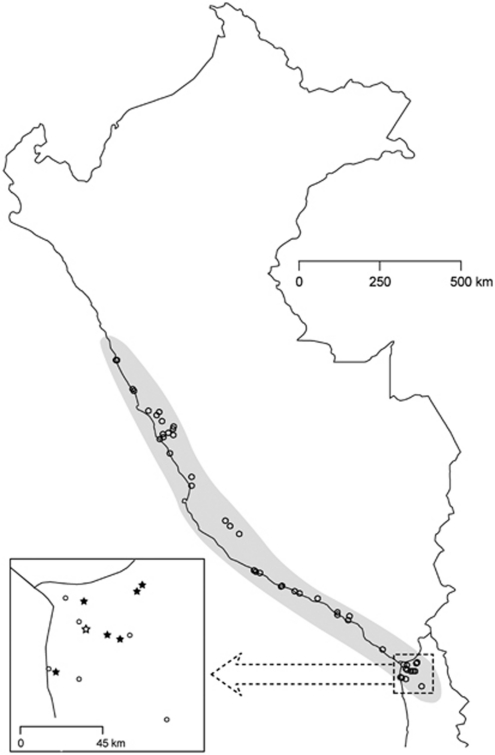
Solanum section Lycopersicon includes the cultivated tomato and twelve wild species that occur natively in western South America (Peralta et al., 2008). Solanum peruvianum is found natively from central Peru to northern Chile (Figure 1; Supplementary Table 1; Peralta et al., 2005, 2008). Populations are typically SI, however, some are apparently SC (see tables in Rick, 1986, Peralta and Spooner, 2001, and Spooner et al., 2005; Peralta et al., 2008). Indeed, at least two populations collected from the Tarapaca region in Chile in the southern region of the species range (∼100 kilometers southeast of the focal location) have been reported as SC (Tomato Genetics Research Center (TGRC), University of California, Davis; LA2955B, latitude -19.317, longitude -69.450; LA4125, latitude -19.306, longitude -69.421). In this study, we evaluate the compatibility status of S. peruvianum using seed collected from near the species' southern border and maintained at the TGRC (Figure 1; Tarapaca, Chile, latitude -18.550, longitude -70.150, elevation 400 m, LA2744). Seeds were germinated and plants grown to flowering in a greenhouse. On flowering, 10–12 styles were collected from each of thirty individuals and preserved in RNAlater (Ambion, Inc., Austin, TX, USA). At the time of style collection, young leaf tissue was also preserved in silica gel desiccant.
Figure 1.
Map of Peru and northern Chile showing the distribution of Solanum peruvianum as determined from collections at the Tomato Genetics Resource Center (open circles; see Supplementary Table 1 for accession numbers and locality information). Inset shows Chilean accessions in the southern part of the range including the collection locality for material used in this study (open star) and nearby accessions for which Genbank sequences were compared (solid stars; Kondo et al., 2002).
Parental generation genotyping
We used the RT-PCR protocol of Richman et al., 1995 to genotype plants, which were used as parentals in controlled pollinations (see below). Five to ten styles were ground in liquid nitrogen and total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Inc., Valencia, CA, USA). We used the First Strand cDNA Synthesis Kit (EMD Biosciences, Inc., Madison, WI, USA) to synthesize cDNAs from the S-RNase encoding mRNAs. A portion of the S-RNase gene (spanning conserved regions C2–C5, see Ioerger et al., 1991) was amplified from cDNA using degenerate primers PR1 and PR3 from Richman et al., 1995. PCRs were performed in 50-μl reactions including 5 μl template, 10 m Tris-HCl buffer, 50 m KCl, 1.5 m MgCl2, 0.2 m dNTPs, 100 ng of each primer, 5 μg BSA and 2 units of Taq polymerase. Because individuals are expected to be heterozygous at the S-RNase locus and as alleles are typically quite divergent, amplification products were cloned into the pT7Blue vector using the Novagen Perfectly Blunt Cloning kit (EMD Biosciences). Positive transformants were amplified using vector-specific primers U19 (5′-GTTTTCCCAGTCACGACGT-3′) and R20 (5′-CAGCTATGACCATGATTACG-3′); 12.5-μl reactions included 2.5 μl template, 10 m Tris-HCl buffer, 50 m KCl, 1.5 m MgCl2, 0.2 m dNTPs, 8 ng of each primer and 0.5 units of Taq polymerase. An average of eight colonies were sequenced for each individual at either the Pennsylvania State University Nucleic Acid Facility (University Park, PA, USA) or Retrogen, Inc. (San Diego, CA, USA).
We developed a PCR-based screening protocol to confirm parental genotypes, as well as genotype offspring arrays following controlled crosses. Following an alignment of the 14 S-RNase sequences isolated using RT-PCR, we designed allele-specific primers to amplify each of the S-RNase sequences recovered in the parental generation. Most primer sets amplified a single allele; however, given sequence similarity two primer sets co-amplified either two (SP14 and SP17) or three (SP1, SP2 and SP3) alleles and these were cloned before sequencing (Supplementary Table 2).
Genomic DNA from parental plants was extracted using the Qiagen DNeasy Mini DNA Extraction Kit (Qiagen). These genotypes contained the full set of 14 S-RNase alleles and were amplified using the battery of allele-specific primers (Supplementary Table 2). These amplifications allowed us to: (1) test the specificity of our primers, (2) confirm parental genotypes and (3) obtain intron sequences for all S-RNase alleles in our sample. PCR amplifications were carried out in either 12.5 or 25 μl volumes containing 0.5 or 1 μl genomic DNA template, 10 m Tris-HCl buffer, 50 m KCl, 0.4 m dNTPs, 3.0 m MgCl2, 0.4 μ of each forward and reverse primer, 1 × Q-Solution (Qiagen) and 0.75 units of Taq polymerase using one of three touchdown PCR programs. Each of the 14 alleles was sequenced following allele-specific amplifications in two to four individuals to confirm the allele sequence and intron. Parental genomic DNAs were used as controls in subsequent offspring genotyping (see below).
Controlled pollinations
To assess the compatibility status of Solanum peruvianum, we compared fruit production following controlled pollinations in an insect-free greenhouse. Because S-RNase genotypes were unknown at the time of pollinations, crosses consisted of either a self-pollen treatment or a cross-pollen treatment. The self-treatment consisted of pollen from other open flowers on the maternal parent, whereas the cross-treatment included pollen collected from a single paternal donor plant. Pollinations were conducted during September–December in 2007 and 2008. As S. peruvianum is buzz pollinated, pollen was collected from paternal donors by removing 1–3 flowers and vibrating the anthers with a handheld electric engraver over a clean petri dish. Pollen was directly applied to the stigmas of flowers on the maternal parent, and marked with the cross identification. Stigmas were coated with donor pollen and received sufficient pollen for seed set. A total of 1249 pollinations were carried out, including 294 self-pollinations (average of 9.8 per maternal parent) and 955 cross-pollinations (including 224 unique parental cross combinations).
Before the analysis of the controlled crosses, genotypes of all parental plants were determined and crosses recorded as either fully compatible (no S-RNase alleles shared between parental plants), semi-compatible (one S-RNase allele shared between parents) or incompatible. Incompatible crosses included both the self-pollen treatment and cross-pollinations between individuals of the same S-RNase genotype, whereas fully or semi-compatible crosses were categorized as compatible. We analyzed fruit set using a general linear model in SAS (SAS Institute, 2002); the model included the effects of plant (random effect), pollination treatment (fixed effect: compatible or incompatible), and the two-way interaction of plant by pollination treatment.
Offspring genotyping
A total of 495 offspring following 17 different controlled crosses were genotyped using allele-specific primers following genomic DNA extractions of offspring. Genomic DNAs from confirmed parental genotypes with the target allele were used as positive controls, whereas individuals without the target allele were used as negative controls. For those primer sets that co-amplified alleles (see Supplementary Table 2), PCR products were cloned and sequenced to confirm S-RNase alleles in progeny.
We used a series of semi-compatible crosses to test the functionality of putative S-RNase allelic sequences. As offspring following a semi-compatible cross can only inherit the non-shared paternal allele, we screened progeny arrays for the unique (and expected) paternal allele. In total, we screened 400 offspring with an average of 26.7 offspring per semi-compatible cross (range, 10–58 offspring per cross). Following Igic et al., 2007, we calculated the probability of the observed progeny array genotypes under the assumption that the cross was fully compatible (P=(0.5)n), where n is the number of progeny genotyped and 0.5 is the probability that offspring share the unique paternal allele. Those probabilities that remained significant following correction for multiple comparisons (that is, 11 alleles tested, P<0.005), allowed rejection of the null hypothesis of full compatibility.
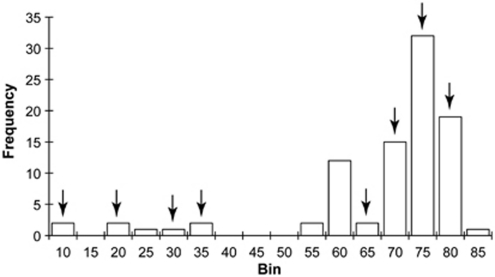
We also determined whether the highly similar alleles in our sample represented unique allelic specificities by analyzing the pattern of S-RNase allele assortment in progeny following three semi-compatible crosses (see above), as well as two additional fully compatible crosses. Specifically, we sought to determine whether highly similar alleles in maternal and paternal plants recognized each other as the same specificity and thus did not pair compared with alternative allele combinations in those parents. A fully compatible cross between a female with the genotype SP2/SP11 and a male with the genotype SP1/SP7 is expected to yield offspring with one of four possible genotypes: SP2/SP1, SP2/SP7, SP11/SP1 or SP11/SP7. However, if alleles SP1 and SP2 are highly similar and are functionally the same specificity, then one might expect an absence of SP2/SP1 offspring. As pairwise divergence values among alleles in our sample were distributed bimodally (ranging from 8 AA differences to 81 AA differences (Figure 2)), we selected crosses that contained potential genotypes in progeny that included both similar (⩽33 AA differences) and alternative (>60 AA differences) allele combinations. We used a Chi-square test to determine whether the number of progeny genotypes in each category (similar vs alternative allele combinations) were different than expected. Expected numbers in each category were determined by the pairwise amino-acid difference between alleles in progeny, and standardized by the total number of similar vs alternative combinations of alleles.
Figure 2.
Distribution of amino-acid differences between S-RNase alleles of Solanum peruvianum in this study. The most similar alleles (SP14 and SP17) differed at 8 AA positions, whereas the most divergent (SP6 and SP12) had 81 AA sites that differed. Arrows indicate those pairwise allele combinations that were present among offspring progeny in selected crosses.
Analysis of S-RNase sequence data
Sequences were edited and assembled using Sequencher version 4.8. (Gene Codes Corp., 1991–2007). Amino-acid alignments were constructed manually using Sequence Alignment Editor (Se-Al) version 2.0 (Rambaut, 2002). Average pairwise distances for S-RNases from Solanum peruvianum were calculated in PAUP* (Swofford, 2002). PAUP* was also used to create a maximum likelihood S-RNase genealogy as a means to assess the extent of ancient lineage diversity. The data set included 64 S-RNase allele sequences from several genera in Solanaceae, including Solanum, Eriolarynx, Lycium, Physalis, Petunia, Nicotiana and Witheringia. Three Antirrhinum sequences were used as outgroups (X96464–66). Maximum likelihood model parameters were determined using Modeltest version 3.7 (Posada and Crandall, 1998). The best-fit model GTR+I+G was used in the maximum likelihood analysis in PAUP*, using the heuristic search option, tree bisection reconnection branch swapping, MulTrees option in effect and a single neighbor-joining tree as a starting topology. Bootstrap support for this topology was determined by a maximum likelihood non-parametric bootstrap analysis in PAUP* on the Amherst College computing cluster. This analysis used the same model parameters as in the maximum likelihood analysis, and 100 full heuristic bootstrap replicates, each with 10 random-addition sequence replicates, tree bisection reconnection branch swapping and the MulTrees option in effect.
We used the codeml package in PAML, version 4.1 to estimate the non-synonymous/synonymous rate ratio (ω=dN/dS) for the data set mentioned above (Yang, 2007). The recommended procedure to investigate positive selection in PAML is to use two sets of models (M1a vs M2a, and M7 vs M8). Where likelihood ratio tests indicate that the models incorporating positive selection (M2a and M8) fit the data significantly better than the corresponding null models (M1a and M7) and the dN/dS rate ratio associated with positive selection (in models M2a and M8) is greater than one, diversifying selection on those sites was inferred. Empirical Bayesian probabilities (Yang et al., 2005) were used to determine the number and identity of sites under positive selection.
Results
S-RNase allele number and identity
We isolated 14 unique partial S-RNase sequences, ranging in length from 354 to 387 base pairs (Genbank accession numbers HM357215–HM357228) from the 30 Solanum peruvianum parental plants. All individuals were heterozygous and there were 25 unique genotypes (Table 1). Average pairwise amino-acid distance among the fourteen sequences was 55% and ranged from 6 to 67%. The average pairwise number of amino-acid differences between sequences was 65.7; however, the range spanned 8–81 AA differences between alleles (Figure 2). The majority of the sequences were extremely divergent from one another, but a few alleles that were similar were identified and confirmed in replicate PCR and sequencing reactions in multiple individuals. Three alleles in our sample (SP6, SP8 and SP17) were identical to previously identified alleles in S. peruvianum obtained from Genbank (see Figure 3 for synonymy). Two additional alleles were nearly identical to previously isolated sequences having either a single synonymous (SP11) or a single non-synonymous (SP13) difference. Many of the Genbank sequences were isolated from nearby populations (solid stars in Figure 1).
Table 1. S-RNase genotypes for individual Solanum peruvianum plants sequenced in this study.
| Individual | Genotype | Individual | Genotype | Individual | Genotype |
|---|---|---|---|---|---|
| PERU1 | SP1/SP6 | PERU11 | SP11/SP13 | PERU21 | SP5/SP17 |
| PERU2 | SP2/SP11 | PERU12 | SP5/SP14 | PERU22 | SP3/SP7 |
| PERU3 | SP7/SP10 | PERU13 | SP10/SP12 | PERU23 | SP1/SP7 |
| PERU4 | SP6/SP7 | PERU14 | SP10/SP16 | PERU25 | SP1/SP13 |
| PERU5 | SP8/SP10 | PERU15 | SP1/SP7 | PERU26 | SP8/SP11 |
| PERU6 | SP5/SP7 | PERU16 | SP5/SP11 | PERU27 | SP3/SP13 |
| PERU7 | SP7/SP11 | PERU17 | SP3/SP10 | PERU28 | SP1/SP3 |
| PERU8 | SP1/SP6 | PERU18 | SP7/SP8 | PERU29 | SP1/SP7 |
| PERU9 | SP8/SP16 | PERU19 | SP8/SP14 | PERU30 | SP1/SP14 |
| PERU10 | SP7/SP12 | PERU20 | SP2/SP11 | PERU31 | SP5/SP7 |
Figure 3.
Maximum likelihood genealogy of S-RNase alleles from Solanum peruvianum (circled and shaded) and other genera in Solanaceae. SP references alleles isolated in the present study, and Genbank accession numbers are given for alleles isolated in previous studies. See Supplementary Table 3 for closely related pairs of S-RNase alleles among Solanum species. Bootstrap support values are at nodes.
S-RNase alleles from Solanum peruvianum are dispersed across many lineages (Figure 3). In particular, there are eight lineages where S. peruvianum alleles cluster with alleles from genera outside Solanum (for example, Eriolarynx, Nicotiana, Petunia, Lycium, Witheringia and Physalis), indicating that much of the polymorphism present in S. peruvianum predates this genus. There are also several additional lineages that include multiple species of Solanum but no alleles isolated from species outside this genus, presumably because of a lack of sampling outside Solanum. There are eight closely related pairs of alleles isolated from the sister species S. peruvianum and S. chilense, and another pair from S. peruvianum and S. neoricki. These nine pairs of sequences are very similar and range from being identical over the region compared to having four amino-acid differences (Supplementary Table 3).
Intron lengths ranged from 76 to 121 base pairs for the 14 S-RNases in our sample and, not surprisingly, given the extreme ages of allelic lineages, we were unable to align introns for most alleles. However, for three sets of sequences belonging to the same lineage ((SP1, SP2, SP3 and SP5), (SP8 and SP10) and (SP14 and SP17); see Figure 3) alignment was possible. An intron was available for Solanum peruvianum Genbank accession number U28795, which was closely related (that is, two amino-acid positions different) to the SP11 allele recovered in this study; the intron sequences for these two sequences differed at only a single base pair.
Selection analyses indicated that models of positive selection (M2a and M8) fit the data significantly better than the models of nearly neutral (M1a and M7) evolution (LRT>54.57, P<0.0001). Not surprisingly, the majority of positively selected sites (9 of 15 sites with P⩾0.95, Table 2) were located in the hypervariable regions of the S-RNase gene. Despite differences in taxon and allelic sampling, as well as in sequence alignments between studies, 14 (of a total of 15 sites, see Table 2) inferred to be under positive selection in this study were also reported as positively selected in one or both previous studies (Igic et al., 2007; Vieira et al., 2007).
Table 2. Amino-acid positions inferred to be positively selected under model M8 as implemented in PAML.
|
Region |
Amino-acid range |
Positively selected amino-acid positions |
|---|---|---|
| 1–12 | 5**, 6**, 8* | |
| Hypervariable region a | 13–32 | 18**, 19*, 20*, 22**, 25**, 30*, 32* |
| 33–43 | None | |
| Hypervariable region b | 44–54 | 47**, 50** |
| 55–61 | 57** | |
| Conserved region 3 | 62–67 | None |
| 68–82 | 68* | |
| Conserved region 4 | 83–90 | None |
| 91–141 | 138* |
Positions in bold identify those sites in which the amino-acid position was also inferred as positively selected in two independent studies (Igic et al., 2007; Vieira et al., 2007); underlined positions indicate convergence with at least one of these studies. Alignment available on request.
*P⩾0.95, **P⩾0.99.
Controlled crosses
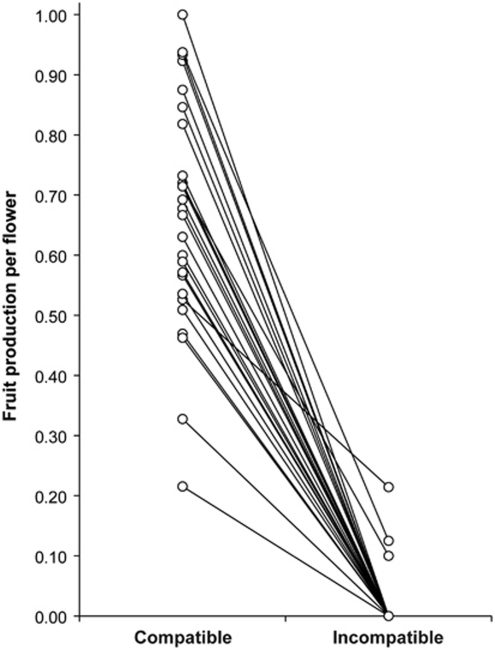
Fruit production did not differ among maternal parents (F29,29=1.01, P=0.49), but was significantly higher following compatible pollinations, indicative of a strong main effect of pollination treatment (59% vs 1%, F1,39.6=278.9, P<0.0001). All plants had higher fruit production following compatible, as compared with incompatible, pollinations (Figure 4); however, there was a significant plant by pollination treatment interaction (F29,1189=2.0, P=0.0018). The interaction is likely the result of two plants with relatively low fruit production in the compatible treatment (⩽33%) and the production of some fruit in the incompatible treatment for three plants (average was ∼15% for plants with non-zero fruit set). Both factors limit treatment differences in these plants as compared with most genotypes, which had high fruit set (⩾50%) in the compatible pollination treatment and zero fruit set following incompatible pollinations (Figure 4).
Figure 4.
Proportion of fruit production per flower for individuals of Solanum peruvianum in this study. Lines connect the same maternal parent following pollination with a single male donor. Incompatible crosses included both the self-pollen treatment and cross-pollinations between individuals of the same S-RNase genotype, whereas fully or semi-compatible crosses were categorized as compatible.
Segregation of S-RNase alleles in offspring
We tested the allelic specificity of 11 putative S-RNase alleles by genotyping 400 offspring following 15 SC crosses, in which parents shared one putative allele (Table 3). The PCR screens were robust and recovered the expected allele in 98.5% of all cases (394 of 400 PCR trials). Not surprisingly, the observed progeny arrays strongly rejected the null hypothesis of full compatibility for each of the 11 tested alleles. Thus, the sequence variants recovered in this study correspond to functional S-RNase alleles. There was no evidence in our sample that highly similar S-RNase sequences represented the same functional allele (χ2=0.966, P=0.326). In fact, in the crosses that potentially produced offspring genotypes with both similar and divergent alleles, offspring genotypes were equally divided between all genotypes (Table 4).
Table 3. We tested S-RNase specificities for 11 putative alleles using semi-compatible crosses and PCR screens of progeny arrays.
| Target allele tested | Cross genotypes (female × male) | Expected allele for PCR | Offspring with allele | Offspring without allele | Probability |
|---|---|---|---|---|---|
| SP1 | SP1/SP6 × SP1/SP3 | SP3 | 58 | 0 | <0.0001 |
| SP3 | SP3/SP1 × SP3/SP10 | SP10 | 10 | 0 | 0.0010 |
| SP5 | SP5/SP11 × SP5/SP14 | SP14 | 18 | 2 | 0.0002 |
| SP6 | SP1/SP6 × SP6/SP7 | SP7 | 18 | 2 | 0.0002 |
| SP7a | SP1/SP7 × SP3/SP7 | SP3 | 45 | 0 | |
| SP7/SP8 × SP3/SP7 | SP3 | 48 | 0 | ||
| SP1/SP7 × SP5/SP7 | SP5 | 45 | 2 | ||
| SP1/SP7 × SP7/SP11 | SP11 | 34 | 0 | ||
| SP7/SP10 × SP7/SP11 | SP11 | 48 | 0 | ||
| 220 | 2 | <0.0001 | |||
| SP8 | SP8/SP11 × SP8/SP16 | SP16 | 20 | 0 | <0.0001 |
| SP10 | SP8/SP10 × SP10/SP12 | SP12 | 10 | 0 | <0.001 |
| SP11 | SP7/SP11 × SP2/SP11 | SP2 | 10 | 0 | <0.001 |
| SP12 | SP10/SP12 × SP7/SP12 | SP7 | 10 | 0 | <0.001 |
| SP13 | SP3/SP13 × SP11/SP13 | SP11 | 10 | 0 | <0.001 |
| SP14 | SP5/SP14 × SP8/SP14 | SP8 | 10 | 0 | <0.001 |
Under GSI and following semi-compatible crosses involving a target allele, all offspring are predicted to have the unique paternal allele (for example, SP3 in the first cross). Numbers of offspring with and without the expected allele are given, as is the probability of the observed progeny array under the assumption of full compatibility (that is, that SP1 was not recognized as self in the first cross). Probabilities that remained significant following correction for multiple comparisons (that is, 11 alleles tested, P<0.005), allowed rejection of the null hypothesis of full compatibility.
Several crosses involving allele SP7 were combined before analysis.
Table 4. Controlled crosses and offspring genotypes as determined by PCR screens using allele-specific primers.
| Female × Male | Offspring genotypes (numbers) | |||
|---|---|---|---|---|
| SP1/SP6 × SP1/SP3 | SP1/SP3 (30) | SP6/SP3 (28) | ||
| SP2/SP11 × SP5/SP7 | SP2/SP5 (13) | SP2/SP7 (15) | SP11/SP5 (10) | SP11/SP7 (8) |
| SP1/SP7 × SP3/SP7 | SP1/SP3 (21) | SP7/SP3 (24) | ||
| SP1/SP7 × SP5/SP7 | SP1/SP5 (29) | SP7/SP5 (16) | ||
| SP2/SP11 × SP1/SP7 | SP2/SP1 (12) | SP2/SP7 (15) | SP11/SP1 (11) | SP11/SP7 (10) |
Offspring genotypes are given for semi-compatible (two genotypes) and fully compatible (four genotypes) crosses. The numbers of progeny with each S-RNase genotype is given in parentheses. Genotypes in bold are those in which the two alleles are similar (⩽33 AA differences), whereas all others were classified as alternative genotypes (see Figure 2).
Discussion
Consistent with gametophytic SI, all 30 parental individuals and 495 genotyped offspring in our sample were heterozygous at the S-RNase locus. In all, 14 unique S-RNase sequences were identified from 30 parental plants. To confirm that the S-RNase sequences were functional alleles, we screened progeny following semi-compatible crosses. For all alleles tested, we rejected the null hypothesis of full compatibility; thus, the S-RNase sequences isolated in this study represent unique specificities. Although allelic diversity in our sample is within the range reported for Solanaceae with GSI (Richman et al., 1995; Wang et al., 2001; Lu, 2002; Stone and Pierce, 2005; Savage and Miller, 2006), we recovered fewer alleles than are typically reported. For example, Igic et al., 2007 report 30 alleles from 34 individuals of the closely related species S. chilense, although several factors likely contribute to this discrepancy. Igic et al., 2007 sampled plants from nine populations (1–10 individuals from each) across the species range of S. chilense, whereas our sample was derived from fruit collected in a single population. Further, the initial collection of this accession likely included a limited number of individuals from the population, and the accession has been regenerated twice since collection (TGRC, personal communication). Consequently, the recovery of 14 alleles in our sample is considerable. Although previous studies (Chung et al., 1994; Kondo et al., 2002) have isolated additional alleles, species-wide estimates of allelic diversity in S. peruvianum may be premature. In total, 21 unique S-RNase alleles have been identified in southern populations of S. peruvianium. Further sampling, particularly across a wider area of the species range, would almost certainly recover novel S-RNase diversity.
Under gametophytic SI, negative frequency-dependent selection functioning over evolutionary time scales results in the maintenance of divergent trans-specific or trans-generic S-RNase lineages (Richman and Kohn, 2000; Igic et al., 2004). S-RNase alleles recovered from S. peruvianum conform to this pattern (Figure 3). Only two exceptions to this pattern have been documented (Richman and Kohn, 2000; Miller et al., 2008), both associated with bottleneck events in the evolutionary histories of these groups. Richman and Kohn, 2000 invoked a reduction in population size (and thus S-RNase diversity) in the ancestor of the sister genera Physalis and Witheringia to explain the presence of a shared, reduced number of TGLs in sampled species in each group. Likewise, Miller et al., 2008 documented a founder event bottleneck in S-RNase diversity for Old World Lycium, following the long-distance dispersal of Lycium from the Americas to the Old World.
Several recovered Solanum peruvianum alleles had highly similar coding sequences (fewer than 4 AA differences; Supplementary Table 3) compared with alleles from related species. Several authors have reported identical (or highly similar) S-RNase alleles across species (Lu, 2001; Savage and Miller, 2006; Surbanovski et al., 2007; Vieira et al., 2007; Sutherland et al., 2008). Similar allele pairs across species could be a result of recently shared evolutionary history (Lu, 2001; Sutherland et al., 2008) or introgression among species (for example, Castric et al., 2008). Solanum peruvianum and S. chilense are certainly recently diverged (⩽0.55 million years, Städler et al., 2008), and analyses of multilocus sequence data suggest historical introgression (Städler et al., 2005, 2008). Castric et al., 2008 reported sets of similar alleles at the SRK (S-locus receptor kinase) pistil SI specificity determining gene in Arabidopsis lyrata and A. halleri (Brassicaceae) and demonstrated that levels of introgression at SRK were higher than background levels. These authors point out that strong negative frequency-dependent selection at mating recognition loci (that is, selection for rare SRK (or S-RNase) alleles) will result in adaptive introgression at such loci, especially in the face of gene flow between diverging species. Similar studies comparing introgression at the S-RNase gene with background levels may be warranted in Solanum.
Comparison of similar allele pairs in different species reveals that, for all allele pairs (with one exception, SP7 vs Solanum chilense S17, which had only a single synonymous difference; Supplementary Table 3), at least 1 AA difference occurs at amino-acid positions also inferred to be under positive selection. Further, the majority of these positively selected sites correspond to amino-acid positions in hypervariable regions, which putatively control allelic specificity. Most comparisons also included positions that were strongly inferred to be under positive selection (that is, those amino-acid positions in Table 2). It is known that differences at a limited number of amino-acid positions can result in changes in allelic specificity (Nunes et al., 2006; Vieira et al., 2007). For example, Matton et al., 1999 altered the S. chacoense S11 allele to the S13 specificity via 4 AA changes. Notably, these four positions were under positive selection in this study supporting the hypothesis that few targeted amino-acid changes can generate new specificities. Our data concur with the observation that few amino-acid changes are responsible for allele specificity differences. Offspring genotyping revealed that highly similar S-RNase alleles (originating from different parental plants) assorted in progeny as novel specificities. Crosses included SP1 and SP2, which share 92% AA similarity and differ at only 10 AA; three of these sites were inferred to be under positive selection. Likewise, Igic et al., 2007 compared alleles with 86% AA similarity in Solanum chilense and found these to encode different allelic specificities. These examples provide clear evidence that allelic specificity can be determined by only a few amino-acid substitutions. That said, we obtained only partial S-RNase sequences, and more extensive sequencing could reveal additional differences. The most similar set of alleles in our sample were SP14 and SP17. Although not tested in this study, these two alleles differ by only 8 AA, five of which are under positive selection. It would be informative to determine whether SP14 and SP17 are functionally distinct; if so, then these five sites that are under positive selection are certainly candidates for specificity determination. Further studies investigating the molecular basis of allelic specificity are warranted, particularly those examining the role of the male determinant, S-locus F-box, and its interaction with S-RNase.
Several studies have used site-specific models to investigate sequence evolution in the S-RNase gene and there is broad convergence in positively selected amino-acid positions across studies. Igic et al., 2007 included 30 S-RNase alleles in Solanum chilense, whereas Vieira et al., 2007 included 64 taxa in three genera of Solanaceae. Despite differences across studies in taxon and allelic sampling, as well as in sequence alignments, 14 of the 15 sites inferred to be under positive selection in this study (Table 2) were also reported as positively selected in one or both of these previous studies. In contrast, there were eight sites reported to be under positive selection in Igic et al., 2007 and Vieira et al., 2007, which were not under positive selection in our analyses. Although not identified as being positively selected in this study, five of these positions had posterior probabilities ranging from 0.729 to 0.946 and thus approached significance. Convergence across studies implies that specific amino-acid positions may be disproportionately important in conferring allelic specificity.
The Solanaceae continues to be a model system for the study of evolution, maintenance and loss of genetic SI. Specifically, GSI is inferred to be the ancestral state in the family (Igic and Kohn, 2001), which has been subsequently lost in multiple lineages (Igic et al., 2008). The transition from SI to SC is one of the most common trends within the angiosperms (Stebbins, 1974), and a recurrent pattern has been the loss of SI on islands or along the edges of species ranges (Rick, 1986; Busch, 2005). This may be at least partially due to pollen limitation in these areas, where the evolution of SC provides reproductive assurance; smaller population size and greater geographical isolation within peripheries also likely contribute to this process (Schoen and Busch, 2008). Although the breakdown of SI in peripheral populations has been documented in Solanaceae, including several wild tomatoes (Rick et al., 1979; Rick and Tanksley, 1981; Rick, 1986; Rick and Chetelat, 1991), our sample of Solanum peruvianum from a population located near the southern range limit retained GSI. Fruit set following compatible pollination was significantly higher than that from incompatible pollination for all individuals in our sample (Figure 4), although it is possible that pseudo-compatible or fully compatible individuals exist but were not sampled. Three plants in our sample (PERU9, PERU13 and PERU19; Table 1) had non-zero fruit set following self pollination. Although two of the three plants shared allele SP8, three additional plants also with this allele had no fruit set following selfing; thus, these data provide little evidence for a ‘leaky' S-RNase allele (Mena-Ali and Stephenson, 2007). Nevertheless, the presence of strong SI in all sampled individuals suggests that the population has maintained functional GSI.
Despite our finding of strong SI in this population of Solanum peruvianum, these data provide an important comparison for species with mating system polymorphism. The wild tomatoes continue to be a model system for understanding the evolution of mating systems (Igic et al., 2006; Moyle, 2008; Städler et al., 2008), and molecular evolutionary studies of species with documented polymorphism in mating system can shed light on the evolutionary fate of the high sequence diversity typical for SI populations. In addition, our work facilitates future efforts directed at studying Solanum and related genera, as allele-specific primers will reduce the cost and effort of identifying S-RNase alleles in additional species and populations. More studies are needed in S. peruvianum, as well as other Solanaceae to continue to characterize GSI and plant-mating system evolution.
Acknowledgments
Funding from NSF DEB-0343735, NSF DEB-0843364 and the Department of Biology at Amherst College (to JSM), and a post baccalaureate Howard Hughes Medical Institute fellowship to JLK supported this work. We thank members of the Biology 32: Evolutionary Biology (Fall 2007, 2008) classes at Amherst College for assistance with data collection, Alan Krause for plant care, and three anonymous reviewers for comments on the manuscript.
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allen AM, Hiscock SJ.2008Evolution and phylogeny of self-incompatibility systemsIn: Franklin-Tong VE (ed).Angiosperms in Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms Springer: NY, USA; 73–101. [Google Scholar]
- Busch JW. The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae) Am J Bot. 2005;92:1503–1512. doi: 10.3732/ajb.92.9.1503. [DOI] [PubMed] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X. Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet. 2008;4:e1000168. doi: 10.1371/journal.pgen.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung IK, Ito T, Tanaka H, Ohta A, Nan HG, Takagi M. Molecular diversity of 3 S-allele cDNAs associated with gametophytic self-incompatibility in Lycopersicon peruvianum. Plant Mol Biol. 1994;26:757–762. doi: 10.1007/BF00013760. [DOI] [PubMed] [Google Scholar]
- Crawford DJ, Archibald JK, Stoermer D, Mort ME, Kelly JK, Santos-Guerraz A. A test of Baker's law: breeding systems and the radiation of Tolips (Asteraceae) in the Canary Islands. Int J Plant Sci. 2008;196:782–791. [Google Scholar]
- Goodwillie C. Multiple origins of self-compatibility in Linanthus Section Leptosiphon (Polemoniaceae): phylogenetic evidence from internal-transcribed-spacer sequence data. Evolution. 1999;53:1387–1395. doi: 10.1111/j.1558-5646.1999.tb05403.x. [DOI] [PubMed] [Google Scholar]
- Ida K, Norioka S, Yamamoto M, Kumasaka T, Yamashita E, Newbigin E, et al. The 1.55Å resolution structure of Nicotiana alata SF11-RNase associated with gametophytic self-incompatibility. J Mol Biol. 2001;314:103–112. doi: 10.1006/jmbi.2001.5127. [DOI] [PubMed] [Google Scholar]
- Igic B, Bohs L, Kohn JR. Historical inferences from the self-incompatibility locus. New Phytol. 2004;161:97–105. [Google Scholar]
- Igic B, Bohs L, Kohn JR. Ancient polymorphism reveals unidirectional breeding system shifts. Proc Natl Acad Sci USA. 2006;103:1359–1363. doi: 10.1073/pnas.0506283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, Lande R, Kohn JR. Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci. 2008;169:93–104. [Google Scholar]
- Igic B, Smith WA, Robertson KA, Schaal BA, Kohn JR. Studies of self-incompatibility in wild tomatoes: I. S-allele diversity in Solanum chilense Dun (Solanaceae) Heredity. 2007;99:553–561. doi: 10.1038/sj.hdy.6801035. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Clark AG, Kao T-h. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc Natl Acad Sci USA. 1990;87:9732–9735. doi: 10.1073/pnas.87.24.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-h. Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod. 1991;4:81–87. [Google Scholar]
- Kohn JR.2008What genealogies of S-alleles tell usIn: Franklin-Tong VE (ed).Angiosperms in Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms Springer: NY, USA; 103–121. [Google Scholar]
- Kohn JR, Graham SW, Morton B, Doyle JJ, Barrett SCH. Reconstruction of the evolution of reproductive characters in Pontederiaceae using phylogenetic evidence from chloroplast DNA restriction-site variation. Evolution. 1996;50:1454–1469. doi: 10.1111/j.1558-5646.1996.tb03919.x. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yamamoto M, Itahashi R, Sato T, Egashira H, Hattori T, et al. Insights into the evolution of self-compatibility in Lycopersicon from a study of stylar factors. Plant J. 2002;30:143–153. doi: 10.1046/j.1365-313x.2002.01275.x. [DOI] [PubMed] [Google Scholar]
- Lu Y. Role of lineage sorting and phylogenetic relationship in the genetic diversity at the self-incompatibility locus of Solanaceae. Heredity. 2001;86:195–205. doi: 10.1046/j.1365-2540.2001.00823.x. [DOI] [PubMed] [Google Scholar]
- Lu Y. Molecular evolution at the self-incompatibility locus of Physalis longifolia (Solanaceae) J Mol Evol. 2002;54:784–783. doi: 10.1007/s00239-001-0080-3. [DOI] [PubMed] [Google Scholar]
- Matton DP, Luu DT, Xike Q, Laublin G, O'Brien M, Maes O, et al. Production of an S-RNase with dual specificity suggests a novel hypothesis for the generation of new S-alleles. Plant Cell. 1999;11:2087–2097. doi: 10.1105/tpc.11.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. S-RNase and SLF determine S-haplotype-specific pollen recognition and rejection. Plant Cell. 2004;16:2840–2847. doi: 10.1105/tpc.104.161130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena-Ali JI, Stephenson AG. Segregation analyses of partial self-incompatibility in self and cross progeny of Solanum carolinense reveal a leaky S-allele. Genetics. 2007;177:501–510. doi: 10.1534/genetics.107.073775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS, Kamath A, Damashek J, Levin RA.2010Out of America to Africa or Asia: Inference of dispersal histories using nuclear and plastid DNA and the S-RNase self-incompatibility locus Mol Biol Evol(e-pub ahead of print 20 September 2010; doi: 10.1093/molbev/msq253 ), http://mbe.oxfordjournals.org/content/early/2010/09/20/molbev.msq253.abstract . [DOI] [PubMed]
- Miller JS, Levin RA, Feliciano NM. A tale of two continents: Baker's Rule and the maintenance of self-incompatibility in Lycium (Solanaceae) Evolution. 2008;62:1052–1065. doi: 10.1111/j.1558-5646.2008.00358.x. [DOI] [PubMed] [Google Scholar]
- Moyle LC. Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon) Evolution. 2008;62:2995–3013. doi: 10.1111/j.1558-5646.2008.00487.x. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Santos RAM, Ferreira SM, Vieira J, Vieira CP. Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wide self-incompatible Prunus spinosa (Rosaceae) population. New Phytol. 2006;172:577–587. doi: 10.1111/j.1469-8137.2006.01838.x. [DOI] [PubMed] [Google Scholar]
- Peralta IE, Knapp S, Spooner DM. New species of wild tomatoes (Solanum section Lycopersicon: Solanaceae) from northern Peru. Syst Bot. 2005;30:424–434. [Google Scholar]
- Peralta IE, Spooner DM. Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon) Am J Bot. 2001;88:1888–1902. [PubMed] [Google Scholar]
- Peralta IE, Spooner DM, Knapp S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae) Syst Bot Monogr. 2008;84:1–186. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A.2002. Se-Al: sequence alignment editor, version 2.0.http://evolvezoo.ox.ac.uk/ .
- Richman AD, Kao T-h, Schaeffer SW, Uyenoyama MK. S-allele sequence diversity in natural populations of Solanum carolinense (Horsenettle) Heredity. 1995;75:405–415. doi: 10.1038/hdy.1995.153. [DOI] [PubMed] [Google Scholar]
- Richman AD, Kohn JR. Evolutionary genetics of self-incompatibility in the Solanaceae. Plant Mol Biol. 2000;42:169–179. [PubMed] [Google Scholar]
- Richman AD, Uyenoyama MK, Kohn JR. S-allele diversity in a natural population of Physalis crassifolla (Solanaceae) (ground cherry) assessed by RT-PCR. Heredity. 1996;76:497–505. doi: 10.1038/hdy.1996.72. [DOI] [PubMed] [Google Scholar]
- Rick CM.1986Reproductive isolation in the Lycopersicon peruvianum complexIn: D'Arcy WG (ed).Solanaceae: Biology and Systematics Columbia University Press: NY, USA; 477–495. [Google Scholar]
- Rick CM, Chetelat RT.1991The breakdown of self-incompatibility in Lycopersicon hirsutumIn: Hawkes JG, Lester RN, Nee M, Estrada N (eds).Solanaceae III: Taxonomy, Chemistry, and Evolution Royal Botanic Gardens: Kew, Richmond, UK; 253–256. [Google Scholar]
- Rick CM, Fobes JF, Tanksley SD. Evolution of mating systems in Lycopersicon hirsutum as deduced from genetic variation in electrophoretic and morphological characters. Plant Syst Evol. 1979;132:279–298. [Google Scholar]
- Rick CM, Tanksley SD. Genetic variation in Solanum pennellii: comparisons with two other sympatric tomato species. Plant Syst Evol. 1981;139:11–45. [Google Scholar]
- Roldán JA, Quiroga R, Goldraij A. Molecular and genetic characterization of novel S-RNases from a natural population of Nicotiana alata. Plant Cell Rep. 2010;7:735–746. doi: 10.1007/s00299-010-0860-6. [DOI] [PubMed] [Google Scholar]
- SAS Institute 2002. JMP, version 5.0.1a. SAS Institute, Cary, NC, USA [Google Scholar]
- Savage AE, Miller JS. Gametophytic self-incompatibility in Lycium parishii (Solanaceae): allelic diversity, genealogical structure, and patterns of molecular evolution at the S-RNase locus. Heredity. 2006;96:434–444. doi: 10.1038/sj.hdy.6800818. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Busch JW. On the evolution of self-fertilization in a metapopulation. Int J Plant Sci. 2008;169:119–127. [Google Scholar]
- Schoen DJ, Johnston M, L'Heureux AM, Marsolais JV. Evolutionary history of the mating system in Amsinckia (Boranginaceae) Evolution. 1997;51:1090–1099. doi: 10.1111/j.1558-5646.1997.tb03956.x. [DOI] [PubMed] [Google Scholar]
- Seavey SR, Bawa KS. Late-acting self-incompatibility in angiosperms. Bot Rev. 1986;52:192–219. [Google Scholar]
- Spooner DM, Peralta IE, Knapp S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes [Solanum L. section Lycopersicon (Mill.) Wettst.] Taxon. 2005;54:43–61. [Google Scholar]
- Städler T, Arunyawat U, Stephan W. Population genetics of speciation in two closely related wild tomatoes (Solanum section Lycopersicon) Genetics. 2008;178:339–350. doi: 10.1534/genetics.107.081810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städler T, Roselius K, Stephan W. Genealogical footprints of speciation processes in wild tomatoes: demography and evidence for historical gene flow. Evolution. 2005;59:1268–1279. [PubMed] [Google Scholar]
- Stebbins GL. Belknap Press: Cambridge, MA, USA; 1974. Flowering plants: evolution above the species level. [Google Scholar]
- Steinbachs JE, Holsinger KE. S-RNase-mediated gametophytic self incompatibility is ancestral in eudicots. Mol Biol Evol. 2002;19:825–829. doi: 10.1093/oxfordjournals.molbev.a004139. [DOI] [PubMed] [Google Scholar]
- Stone JL, Pierce SE. Rapid recent radiation of S-RNase lineages in Witheringia solanacea (Solanaceae) Heredity. 2005;94:547–555. doi: 10.1038/sj.hdy.6800657. [DOI] [PubMed] [Google Scholar]
- Surbanovski N, Tobutt KR, Konstantinovic M, Maksimovic V, Sargent DJ, Stevanovic V, et al. Self-incompatibility of Prunus tenella and evidence that reproductively isolated species of Prunus have different SFB alleles coupled with an identical S-RNase allele. Plant J. 2007;50:723–734. doi: 10.1111/j.1365-313X.2007.03085.x. [DOI] [PubMed] [Google Scholar]
- Sutherland BG, Tobutt KR, Robbins TP. Trans-specific S-RNase and SFB alleles in Prunus self-incompatibility haplotypes. Mol Genet Genomics. 2008;279:95–106. doi: 10.1007/s00438-007-0300-7. [DOI] [PubMed] [Google Scholar]
- Swofford DL.2002PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods)Version 4.Sinauer, Sunderland, MA, USA [Google Scholar]
- Takebayashi N, Brewer PB, Newbigin E, Uyenoyama MK. Patterns of variation within self-incompatibility loci. Mol Biol Evol. 2003;20:1778–1794. doi: 10.1093/molbev/msg209. [DOI] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. J Mol Evol. 2008;67:179–190. doi: 10.1007/s00239-008-9137-x. [DOI] [PubMed] [Google Scholar]
- Vieira J, Ferreira PG, Aguiar B, Fonseca NA, Vieira CP.2010Evolutionary patterns at the RNase based gametophytic self - incompatibility system in two divergent Rosaceae groups (Maloideae and Prunus) BMC Evol Biol 10200doi:10.1186/1471–2148–10–200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Morales-Hojas R, Santos RAM, Vieira CP. Different positively selected sites at the gametophytic self-incompatibility pistil S-RNase gene in the Solanaceae and Rosaceae (Prunus, Pyrus, and Malus) J Mol Evol. 2007;65:175–185. doi: 10.1007/s00239-006-0285-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Hughes AL, Tsukamoto T, Ando T, Kao T-h. Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 2001;125:1012–1022. doi: 10.1104/pp.125.2.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller SG, Donoghue MJ, Charlesworth D.1995The evolution of self-incompatibility in flowering plants: A phylogenetic approachIn: Hoch PC, Stephenson AG (eds).Experimental and Molecular Approaches to Plant Biosystematics Missouri Botanical Garden Press: St Louis, MO, USA; 355–382. [Google Scholar]
- Wright S. The Distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under diversifying selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.