Abstract
Toxin production in algal blooms presents a significant problem for the water industry. Of particular concern is microcystin, a potent hepatotoxin produced by the unicellular freshwater species Microcystis aeruginosa. In this study, the proteomes of six toxic and nontoxic strains of M. aeruginosa were analyzed to gain further knowledge in elucidating the role of microcystin production in this microorganism. This represents the first comparative proteomic study in a cyanobacterial species. A large diversity in the protein expression profiles of each strain was observed, with a significant proportion of the identified proteins appearing to be strain-specific. In total, 475 proteins were identified reproducibly and of these, 82 comprised the core proteome of M. aeruginosa. The expression of several hypothetical and unknown proteins, including four possible operons was confirmed. Surprisingly, no proteins were found to be produced only by toxic or nontoxic strains. Quantitative proteome analysis using the label-free normalized spectrum abundance factor approach revealed nine proteins that were differentially expressed between toxic and nontoxic strains. These proteins participate in carbon-nitrogen metabolism and redox balance maintenance and point to an involvement of the global nitrogen regulator NtcA in toxicity. In addition, the switching of a previously inactive toxin-producing strain to microcystin synthesis is reported.
Blooms of cyanobacteria occur worldwide in nutrient-rich waters and can pose a public health threat when toxin-producing species are involved. One of the most significant cyanotoxins is microcystin, produced by the genera Microcystis, Anabaena, Anabaenopsis, Aphanizomenon, Fischerella, Planktothrix, and the terrestrial Hapalosiphon (1–3). Microcystins are cyclic heptapeptides with the general structure cyclo(-d-Ala-l-X-d-MeAsp-l-Z-Adda-d-Glu-Mdha), where Adda is 3-amino-9-methoxy-2,6,8,-trimethyl-10-phenyl-4,6,-decadienoic acid, MeAsp is 3-methylaspartic acid, and Mdha is N-methyl-dehydroalanine (4). The most common isoform produced is microcystin-LR (MC-LR)1 with a molecular weight of 995 Da. Incorporation of variable amino acids at the X and Z positions, together with differences in the peptide backbone, account for the generation of close to 90 microcystin isoforms that have been identified to date, each with differing polarity and toxicity (2, 5, 6).
Microcystins exert their hepatotoxicity in mammals by inhibiting protein phosphatases in the liver (PP1 and PP2a) (7, 8). Prolonged exposure to microcystin and the ensuing reactive oxygen species (ROS)-induced damage to DNA have been shown to be a powerful tumor promoter in rodents, but its impact as a carcinogen is yet to be established in humans (6, 8, 9). The effects of microcystin are not limited to mammals, with the toxin also able to cause oxidative stress damage, via the production of ROS, loss of mitochondrial potential, and apoptosis in macrophytes (10).
Numerous studies have been published on the putative role of microcystin, suggesting the possibility of the toxin to be a molecule involved in quorum-sensing, light sensitivity, or the acquisition of iron (11–14). However, the essential contribution of microcystin to the metabolism of cyanobacteria has been questioned by the existence of nontoxic strains within all species capable of microcystin production. In fact, under certain environmental conditions, nontoxic strains in both Planktothrix and Microcystis spp. are capable of dominating their environment and most blooms contain a mixture of toxic and nontoxic strains (5, 15–17).
In addition, the currently available nontoxic strains generated by insertional inactivation of mcyS genes show altered morphology, compared with the wild-type toxic strain Microcystis aeruginosa PCC 7806. Some of these changes are readily observable, such as the presence of gas vesicles, changes in pigmentation and thylakoid organization, reduced cell size, as well as the tendency of mutant cells to form aggregates (18, 19). Such changes have been suggested to be influenced by the association of microcystin with the thylakoid membranes in toxic cells, observed by electron microscopy and possibly, the binding of microcystin to phycobilin proteins (20, 21). However, a study by Vela and colleagues (22) considered this association to be nonspecific.
Proteomic studies in several bacterial species have opened the possibility to assess the role of various metabolites and the effect of stress-inducing conditions on cellular function. On the other hand, proteomic research of toxic cyanobacteria has been limited because of the relative scarcity of sequenced genomes. The first proteome studies in cyanobacteria were performed in the model photosynthetic prokaryote Synechocystis sp. PCC 6803 following the sequencing of its genome (23). As the wealth of cyanobacterial genomic information increased, proteomic investigations into the response of cyanobacteria under various stress conditions, the composition of the thylakoid membranes, and the processes of heterocyst formation and nitrogen fixation, among others have been carried out (24–27). These initial studies reported optimized extraction and separation protocols for cyanobacterial proteomes, which contain large amounts of phycobilisomes that can mask less-abundant proteins, in particular when two-dimensional electrophoresis (2DE) is used (28). Although the majority of studies have used 2DE, several groups have also reported comparable results with gel-free shotgun approaches (29, 30).
In 2001, Dittmann et al. performed the only proteomic study in M. aeruginosa using 2DE to compare the wild-type strain PCC 7806 and the mcyB− mutant strain (11). Using the partially sequenced genome, three proteins, showing some homology to quorum-sensing and light-regulated proteins, were identified as differentially displayed and were assigned as microcystin-related proteins (MrpA-C) (11). However, these proteins were not present in the genome of Microcystis aeruginosa NIES-843, indicating that they may not be necessarily linked to toxicity, but rather represent a strain-specific feature (31).
The recent publication of two Microcystis aeruginosa genomes has opened the possibility of studying the proteome expression of this toxin-producing species in more detail (31, 32). Comparison of the two genomes has revealed that they are both highly plastic and contain regions that could be the result of recent horizontal gene transfer (31). It was found that 16% of the M. aeruginosa PCC 7806 genome and 28% of the NIES-843 genome coded for proteins not present in the other strain, showing many strain-specific differences, despite the high 16S rRNA gene sequence conservation reported for the M. aeruginosa species cluster (31). In the past, phylogenetic comparisons have led to the belief that the Microcystis genus should be unified and strains should be defined as ecotypes adapted to a particular environment, rather than be divided into species based on morphological characteristics (33). Although most comparative proteomic studies have been performed on the same organism grown in different conditions, there is a general lack of understanding of what constitutes the proteomic diversity within cyanobacterial species.
In this study, the proteome of the model microcystin-producing organism M. aeruginosa PCC 7806 and five other morphologically distinct strains of M. aeruginosa isolated from various geographic locations was determined under nutrient-replete conditions. The aim of these studies was to see if the genome plasticity characteristic of the Microcystis species is observed at the protein expression level. In addition, protein expression differences between toxic and nontoxic strains grown in nutrient-replete conditions may be the underlying reason for the reported differential response of these organisms to nutrient stress (14, 16, 17). To test this hypothesis, protein expression was quantified using the label-free normalized spectral abundance factor (nSAF) method in an attempt to expand the current knowledge on the putative metabolic role of microcystin in the cyanobacterial cells. Several proteins involved in carbon-nitrogen metabolism and redox balance were found to be differentially expressed in toxic and nontoxic strains suggesting a link between microcystin synthesis and metabolic control by the global nitrogen regulator NtcA.
EXPERIMENTAL PROCEDURES
Strains and Culturing
M. aeruginosa cultures were grown in BG-11 media (Fluka) without shaking under continuous light (25 μm photons m−2 s−1) supplied by cool light fluorescent lamps at 28 °C. The strains used in this study are listed in Table I. Cells were filtered through a 3 μm pore-size membrane (Millipore) to remove contaminating bacteria and maintained in 100 μg/ml cyclohexamide to achieve axenic culture.
Table I. Strains used in this study.
| Strain | Isolate location | Isolate date | Toxicitya | Reference |
|---|---|---|---|---|
| PCC 7806 | Braakman Reservoir, The Netherlands | 1972 | + | [87] |
| UWOCC MRC | Malpas Dam, Armidale, Australia | 1973 | + | [88] |
| UWOCC MRD | Malpas Dam, Armidale, Australia | 1973 | + | [88] |
| UWOCC CBS | Lake Mendota, WI, USA | Pre 1983 | − | |
| PCC 7005 | Florida, USA | 1948 | − | [87] |
| HUB5.3 | Lake Pehlitzsee, Germany | 1977 | − | [89] |
a As determined by this study.
Toxicity Assays
Microcystin Extraction
To verify the toxicity of the strains chosen for proteomic studies, 2 ml of late exponential culture were pelleted by centrifugation, the cells were resuspended in 70% methanol and lysed by four 30-s cycles of bead-beating. Cell debris was removed by centrifugation. The cell lysate was dried completely under vacuum, resuspended, and mixed vigorously in 750 μl of chloroform and an equal volume of 20% methanol. After centrifugation at 13,400 × g for 20 min, the supernatant was collected and stored at −20 °C for further analysis.
Protein Phosphatase Inhibition Assay
Microcystin extracts were tested for inhibition of protein phosphatase type 2a (PP2a) (Promega, Madison, WI) as described in the study by Carmichael and An (34). Samples or standards (0–33 nm MCYST-LR in 20% methanol) were incubated for 5 min at 37 °C with 0.05 U PP2a diluted in 50 mm Tris pH 7.0, 2 mm MnCl2, 1 mg/ml bovine serum albumin (BSA) and 2 mm dithiotreitol. A reaction mixture of 25 mm p-NPP substrate in 0.2 m Tris pH 8.1, 80 mm MgCl2, 0.4 mm MnCl2, 2 mg/ml BSA and 4 mm dithiotreitol was added and the reaction was allowed to proceed at 37 °C for a further 80 min. Sample absorbance was read in a BioRad microplate reader at 405 nm and the percent inhibition was calculated.
High Performance Liquid Chromatography (HPLC) Detection of Microcystins
Samples showing inhibition of PP2a were further tested by HPLC according to the procedure described by (35). Briefly, microcystin extracts were loaded on an Alltech 5 μm Nucleosil C18 column (250 × 4.6 mm) and separated using a linear gradient of 30–70% Solvent B (acetonitrile containing 0.05% trifluoroacetic acid) for 30 min. Solvent A was 0.05% trifluoroacetic acid in water with a flow rate of 1 ml/min. Fractions eluting between 10 and 30 min were collected, concentrated under vacuum to 50 μl and were tested again for PP2a inhibition as described above.
Electrospray Ioization Quadrupole Time-of-Flight Tandem MS (ESI Q-TOF MS/MS)
To distinguish the isoforms of the toxins produced by M. aeruginosa PCC 7806, UWOCC MRC and UWOCC MRD, fractions collected by HPLC were subjected to mass spectrometry in a Q-TOF Ultima (Micromass) fitted with nanospray needles prepared in-house at the Biological Mass Spectrometry Facility, UNSW. Tandem mass spectra were acquired using Ar as the collision gas at different collision energies (8–50 eV). Capillary voltage was set at 1.3 kV. Cone voltage was 50 V. Microcystin-LR (Sigma) was used as a standard.
Proteome Analysis
Protein Extraction
Before protein extraction, cells were grown to mid-exponential phase (OD730 nm 0.75–0.85) and synchronized using the block-release method (48 h dark/72 h light) as described in Yoshida et al. (36). The cultures were grown for further 5 days with a 12:12 light:dark cycle and 100 ml of cells were harvested by centrifugation. The pellets were washed twice with sterile Milli-Q water before resuspending in a final volume of 500 μl MilliQ. The cells were partially lysed by four freeze-thaw cycles in liquid nitrogen and 37 °C in the presence of 15 μm PMSF (Sigma) and the lysates were treated with endonuclease (Sigma). The cells were resuspended with an equal volume of acid extraction buffer containing 7 m urea, 2 m thiourea, 2% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), 2% sulfobetaine 3–10 (SB3–10) and 80 mm citric acid, pH 4 and proteins were extracted according to the method in Herbert and colleagues (37). The proteins were resuspended in 150 μl 2DE buffer (7 m urea, 2 m thiourea, 2% CHAPS, 2% SB3–10) and quantified by a Bradford assay (Sigma), as well as by serial dilutions of the sample using one-dimensional SDS-PAGE against known BSA standards. Proteome analysis for all strains was performed with biological triplicates.
One-dimensional SDS-PAGE
One hundred micrograms of protein were buffer-exchanged in one-dimensional SDS-PAGE buffer and boiled for 10 min. Samples were loaded on a 10–20% Criterion gel (BioRad) and electrophoresis was performed at 5 mA/gel for 15 min followed by 200 V until the dye front had reached the end of the gel. The gels were fixed in 40% methanol and 10% acetic acid and stained with Coomassie G250 overnight. The gels were destained with Milli-Q water before visualization. A standard containing 1 μg BSA (Sigma) was included in all electrophoresis runs.
In-gel Digestion
For nanoLC/MS-MS analysis, each lane of a Coomassie-stained gel was cut into 16 segments and these were sliced to 1 mm3 pieces. Destaining was achieved by washing the gel pieces with 50% acetonitrile/50 mm NH4HCO3 until the stain was no longer visible. After dehydrating with 100% acetonitrile, and air-drying to remove residual acetonitrile, proteins were reduced in 10 mm dithiotreitol dissolved in 100 mm NH4HCO3 for 1 h at 55 °C and alkylated in 55 mm iodoacetamide in 100 mm NH4HCO3 for 45 min in the dark at room temperature. The washing steps described above were repeated twice and gel pieces were dehydrated with 100% acetonitrile.
The gel pieces were covered with 15 μg ml−1 sequencing-grade trypsin solution (Promega) and rehydrated at 4 °C for 30 min. Samples were then covered with an additional 25 μl NH4HCO3 buffer and digested overnight at 37 °C.
The digest supernatant was transferred to a clean tube and 30 μl of 50% acetonitrile/2% formic acid was added to the gel pieces, which were vortexed over 20 min to extract the remaining peptides. The supernatant was combined with the initial digest solution and the step was repeated to give a final volume of ∼60 μl. The volume of the extracted peptides was reduced to 10 μl under vacuum and the solution was desalted using C18 tips (Eppendorf). The extracted peptides were spun at 16,000 × g for 10 min to remove particulate matter and diluted to 10 μl with 1% formic acid if necessary.
nanoLC-Tandem Mass Spectrometry
The tryptic digest extracts from 1DE gel slices were analyzed by nanoLC-MS/MS using a LTQ-XL ion-trap mass spectrometer (Thermo) according to Hattrup and colleagues (38). Reversed phase columns were packed in-house to ∼7 cm (100 mm i.d.) using 100 Å, 5 mm Zorbax C18 resin (Agilent Technologies, Santa Clara, CA) in a fused silica capillary with an integrated electrospray tip. A 1.8 kV electrospray voltage was applied via a liquid junction up-stream of the C18 column. Samples were injected onto the C18 column using a Surveyor autosampler (Thermo). Each sample was loaded onto the C18 column followed by an initial wash step with buffer A (5% (v/v) acetonitrile, 0.1% (v/v) formic acid) for 10 min at 1 ml/min. Peptides were subsequently eluted from the C18 column with 0–50% Buffer B (95% (v/v) acetonitrile, 0.1% (v/v) formic acid) over 58 min at 500 nL min−1, followed by 50–95% Buffer B over 5 min at 500 nL/min. The column eluate was directed into a nanospray ionization source of the mass spectrometer. Spectra were scanned over the range 400–1500 m/z. Automated peak recognition, dynamic exclusion, and tandem MS of the top six most intense precursor ions at 35% normalization collision energy were performed using the Xcalibur software (version 2.06) (Thermo).
Protein Identification
Raw files were converted to mzXML format and MS/MS spectra from all fractions were processed using the Global Proteome Machine Tandem software (version 2007.08.29) against the proteome of M. aeruginosa NIES-843 containing 6312 open reading frames (database derived from National Center for Biotechnology Information (NCBI)), and the common repository for abundant peptides (39, 40). This proteome was chosen so that only proteins universal to the M. aeruginosa species rather than strain-specific proteins were identified. Reverse database searching was used for estimating false discovery frequencies (41). To ensure the specificity of the database, mass spectra were also searched against proteome databases of known bacterial contaminants in M. aeruginosa cultures, such as Staphylococcus aureus and Pseudomonas aeruginosa, but no significant matches were found. Peptide identification was determined using a 0.4 Da fragment ion tolerance and a parent ion tolerance of 1.4 Da. Carbamidomethyl was considered as a complete modification and partial modifications, including the oxidation of methionine and deamidation of asparagine and glutamine, were also considered. Up to one missed cleavage was permitted. Peptide matches and proteins identified by a single protein are listed in supplemental Data 1 and 2.
The mass spectrometry data and identification files were processed with PRIDE Converter and submitted to the Proteomics Identifications database (PRIDE).
Data Analysis
Protein function was assigned according to the categories used in CyanoBase (bacteria.kazusa.or.jp/cyanobase/). Hierarchical multivariate cluster analysis with the chi-square method of cluster distance calculation was performed in SYSTAT on proteins that were present in all biological replicates for each strain.
The nSAF quantitation method developed by Zybailov et al. (2006) and Chick et al. (2008) (42–43) was used to quantify changes in the expression levels of differentially displayed proteins between toxic and nontoxic strains, as well as pairwise comparison of proteins in UWOCC CBS, UWOCC MRC, and UWOCC MRD. For each protein k, the number of assigned spectral counts SpC, was corrected for the predicted molecular weight MW of the protein in kDa. The (SpC/MW), values were divided by the sum of (SpC/MW) for all proteins to give the nSAF values used for further analysis. A spectral fraction of 0.5 was added to all nSAF values to achieve normal distribution of the data. To determine differential protein expression in toxic and nontoxic M. aeruginosa, ANOVA with no multiple adjustments was performed on the log-transformed data using the statistical package R (44). Only proteins present in at least eight of the total nine replicates for the toxic or nontoxic strain group were considered for this analysis.
The full log nSAF abundance data (242 proteins × 18 samples, supplemental Data 5) was also examined for patterns in the overall protein abundance profile that could be associated with the synthesis of particular toxin isoforms. To this end, the Pearson correlation matrix for all samples was generated and then visualized as an image using basic functionality from the R stats package. Also a principal component analysis was carried out, and the different samples were plotted in the space of the first three principal components. All computations were carried out using the R statistical programming environment (44).
Transcription Analysis
RNA Extraction
Parallel to protein extraction, RNA was extracted from the same cultures of M. aeruginosa. Fifty milliliters of cells were pelleted by centrifugation, washed with Milli-Q water, and resuspended in 1 ml of Trizol reagent (Invitrogen). The cells were snap-frozen in liquid nitrogen and all subsequent steps were performed at 4 °C where possible. Cell lysis was achieved by vigorous pipetting and 400 μl chloroform was added to the lysate. After centrifugation at 13,400 × g for 15 min, the aqueous layer was collected and RNA was precipitated in ice-cold isopropanol. The extracts were centrifuged at 13,400 × g for 30 min and the pellets were washed twice with 75% ethanol, before being resuspended in DEPC-treated water (Invitrogen).
DNase Treatment
RNA extracts were treated with 3 U of Turbo DNase (Ambion) for 4 h at 37 °C. This was followed by a second extraction in Trizol as outlined above. The success of DNA removal was assessed by PCR targeting the 16S rDNA gene. The purity and quantity of RNA were estimated using a Nanodrop spectrophotometer (Nanodrop Technologies).
cDNA Synthesis
The Marligen random cDNA synthesis kit (Marligen Bioscience) was used for reverse transcription of 500 ng RNA following the manufacturer's instructions. The reaction conditions were 22 °C for 5 min, 42 °C for 90 min, and 85 °C for 5 min.
Quantitative Real-time PCR (qRT-PCR)
qRT-PCR was used to quantify transcription levels for genes that were found to be differentially expressed by the nSAF proteomic analysis. Primer sequences are listed in Table II. Both the 16S rRNA and the RNA polymerase subunit C (rpoC1) genes were considered as possible reference genes and rpoC1 was chosen for further studies after validation. Transcript levels were quantified by qPCR using the Rotor-Gene 3000 system (Corbett). Reactions were performed in a total volume of 25 μl using 1 μg cDNA, 10 pmol of forward and reverse primer and the Platinum SYBR Green qPCR supermix UDG kit (Invitrogen). Two-step cycling was performed with an initial hold of 60 °C for 2 min and 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s. The efficiency of amplification for each primer set was determined using standard cDNA curves and calculated according to the equation E = 10 [-1/slope] (45). Transcript levels were normalized to rpoC transcription and calculated relative to values for toxic strains using the 2−ΔΔCt method as described elsewhere (45). All analyses were performed using biological and technical triplicates.
Table II. Primers used for the quantitative real-time PCR (qRT-PCR) analysis. The rpoC gene was chosen as a reference [56].
| Primer name | Sequence (5′-3′) | Target |
|---|---|---|
| trxMF | TCAGGAACTGTTGCAATCCA | thioredoxin trxM |
| trxMR | AGAATCGGAGCCATCATTTG | |
| MAE27590F | AATCGAACCCGATAAACCCT | hypothetical protein MAE27590 |
| MAE27590R | TCACAACCGACAACAGAAGC | |
| ccmKF | ACGGATCACGATTGTTGGTT | carboxysome shell subunit ccmK3 |
| ccmKR | TCGGTTTTATTGACGGCTTC | |
| PIIF | AAGCGATTATCCGACCCTTT | nitrogen regulatory protein PIIglnB |
| PIIR | ATTGACCTTTCTGACGACCG | |
| ccmLF | TCTGCTTTTGCAATTCATCG | carboxysome shell subunit ccmL |
| ccmLR | CAATAATCCCCACCACCATC | |
| MAE06820F | AGTGGTAGCCGAAAGCGATA | hypothetical protein MAE06820 |
| MAE06820R | TCCCTTCCAAGAACAAATGG | |
| nrtAF | TGATGGTCGCAAAATTGAAA | nitrate transporter nrtA |
| nrtAR | GGAATATAGCCCCAACGGAT | |
| ndhKF | ACTACCCACAAAATGCAGGC | NADH dehydrogenase subunit K ndhK |
| ndhKR | CTCTTTCGGAGGTGCTTGAC | |
| MAE06360F | GCACGATCGGTTTTTGTTTT | carboxymethylenebutenolidase |
| MAE06360R | ATGCGTAGATTGTCCCCTTG | |
| rpoC1F | CCTCAGCGAAGATCAATGGT | RNA polymerase gamma subunit rpoC1 |
| rpoC1R | CCGTTTTTGCCCCTTACTTT | |
| ntcArealF | CATTTCCGTTTGCAGAATCC | global nitrogen regulator ntcA |
| ntcArealR | TGTTTTTGGGGTGCTATCCT |
Reverse-transcription PCR
To assess the cotranscription of trxM and the neighboring ORF MAE06820, 1 μg of RNA extracted from PCC 7806 was reverse-transcribed using the Marligen random cDNA synthesis kit as described above. A 470 bp fragment spanning both the trxM and MAE06820 gene was amplified with the primers trxMF and MAE06820R and the successful amplification was visualized after electrophoresis in a 1% agarose gel.
RESULTS
The Core Proteome of M. aeruginosa Strains
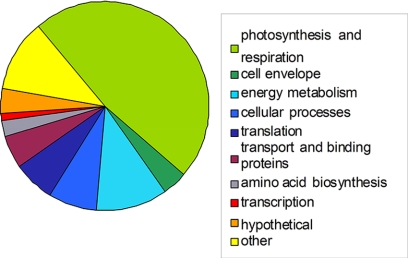
Proteome analysis of the six strains in this study identified 475 proteins reproducible in biological triplicates, which were considered for further statistical analysis (supplemental Data 3). Of the compiled proteins, only 82 were found to be expressed in all strains (Fig. 1 and Table III), constituting between 29 and 43% of the identified proteins. This interstrain variability in identified proteins confirms the observed differences in SDS-PAGE migration patterns (supplemental Data 4).
Fig. 1.
Functional class distribution in the core M. aeruginosa proteome. Functional class categories were assigned according to CyanoBase (http://genome.kazusa.or.jp/cyanobase).
Table III. Functional categories of proteins in M. aeruginosa strains. The proteins universally expressed for all strains are shown in the first column (M. aeruginosa). The number of proteins identified is in black and the % of the proteome is below in italics. Functional categories were assigned according to CyanoBase (http://genome.kazusa.or.jp/cyanobase).
| Functional category | Protein % |
||||||
|---|---|---|---|---|---|---|---|
| M. aeruginosa | PCC 7806 | UWOCC MRC | UWOCC MRD | UWOCC CBS | HUB5.3 | PCC 7005 | |
| Amino acid biosynthesis | 4 | 20 | 13 | 11 | 12 | 11 | 17 |
| 4.8 | 7.6 | 6.0 | 4.1 | 6.2 | 5.2 | 6.1 | |
| Biosynthesis of cofactors, prosthetic groups and carriers | - | 4 | 2 | 3 | 3 | 4 | 9 |
| - | 1.5 | 0.9 | 1.1 | 1.5 | 1.9 | 3.2 | |
| Cell envelope | 3 | 5 | 7 | 5 | 5 | 4 | 3 |
| 3.6 | 1.9 | 3.2 | 1.9 | 2.6 | 1.9 | 1.1 | |
| Cellular processes | 5 | 10 | 10 | 11 | 7 | 9 | 10 |
| 6.1 | 3.8 | 4.6 | 4.1 | 3.6 | 4.2 | 3.6 | |
| Central intermediary metabolism | - | 3 | 2 | 2 | - | 1 | 4 |
| - | 1.1 | 0.9 | 0.7 | - | 0.4 | 1.4 | |
| Energy metabolism | 5 | 18 | 15 | 15 | 12 | 12 | 22 |
| 6.1 | 6.9 | 6.9 | 5.7 | 6.2 | 5.7 | 7.8 | |
| Fatty acid, phospholipids and sterol metabolism | - | - | 1 | 1 | 1 | 1 | 2 |
| - | - | 0.4 | 0.3 | 0.5 | 0.4 | 0.7 | |
| Photosynthesis and respiration | 32 | 53 | 54 | 56 | 41 | 53 | 52 |
| 39 | 20.3 | 25 | 21.3 | 21.3 | 25.2 | 18.5 | |
| Purines, pyrimidines, nucleosides and nucleotides | - | 1 | 1 | 2 | - | 1 | 2 |
| - | 0.3 | 0.4 | 0.7 | - | 0.4 | 0.7 | |
| Regulatory functions | - | 2 | - | 5 | 2 | 2 | 4 |
| - | 0.7 | - | 1.9 | 1.0 | 0.9 | 1.4 | |
| DNA replication, restriction, modification, recombination and repair | - | 2 | 2 | 1 | 1 | 1 | 4 |
| - | 0.7 | 0.9 | 0.3 | 0.5 | 0.4 | 1.4 | |
| Transcription | - | 4 | 1 | 2 | - | 2 | 2 |
| - | 1.5 | 0.4 | 0.7 | - | 0.9 | 0.7 | |
| Translation | 7 | 27 | 17 | 17 | 10 | 17 | 40 |
| 8.5 | 10.3 | 7.9 | 6.4 | 5.2 | 8.1 | 14.3 | |
| Transport and binding proteins | 6 | 10 | 9 | 17 | 14 | 9 | 8 |
| 7.3 | 3.8 | 4.1 | 6.4 | 7.3 | 4.2 | 2.8 | |
| Other | 13 | 53 | 41 | 55 | 47 | 44 | 61 |
| 15.8 | 20.3 | 18.9 | 20.9 | 24.4 | 20.9 | 21.7 | |
| Hypothetical | 7 | 46 | 37 | 56 | 35 | 36 | 37 |
| 8.5 | 17.7 | 17.1 | 21.3 | 18.2 | 17.1 | 13.2 | |
| Unknown | - | 2 | 4 | 4 | 2 | 3 | 3 |
| - | 0.7 | 1.8 | 1.5 | 1.0 | 1.4 | 1.1 | |
| Total proteins | 82 | 260 | 216 | 263 | 192 | 210 | 280 |
Ten functional categories were represented in the core proteome, the highest fraction being comprised of proteins involved in photosynthesis and respiration. Seven hypothetical proteins were identified in all strains (Table III, supplemental Data 2), including MAE06270, which shows homology to slr1623 and cce 4330, a novel NADH dehydrogenase subunit in Synechocystis sp. PCC 6803 and Cyanothece sp. ATCCC 51142, respectively. No unknown proteins were present in the M. aeruginosa core proteome.
Functional Categories of Proteins Expressed in M. aeruginosa Strains
The total number of proteins identified for each strain varied, with the most found in PCC 7005 (280 proteins) and the least in UWOCC CBS (192 proteins). However, in the six strains proteins from all functional categories were identified (Table III). Similar to the results obtained for the core proteome, the photosynthetic, respiratory and hypothetical proteins, as well as proteins involved in diverse functions (the category designated “other” in CyanoBase) were predominant.
A total of 90 hypothetical proteins and nine unknown proteins were identified reproducibly, confirming the correct annotation of these open reading frames and the expression of these proteins in nutrient-replete conditions in the exponential growth phase. Two of the unknown proteins (MAE60260 and MAE60250) were in adjacent open reading frames and may form part of an operon, expressed only in strain PCC 7005. Similarly, two putatively cotranscribed clusters of hypothetical proteins were found in PCC 7005 - MAE36710 and MAE36720, and MAE45800 and MAE45790, the latter pair also being expressed in HUB5.3. The fact that these proteins were only expressed in a small number of the strains suggests that they may be involved in adaptation processes specific for a particular environmental niche.
Diversity of Protein Expression in M. aeruginosa Strains
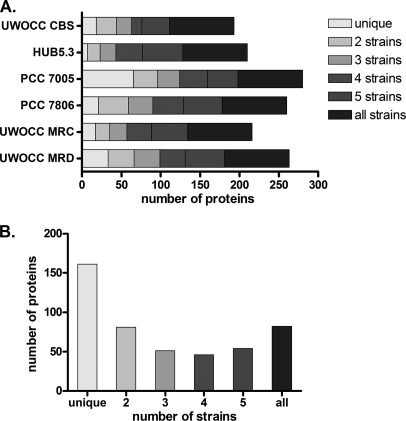
Genome analysis of M. aeruginosa strains has revealed that these organisms are genetically highly plastic and contain a large number of transposases and loci that could have been the result of horizontal gene transfer (31, 32). We were interested to see whether this diversity was also present at the proteome level. For each strain, the larger part of the proteome consisted of proteins that were present in the M. aeruginosa core proteome (Fig. 2). Strain PCC 7005 was the most divergent, expressing 65 strain-specific proteins (23% of the strain proteome). The majority of the proteins expressed uniquely in this strain were involved in carbon and nitrogen metabolism, as well as cofactor synthesis (supplemental Data 3). In contrast, in HUB5.3 only seven strain-specific proteins were identified (3% of the proteome). When the number of proteins shared among strains was considered, it became evident that proteins that were present in a single strain (unique) comprised the largest category, followed by proteins in the core proteome (Fig. 2, Table IV). Most of the proteins expressed in a single strain were hypothetical or proteins in the other category. This suggests that the M. aeruginosa strains differ mostly in processes involved in adaptation to a particular environment or a growth condition, rather than in essential metabolic reactions. No protein was expressed uniquely in either the toxic or the nontoxic strains, which could serve as potential markers for toxicity.
Fig. 2.
Protein distribution among the six strains in this study. A, Distribution of unique and shared proteins for each strain B. Total number of proteins involved in each combination (1 being unique and 6 being present in all strains).
Table IV. Protein distribution in strains of M. aeruginosa. Combinations of proteins expressed by different strains.
| Strain | Number of proteins |
||||||
|---|---|---|---|---|---|---|---|
| Unique | 2 Strains | 3 Strains | 4 Strains | 5 Strains | All Strains | Grand Total | |
| PCC 7005 | 65 | 31 | 28 | 35 | 39 | 82 | 280 |
| PCC 7806 | 21 | 38 | 31 | 39 | 49 | 82 | 260 |
| UWOCC CBS | 18 | 26 | 19 | 13 | 35 | 82 | 192 |
| HUB5.3 | 7 | 16 | 20 | 34 | 51 | 82 | 210 |
| UWOCC MRC | 17 | 18 | 22 | 31 | 46 | 82 | 216 |
| UWOCC MRD | 33 | 33 | 33 | 32 | 50 | 82 | 263 |
| Total proteins | 161 | 81 | 51 | 46 | 54 | 82 | 475 |
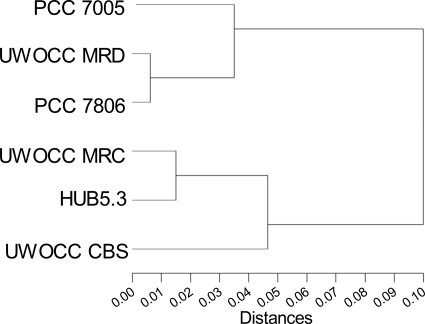
Cluster analysis was also performed based on the distribution pattern of proteins expressed reproducibly in M. aeruginosa strains, and all strains were found to be closely related with the largest distance between them being only 10% (Fig. 3). Two large clades were observed, each containing both toxic and nontoxic strains. Interestingly, UWOCC MRD and UWOCC MRC, which are identical in 16S rRNA gene phylogeny and two intergenic spacer region sequences, studied so far, were separated in different clades of the tree. UWOCC MRC, which has recently reverted to toxin production, was grouped with two nontoxic strains, whereas the protein expression profile of the toxic UWOCC MRD was closest to that of the model toxic strain PCC 7806.
Fig. 3.
Cluster dendrogram of protein expression in strains of M. aeruginosa. Hierarchical multivariate cluster analysis (n = 3475 proteins) with chi-square method of cluster distance calculation.
Differentially Expressed Proteins in M. aeruginosa
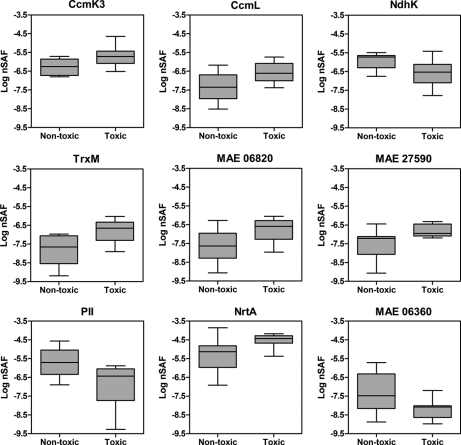
The expression levels of nine proteins were found to differ significantly between toxic and nontoxic strains when ANOVA was performed on nSAF calculated for proteins in the whole proteome data set (Table V and Fig. 4, supplemental Data 5). Of these, two proteins were involved in nitrogen uptake and metabolism (PII and NrtA), two comprised components of the carboxysome shell (CcmK3 and CcmL) and two are involved in maintaining the cellular redox balance (NdhK and TrxM). The two hypothetical proteins with a putative role in redox balance maintenance were also identified as differentially expressed. The majority of the proteins were down-regulated in the nontoxic strains, with the exception of PII, NdhK and the carboxymethylenebutenolidase. Quantitative real-time PCR was used to measure differences at the transcript level and the patterns predicted at the protein level were observed at the RNA level for nrtA, ndhK, and ccmL (Table V). In general, greater differences were observed at the protein expression level compared with gene transcription.
Table V. Proteins identified as significantly different between the M. aeruginosa toxic and nontoxic group after nSAF analysis and ANOVA. Expression patterns determined by nSAF and transcription values from qRT-PCR are shown for nontoxic strains relative to those for the toxic strains. Standard deviation values are in brackets and italicized. Accession numbers are shown as they appear in CyanoBase.
| Protein GI | p value | Accession | Function | Expression | Transcription |
|---|---|---|---|---|---|
| gi 166085797 | 0.01 | MAE06830 | thioredoxin (trxM) | −2.79 | 1.47 (1.12) |
| gi 166087873 | 0.02 | MAE27590 | hypothetical protein | −1.76 | 1.02 (1.98) |
| gi 166090653 | 0.02 | MAE55390 | carbon concentrating mechanism protein (ccmK3) | −1.94 | 2.52 (1.08) |
| gi 166090860 | 0.02 | MAE57460 | nitrogen regulatory protein PII (glnB) | 2.87 | −1.85 (0.95) |
| gi 166089906 | 0.02 | MAE47920 | carbon concentrating mechanism protein (ccmL) | −2.02 | −1.94 (0.92) |
| gi 166085796 | 0.03 | MAE06820 | hypothetical protein | −2.14 | 3.65 (1.36) |
| gi 166086594 | 0.03 | MAE14800 | nitrogen transport protein (nrtA) | −1.71 | −2.58 (0.93) |
| gi 166086291 | 0.04 | MAE11770 | NADH dehydrogenase subunit (ndhK) | 1.67 | 1.95 (1.31) |
| gi 166085750 | 0.05 | MAE06360 | carboxymethylenebutenolidase | 3.84 | −1.02 (1.71) |
Fig. 4.
Box-plots of nSAF values for the proteins identified by ANOVA as significantly different (p < 0.05) between the toxic (PCC7806, UWOCC MRC, UWOCC MRD) and nontoxic (PCC7005, HUB5–3, UWOCC CBS) strain group. nSAF values were calculated according to Zybailov et al. (42).
Because of the possible involvement of the transcriptional regulator NtcA in this differential display (see Discussion), the transcription of this gene was also measured and found to be 2.61-fold up-regulated in nontoxic strains.
Coexpression of MAE06820 and TrxM
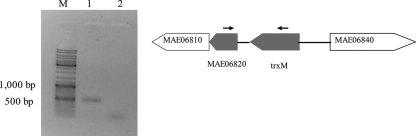
The similar expression profiles of TrxM and the hypothetical protein MAE06820 observed in Fig. 4, prompted us to perform reverse transcription in order to evaluate whether the two open reading frames were cotranscribed. The amplification of a fragment of the expected size (470 bp) spanning both genes was successful, confirming that both genes can be transcribed from a single promoter (Fig. 5).
Fig. 5.
Reverse-transcription of trxM and MAE06820. After amplification with primers spanning a 470 bp region of the two open reading frames shown in the right panel, amplicons were electrophoresed in a 1% agarose gel. M, Gene Ruler DNA ladder (MBI Fermentas, Burlington Canada); 1, reverse-transcribed PCC 7806 RNA; 2, negative control. The binding sites of the primers used in the reverse transcription reaction are marked with black arrows and the cotranscribed region is in gray. The open reading frames are shown as their accession numbers in CyanoBase: MAE06810, phosphopantheine adenyltransferase; MAE06820, hypothetical protein; trxM, thioredoxin M; MAE06840, methionine aminopeptidase.
Toxicity Switching in M. aeruginosa UWOCC MRC
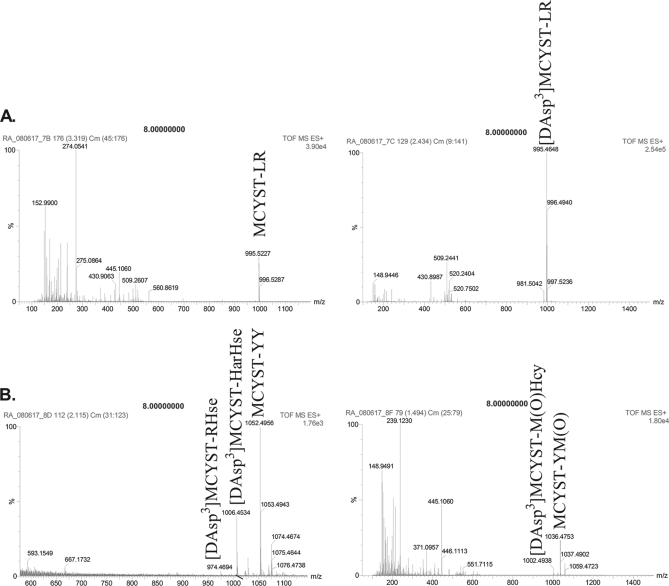
A toxicity screen of the cultures used in this study revealed that UWOCC MRC, a strain that was expected to be nontoxic, was producing MCYST-LR and [DAsp3] MCYST-LR (Fig. 6 and Table VI, supplemental Data 6). These isoforms are found in M. aeruginosa PCC 7806 and are different from the microcystin species produced by the toxic strain UWOOC MRD, which produced several aromatic and homoamino acid isoforms of the toxin (Table VI). Contamination by other Microcystis spp. was ruled out by sequencing the 16S rRNA gene and PCR of the regions of the mcy cluster characteristic for MRC and MRD (46). Several subcultures were tested over a period of two years, and the toxicity of MRC appears to be a stable trait.
Fig. 6.
MS spectra showing microcystin production by M. aeruginosa UWOCC MRC (A) and UWOCC MRD (B). Labeled peaks were identified by MS/MS spectra as microcystin isoforms.
Table VI. Microcystin isoforms identified by Q-TOF MS/MS. Purified MCYST-LR was used as a standard for both HPLC and MS studies.
| Strain | Toxin isoformsa | Molecular weight (Da) | HPLC elution time (min) | Reference |
|---|---|---|---|---|
| PCC 7806 | MCYST-LR | 995.45 | 19.4–20.4 | Phelan and Downing, 2007 [90] |
| [DAsp3]MCYST-LR | 995.5 | 18.7–19.4 | ||
| UWOCC MRD | [DAsp3] MCYST-RHse | 974 | 25.0–25.6 | This study |
| [DAsp3] MCYST-M(O)Hcy | 1002 | 30.1–30.7 | ||
| [DAsp3] MCYST-HarHse | 1006 | 25.0–25.6 | ||
| MCYST-YY | 1052.3 | 25.0–25.6 | ||
| MCYST-YM(O) | 1036.5 | 30.1–30.7 | ||
| UWOCC MRC | MCYST-LR | 995.5 | 17.2–19.7 | This study |
| [DAsp3]MCYST-LR | 995.5 | 18.4–19.7 |
a Har, homoarginine; Hse, homoserine; Hcy, homocysteine; M(O), methionine-S-oxide.
Effects of Toxin Isoform Synthesis on Protein Expression
The different isoforms of microcystin synthesized by the three toxic strains in this study, prompted us to consider the possibility that the toxin variant produced has an effect on the protein expression profile of the producing cells. However, significant separation of UWOCC MRD from UWOCC MRC and PCC 7806 could not be achieved when the protein expression patterns in these strains were considered (supplemental Data 7). The protein phosphatase inhibition assay toxin analysis revealed that the microcystin concentration was similar in all toxic strains used here, excluding the possibility that quantitative differences in the intracellular microcystin content influence protein expression in M. aeruginosa. PCC 7806, UWOCC MRC, and UWOCC MRD contained 245.28 ± 0.20 nm, 245.94 ± 0.11 nm, and 245.75 ± 0.37 nm microcystin, respectively. Correlation analysis of the expressed proteins in all strains also did not achieve distinct separation between the toxic and nontoxic strain group (supplemental Data 7).
DISCUSSION
Diversity of Protein Expression in Strains of M. aeruginosa
The proteome studies presented here indicate that protein expression in strains of M. aeruginosa is highly variable, with only a third of the expressed proteins in each strain constituting the core proteome of the species. Such protein diversity was also observed when the protein samples were run on SDS-PAGE (supplemental Data 4). Such inter-strain variability in protein expression was unexpected, given the close 16S rRNA gene homology shared within strains of this species. The observed proteome diversity supports previous suggestions that M. aeruginosa strains should be considered as ecotypes adapted to survival in a particular environmental niche (33). It also highlights the dangers of using predominantly M. aeruginosa PCC 7806 as a model organism for the entire species. In particular, under stress-inducing conditions, such as nutrient starvation or changes in light quality and intensity, many strain-specific acclimation processes would be expected to take place and differences in protein expression are expected.
The use of a single model strain also seems an inappropriate approach to study toxicity and the effects of microcystin production on cyanobacterial metabolism because proteome diversity was high within the toxic and nontoxic strain groups studied here as well. This is evident from the cluster analysis (Fig. 3), which failed to separate the strains based on their ability to produce microcystin, geographical origin or period of culture in the laboratory. No proteins were expressed solely in toxic or nontoxic strains, including the McyS proteins involved in microcystin analysis. The mixed toxic and nontoxic strains in the same clade of the cluster tree present a similar situation to what has been observed on a genetic level, where phylogeny-based methods have not been able to discriminate between toxic and nontoxic, but toxigenic strains. The existing proteome and genome diversity in M. aeruginosa should therefore be taken into consideration, in particular in the study of physiological responses to particular conditions. Ensuring that several strains are used for such comparative studies would avoid confusing conclusions as illustrated by the study of PCC 7806 and its knock-out mutant by Dittmann and colleagues in 2001 (11) and the more recent comparative genomic analysis of PCC 7806 and NIES-843 (31). The authors reported the identification of two light-sensitive proteins in the mcyB- mutant strain and proposed their use as a suitable marker for toxin production (11). However, these proteins were not found in the genome of the toxic NIES-843 and therefore are not universal for the M. aeruginosa genus.
An interesting observation was the unusually large number of proteins expressed solely in PCC 7005 (Table I). In the environment, strains from a dense culture in the late exponential growth stage such as the ones used here, are likely to be starved for micronutrients, such as iron. One possibility for the increased expression of proteins in this strain, may be its ability to synthesize cofactors needed for the utilization and storage of the excess carbon and nitrogen provided in the laboratory media (supplemental Data 3). Because this strain has been maintained in laboratory culture for several decades longer than the other strains in this study, its proteome may reflect an ability to adapt to the nutrient-rich environment of laboratory media and make use of the excess macronutrients provided. Such changes have been observed previously in M. aeruginosa with the loss of gas vesicles because of transposition events and the dispersal of colonies after prolonged culturing (18, 47).
The M. aeruginosa strain PCC 7005 also contained most of the hypothetical and unknown proteins found in adjacent reading frames, the majority of which were absent in the remaining strains of the cohort. In fact, only nine unknown proteins, out of 1404 predicted, were identified in the proteome of the strains studied here and none of these were present in the core proteome. It is likely that the majority of proteins designated as unknown are involved in adaptation or growth-stage specific processes in M. aeruginosa.
The Core Proteome of M. aeruginosa
The majority of proteins shared among all strains of M. aeruginosa were involved in photosynthesis and respiration. This distribution of protein expression is typical for cyanobacteria, where phycobilisome components are regularly found in high abundance and obscure less-abundant proteins (48). The number of proteins that were identified, as well as the pI and molecular range covers are also similar to other gel-based studies of total cell extracts of cyanobacteria, such as the unicellular Synechocystis sp. PCC 6803 and the filamentous nitrogen-fixing Anabaena variabilis ATCC 29413 (49). It is likely that other proteins remained undetected because only one genomic database was used for identification and there may be proteins expressed in certain strains but not encoded in the NIES-843 genome. However, the aim of this study was to find proteins shared across M. aeruginosa species, rather than perform an exhaustive proteome study on one strain, for which sub-cellular protein fraction analysis and multiple analytical approaches would be required.
The large differences in protein expression among strains of M. aeruginosa, although unexpected, are not unique for bacteria. A comparative analysis of two strains of E. coli found that only 38% of the secreted proteins were conserved and a similar fraction were strain-specific (50). Similarly, a study of strains of the marine cyanobacteria Prochlorococcus spp. found that the elemental composition of their proteomes was strongly influenced by their site of isolation and the nutritional availability at that site (51). However, most proteome studies comparing bacterial strains have regularly found the core proteome to consist of 70 to 90% of the identified proteins (52–55).
It is important to note that the proteomes presented here reflect the state of the cells during nutrient-replete, exponential growth. Although no protein with the potential to be used as a biomarker for toxicity was detected, the possibility that one would be expressed during other growth stages or under different environmental conditions cannot be excluded.
Differential Display Between Toxic and Nontoxic Strains of M. aeruginosa
The nSAF approach was used as a label-free method to quantify expression of proteins in the toxic and nontoxic subsets of M. aeruginosa strains. Differences between toxic and nontoxic strains were revealed in proteins involved in maintaining the cellular redox balance and integration of carbon-nitrogen (C-N) metabolism, as well as two hypothetical proteins MAE06820 and MAE27590 (Table V and Fig. 4).
Suitability of the nSAF Approach for Comparison of Bacterial Strains
The nSAF approach was initially used to compare the expression profiles of a single organism grown under different conditions (42). In this study, this label-free quantitation method was applied to analyze protein expression in several strains of a bacterial species. The M. aeruginosa strains studied here showed large diversity in their proteomes. The analysis of similarly diverse proteomes is not possible with image-based methods such as 2DE, which has been recently attempted unsuccessfully in cylindrospermopsin-producing cyanobacteria (28). However, the reciprocal expression profiles of PII and NrtA, the similar expression of the CcmL/CcmK3 and TrxM/MAE06820 pairs, as well as the up-regulation of the gas vesicle and phycocyanin proteins in UWOCC MRC, that were seen here, all suggest that biologically meaningful data can be extracted from even highly diverse datasets using nSAF.
Nitrogen Uptake and Metabolism Proteins
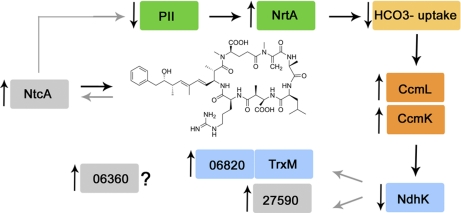
The expression of the signal transduction protein, PII, encoded by glnB, was altered to a significant extent by the presence of microcystin in M. aeruginosa cells (Table V). This protein is central to coordinating photosynthetic activity, carbon metabolism and nitrogen assimilation, because of its dependence on the availability of ATP, 2-oxoglutarate (2OG) levels and competition with the global nitrogen regulator NtcA for binding to the PII interaction protein X (PipX) (56, 57) (Fig. 7). This complex post-transcriptional regulation of PII may be the reason for the lack of correlation between the expression and transcription data for glnB observed here (Table V). Nevertheless, its decreased abundance in the toxic M. aeruginosa was consistent with the observed up-regulation of the nitrate transporter NrtA in the same strains (Table V, Fig. 7). Such reciprocal relationship between PII and NrtA is the result of the regulation of both the glnB and nrtA genes by NtcA, and has been reported previously (58). The differential expression of PII between toxic and nontoxic strains is interesting, given the fact that the mcy gene cluster also appears to be regulated by ntcA (59), and may suggest a regulatory process of microcystin synthesis, involving the PII-NtcA couple. The observed expression patterns of PII and NrtA are indicative of a high 2OG level in the cell as a result of a high C:N metabolic ratio, and the following activation of NtcA and up-regulation of nitrogen assimilation (60). Unfortunately, NtcA expression was at a level below the detection limit of the proteomic approach used here and the levels of the NtcA protein could not be determined directly, but ntcA transcription did not change significantly. Thus, the differential regulation of these proteins may be the result of increased NtcA activity, rather than levels of expression, being modulated by microcystin (Fig. 7).
Fig. 7.
Proposed model for the involvement of microcystin and NtcA regulation in the differential expression of proteins in M. aeruginosa. Boxes in gray have a putative function in this proposed model and have not been shown to be differentially expressed experimentally.
The nitrate ABC transporter, of which NrtA is a subunit has an unusual structure that is shared with members if the cyanobacterial cyanate (Cyn) and bicarbonate (Cmp) transporters (61). Both NrtA and CmpA are inhibited by high levels of nitrate, providing one of the links between carbon and nitrogen metabolism (61). In addition, PII has been shown to regulate carbon transport and respond to the carbon fixation rate in the model cyanobacterium Synechocystis sp. PCC 6803 (62, 63). In this microorganism, PII control results in immediate decrease of nitrate transport, when CO2 fixation is inhibited or CO2 supply is limited (62). However, a ΔPII mutant expressed high-affinity carbon transporters, even in the presence of inorganic carbon (63). Thus, the decreased abundance of the PII protein in the toxic strains studied here, is expected to affect both nitrate and carbon metabolism, resulting in enhanced carbon fixation in microcystin-producing cells.
Carboxysome Proteins
Carbon fixation in cyanobacteria occurs in specialized compartments, the carboxysomes. In β-cyanobacteria these are formed by a multifaceted protein shell, comprised of repeated hexamers of the homologous carbon concentrating mechanism proteins CcmK1–4, as well as CcmL, a pentamer that forms pores at the vertices of the icosahedral shell and allows flux of metabolites in and out of the carboxysome (64, 65). The protein shell encases the enzymes Rubisco and carbonic anhydrase, which are involved in extraction of CO2 from HCO3−, and thus, the final stages of carbon fixation (66). From the data presented here, it appears that the nontoxic strains down-regulated the expression of two shell subunits, CcmK3 and CcmL. It is likely that this was the result of an increase in carboxysome numbers in toxic strains, rather than a change in the carboxysome size (in which case the expression level of CcmL would remain constant). However, this was not reflected by a change in expression of the internal components of the carboxysome, such as Rubisco and carbonic anhydrase, suggesting that at the time of harvesting the cells, the toxic strains contained several empty carboxysomes.
In cyanobacteria, such anomalies in carboxysome structure have been observed previously by electron microscopy in high-carbon requiring mutants (67). This hypothesis is consistent with the observed down-regulation of PII, causing an increase in NrtA, and nitrate uptake, with subsequent inhibition of bicarbonate uptake in microcystin producers. Interestingly, of the four CcmK homologs, which were all expressed in the strains studied here, only CcmK3 was differentially regulated based on toxicity. The contribution of CcmK3 to the structure of the shell has not been studied. However, changes in the carboxysome shell composition have been proposed to influence the permeability of this compartment to metabolites and could be dependent on the carbon status of the cell (68). At the transcript level, ccmL was down-regulated in nontoxic strains, consistent with expression data, but unexpectedly, ccmK transcription appeared to be up-regulated. Such discrepancies in the transcription and expression of carboxysome genes have been reported previously in carbon-limited cells of Synechocystis sp. PCC 6803, and have been accounted to a yet unknown mechanism of post-transcriptional regulation (68). It is possible that NtcA is involved in the expression of both ccmK and ccmL genes to some extent (69).
In terms of the relationship between carbon-fixation and microcystin synthesis, immunogold labeling experiments have shown that the toxin is sometimes localized in proximity to carboxysomes and the possibility of the molecule acting as a inhibitor for Rubisco in carbon-limiting conditions has been discussed (20). However, the relationship between carboxysomes and microcystin is not exclusive, as the toxin is also found close to the thylakoid membranes, polyphosphate bodies and in the nucleoplasm (20, 70). The association of microcystin with the thylakoids, in particular under different light regimes, as well as its proposed ability to bind divalent metal ions have lead to hypotheses that it could be involved in the general oxidative stress response of M. aeruginosa (20, 71).
Proteins Involved in Redox Balance Maintenance
NdhK is an essential subunit of the NADH dehydrogenase complex, similar in function to the NAD(P)H: quinine oxidoreductase (NDH-1) in mitochondria and eubacteria. This enzyme is involved in accepting electrons from reduced plastoquinone, respiration and CO2 uptake (72). This multitude of functions is possible because of the presence of several forms of the complex containing alternative D and F subunits, as well as an accessory carbon uptake (CUP) domain (73, 74). The substrate specificity of the cyanobacterial NDH-1 complex is still unclear, with NADH, NADPH, and reduced ferredoxin being considered as possibilities (72). The NdhK subunit is a soluble protein involved in connecting the membrane and peripheral domains of all NDH complexes and is present both in the thylakoid and plasma membranes (75). It is unclear how the changed expression of this particular NDH subunit would influence the metabolism of M. aeruginosa, but an ndhK knock-out strain of Synechocystis sp. PCC 6803 demonstrated a decreased ability of cells to accumulate inorganic carbon (76). Such a response, where NdhK expression is low in toxic strains, but CcmK and CcmL proteins are highly expressed is consistent with the expected high-carbon requiring phenotype of microcystin-producers discussed above.
Thioredoxin M expression was found to be elevated in toxic cells and further supports a difference in the mechanism for coping with alteration in the redox status of microcystin-producing and nontoxic cells. Thioredoxins are small redox-active proteins, which in cyanobacteria receive electrons from the photosynthetic chain via ferredoxin and ferredoxin-dependent thioredoxin reductase. In high light conditions, NADPH levels increase, leading to an increase in reduced thioredoxin and oxidized target proteins, which are then deactivated and thus protected from oxidative stress (77). In Synechocystis sp. PCC 6803 thioredoxins interact with and modulate the activity of proteins involved in CO2 fixation, glycolysis, nitrogen metabolism and the oxidative stress response (78, 79). These processes are all tightly linked to photosynthetic activity, and accordingly, thioredoxin is placed under transcriptional regulation in response to light, where dark or photoinhibitory light intensities inhibit thioredoxin transcription (80).
The hypothetical protein MAE06820 encoded by the open reading frame adjacent to thioredoxin M was found to follow the TrxM expression pattern. The two open reading frames are separated by a conserved region of 34 nucleotides in both published genomes of M. aeruginosa PCC 7806 and NIES-843 and are cotranscribed, as shown on Fig. 4. This apparent coregulation at both the transcriptional and expression level suggests a thioredoxin or redox-related function of MAE06820. Interestingly, none of the homologs of MAE06820 in other cyanobacteria are encoded in open reading frames proximal to thioredoxin, suggesting that this protein may have a more divergent function in M. aeruginosa. Another hypothetical protein (MAE27590) was also found to be down regulated in nontoxic strains. The role of this protein has not been established yet, although it contains a putative flavin mononucleotide-binding domain and as such, may also be involved in redox balance or photosynthesis. The differential expression of TrxM and the TrxM-associated hypothetical protein in the toxic and nontoxic strains studied here may be the result of altered NdhK activity and thus, changes in photosynthetic electron flow. The increase in TrxM levels in toxic strains would lead to oxidation of its target proteins and protection in oxidative stress, which may explain why toxic strains seem to be better competitors in iron limitation or high light, processes that lead to the formation of reactive oxygen species.
NtcA Regulation and Microcystin Synthesis
Five of the proteins that were differentially expressed, have been identified as targets of the global nitrogen regulator NtcA in cyanobacteria. These include NrtA, PII, CcmK, CcmL, and TrxM (69). The expression of MAE06820 is also likely to be influenced by NtcA activity when it is cotranscribed with TrxM. In conjunction with PII, NtcA is able to either activate or suppress transcription and links photosynthesis and nitrogen metabolism in cyanobacteria (69). The general response of the toxic strains observed here, with an increase in nitrate transporter expression because of lower levels of PII, increased carboxysome shell subunits, low NdhK expression and higher thioredoxin/MAE06820 expression suggest that toxic strains would have higher carbon requirements and increased NtcA activity (Fig. 7).
NtcA generally acts on its own promoter as a positive regulator, hence it was surprising to find that ntcA transcription was lower in toxic strains, relative to nontoxic M. aeruginosa. This may be explained by interaction between NtcA with PII, 2-OG levels in the cell and PipX availability. The lower levels of PII in the microcystin-producing cells would make more PipX available for binding to NtcA-DNA complexes, stimulating NtcA activity on promoters recognized by the global nitrogen regulator. On the other hand, the higher NrtA levels and putatively, nitrate uptake, would lead to low levels of 2-OG, inhibiting ntcA transcription. In fact, high 2-OG during nitrogen starvation has been shown to stimulate binding of pre-existing low levels of NtcA to one of its own promoters, P1 in the nontoxic Anabaena sp. PCC 7120 (81).
As discussed, the NtcA-binding sites present in the microcystin promoter and the differential expression of several NtcA-regulated genes in toxic and nontoxic strains of M. aeruginosa strongly suggests an interaction of the toxin molecule with NtcA. The mcyS gene cluster promoter contains binding sites for two other transcription factors, the ferric uptake regulator Fur and the light-responsive Rca. However, the results presented here did not include genes regulated by either Fur or Rca, although interaction and cross-regulation of all three transcription factors with the toxin cluster cannot be excluded. Instead, the data points to an involvement of microcystin in processes that ultimately influence the integration of photosynthesis with carbon and nitrogen metabolism. These could explain previous observations of toxic and nontoxic strains behaving differently under varying light intensities or in nutrient starvation (11, 16, 82, 83). It is important to consider that the results from this study reflect protein expression in the exponential growth phase and in nutrient-replete conditions. It is highly likely that many more proteins would be differentially expressed under different nutrient and light regimes, in addition to proteins that could contribute to strain-specific adaptation in a particular environment regardless of the ability to synthesize microcystin.
Induction of Toxicity in an Inactive Microcystin Producing Strain
Two of the strains used in this analysis, UWOCC MRC and UWOCC MRD were chosen because of their 16S rRNA gene sequence identity and the reported inability of UWOCC MRC to produce microcystin (84). However, during this study UWOCC MRC was found to have reverted to toxicity and was able to produce the commonly identified MCYST-LR and [DAsp3]MCYST-LR not found in its parent strain UWOCC MRD (Fig. 6 and Table VI). The rare microcystin isoforms produced by UWOCC MRD (Table VI), do not appear to correlate with the protein expression profile in M. aeruginosa strains (supplemental Data 7). In addition, pair-wise nSAF analysis of UWOCC MRC and UWOCC MRD did not reveal differentially expressed proteins that could account for the reversal to toxin synthesis in UWOCC MRC (supplemental Data 8). These findings support previous reports that the toxin isoform is genetically predetermined by the relaxed specificity of the microcystin synthetase aminoacyl adenylation domains, and the activity and presence of the various tailoring enzymes. Nevertheless, a post-transcriptional regulatory process of microcystin synthesis, cannot be excluded in the case of inactive toxigenic strains such as UWOCC MRC, which represent ∼3% of the bloom population in Microcystis and Planktothrix spp (85). This is suggested by the ability of UWOCC MRC to revert back to toxicity, without any associated mutations in the mcy cluster or other phenotypic changes. It is possible that point mutations within the cluster may cause premature stop codons. Indeed, several insertions in the mcy gene cluster have been reported in M. aeruginosa strains, including UWOCC MRC and may affect the transcription of toxin genes (46). These insertions, identified in the promoter region and mcyB, could be the result of past transposition events but do not seem to correlate with toxin production in the strain.
A study by Wilson and colleagues (86) speculated that solid media selects for nontoxic strains, whereas liquid media favors toxin production. Because our strains were maintained in liquid BG11, prolonged culturing under these conditions may have induced the toxicity in UWOCC MRC over time, although the actual event that has caused the reversal of UWOCC MRC to toxicity remains unclear. Unfortunately, we have not been able to acquire a nontoxic UWOCC MRC strain, which would enable us to compare its protein expression to its presently toxic counterpart in our laboratory and the parental UWOCC MRD. This would certainly shed light on the regulatory mechanisms governing toxin biosynthesis.
Proteome analysis of M. aeruginosa revealed that the strains studied here have highly variable protein expression profiles, which may be a result of the adaptation of these organisms to the particular environmental niche that they occupy. No qualitative differences were found between toxic and nontoxic strains, and cluster analysis of protein expression did not group strains according to their toxicity. The literature reports that production of microcystin isoforms shifts with changing nutrient conditions (16), and toxic strains have an advantage in low-iron or high light environments. Thus, it can be expected that the culture conditions used here may not be optimal for the segregation of toxic and nontoxic strains based on their protein expression profile. However, differences in the expression of proteins involved in C-N metabolism and cellular redox status identified in the nSAF analysis, suggest a link between toxin production and cyanobacterial primary metabolism. The expression of several of these proteins, as well as microcystin, is regulated by the global nitrogen transcriptional regulator NtcA, and may reflect a cross-regulatory mechanism linking toxin production to NtcA-controlled metabolic processes under nutrient limitation.
Acknowledgments
Ralitza Alexova was supported by an Environmental Biotechnology CRC PhD scholarship. BAN is a fellow of the Australian Research Council. We thank Dana Pascovici for assistance with the nSAF analysis.
Footnotes
* This work was supported by the Environmental Biotechnology Cooperative Research Center and the Australian Research Council.
 This article contains supplemental Data S1 to S8.
This article contains supplemental Data S1 to S8.
The data associated with this manuscript has been submitted to PRIDE (www.ebi.ac.uk/pride) under accession numbers 13291–13308.
1 The abbreviations used are:
- MC-LR
- microcystin-LR
- Adda
- 3-amino-9-methoxy-2,6,8,-trimethyl-10-phenyl-4,6,-decadienoic acid
- nSAF
- normalized spectrum abundance factor
- PP2a
- protein phosphatase 2a
- qRT-PCR
- quantitative real-time PCR
- 2DE
- two-dimensional electrophoresis
- PRIDE
- Proteomics Identifications Database.
REFERENCES
- 1. Fiore M. F., Genuario D. B., da Silva C. S. P., Shishido T. K., Moraes L. A. B., Cantusio Neto R., Silva-Stenico M. E. (2009) Microcystin production by a freshwater spring cyanobacterium of the genus. Fischerella. Toxicon 53, 754–761 [DOI] [PubMed] [Google Scholar]
- 2. Wiedner C., Visser P. M., Fastner J., Metcalf J. S., Codd G. A., Mur L. R. (2003) Effects of light on the microcystin content of Microcystis strain PCC 7806. Appl. Environ. Microbiol. 69, 1475–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rantala A., Fewer D. P., Hisbergues M., Rouhiainen L., Vaitomaa J., Börner T., Sivonen K. (2004) Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. U.S.A. 101, 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaebernick M., Neilan B. A. (2001) Ecological and molecular investigations of cyanotoxin production. FEMS Microbiol. Ecol. 35, 1–9 [DOI] [PubMed] [Google Scholar]
- 5. Tonk L., Visser P. M., Christiansen G., Dittmann E., Snelder E. O. F. M., Wiedner C., Mur L. R., Huisman J. (2005) The microcystin composition of the cyanobacterium Planktothrix agardhii changes toward a more toxic variant with increasing light intensity. Appl. Environ. Microbiol. 71, 5177–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babica P., Blaha L., Marsalek B. (2006) Exploring the natural role of microcystins - a review of effects on photoautotrophic organisms. J. Phycol. 42, 9–20 [Google Scholar]
- 7. Bagu J. R., Sykes B. D., Craig M. M., Holmes C. F. B. (1997) A molecular basis for different interactions of marine toxins with protein phosphatase-1. J. Biol. Chem. 272, 5087–5097 [DOI] [PubMed] [Google Scholar]
- 8. Soares R. M., Yuan M., Servaites J. C., Delgado A., Magalhães V. F., Hilborn E. D., Carmichael W. W., Azevedo S. M. (2006) Sublethal exposure from microcystins to renal insufficiency patients in Rio de Janeiro, Brazil. Environ. Toxicol. 21, 95–103 [DOI] [PubMed] [Google Scholar]
- 9. Mikalsen B., Boison G., Skulberg O. M., Fastner J., Davies W., Gabrielsen T. M., Rudi K., Jakobson K. S. (2003) Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185, 2774–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pflugmacher S. (2002) Possible allelopathic effects of cyanotoxins, with reference to microcystin-LR, in aquatic ecosystems. Environ. Toxicol. 17, 407–413 [DOI] [PubMed] [Google Scholar]
- 11. Dittmann E., Erhard M., Kaebernick M., Scheler C., Neilan B. A., von, Dohren H., Borner T. (2001) Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiology 147, 3113–3119 [DOI] [PubMed] [Google Scholar]
- 12. Kaebernick M., Neilan B. A., Borner T., Dittmann E. (2000) Light and the Transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 66, 3387–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schatz D., Keren Y., Vardi A., Sukenik A., Carmeli S., Börner T., Dittmann E., Kaplan A. (2007) Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ. Microbiol. 9, 965–970 [DOI] [PubMed] [Google Scholar]
- 14. Humble A. V., Gadd G. M., Codd G. A. (1997) Binding of copper and zinc to three cyanobacterial microcystins quantified by differential pulse polarography. Water Res. 31, 1679–1686 [Google Scholar]
- 15. Briand E., Yéprémian C., Humbert J. F., Quiblier C. (2008) Competition between microcystin- and non-microcystin-producing Planktothrix agardhii (cyanobacteria) strains under different environmental conditions. Environ. Microbiol. 10, 3337–3348 [DOI] [PubMed] [Google Scholar]
- 16. Oh H. M., Lee S. J., Jang M. H., Yoon B. D. (2000) Microcystin production by Microcystis aeruginosa in a phosphorus-limited chemostat. Appl. Environ. Microbiol. 66, 176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vézie C., Rapala J., Vaitomaa J., Seitsonen J., Sivonen K. (2002) Effect of nitrogen and phosphorus on growth of toxic and nontoxic Microcystis strains and on intracelular microcystin concentrations. Microb. Ecol. 43, 443–454 [DOI] [PubMed] [Google Scholar]
- 18. Kehr J. C., Zilliges Y., Springer A., Disney M. D., Ratner D. D., Bouchier C., Seeberger P. H., de Marsac N. T., Dittmann E. (2006) A mannan binding lectin is involved in cell–cell attachment in a toxic strain of Microcystis aeruginosa. Mol. Microbiol. 59, 893–906 [DOI] [PubMed] [Google Scholar]
- 19. Schatz D., Keren Y., Hadas O., Carmeli S., Sukenik A., Kaplan A. (2005) Ecological implications of the emergence of non-toxic subcultures from toxic Microcystis strains. Environ. Microbiol. 7, 798–805 [DOI] [PubMed] [Google Scholar]
- 20. Gerbersdorf S. U. (2006) An advanced technique for immuno-labelling of microcystins in cryosectioned cells of Microcystis aeruginosa PCC 7806 (cyanobacteria): Implementations of an experiment with varying light scenarios and culture densities. Toxicon 47, 218–228 [DOI] [PubMed] [Google Scholar]
- 21. Jüttner F., Lüthi H. (2008) Topology and enhanced toxicity of bound microcystins in Microcystis PCC 7806. Toxicon 51, 388–397 [DOI] [PubMed] [Google Scholar]
- 22. Vela L., Sevilla E., Gonzalez C., Bes M. T., Fillat M. F., Peleato M. L. (2008) Exploring the interaction of microcystin-LR with proteins and DNA. Toxicol. in Vitro 22, 1714–1718 [DOI] [PubMed] [Google Scholar]
- 23. Sazuka T., Ohara O. (1997) Towards a proteome project of cyanobacterium Synechocystis sp. strain PCC6803: linking 130 protein spots with their respective genes. Electrophoresis 18, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 24. Castielli O., de la Cerda B., Navarro J. A., Hervás M., de la Rosa M. A. (2009) Proteomic analyses of the the response of cyanobacteria to different stress conditions. FEBS Lett. 583, 1753–1758 [DOI] [PubMed] [Google Scholar]
- 25. Srivastava R., Pisareva T., Norling B. (2005) Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5, 4905–4916 [DOI] [PubMed] [Google Scholar]
- 26. Ow S. Y., Cardona T., Taton A., Magnuson A., Lindblad P., Stensjö K., Wright P. C. (2008) Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC7120 using 8-plex isobaric peptide tags. J. Proteome Res. 7, 1615–1628 [DOI] [PubMed] [Google Scholar]
- 27. Ow S. Y., Noirel J., Cardona T., Taton A., Lindblad P., Stensjö K., Wright P. C. (2009) Quantiative overview of N2 fixation in Nostoc punctiforme ATCC 29133 through cellular enrichments and iTRAQ shotgun proteomics. J Proteome Res. 8, 187–198 [DOI] [PubMed] [Google Scholar]
- 28. Plominsky A. M., Soto-Liebe K., Vásquez M. (2009) Optimization of 2D-PAGE protocols for proteomic analysis of two nonaxenic toxin-producing freshwater cyanobacteria: Cylindrospermopsis raciborskii and Raphidiopsis sp. Lett. Appl. Microbiol. 49, 332–337 [DOI] [PubMed] [Google Scholar]
- 29. Barrios-Llerena M. E., Chong P. K., Gan C. S., Snijders A. P., Reardon K. F., Wright P. C. (2006) Shotgun proteomics of cyanobacteria applications of experimental and data-mining techniques. Brief Funct. Genomic Proteomic 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 30. Stensjö K., Ow S. Y., Barrios-Llerena M. E., Lindblad P., Wright P. C. (2007) An iTRAQ-Based Quantitative Analysis To Elaborate the Proteomic Response of Nostoc sp. PCC 7120 under N2 Fixing Conditions. J. Proteome Res. 6, 621–635 [DOI] [PubMed] [Google Scholar]
- 31. Frangeul L., Quillardet P., Castets A. M., Humbert J. F., Matthijs H. C., Cortez D., Tolonen A., Zhang C. C., Gribaldo S., Kehr J. C., Zilliges Y., Ziemert N., Becker S., Talla E., Latifi A., Billault A., Lepelletier A., Dittmann E., Bouchier C., de Marsac N. T. (2008) Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9, 274–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaneko T., Nakajima N., Okamoto S., Suzuki I., Tanabe Y., Tamaoki M., Nakamura Y., Kasai F., Watanabe A., Kawashima K., Kishida Y., Ono A., Shimizu Y., Takahashi C., Minami C., Fujishiro T., Kohara M., Katoh M., Nakazaki N., Nakayama S., Yamada M., Tabata S., Watanabe M. M. (2007) Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 14, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otsuka S., Suda S., Shibata S., Oyaizu H., Matsumoto S., Watanabe M. M. (2001) A proposal for the unification of five species of the cyanobacterial genus Microcystis Kutzing ex Lemmermann 1907 under the rules of the Bacteriological Code. Int. J. Syst. Evol. Microbiol. 51, 873–879 [DOI] [PubMed] [Google Scholar]
- 34. Carmichael W. W., An J. (1999) Using an enzyme linked immunosorbent assay (ELISA) and a protein phosphatase inhibition assay (PPIA) for the detection of microcystins and nodularins. Nat. Toxins 7, 377–385 [DOI] [PubMed] [Google Scholar]
- 35. Lawton L. A., Edwards C., Codd G. A. (1994) Extraction and high-performance liquid chromatographic method for the determination of microcystins in raw and treated waters. Analyst 119, 1525–1530 [DOI] [PubMed] [Google Scholar]
- 36. Youshida T., Maki M., Okamoto H., Hiroshi S. (2005) Coordination of DNA replication and cell division in cyanobacteria. M. aeruginosa. FEMS Microbiol. Lett. 251, 149–154 [DOI] [PubMed] [Google Scholar]
- 37. Herbert B. R., Grinyer J., McCarthy J. T., Isaacs M., Harry E. J., Nevalainen H., Traini M. D., Hunt S., Schulz B., Laver M., Goodall A. R., Packer J., Harry J. L., Williams K. L. (2006) Improved 2-DE of microorganisms after acidic extraction. Electrophoresis 27, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 38. Hattrup E., Neilson K. A., Breci L., Haynes P. A. (2007) Proteomic analysis of shade-avoidance response in tomato leaves. J. Agric. Food Chem. 55, 8310–8318 [DOI] [PubMed] [Google Scholar]
- 39. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 40. Craig R., Beavis R. C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 [DOI] [PubMed] [Google Scholar]
- 41. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 42. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in. Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 43. Chick J. M., Haynes P. A., Bjellqvist B., Baker M. S. (2008) A combination of immobilised pH gradients improves membrane proteomics. J. Proteome Res. 7, 4974–4981 [DOI] [PubMed] [Google Scholar]
- 44. Team R. D. C. (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria [Google Scholar]
- 45. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roberts A. A. (2008) Unnatural production of natural products: Heterologous expression and combinatorial biosynthesis of cyanobacterial-derived compounds, in School of Biotechnology and Biomolecular Sciences. PhD thesis, The University of New South Wales [Google Scholar]
- 47. Mlouka A., Comte K., Castets A. M., Bouchier C., Tandeau, de Marsac N. (2004) The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to the loss of cell buoyancy. J. Bacteriol. 186, 2355–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anderson D. C., Campbell E. L., Meeks J. C. (2006) A soluble 3D LC/MS/MS Proteome of the filamentous cyanobacterium. Nostoc punctiforme. J. Proteome Res. 5, 3096–3104 [DOI] [PubMed] [Google Scholar]
- 49. Barrios-Llerena M. E., Reardon K. F., Wright P. C. (2007) 2-DE proteomic analysis of the model cyanobacterium Anabaena variabilis. Electrophoresis 28, 1624–1632 [DOI] [PubMed] [Google Scholar]
- 50. Xia X. X., Han M. J., Lee S. Y., Yoo J. S. (2008) Comparison of the extracellular proteomes of Escherichia coli B and K-12 strains during high cell density cultivation. Proteomics 8, 2089–2103 [DOI] [PubMed] [Google Scholar]
- 51. Lv J., Li N., Niu D. K. (2008) Association between the availability of environmental resources and the atomic composition of organismal proteomes: Evidence from Prochlorococcus strains living at different depths. Biochem. Biophys. Res. Commun. 375, 241–246 [DOI] [PubMed] [Google Scholar]
- 52. Dumas E., Desvaux M., Chambon C., Hébraud M. (2009) Insight into the core and variant exoproteomes of Listeria monocytogenes species by comparative subproteomic analysis. Proteomics 9, 3136–3155 [DOI] [PubMed] [Google Scholar]
- 53. Wang R., Zhang H., Qiu H., Gao S., Kan B. (2009) Proteins involved in difference of sorbitol fermentation rates of the toxigenic and nontoxigenic Vibrio cholerae El Tor strains revealed by comparative proteome analysis. BMC Microbiol. 9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patrauchan M. A., Sarkisova S. A., Franklin M. J. (2007) Strain-specific proteome responses of Pseudomonas aeruginosa to biofilm-associated growth and to calcium. Microbiology 153, 3838–3851 [DOI] [PubMed] [Google Scholar]
- 55. Donaldson J. R., Nanduri B., Burgess S. C., Lawrence M. L. (2009) Comparative proteomic analysis of Listeria monocytogenes strains F2365 and EGD. Appl. Environ. Microbiol. 75, 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Espinosa J., Frochammer K., Burillo S., Contreras A. (2006) Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61, 457–469 [DOI] [PubMed] [Google Scholar]
- 57. Fadi, Aldehni M., Sauer J., Spielhaupter C., Schmid R., Forchhammer K. (2003) Signal transduction protein PII is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J. Bacteriol. 185, 2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Takatani N., Omata T. (2006) Effects of PII deficiency on expression of the genes involved in ammonium utilization in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 47, 679–688 [DOI] [PubMed] [Google Scholar]
- 59. Ginn H. P., Pearson L. A., Neilan B. A. (2010) NtcA from Microcystis aeruginosa PCC 7806 is autoregulatory and binds to the microcystin promoter. Appl. Environ. Microbiol. 76, 4362–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Muro-Pastor M. I., Reyes J. C., Florencio F. J. (2005) Ammonium assimilation in cyanobacteria. Photosyn. Res. 83, 135–150 [DOI] [PubMed] [Google Scholar]
- 61. Koropatkin N. M., Pakrasi H. B., Smith T. J. (2006) Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc. Natl. Acad. Sci. U.S.A. 103, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Forchhammer K. (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28, 319–333 [DOI] [PubMed] [Google Scholar]
- 63. Hisbergues M., JeanJean R., Joset F., Tandeau de Marsac N., Bedu S. (1999) Protein PII regulates both inorganic carbon and nitrate uptake and is modified by redox signal in Synechocystis PCC 6803. FEBS Lett. 463, 216–220 [DOI] [PubMed] [Google Scholar]
- 64. Kerfeld C. A., Sawaya M. R., Tanaka S., Nguyen C. V., Phillips M., Beeby M., Yeates T. O. (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938 [DOI] [PubMed] [Google Scholar]
- 65. Tanaka S., Kerfeld C. A., Sawaya M. R., Cai F., Heinhorst S., Cannon G. C., Yeates T. O. (2008) Atomic-level models of the bacterial carboxysome shell. Science 318, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 66. Badger M. R., Price G. D. (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54, 609–622 [DOI] [PubMed] [Google Scholar]
- 67. Orús M. I., Rodríguez-Buey M. L., Marco E., Fernández-Valiente E. (2001) Changes in carboxysome structure and grouping and in photosynthetic affinity for inorganic carbon in Anabaena strain PCC 7119 (Cyanophyta) in response to modification of CO2 and Na+ supply. Plant Cell Physiol. 42, 46–53 [DOI] [PubMed] [Google Scholar]
- 68. Eisenhut M., von Wobeser E. A., Jonas L., Schubert H., Ibelings B. W., Bauwe H., Matthijs H. C., Hagemann M. (2007) Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 144, 1946–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Su Z., Olman V., Mao F., Xu Y. (2005) Comparative genomics analysis of NtcA regulons in cyanobacteria: regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res. 33, 5156–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Young F. M., Thomson C., Metcalf J. S., Lucocq J. M., Codd G. A. (2005) Immunogold localisation of microcystins in cryosectioned cells of. Microcystis. J. Struct. Biol. 151, 208–214 [DOI] [PubMed] [Google Scholar]
- 71. Utkilen H., Gjølme N. (1995) Iron-stimulated toxin production in. Microcystis aeruginosa. Appl. Environ. Microbiol. 61, 797–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Prommeenate P., Lennon A. M., Markert C., Hippler M., Nixon P. J. (2004) Subunit Composition of NDH-1 Complexes of Synechocystis sp. PCC6803 Identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J. Biol. Chem. 279, 28165–28173 [DOI] [PubMed] [Google Scholar]
- 73. Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E. M. (2004) Towards Functional Proteomics of Membrane Protein Complexes in Synechocystis sp. PCC 68031 Plant Physiol. 134, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ogawa T., Mi H. (2007) Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93, 69–77 [DOI] [PubMed] [Google Scholar]
- 75. Battchikova N., Aro E. M. (2007) Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol. Plant 131, 22–32 [DOI] [PubMed] [Google Scholar]
- 76. Pieulle L., Guedeney G., Cassier-Chauvat C., Jeanjean R., Chauvat F., Peltier G. (2000) The gene encoding the NdhH subunit of type 1 NAD(P)H dehydrogenase is essential to survival of Synechocystis PCC 6803. FEBS Lett. 487, 272–276 [DOI] [PubMed] [Google Scholar]
- 77. Schuermann P., Buchanann B. B. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal, 10, 1235–1273 [DOI] [PubMed] [Google Scholar]
- 78. Florencio F. J., Perez-Perez M. E., Lopez-Maury L., Mata-Cabana A., Lindahl M. (2006) The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 89, 157–171 [DOI] [PubMed] [Google Scholar]
- 79. Lindahl M., Kieselbach T. (2009) Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J. Proteomics. 72, 416–438 [DOI] [PubMed] [Google Scholar]
- 80. Navarro F., Martın-Figueroa E., Florencio F. J. (2000) Electron transport controls transcription of the thioredoxin gene (trxA) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 43, 23–32 [DOI] [PubMed] [Google Scholar]
- 81. Olmedo-Verd E., Valladares A., Flores E., Herrero A., Muro-Pastor A. M. (2008) Role of two NtcA-binding sites in the complex ntcA gene promoter of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 190, 7584–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kardinaal W. E., Tonk L., Janse I., Hol S., Slot P., Huisman J., Visser P. M. (2007) Competition for light between toxic and non-toxic strains of the harmful cyanobacterium Microcystis. Appl. Environ. Microbiol. 73, 2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sevilla E., Martin-Luna B., Vela L., Bes M. T., Fillat M. F., Peleato M. L. (2008) Iron availability affects mcyD expression and microcystin-LR synthesis in Microcystis aeruginosa PCC7806. Environ. Microbiol. 10, 2476–2483 [DOI] [PubMed] [Google Scholar]
- 84. Kaebernick M., Rohrlack T., Christoffersen K., Neilan B. A. (2001) A spontaneous mutant of microcystin biosynthesis: genetic characterisation and effect on Daphnia. Environ. Microbiol. 3, 669–679 [DOI] [PubMed] [Google Scholar]
- 85. Kurmayer R., Christiansen G., Fastner J., Börner T. (2004) Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6, 831–841 [DOI] [PubMed] [Google Scholar]
- 86. Wilson A. E., Sarnelle O., Neilan B. A., Salmon T. P., Gehringer M. M., Hay M. E. (2005) Genetic Variation of the bloom-forming cyanobacterium Microcystis aeruginosa within and among lakes: Implications for harmful algal blooms. Appl. Environ. Microbiol. 71, 6126–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rippka R., Herdman M. (1992) Pastuer culture collection (PCC) of cyanobacterial strains in axenic culture. Vol. 1 Paris: Institut Pasteur [Google Scholar]
- 88. Jackson A. R., McInnes A., Falconer I. R., Runnegar M. T. (1984) Clinical and pathological changes in sheep experimentally poisoned by the blue-green alga Microcystis aeruginosa. Vet. Pathol. 21, 102–113 [DOI] [PubMed] [Google Scholar]
- 89. Schwabe W., Weihe A., Börner T., Henning M., Kohl J. G. (1988) Plasmids in toxic and nontoxic strains of the cyanobacterium Microcystis aeruginosa. Curr. Microbiol. 17, 133–137 [Google Scholar]
- 90. Phelan R. R., Downing T. G. (2007) Optimisation of laboratory scale production and purification of microcystin-LR from pure cultures of Microcystis aeruginosa. Afr. J. Biotechnol. 6, 2451–2457 [Google Scholar]