Abstract
Crop-plant-yield safety is jeopardized by temperature stress caused by the global climate change. To take countermeasures by breeding and/or transgenic approaches it is essential to understand the mechanisms underlying plant acclimation to heat stress. To this end proteomics approaches are most promising, as acclimation is largely mediated by proteins. Accordingly, several proteomics studies, mainly based on two-dimensional gel-tandem MS approaches, were conducted in the past. However, results often were inconsistent, presumably attributable to artifacts inherent to the display of complex proteomes via two-dimensional-gels. We describe here a new approach to monitor proteome dynamics in time course experiments. This approach involves full 15N metabolic labeling and mass spectrometry based quantitative shotgun proteomics using a uniform 15N standard over all time points. It comprises a software framework, IOMIQS, that features batch job mediated automated peptide identification by four parallelized search engines, peptide quantification and data assembly for the processing of large numbers of samples. We have applied this approach to monitor proteome dynamics in a heat stress time course using the unicellular green alga Chlamydomonas reinhardtii as model system. We were able to identify 3433 Chlamydomonas proteins, of which 1116 were quantified in at least three of five time points of the time course. Statistical analyses revealed that levels of 38 proteins significantly increased, whereas levels of 206 proteins significantly decreased during heat stress. The increasing proteins comprise 25 (co-)chaperones and 13 proteins involved in chromatin remodeling, signal transduction, apoptosis, photosynthetic light reactions, and yet unknown functions. Proteins decreasing during heat stress were significantly enriched in functional categories that mediate carbon flux from CO2 and external acetate into protein biosynthesis, which also correlated with a rapid, but fully reversible cell cycle arrest after onset of stress. Our approach opens up new perspectives for plant systems biology and provides novel insights into plant stress acclimation.
Crop plant breeding in the past was mainly directed toward high-yield potential and quality. However, in light of the forecast increase in global temperature, yield safety is jeopardized as drought and high temperature are considered as key stress factors with high potential impact on crop yield (1). To improve the resistance of crop plants to abiotic stress it is crucial to understand the fundamental mechanisms underlying the stress response and stress acclimation in plants. As stress response and acclimation are mediated by proteins they can best be studied by quantitative proteomics. Although transcriptomic approaches are more comprehensive (2), they cannot give insights to protein abundance, which may be regulated at the levels of turnover and translation initiation. Accordingly, several studies on the effects of heat stress at the protein level in various plant species have been performed that were mainly based on differential display of proteins by two-dimensional gels and mass spectrometry-based protein identification (3). Proteins like members of the five major chaperone classes (Hsp100, Hsp90, Hsp70, Hsp60, and sHsps) were consistently reported to be up-regulated and others, like methionine synthase, to be down-regulated during heat stress (4–14). However, several proteins were reported only in single studies to be differentially expressed and even worse, for many proteins contradictory reports exist on the direction of differential expression during heat stress. These results suggest that the plant heat stress response may consist of general and of species- or tissue-specific components. Alternatively, seemingly differential expression of some proteins may be because of experimental artifacts inherent to two-dimensional gel-tandem MS (MS/MS) approaches—the approach almost exclusively used for earlier proteomics studies on the plant stress response (4–7, 9–15). For example, if several proteins comigrate in a single spot of a two-dimensional gel differential expression may be attributed to the wrong protein. Or post- translational modifications induced by stress may shift proteins to different pIs/MWs, thus leading to reduced levels of the unmodified protein. Moreover, differential expression of many proteins is likely to be concealed by the high complexity of the plant proteome.
To reveal highly conserved and thus fundamental components of the heat shock response in plants it is imperative to study the response in plant models that are evolutionarily apart. As proteomic studies on the heat stress response in the green lineage have yet only been carried out on higher plants (3), we decided to perform a study on the unicellular green alga Chlamydomonas reinhardtii. Chlamydomonas is well suited for such an analysis for the following reasons: (1) heat stress can be applied homogeneously and rapidly to all cells in a liquid culture. (2) Vegetative cells are undifferentiated, thus ruling out varying responses because of cell differentiation. (3) All genetic compartments of Chlamydomonas are sequenced and therefore peptides can be identified by database searches (16). (4) (Stress) gene families in Chlamydomonas are much smaller than in higher plants, thus facilitating the interpretation of results (17). This is particularly important for bottom-up proteomic studies, where peptides need to be mapped to single proteins (8). However, annotation of Chlamydomonas gene models is still unsatisfactory, thus complicating spectra-to-peptide mapping. (5) Chlamydomonas tolerates a temperature range between ∼15 and ∼43 °C and, quite similar to higher plants (3), upon shifting from 20–25 °C to temperatures above ∼35 °C induces a stress response (18–20). The stress response in Chlamydomonas is mediated largely by a single heat shock transcription factor sharing all features characteristic of class A heat shock factors of higher plants (21, 22). Hence, the stress response appears to be conserved from Chlamydomonas to higher plants.
For a thorough quantitative proteomic analysis we were looking for a method that allowed for an untargeted identification and accurate quantification of as many proteins as possible. As we wanted to follow proteome dynamics in time courses we disfavored gel-based assays because of the high work load and therefore chose a shotgun approach. Out of the current quantification methods, metabolic labeling with stable isotopes allows for the highest accuracy (23). Here, unlabeled and heavy isotope-labeled cells are mixed and proteins are extracted, precipitated, digested, and analyzed by nano liquid chromatography (LC)-MS/MS in parallel. As losses during processing will affect proteins and peptides from both, light and heavy labeled samples, the ratios of light to heavy peptides will remain constant. Metabolic labeling with 15N for quantitative proteomics has been successfully applied to several eukaryotic model organisms including Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana (24–28). In Chlamydomonas, metabolic labeling for quantitative proteomics has yet been carried out only by stable isotope labeling by amino acids in cell culture (SILAC), where arginine-auxotrophic strains were fed with 13C-arginine (29–32). However, the disadvantages of using 13C-arginine are that it is costly, that quantification of tryptic lysine peptides is precluded, and that quantification is tedious because of significant arginine-to-proline interconversion (31). We therefore decided to label Chlamydomonas proteins with 15N by growing cells in medium containing comparably cheap 15NH4Cl as the nitrogen source. All 15N-labeled tryptic peptides can be quantified, however, currently available evaluation tools either do not support 15N-based quantification (33, 34), or do not support batch job mediated identification, quantification and result data assembly to process the huge amounts of data produced by shotgun proteomics on time course experiments done with multiple biological replicates (35, 36). Hence, the evaluation of shotgun proteomics data based on 15N-labeling required new software solutions.
In this study, we demonstrate the applicability of 15N-labeling based quantitative shotgun proteomics to monitor proteome dynamics during heat stress in Chlamydomonas. Using our in-house developed software framework ‘IOMIQS’ for the management and evaluation of 15N shotgun proteomics data we could identify 3433 Chlamydomonas proteins and out of these quantify 1116 in a heat stress time course experiment. We found levels of 244 proteins to change significantly during heat stress and therefore gained novel, deep insights into how plant cells adapt to heat stress.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions
Chlamydomonas reinhardtii strain CF185 (37) was used in all experiments. Cells were grown photomixotrophically in Tris acetate-phosphate (TAP1) medium (38) in an incubation shaker (INFORS HT, Multitron) at 120 rpm, 25 °C and constant illumination at ∼40 μE m−2 s−1. Cells were counted using a Z2 Coulter Counter (Beckman Coulter, Fullerton, CA).
Heat Shock Kinetics
CF185 cells were grown at constant illumination in TAP medium to a density of ∼5 × 106 cells/ml, harvested by centrifugation at room temperature and 1950 × g for 4 min, resuspended in an equal amount of prewarmed (42 °C) TAP medium and incubated at 42 °C and constant illumination in a water bath. Samples were taken just before and 30, 60, 120, and 180 min after transfer to 42 °C and directly stored on ice. The cells were harvested by centrifugation at 4 °C and 3220 × g for 2 min, washed with 5 mm Hepes-KOH pH 7.4, transferred to Eppendorf tubes, centrifuged again at 4 °C and 21,500 × g for 2 min, resuspended in lysis buffer (50 mm NH4HCO3, 1 mm DTT, 1 mm NaCl), frozen in liquid nitrogen and stored at −80 °C.
15N-Reference Cells
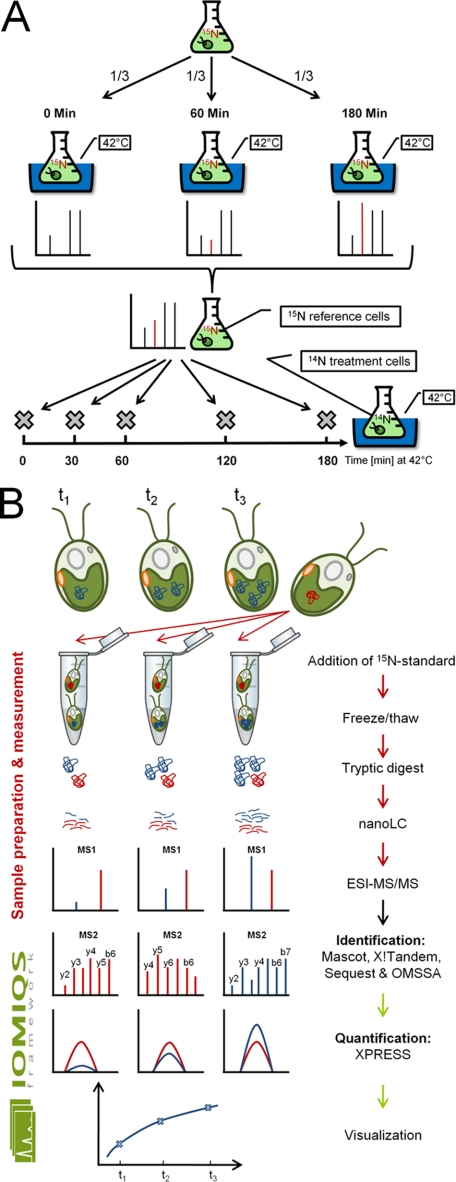
CF185 cells were grown at constant illumination in 50 ml TAP medium containing 7.5 mm 15NH4Cl (≥98%; Cambridge Isotope laboratories, Andover, MA; NLM-467) as nitrogen source to a density of ∼5 × 106 cells/ml. The preculture was diluted into 1 L of 15NH4Cl-containing TAP medium and again grown to a density of ∼5 × 106 cells/ml (overall ≥10 generations). Labeling efficiency was determined to be >98%. Cells were exposed to heat stress as described above and samples taken just before, 60, and 180 min at 42 °C were pooled, harvested, and stored also as described above (see Fig. 1A).
Fig. 1.
Schemes for the preparation of 15N-labeled reference cells, heat stress sampling, sample processing, and data evaluation. A, To prepare a 15N-labeled reference, a Chlamydomonas cell culture was grown for at least 10 generations in medium containing 15NH4Cl as nitrogen source and incubated in a 42 °C water bath. One thirds of the culture were harvested just before and 60 min and 180 min after shifting to 42 °C and pooled. This procedure ensured that proteins absent before or after heat stress were present in the final reference (red spike). Heat stress kinetics were performed by incubating cells grown in medium containing 14NH4Cl as nitrogen source in a 42 °C water bath and sampling shortly before and 30, 60, 120, and 180 min after shifting to 42 °C. B, 15N-labeled reference cells (indicated by the red protein) were mixed with unlabeled samples (indicated by the blue protein) before protein extraction. Soluble proteins prepared by freeze/thaw cycles were acetone-precipitated, tryptically digested and analyzed by nanoLC-ESI-MS/MS. Depending on which protein was more abundant in the sample (heavy or light) peptide identification by MS2 was preferentially based on heavy (red) or light (blue) peptides. Peptide identification by four search engines in parallel, quantification of heavy and light peptides by XPRESS, integration of quantification values at the protein level and export to R! for visualization was managed by the IOMIQS framework.
Gel Blot Analyses
Resuspended cells were ruptured by two freeze/thawing cycles. Broken cells were centrifuged in a table lab centrifuge at 4 °C and 21,500 × g for 35 min and the supernatant, containing soluble proteins, was recovered. Protein concentrations were determined using the bicinchoninic acid protein assay (Thermo Scientific) and 8 μg proteins were separated by SDS-PAGE (39) and transferred to nitrocellulose membranes (GE-Healthcare) by semidry blotting. Blocking and immunodecorations were performed in phosphate-buffered saline containing 3% nonfat dry milk and membranes were immunodetected by enhanced chemiluminesecence (ECL). Antisera used were against cytosolic HSP70A and HSP90A (21), plastidic HSP70B (37), plastidic HSP90C (40), and ATPase subunit CF1β (41). ECL signals were detected with Hyperfilm-ECL (Amersham Biosciences). Films were scanned using an Image Scanner III (GE Healthcare) and signals were quantified using Quantity One-4.6.3 (Bio-Rad). Signal intensities were corrected for unequal loading by using CF1β signals obtained for the respective lanes.
RNA Isolation, Synthesis of cDNA, and qRT-PCR
Samples for quantitative real-time PCR (qRT-PCR) were taken directly, 30 and 180 min after transfer to 42 °C and quenched with ice. Approximately 7 × 107 cells were harvested by centrifugation at 4 °C and 3220 × g for 2 min and total RNA isolation was performed immediately by the trizol extraction method. Harvested cells were resuspended in 1 ml trizol (TRIzol, Invitrogen), 200 μl chloroform were added and the composite was mixed by shaking for 1 min. After centrifugation the supernatant was transferred into a fresh tube and extracted twice with chloroform and isoamylalcohol (24:1). RNA was precipitated by the addition of 1 vol. isopropanol on ice for ∼1 h, collected by centrifugation, washed with 80% ethanol and resuspended in water. RNA concentration and purity were determined spectrophotometrically (NanoDrop-1000). RNA samples were treated with DNase according to the manufacturer's instructions (Turbo DNA-free; Ambion, Austin, TX). cDNA was synthesized using (dT)18 oligonucleotides following the M-MLV reverse transcriptase protocol (Promega, Madison, WI). qRT-PCR was performed using the StepOnePlus RT-PCR System (Applied Biosystems, Foster City, CA) and the SYBR Green kit from Fermentas (Hanover, MD) according to the manufacturer's guidelines. Primer sequences, efficiencies and amplicon lengths for the targets used in this analysis are listed in supplemental Table S5. Fold changes were calculated based on the relative 2−ΔΔCt method using CBLP2 as internal standard.
Sample Preparation for 15N-Labeling Based Quantitative Shotgun Proteomics
15N-labeled reference cells were mixed with nonlabeled samples from the heat stress time course at a 15N/14N-ratio of 0.8 and 0.4 based on chlorophyll content. Mixed cells were ruptured by freeze/thawing cycles and soluble proteins were recovered as described above. Two hundred micrograms of soluble proteins as determined by the bicinchoninic acid assay were precipitated in 80% acetone over night at −80 °C. Precipitated proteins were resuspended in 6 m urea, 2 m thiourea, reduced with 0.24 mm dithiotreitol at room temperature for 30 min and carbamidomethylated with 1 mm iodacetamide at 25 °C for 20 min in the dark. Samples were diluted with 40 mm ammonium hydrogen carbonate to 4 m urea and digested with 0.5 μg endoproteinase Lys C (Roche) on a rotating wheel over night at 37 °C. The digest was diluted to 2 m urea with 20 mm ammonium hydrogen carbonate and 5% acetonitrile and digested with 10 μl of immobilized trypsin (Poroszyme immobilized trypsin, 2–3127-00, Applied Biosystems) for 16 h at 37 °C on a rotating wheel. The resulting peptide mixture was diluted with 20 mm ammonium hydrogen carbonate to a final acetonitrile concentration of 2.5%, desalted using SPEC-C18 solid phase extraction plates (Varian) and dried in a speed vac. The extracted peptides were resuspended in 24 μl of 2% acetonitrile and acidified with 0.5% acetic acid.
nanoLC-ESI-MS/MS
Three times 4 μl of extracted peptides for each sample were subjected to reverse phase separation by nanoLC (EASY-nLC; Proxeon Biosystems) using a monolithic capillary column (RP-18e; 150 - 0.1 mm; Merck) and a linear acetonitrile gradient at a flow rate of 350 nl/min. The gradient was directly initiated after sample application ramping to 65% A, 35% B (A, 2% acetonitrile and 0.5% acetic acid; B, 80% acetonitrile and 0.5% acetic acid) after 100 min, 60% B after 110 min, and 100% B after 115 min. After each run an intermediate wash step consisting of a 20-min linear gradient from 0 to 80% B was performed. Separated peptides were directed to the linear trap quadrupole-Orbitrap mass spectrometer (Thermo Scientific) through a noncoated silica electrospray emitter (PicoTip, FS360–20-10-N-20; New Objective, Woburn, MA) and ionized at ∼2 kV. Spectra were recorded in positive ionization mode. The mass spectrometer was operated in a cycle of one full scan mass spectrum (Orbitrap, 300–1800 m/z) at a set resolution of 30,000 at 400 m/z followed by consecutive data-dependent MS2 scans of the five most intense peaks. MS2 were taken in the linear ion trap at 35% normalized collision energy, using wideband activation and a dynamic exclusion of parent ion masses for 20 s. All single-charged ions were excluded. The raw data may be downloaded from ProteomeCommons.org Tranche using the following hash:/CJRJNPpKfUDFDiXNVFcnq5a5N6uiF1/1VO6gN+TZVxgwDlpEUPJIOCnIkhcuCCU3WuxvsFW8E78fnSPQDiHOmUp0BUAAAAAAABOYg = =
Protein Identification and Quantification
Proteins were identified and quantified using the in-house developed IOMIQS framework (Integration Of Mass spectrometry Identification and Quantification Software). Raw spectra data coming in RAW file format (Thermo Scientific) were converted into mzXML by using the ReAdW program (http://sashimi.sourceforge.net/software_glossolalia.html) and spectra lists were extracted and written into mgf and dta files with the MzXML2Search program (http://tools.proteomecenter.org/wiki/index.bpphp?title = Software:MzXML2Search). The ReAdW and MzXML2Search programs both are part of the Trans-Proteomic Pipeline (36).
For peptide identification the search engines Mascot [version 2.2.04] (42), Sequest [version 28] (43), OMSSA [version 2.1.4] (44), and X!Tandem [version 2009.04.01.1] (45) were used. For the searches the precursor mass tolerance was set to 10 ppm (for OMSSA it was set to 0.015 Da), whereas the fragment ion tolerance was set to 0.8 Da. Up to three missed cleavages were allowed for tryptic peptides. These settings were used for all four search engines. As variable modifications, carbamidomethylation of cysteine, oxidation of methionine and N-terminal acetylation were selected for Mascot, OMSSA, and X!Tandem. To identify 15N-labeled peptides, the searches were repeated with exactly the same settings, but in 15N mode. This mode causes the search engines to consider the heavy isotope when calculating the peptide masses. As our version of Sequest did not have such a 15N mode, we searched labeled peptides with Sequest by assigning a fixed modification to each amino acid with the mass it would have if it was fully 15N-labeled. Combining this approach with the use of variable modifications did not work with our Sequest version, which is why we decided to forgo searching for modifications in favor of being able to identify heavy labeled peptides with Sequest.
The search database was built as follows: all protein sequences released in version 3.1 of the Chlamydomonas genome sequence (16) were combined with all sequences from Chlamydomonas mitochondrial and chloroplast proteins (downloaded from http://chlamycyc.mpimp-golm.mpg.de/files/sequences/protein/). A copy of all target sequences was shuffled and added to build the final FASTA file.
As peptide identification was done within IOMIQS, all output files of the different search engines were automatically converted into pepXML file format by our wrapper program, which makes use of tools of the Trans-Proteomic Pipeline (Mascot2XML, Sequest2XML and Tandem2Xml) (http://tools.proteomecenter.org/TPP.php). In addition, the wrapper program uses the corresponding mzXML file to supplement data like the retention times to ensure that all pepXML files resulting from different search engines contain the same information.
For peptide quantification the algorithm XPRESS [version 2.1] (46) was used with peptide mass tolerance set to 0.8 Da. As we searched both, unlabeled and labeled peptides, for each raw file and search engine two pepXML files were processed by XPRESS. This approach yielded 8 pepXML files for each raw file containing quantitative peptide data.
Data Processing with IOMIQS
To implement the batch job processing, IOMIQS incorporates a job management system, which is based on a database. This database is connected to the IOMIQS web interface and stores all jobs with the corresponding parameter settings that the user selects via the web interface. In addition, the database holds information about the dependences between different processes as well as the computing capacity each process can employ. Using this information the job management system is able to start independent processes in parallel and exploit the full computing capacity that is available.
Data Analysis and Significance Tests
The data contained in all obtained pepXML files as well as the corresponding MS2 spectra were uploaded into a MSSQL database via IOMQS. In the first analysis step, a threshold is calculated for each search engine and raw file at which the false positive rate is at most 1%. For the false positive calculation we used the approach of Elias and Gygi (47). The basic assumption of this approach is that after peptide identification, using a database containing target and shuffled sequences, the number of false positive hits with a score above a certain threshold is two times the number of hits above this threshold where spectra are matched to shuffled sequences. From this the fraction of incorrect assignments above a certain score can be calculated. We determined for each sample and each search engine the threshold that ensures a false positive rate of at most 1%. For this threshold the following scores are used: ion score (Mascot), xcorr (Sequest), expect (OMSSA), and expect (X!Tandem). All peptides that received a score above (Mascot, Sequest)/below (OMSSA, X!Tandem) this threshold from at least one search engine were selected for further analyses. Furthermore, only peptides that map to exactly one protein were taken into account, which, however, leads to the preclusion of isoforms and splice variants represented by individual gene models, like RBCS1/2 (48) and CGE1a/b (49). Annotated, mass labeled spectra for single peptide identifications and all post-translationally modified peptides may be downloaded from ProteomeCommons.org Tranche using the following hash:
Qf8fXTJrSNyauoJ//IuGIMZxq0nMBtE5f0W2Ns6A0JIfMKxdBSWILEIXR0kir5uGu2CYNdiBScEE7xpdlcyhIfC+NxUAAAAAAAAHIw = =. The “light-to-heavy” ratios calculated by XPRESS for all unique peptides that passed the threshold were used to determine the relative abundance of the corresponding proteins. Unequal mixing of labeled and unlabeled proteins in each sample was corrected for by normalizing the light-to-heavy ratio determined for each peptide in a sample by the geometric median of light-to-heavy ratios of all peptides in that sample. The median of the normalized light-to-heavy ratios of all peptides from one protein was taken as the relative abundance of that protein.
To assess, whether changes of protein abundance during the heat stress time course were significant, quantified protein data were transformed to Z-score and tested by one-way ANOVA over the time points. The Z-score transformation was performed to eliminate differences in the kinetic behavior of the proteins caused by the addition of different amounts of 15N standard to the biological and technical replicates. After p value adjustment according to the Benjamini and Hochberg procedure (50) the significance threshold was set to a p value ≤ 0.05. For the visualization of original (untransformed) quantitative protein data, significantly changing proteins were transformed to the percentage of maximum change for a biologically more intuitive presentation. The mean and standard deviation calculated from three biological and two technical replicates were plotted with R! (http://www.r-project.org/).
Protein Binning and Bin Enrichment
We created 47 bins and sub-bins to group all quantified proteins according to their function (see Table I). Bins were mainly based on MapMan binning (51) and on the assignments and annotations made by the Chlamydomonas community (16, 38) and curated manually. Proteins were placed only into single bins, although they might participate in more than one function (e.g. alanine-glyoxylate transaminase participates in photorespiration and glycine biosynthesis, but was placed only into bin “Proteins of photosynthetic and photorespiratory carbon metabolism”). To elucidate whether changing proteins were enriched within an annotation category with respect to the total protein population, we used hypergeometric testing. The total protein population for which we have kinetic information in our experiment is 1116 and the number of differential changing proteins 244 (Table I). The null hypothesis to test is that the change in protein abundance and its belonging to an annotation category is statistically independent. Hypergeometric formulation is directly derived from the problem statement and was shown to be appropriate even for a small number of proteins (52). Again, p values were adjusted according to the Benjamini and Hochberg procedure and the significance threshold was set to a p value ≤ 0.01. Calculations were performed with R!.
Table I. List of the 244 proteins found among the 1116 quantified to significantly change during heat stress. Protein IDs are derived from gene models from version 3.1 of the Chlamydomonas genome sequence (16).
| Bin | Functional group | No of proteins in bin | Enrichment (p value)a | Ups (Cluster; maximum fold increase)b | Downs (Cluster; maximum percent decrease)c |
|---|---|---|---|---|---|
| 1 | Flagellar and basal body proteins | 38 | 1,0E+00 | - | FAP24(C;40.1), FAP103(D;11.3), FAP278(E;21.5), PF9(-;66.4) |
| 2.0 | Other proteins associated with light reactions | 19 | 6,1E-01 | LHCSR3(A;1.2), ULP1(B;2.7) | FNR1(E;9.4), TEF8(C;45.0) |
| 2.1 | Proteins associated with PS I | 11 | 4,0E-01 | PSAN(B;2.1), LHCA3(B;1.2) | LHCA5(E;16.9) |
| 2.2 | Proteins associated with PS II | 10 | 1,3E-01 | - | PSBO(D;19.1), PSBP1(C;30.7), LHCBM1(C;45.6), LHCBM3(C;35.5) |
| 3 | Proteins of photosynthetic and photorespiratory carbon metabolism | 30 | 2,2E-7 | - | SHMT1(D;25.9), SHMT2(C;13.8), SHMT3(E;9.8), GCST(C;7.1), GCSP(E;11.3), rbcL(D;11.4), RCA1(C;29.6), CP12(D;22.2), FBA3(E;8.1), TRK1(E;9.7), FBP1(E;10.9), SBP1(E;8.7), RPE1(C;26.5), TPIC(E;14.8), PRK1(E;5.9), LCIB(D;8.2), LCIC(D;10.3), LCI23(-;87.2), CAH4(D;12.9), PPD2(C;31.9) |
| 4 | Starch synthesis & degradation | 8 | 4,4E-01 | - | PHOA(D;17.5), PHOB(D;14.6) |
| 5 | Porphyrin biogenesis | 14 | 1,7E-01 | - | GSA(D;17.8), ALAD(D;12.7), UROD1(E;30.9), UROD2(-;82.0), CHLI1(C;18.8) |
| 6 | Carotenogenic enzymes | 5 | - | - | |
| 7 | Glycolysis and gluconeogenesis | 13 | 7,9E-01 | - | PGH1(D;10.0), PYK1(E;27.0) |
| 8 | TCA | 14 | 3,7E-01 | - | PDC1(D;19.1), IDH3(E;7.2), OGD1(D;19.7), OGD2(D;32.1) |
| 9 | Oxidative pentose phosphate pathway | 6 | 6,0E-01 | - | 101528(E;19.8) |
| 10 | Acetate metabolism/glyoxylate cycle | 10 | 6,4E-03 | - | ACS1(D;15.1), ACS3(D;10.4), ACH1(D;9.2), ICL1(D;21.0), MAS1(D;11.4), MDH1(D;10.8) |
| 11 | Fermentation | 6 | 2,8E-01 | - | PDC3(C;59.1), PFL1(C;67.2) |
| 12 | Glycerolipid metabolism | 22 | 5,8E-01 | - | BCR1(C;20.2), KAR1(E;18.2), ENR1(C;26.1), 190684(C;60.3), ATO1(D;29.7) |
| 13 | Mitochondrial electron transport chain and ATP synthase | 12 | 9,6E-01 | - | CYC(D;36.6) |
| 14 | N-metabolism | 7 | 3,3E-03 | - | GSF1(D;14.2), GSN1(C;32.8), GLN1(E;25.5), 194028(C;22.0), MDH5(E;20.4) |
| 15 | Amino acid metabolism | 47 | 1,8E-01 | - | 136279(E;42.5), 162449(D;21.3), 126232(E;29.0), HIS1(D;43.1), 140252(E;14.0), ASSD1(C;24.2), METE(C;58.6), THS1(D;19.8), AAD1(D;16.1), AAI1(C;22.5), ALSS1(C;17.6), LEU2(E;25.4), AGS1(D;12.3), CMPL1(D;16.7) |
| 16 | S-assimilation | 5 | 5,2E-01 | - | ATS1(C;60.5) |
| 17 | Metal handling | 3 | 2,8E-01 | - | SBD1(E;16.9) |
| 18 | C1 metabolism | 6 | 8,7E-03 | - | FDH1(D;80.2), 111330(C;15.0), METM(C;12.3), SAH1(D;21.6) |
| 19 | Vitamin biosynthesis | 7 | 3,7E-01 | THI4a(A;1.5) | PDX1(C;41.5) |
| 20.0 | Protein folding and abiotic stress | 8 | 7,9E-01 | 154979(A;2.5) | - |
| 20.1 | Small heat shock proteins | 3 | 0,0E+00 | HSP22A(B;21.4), HSP22C(B;26.0), HSP22E(B;13.2) | - |
| 20.2 | HSP60s | 15 | 6,4E-03 | CPN60A(B;4.1), CPN60B1(B;2.1), CPN60B2(B;4.0), CPN60C(A;1.3), CPN23(B;2.5), CPN20(B;2.5), CPN10(B;3.0), CPN11(B;2.5) | - |
| 20.3 | HSP70 and co-chaperones | 11 | 3,3E-04 | HSP70A(B;2.9), HSP70B(B;2.4), HSP70C(B;1.3), HSP70E(B;2.0), 150437(A;6.4), MOT53(-;3.2), DNJ1(A;2.3) | HSP70G(E;56.9) |
| 20.4 | HSP90 and co-chaperones | 5 | 3,0E-03 | HSP90A(B;5.2), HSP90B(B;1.4), HSP90C(B;3.4), HOP1(A;2.6) | - |
| 20.5 | HSP100 | 4 | 8,7E-03 | CLPB1(A;22.1), CLPB3(A;5.7), CLPB4(A;6.2) | - |
| 20.6 | FKBPs/Cyclophilins | 10 | 1,1E-03 | FKB62(B;11.5) | FKB12(D;17.7), FKB53(E;10.7), CYN19–2(C;43.4), CYN20–1(E;9.0), CYN20–3(C;46.7), CYN38(D;29.3) |
| 21 | Proteases/Peptidases | 53 | 1,0E+00 | - | 82208(E;7.5), 139416(D;18.0), UBC2(D;23.1), CDC48(D;16.8) |
| 22 | Redox regulation/oxidative stress response | 35 | 4,1E-01 | - | TRXH(C;15.4), CITRX(E;8.4), RB60(D;15.3), CCPR1(C;53.4), 165193(E;9.4), APX1(C;36.5), GPX5(E;16.8), FSD1(E;4.9), MSRA4(-;39.2) |
| 23 | Nucleotide metabolism | 26 | 1,0E+00 | - | 175039(D;28.7) |
| 24 | Sugar nucleotide metabolism | 10 | 1,3E-01 | - | 158152(C;23.6), SNE1(E;15.4), SNE6(C;10.8), UPTG1(D;17.6) |
| 25 | RNA processing/binding | 43 | 1,0E+00 | - | RSZ22(C;46.1), UAP56(E;6.0), NOP1(D;43.6), 184151(D;15.9), RB47(D;13.0), RB38(D;16.9) |
| 26 | DNA binding/transcription/chromatin remodeling | 65 | 1,0E+00 | 153995(-;3.3), 24073(B;1.5) | GBP1(E;27.1), 186410(C;33.5), 154370(E;16.5) |
| 27.1 | Protein biosynthesis (not ribosome) | 50 | 6,0E-01 | - | 186253(E;23.6), 82920(D;32.6), 38643(E;19.9), 145770(D;14.0), EFG2(D;13.6), EFT1a(C;64.4), tufA(D;13.2), EIF3K(D;19.7), 188942(C;13.9), EIF4E(C;17.1), EIF6A(D;23.4) |
| 27.2.0 | Protein biosynthesis | 4 | - | - | |
| 27.2.1 | Protein biosynthesis (LSU plastid) | 21 | 9,4E-01 | - | PRPL1(D;37.0), rpl2(E;10.7), PRPL9(E;10.2) |
| 27.2.2 | Protein biosynthesis (SSU plastid) | 15 | - | - | |
| 27.2.3 | Protein biosynthesis (LSU cytosol) | 40 | 1,4E-06 | - | RPP0(E;7.3), RPL3(D;21.1), RPL4(D;10.5), RPL5(E;5.9), RPL6(E;14.3), RPL7(D;20.6), RPL9(E;6.3), RPL10(D;16.9), RPL10a(D;11.4), RPL12(C;10.4), RPL13(D;13.3), RPL14(D;12.4), RPL18a(D;11.5), RPL19(E;10.3), RPL21(E;8.3), RPL22(E;11.1), RPL27(D;7.9), RPL28(C;14.9), RPL32(D;28.6), RPL35(D;14.1), RPL35a(D;18.9), RPL36(E;15.5), RPL40(E;31.2) |
| 27.2.4 | Protein biosynthesis (SSU cytosol) | 31 | 9,4E-09 | - | RPS2(D;26.9), RPS3(E;13.8), RPS3a(E;15.0), RPS4(D;31.4), RPS6(D;29.5), RPS7(D;26.2), RPS8(D;17.3), RPS9(E;27.1), RPS10(E;14.2), RPS14(C;17.4), RPS15(D;12.1), RPS15a(C;27.6), RPS16(D;18.8), RPS18(D;10.2), RPS19(E;9.2), RPS20(D;15.0), RPS23(D;35.4), RPS24(E;22.5), RPS25(D;17.0), RPS27-A(D;18.0), RPS28(D;13.3), RPS29(D;27.3) |
| 28 | Signal transduction | 58 | 1,0E+00 | CRT2(B;1.1), AKC2(-;2.0) | 184895(D;6.7), RACK1(E;7.3), PP2A-2r(E;21.5), RABC2(-;25.1), FTT1(D;15.2) |
| 29 | Pumps and transporters | 16 | 9,4E-01 | - | ATPvA1(D;16.2), ATPvB(D;10.4) |
| 30 | Vesicle transport | 22 | 9,5E-01 | - | COPE1(E;30.6), COPG1(C;17.1), VPS35(C;18.3) |
| 31 | Cytoskeleton | 9 | - | - | |
| 32 | Miscellaneous | 145 | 1,0E+00 | 193531(A;43.8), 148088(B;1.6) | SPS1(E;21.1), CPLD48(-;35.9), 190138(C;22.5), 190503(C;23.6), RCD2(C;26.0), 194623(C;20.0), 179100(D;54.5) |
| 33 | Proteins without functional annotation | 115 | 1,0E+00 | - | 144849(C;15.6), 178937(C;97.1), 95661(-;30.0), 192803(D;12.6), 137973(-;44.3), CGL13(-;20.7), 166509(-;33.5) |
| 34 | Protein sorting | 9 | 9,4E-02 | - | TIC40(D;46.6), TIC110(-;19.1), SRP19(C;32.6), IPB2(E;23.7) |
a Significant bin enrichment is given at p values below 1,0E-02.
b Cluster A = UP fast; B = UP delayed; C = DOWN fast; D = DOWN delayed; E = DOWN delayed, into steady state. Maximum fold increase was calculated by dividing protein quantification values for all time points with the value of the first available time point and selecting the quotient with the highest value for display.
c Maximum percent decrease was calculated by subtracting the quotients of protein quantification values from all time points and the first available time point from 1 and selecting the difference with the highest value for display.
Cluster Analysis
From the heat stress time course experiment we expected eight cluster types, four each for up- and down regulated proteins: 1-fast, 2-fast into steady-state, 3-slow, 4-slow into steady-state. Using hierarchical clustering, based on Manhattan distance and Ward's linkage (53), clusters were reduced until they contained not less than three observations, which eventually resulted in five clusters. Clustering was performed on Z-score transformed data using R!.
RESULTS
Chlamydomonas lives in diverse freshwater environments like lakes, ponds, puddles, or water-filled cavities in the soil (38). In particular smaller liquid volumes may experience rapid temperature shifts, e.g. by sudden exposure to sunlight on a cloudy day. Hence, to cope with sudden temperature stress, algae dwelling such environments require fast acclimation responses. To monitor how acclimation to a rapid temperature increase is realized in Chlamydomonas at the proteome level, we rapidly transferred mixotrophically grown cells from 25 °C to 42 °C and took samples just before and 30, 60, 120, and 180 min after transfer to 42 °C (Fig. 1A). This rapid temperature shift has been frequently used to elicit a physiological heat stress response in Chlamydomonas (18–21). Cells grown with 15NH4Cl as nitrogen source for at least ten generations were subjected to the same treatment and samples taken shortly before and 60 and 180 min after onset of heat stress were pooled to generate a uniform 15N-labeled reference proteome. Cells taken during the heat shock time course were then mixed with 15N-labeled reference cells at ratios of 0.8 and 0.4. The on average lower abundance of heavy peptides ensures that light peptides would preferentially be selected for MS2, thus increasing the number of MS2 events on different (light) peptides. Soluble proteins were extracted after disruption of cells by freeze/thawing cycles, precipitated in acetone and digested with LysC and trypsin (Fig. 1B). After desalting, peptides were separated on a reversed phase column by nanoLC and injected online into a linear trap quadrupole-Orbitrap mass spectrometer. The experiment was done with three biological and two technical replicates (0.8 and 0.4 ratios of heavy to light proteins). As each sample was measured three times to increase peptide coverage, the heat shock time course generated 90 raw files containing a total of 2,035,419 spectra, 419,739 MS1 full scans, and 1,615,680 MS2 spectra.
Identification and Quantification of Peptides and Proteins
To evaluate these huge raw data sets, we developed IOMIQS (Integration Of Mass spectrometry Identification and Quantification Software). IOMIQS integrates different published identification and quantification algorithms to allow for an automated evaluation of large raw data sets from shotgun proteomics based on 15N labeling (Fig. 1B).
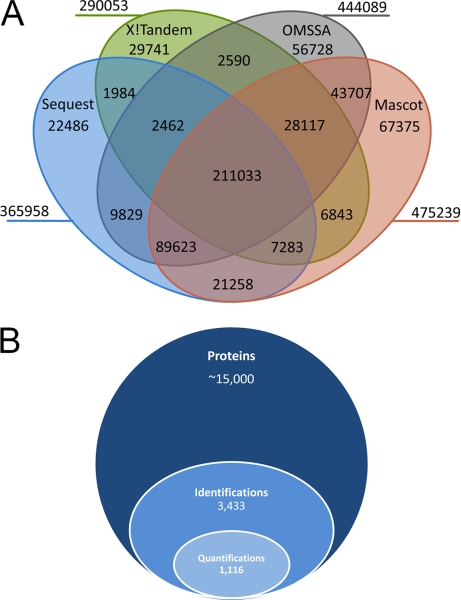
As shown in Fig. 2A, the combined use of the four search engines Mascot (42), Sequest (43), OMSSA (44), and X!Tandem (45) enhanced peptide identification rates significantly: at a false discovery rate of 1%, most spectra-to-peptide mappings (SPMs) were achieved by Mascot (475, 239), followed by OMSSA (444, 089), Sequest (365, 958) and X!Tandem (290, 053). Peptides of highest confidence were the 211,033 peptides identified by all four search engines. Sensitivity increased at the expense of confidence when peptides were required to be identified by only three (338,518 SPMs), two (424,729 SPMs), or a single search engine (601,059 SPMs). Thus, although the apparently best search engine Mascot allowed SPMs for 29% of all 1,615,680 spectra, this value was 37% when SPMs of all four search engines were combined, thus corroborating previous results on samples of low complexity (54).
Fig. 2.
Overview of peptides and proteins identified and quantified in this study. A, Venn diagram of spectra-to-peptide mappings by search engines Sequest, X!Tandem, OMSSA and Mascot. MS2 spectra were taken from 90 raw files. B, Stacked Venn diagram showing the fractions of identified and quantified proteins from the total protein-coding gene models present in V3.1 of the Chlamydomonas genome.
We have three likely explanations for why only about one third of all spectra were mapped to peptide sequences: first, low quality fragment spectra e.g. from short peptides will not be mapped to peptide sequences with sufficiently high scores. Second, peptides containing post-translational modifications other than methionine oxidation and N-terminal acetylation are not included in the database search. Third, as full cDNA coverage is lacking for many of the gene models in version 3.1 of the Chlamydomonas genome sequence (16), many exon-intron borders are not correctly annotated and, therefore, many sequence templates are lacking for spectra mapping.
In addition to peptide identification, IOMIQS features automated peptide quantification by passing data on identified peptides from the search engines over to the quantification algorithm XPRESS (46). Information on peptide sequences, the search engines by which they were identified, and XPRESS quantification values for all three biological replicates and time points of the heat stress time course are compiled in supplemental Table S1. Of the estimated ∼15,000 protein coding genes in the Chlamydomonas genome (16), 3433 proteins were identified and are listed in supplemental Table S2. Quantitative data for at least three of the five time points within the heat shock time course was obtained for 1116 proteins (Fig. 2B and supplemental Table S3).
Verification of Quantitative Shotgun Proteomics Results by Immunoblot Analysis
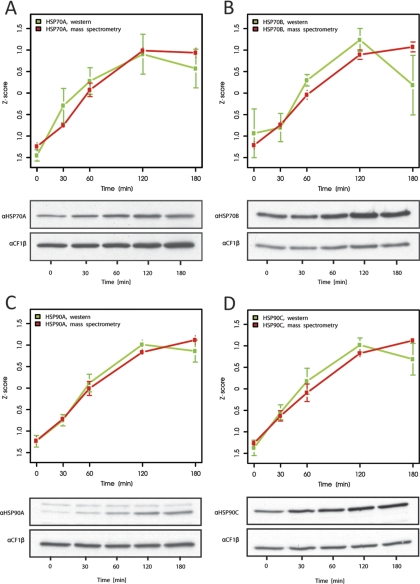
To verify that quantification by our approach was accurate we compared results from quantitative shotgun proteomics using a uniform 15N standard with results obtained by quantitative immunoblotting. For this, aliquots taken from the unlabeled heat stress time courses before mixing with the 15N-labeled reference proteome were analyzed using antisera against cytosolic HSP70A and HSP90A, and chloroplast HSP70B and HSP90C. These proteins were previously shown to increase during heat stress (21, 40, 55). As shown in Fig. 3, both quantification methods gave very similar results for the kinetics of increase of the four chaperones. As judged from the much larger error bars obtained for the quantification by immunoblotting as compared with quantitative shotgun proteomics, the latter appears to be even more accurate than quantitative immunoblotting, but is not restricted to proteins against which antisera are available.
Fig. 3.
Comparison of chaperone expression kinetics by quantitative immunoblotting and quantitative shotgun proteomics. Accumulation of cytosolic HSP70A (A), chloroplast HSP70B (B), cytosolic HSP90A (C), and chloroplast HSP90C (D) during heat shock as measured by immunoblot analysis (green curves) or quantitative shotgun proteomics (red curves). Values from immunoblot analyses were derived from ECL signals from the chaperones normalized to the signals from CF1β. Mean values and standard deviations derive from 3 biological and 2–4 technical replicates. Shown are Z-score transformed values to facilitate comparison of protein kinetics.
We sorted 856 of the 1116 quantified proteins into 47 functional categories (Table I). For 260 proteins a functional annotation was either not available at all (bin “Proteins without functional annotation”), or a putative function was not covered by one of the 47 functional bins (bin “Miscellaneous”). According to ANOVA testing based on a Z-score representation of a protein's heat stress time course curve shape, 244 of the 1116 quantified proteins changed significantly during heat stress: 38 proteins were up- and 206 were down-regulated. The remaining 872 proteins either did not change during heat stress, or changes in protein levels based on the ANOVA test were not significant. A full list of all 1116 quantified proteins including protein IDs, bin assignment, % coverage, annotation, links to the gene models and protein quantification values is provided in supplemental Table S3. In the following, we will give an overview to the scope of the 1116 quantified proteins in each bin and describe the 244 significantly changing proteins in more detail.
Proteins Involved in Protein Folding and Abiotic Stress
Fifty-six of the 1116 quantified proteins have functions in protein folding and abiotic stress. Thirty-eight of them belong to one of the five major classes of molecular chaperones, termed Hsp100, Hsp90, Hsp70, Hsp60/chaperonins and sHsps, or are cochaperones of Hsp90s, Hsp70s, or Hsp60s (Table I, bins 20.1–20.5). 25 of these 38 (co-)chaperones increased during heat shock. Most dramatic increases (13 to 26-fold) were observed for cytosolic HSP22A, HSP22E, and CLPB1, and for chloroplast HSP22C (Figs. 4A and 4B). Strong, two- to sevenfold increases in protein concentrations were observed for cytosolic HSP90A/HOP1, HSP70A/DNJ1/MOT53 (= HSPBP1), HSP70E, and BAG6-like protein with ID 150437, and for chloroplast CLPB3/4, HSP90C, HSP70B, CPN60A, CPN60B1, CPN60B2, CPN23, CPN20, and CPN11 (Figs. 4B–4E). The 3.4- and 2.4-fold increases in levels of HSP90C and HSP70B, respectively, closely matched the values reported previously (3.9- and ∼3-fold, respectively) (40, 55). Interestingly, concentrations of mitochondrial HSP70C and CPN60C increased only moderately by 1.3-fold, whereas those of the putative CPN60C co-chaperone CPN10 increased threefold during heat stress. Similarly, among the quantified ER-targeted chaperones a modest increase by 1.4-fold was observed only for HSP90B, whereas Hsp110 family member HSP70G even declined by 57%. Levels of (co-)chaperones HSP22A/E, HSP90A/B/C, HSP70B, HSPBP1, CPN60A/B1/B2, and CPN23/20/11/10 were still slightly increasing between 120 and 180 min of heat stress, whereas those of HSP22C, CLPB1/3/4, HOP1, HSP70A/C/E, DNJ1, BAG6-like protein, and CPN60C had reached a plateau or were slightly declining after 120 min of stress. Levels of chloroplast Hsp100 member CLPC, which is involved in chloroplast protein import and protein quality control when associated with the ClpP protease (56), did not change during heat stress (supplemental Table S3). Significant changes in levels of the seven quantified subunits of the cytosolic CCT chaperonins and of the four putative subunits of prefoldin—both are mainly involved in folding of actin and tubulin (57, 58)—could not be detected. In addition to the quantified 38 members of the five main chaperone families, few peptides were identified for 9 more members. An overview to all currently annotated family members and to which extent they were detected in this study is provided in supplemental Table S4.
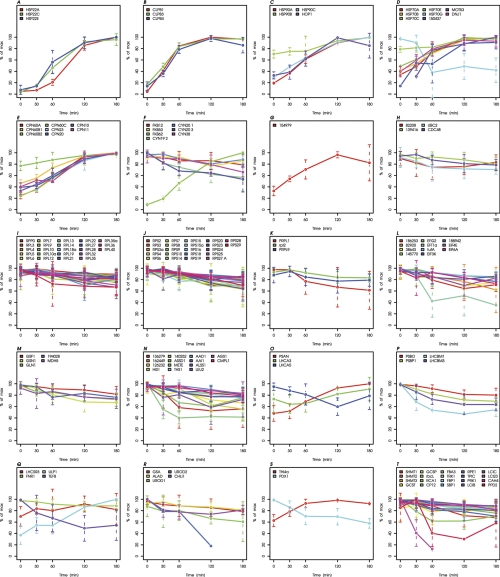
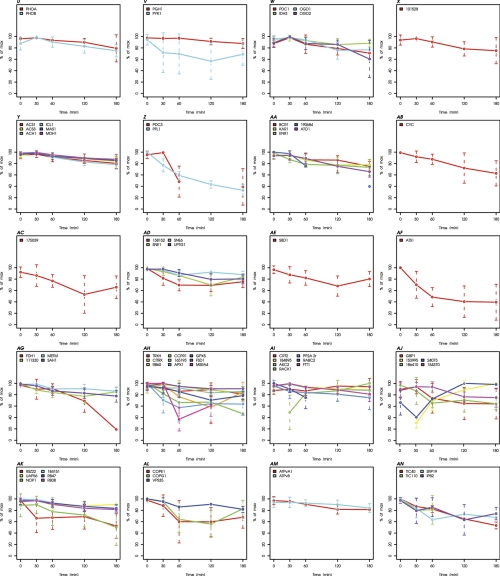
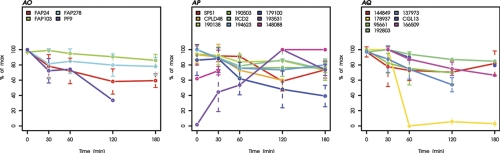
Fig. 4.
Kinetics of proteins exhibiting significant changes during heat stress. Kinetics of all 244 proteins significantly changing during heat stress are plotted bin-wise as percent of the maximal value in the time course. Values and standard deviations derive from three biological and two technical replicates (listed in supplemental Table S3). A, Small heat shock proteins. B, HSP100s. C, HSP90 and co-chaperones. D, HSP70 and co-chaperones. E, HSP60s. F, FKBPs/Cyclophilins. G, Protein folding and abiotic stress. H, Proteases/Peptidases. I, Protein biosynthesis (LSU cytosol). J, Protein biosynthesis (SSU cytosol). K, Protein biosynthesis (LSU plastid). L, Protein biosynthesis (not ribosome). M, N-metabolism. N, Amino acid metabolism. (O) Proteins associated with PS I. P, Proteins associated with PS II. Q, Other proteins associated with light reactions. R, Porphyrin biogenesis. S, Vitamin biosynthesis. T, Proteins of photosynthetic and photorespiratory carbon metabolism. U, Starch synthesis and degradation. V, Glycolysis and gluconeogenesis. W, TCA cycle. X, Oxidative pentose phosphate pathway. Y, Acetate metabolism/glyoxylate cycle. Z, Fermentation. AA, Glycerolipid metabolism. AB, Mitochondrial electron transfer chain and ATP synthase. AC, Nucleotide metabolism. AD, Sugar nucleotide metabolism. AE, Metal handling. AF, S-assimilation. AG, C1-metabolism. AH, Redox regulation/oxidative stress response. AI, Signal transduction. AJ, DNA binding/transcription/chromatin remodeling. AK, RNA processing/binding. AL, Vesicle transport. AM, Pumps and transporters. AN, Protein sorting. AO, Flagellar and basal body proteins. AP, Miscellaneous. AQ, Proteins without functional annotation.
Changes in protein concentrations were also observed for seven out of 10 quantified peptidyl-prolyl cis-trans isomerases of the FK506-binding protein (FKB) and cyclophilin (CYN) families (59) (Table I): whereas cytosolic, TPR-repeat containing FKB62 increased 11.5-fold, 9–47% decreases were observed for cytosolic FKB12, FKB53, and CYN19–2, ER-localized CYN20–1, stromal CYN20–3, and thylakoidal CYN38 (Fig. 4F). Two of the 56 quantified stress-related proteins are homologs of universal stress proteins (USP) (Table I, bin 20.0). Of these, only UspA-like protein with ID 154979 increased by 2.5-fold (Fig. 4G).
Protein Quality Control and Degradation
Fifty-three of the 1116 quantified proteins have functions in protein quality control/degradation. These comprise nine amino-, endo-, and carboxypeptidases, a presequence protease (PREP1), nine subunits of chloroplast proteases of the Clp-, Deg-, or FtsH-type, 10 proteins involved in (de-)ubiquitylation, 21 subunits of the 26S-proteasome, and three other proteases (Table I, bin 21). Surprisingly, changes in protein levels were observed only for two aminopeptidases (IDs 82208 and 139416) and two proteins involved in protein ubiquitylation (UBC2 and CDC48), which decreased by 7.5–23% during heat stress (Fig. 4H).
Protein Biosynthesis
One hundred and sixty one proteins involved in protein biosynthesis were among the 1116 quantified proteins and therefore represent the largest group of quantified proteins (Table I, bin 27). Among these, 40 belong to the large subunit of cytosolic ribosomes and 31 to the small subunit. Another 21 proteins belong to the large subunit of chloroplast ribosomes (four chloroplast- and 17 nuclear-encoded) and 15 proteins belong to the small subunit (five chloroplast- and 10 nuclear-encoded). The fact that we could not quantify mitochondrial ribosomal proteins in the time course (supplemental Table S1) suggests that mitochondrial ribosomes are of low abundance in Chlamydomonas cells. 15 of the 161 proteins involved in protein synthesis are aminoacyl-tRNA ligases, 22 are translation initiation, and 10 are elongation factors. Strikingly, the cytosolic ribosome content appeared to decrease during heat stress: 23 proteins of the large subunit and 22 of the small subunit declined, on average by 15–20% (Figs. 4I and 4J). A reduction in levels of chloroplast ribosomal subunits was observed for 3 proteins of the large subunit, among them chloroplast-encoded rpl2 (Fig. 4K). Heat stress also resulted in decreases of three aminoacyl-tRNA ligases, four initiation, and four elongation factors (by 13–64%) (Fig. 4L).
Nitrogen and Amino Acid Metabolism
Seven of the 1116 quantified proteins are involved in nitrogen metabolism (Table I, bin 14). Protein levels of 5 of them were declining by 14–33% during heat stress, namely of two glutamate synthases (GSF1, GSN1), one glutamine synthetase (GLN1), a protein with ID 194028 similar to the PII regulatory subunit of glutamine synthetase and NADP-malate dehydrogenase (MDH5) (Fig. 4 M). Forty-seven quantified proteins are involved in amino acid metabolism, 46 of them in amino acid biosynthesis and one in valine degradation (Table I, bin 15). The former catalyze reactions for the biosynthesis of the following amino acids: aspartate (3), serine (3), cysteine (1), histidine (4), asparagine (1), lysine/methionine/threonine (9), valine/leucine/isoleucine (8), ornithine/arginine (11), and aromatic amino acids (5). Fourteen of these 47 proteins show reductions in protein levels during heat stress (Fig. 4N). Affected are two proteins involved in serine biosynthesis (two phosphoglycerate dehydrogenases (protein IDs 136279 and 162449), declining by 42 and 21%, respectively); two proteins of the histidine biosynthesis pathway (histidinol dehydrogenase, protein ID 126232, and N-5-phosphoribosyl-ATP transferase (HIS1), declining by 29 and 43%, respectively); asparagine synthase (ID 140252), declining by 14%; 3 proteins leading to lysine/methionine/threonine synthesis (aspartate semialdehyde dehydrogenase (ASSD1), cobalamine-independent methionine synthase (METE), and threonine synthase (THS1), declining by 24%, 59%, and 20%, respectively); 4 proteins involved in valine/leucine/isoleucine biosynthesis (acetohydroxyacid dehydratase (AAD1), acetohydroxyacid isomeroreductase (AAI1), acetolactate synthase (ALSS1), and isopropylmalate synthase (LEU2), declining by 16–25%); and finally, argininosuccinate synthase (AGS1) and carbamoyl phosphate synthase (CMPL1), two enzymes involved in ornithine/arginine biosynthesis, which declined by 12–17%.
Light Harvesting and Photosynthesis
Although we focused on soluble proteins in this study, the kinetic behavior of 31 proteins directly associated with light reactions at the thylakoid membranes could be quantified (Table I, bin 2). 12 of these are peripheral proteins of photosystem II (PSBO, PSBP1/3/6, PSBQ,), photosystem I (PSAN), and the ATP synthase (atpA, atpB, ATPC, ATPD), or soluble proteins (plastocyanin, FNR1). However, 19 of these proteins, associated with one of the photosystems or the cytochrome b6/f complex, do contain membrane-spanning domains (PSBR, PSBW, LHCB4/M1/M3, PSAF, PSAG, LHCA1–3/5–9, LHCSR1/3, petA/O). Within the latter, levels of LHCA3 and LHCSR3 increased 1.2-fold during heat stress, while LHCA5, LHCBM1, and LHCBM3 declined by 17–46% (Figs. 4O–Q). That these membrane proteins are well quantifiable suggests that they are abundant and extracted at constant percentages from light and heavy labeled membranes in the individual samples. However, their apparent differential expression might also be caused by an altered extractability as a consequence of e.g. increased membrane fluidity at elevated temperatures. Or we might detect precursors that are differentially expressed, or unequally well incorporated into the thylakoid membranes during heat stress.
Within the extrinsic membrane proteins, levels of PSAN increased 2.1-fold during heat stress, whereas we observed a 19–31% decline in levels of PSBO and PSBP1 (Figs. 4O and 4P). Moreover, another 8 proteins found to be enriched in thylakoid fractions were quantified (Table I). Among these, a 2.7-fold increase was measured for uncharacterized lumenal polypeptide1 (ULP1), while a protein of the thylakoid enriched fraction (TEF8) and FNR1 decreased by 45 and 9%, respectively (Fig. 4Q).
Interestingly, we observed a decline under heat stress in levels of five out of 14 quantified proteins that are involved in tetrapyrrole biosynthesis (Table I, bin 5): glutamate-1-semialdehyde aminotransferase (GSA, by 18%), delta-aminolevulinic acid dehydratase (ALAD, by 13%), uroporphyrinogen decarboxylases UROD1 and UROD2 (by 31 and 82%, respectively), and Mg-chelatase subunit chlI (CHLI, by 19%) (Fig. 4R). The 41% decrease of the pyridoxine (vitamin B6) biosynthesis protein (PDX1) (Fig. 4S) is likely to result in reduced levels of pyridoxal phosphate, which is an essential cofactor for GSA. In contrast to the apparent decline of proteins generally involved in tetrapyrrole biosynthesis (GSA, ALAD, UROD1, UROD2, and PDX1) and specifically involved in chlorophyll biosynthesis (CHLI), we could not detect significant changes in levels of the five quantified carotenogenic enzymes during heat stress (supplemental Table S3).
Thirty proteins participating in photosynthetic carbon fixation were quantified, nine of which are involved in photorespiration, 13 in the Calvin cycle, and eight in carbon concentration (Table I, bin 3). Heat stress triggered a decline in levels of four of the nine proteins involved in photorespiration, of 10 of the 13 Calvin cycle enzymes, and of five of the 10 proteins participating in carbon concentration. Although this decline was modest in the range of 6–15% for most proteins, it was more dramatic for serine hydroxymethyltransferase SHMT1 (26%), Rubisco activase RCA1 (30%), the small CP12 protein associating with GAPDH and PRK (22%), ribulose phosphate epimerase RPE1 (26%), low CO2 inducible protein LCI23 (87%) and pyruvate phosphate dikinase PPD2 (32%) (Fig. 4T). Heat stress also affected levels of two of eight quantified proteins involved in starch metabolism, namely starch phosphorylases PHOA and PHOB, which declined by 15–17% (Table I, bin 4; Fig. 4U).
Glycolysis and Gluconeogenesis, TCA, OPP, Acetate Metabolism, Fermentation, Glycerolipid Metabolism and Mitochondrial Respiration
Two out of 13 quantified enzymes participating in glycolysis were affected by heat stress (Table I, bin 7): these were 2-phosphoglycerate dehydratase (PGH1) and pyruvate kinase (PYK1), which catalyze the final steps of glycolysis and declined by 10 and 27%, respectively (Fig. 4V). Similarly, levels of four of 14 quantified proteins involved in the TCA cycle declined by 7–32% during heat stress. The affected proteins are E1 subunits of pyruvate and 2-oxoglutarate dehydrogenase (PDC1 and OGD1, respectively), the E2 component of oxoglutarate dehydrogenase (OGD2), and isocitrate dehydrogenase (IDH3) (Table I, bin 8; Fig. 4W). One of the six quantified proteins acting in the oxidative pentose phosphate pathway, 6-phosphogluconolactonase with protein ID 101528, decreased by 20% during heat stress (Table I, bin 9; Fig. 4X).
Ten proteins attributed to acetate metabolism/glyoxylate cycle were quantified (Table I, bin 10). In this group six proteins exhibited modest declines by 9–21% during heat stress (Fig. 4Y). Six of the 1116 quantified proteins are linked to fermentation processes, including four putative alcohol dehydrogenases, a pyruvate dehydrogenase (PDC3), and pyruvate-formate lyase (PFL1). The latter two decreased by 59–67% during heat stress (Table I, bin 11; Fig. 4Z). The presence of enzymes involved in fermentation under aerobic conditions was surprising, but has been observed by others as well (32, 60) and suggested that they only need to be activated under anaerobic conditions.
Twenty-two proteins involved in glycerolipid metabolism were quantified. Four in lipid oxidation (Table I, bin 12). In this group, decreases in protein levels by 18–60% were observed for lipid biosynthesis enzymes biotin carboxylase (BCR1), 3-oxoacyl-[acyl-carrier protein] reductase (KAR1), enoyl-ACP-reductase (ENR1), and acyl-coA-binding protein (ACBP, ID 190684), and for fatty acid oxidation enzyme 3-oxoacyl coA thiolase (ATO1) (Fig. 4AA).
Finally, 12 proteins of the mitochondrial electron transfer chain were quantified (Table I, bin 13). Among these a decrease by 37% during heat stress was observed only for cytochrome c (CYC) (Fig. 4AB).
Nucleotide and Sugar Nucleotide Metabolism
Twenty-six proteins involved in nucleotide metabolism were quantified (Table I, bin 23), but a decline by 29% was detected only for glycinamide ribonucleotide synthetase (ID 175039) (Fig. 4AC). Moreover, four out of 10 quantified proteins involved in sugar nucleotide metabolism exhibited reductions during heat stress. These were dTDP-4-dehydrorhamnose 3,5-epimerase (protein ID 158152; by 24%), GDP-mannose 3,5-epimerase (SNE1; by 15%), UDP-glucose 4-epimerase (SNE6; by 11%), and UDP-glucose:protein transglucosylase (UPTG1; by 18%) (Table I, bin 24; Fig. 4AD).
Metal Handling, S-assimilation, C1-metabolism and Vitamin Biosynthesis
Of the 1116 quantified proteins, five play roles in S-assimilation and three in metal handling (Table I, bins 16 and 17). Proteins declining during heat stress were observed in both categories: an ATP-sulfurylase (ATS1) declined by 60% and a selenium binding protein (SBD1) by 17% (Figs. 4AE and 4AF).
Of the six quantified proteins involved in C1-metabolism, levels of four decreased during heat stress (Table I, bin18): an 80% decrease was observed for formaldehyde dehydrogenase (FDH1) and 12–22% decreases for a putative methylenetetrahydrofolate reductase (protein ID 111330), S-adenosylmethionine synthetase (METM), and S-adenosyl homocysteine hydrolase (SAH1) (Fig. 4AG).
Seven quantified proteins are involved in the biosynthesis of thiamine, riboflavin, and pyridoxamine (Table I, bin 19). We observed a 1.5-fold increase in THI4a protein levels during heat stress, while the pyridoxin biosynthesis protein PDX1 declined by 41% (Fig. 4S).
Redox Regulation and Oxidative Stress Control
Thirty-five proteins related to redox regulation and oxidative stress control were quantified (Table I, bin 22). These include nine proteins of the thioredoxin system, two protein-disulfide isomerases, seven ascorbate oxidases/reductases, seven proteins involved in glutathione metabolism, five peroxiredoxins, three superoxide dismutases, one catalase, and a methionine sulfoxide reductase. Nine of these 35 quantified proteins changed during heat stress, but surprisingly all of them declined (Fig. 4AH). These were cytosolic thioredoxin h (TRXH, by 15%), thioredoxin-related CITRX1 (by 8%), protein disulfide isomerase RB60 (by 15%), three ascorbate peroxidases (CCPR1 by 53%; protein with ID 165193 by 9%; APX1 by 36%), glutathione peroxidase GPX5 (by 17%), Fe-superoxide dismutase FSD1 (by 5%), and methionine sulfoxide reductase MSRA4 (by 39%).
Signal Transduction
Fifty-eight of the 1116 quantified proteins are likely to play roles in signal transduction processes (Table I, bin 28). They comprise nine calcium responsive proteins, 20 protein kinases, a receptor of activated protein kinase, six protein phosphatase components, 10 small G-proteins or associated factors, six nucleotide cyclases and a nucleotide phosphodiesterase, two 14–3-3 proteins, a PAS/PAC sensor domain protein, a phospholipase C, and the blue light receptor phototropin. Changes in protein levels during heat stress were observed for eight of the 58 proteins. Increasing were the calcium-binding protein calreticulin 2 (CRT2; 1.1-fold) and a putative ABC1-like ser/thr kinase (AKC2; 2-fold) (Fig. 4AI). Proteins decreasing were the gamma subunit of a 5′ activated protein kinase (ID 184895; by 7%); the receptor of activated protein kinase C1 (RACK1; by 7%); the regulatory subunit of protein phosphatase 2A (PP2A-2r; by 21%); a small Rab-related GTPase (RABC2; by 25%); and a 14–3-3 protein (FFT1; by 15%).
Proteins Involved in DNA Maintenance, Transcription, and Chromatin Remodeling
Sixty-five of the quantified proteins have putative functions in DNA maintenance, transcription, or chromatin remodeling (Table I, bin 26). Among these, 10 are predicted to play roles in replication/recombination/repair, five in chromosome organization, eight in histone modification, nine in nucleosome assembly, and five in the read-out of histone modifications. Moreover, two are high-mobility group proteins, 14 are putative transcription factors, and six are polymerase subunits. Changes in levels of five of the 65 quantified proteins were observed, where two increased and three decreased during heat stress (Fig. 4AJ). Protein with ID 153995 increased 3.3-fold. It may contain a bacterial SMC domain (structural maintenance of chromosomes). And protein with ID 24073, increasing 1.5-fold during heat stress, is a TATA-binding protein interacting protein with an ATP-dependent DNA helicase domain. The three proteins declining during heat stress are a single-stranded G-strand telomeric DNA-binding protein (GBP1; by 27%), a nucleosome assembly protein (ID 186410; by 33%) and a DEK-like chromatin associated protein (ID 154370; by 16%).
RNA-binding Proteins
Forty-three of the 1116 quantified proteins are putative RNA-binding proteins (Table I, bin 25). They comprise among several others nine splicing factors, five RNA helicases, five nucleolar rRNA processing proteins, the two CRB subunits of the circadian RNA-binding protein CHLAMY1 (61), and two chloroplast proteins controlling translation initiation (RB38 and RB47). Changes during heat stress were detected for six RNA-binding proteins, which all declined (Fig. 4AK). These are putative mRNA splicing factors RSZ22 (by 46%) and UAP56 (by 6%), nucleolar protein NOP1 (by 44%), a glycine-rich RNA binding protein (ID 184151; by 16%) and RB38 and RB47 (by 13–17%), which regulate translation of psbD and psbA transcripts, respectively (62, 63).
Vesicle Traffic, Pumps and Transporters, Protein Traffic, and Cytoskeleton
Twenty-two proteins with functions in vesicle traffic were quantified, among them seven subunits of coat protein I (COP), six of coat protein II (Sec), three involved in vacuolar protein sorting (VPS), two dynamins, clathrin heavy chain and an adaptor protein (Table I, bin 30). Of these 22 proteins, only COPE1 and COPG1 decreased by 31 and 17%, respectively, during heat stress (Fig. 4AL). This is somewhat surprising in light of the heptameric stoichiometry of the coat protein (64). A slight decrease by 18% was also observed for retromer subunit VPS35.
Sixteen proteins being parts of pumps and transporters were quantified, comprising six subunits of the vacuolar ATPase (ATPv), seven putative ABC transporters, and three channels/carriers. In this group, only levels of the A1 and B1 subunits of vacuolar ATPase declined by 10–16% during heat stress (Table I, bin 29; Fig. 4AM).
Nine proteins participating in the sorting of proteins to the chloroplast, mitochondria, ER and nucleus were quantified (Table I, bin 34). Four proteins in this category declined during heat stress: two subunits of the translocon across the inner chloroplast membrane (TIC110 and TIC40; by 19 and 47%, respectively), a signal recognition particle protein (SRP19; by 33%) and the beta subunit of nuclear importin (IPB2; by 24%) (Fig. 4AN). None of the nine quantified cytoskeleton proteins changed significantly during heat stress.
Flagellar and Basal Body Proteins
Thirty-eight of the 1116 quantified proteins are annotated as flagellar or basal body proteins (Table I, bin 1). Given that we have used a strain lacking flagella the identification of the flagella associated proteins appears surprising. Possibly our strain contains rudimentary flagellar structures, or the proteins found belong to stable complexes that are preassembled in the cytosol, but cannot be further processed because of an early block in flagellar biogenesis. Preassembly of the outer dynein arms (ODA) in the cytosol has been reported (65), and accordingly ODA components are among those quantified (ODA2/4/11, FLA14). Alternatively, the proteins found might serve functions at multiple intracellular locations, among them in the flagella. Only four of the 38 proteins associated with flagella or basal bodies were found to decrease during heat stress, which are FAP24 (by 40%), FAP103 (by 11%), FAP278 (by 21%), and PF9 (by 66%) (Fig. 4AO).
Miscellaneous and Nonannotated Proteins
One hundred and forty-five quantified proteins were sorted into the category “miscellaneous,” because they carry out putative functions that do not fit into one of the 47 functional categories, or because annotation was ambivalent or too weak to allow for a clear functional assignment (Table I, bin 32). Of the 145 proteins in this category, two proteins increased and seven proteins decreased during heat stress. The first of the former with protein ID 193531 dramatically increased during heat stress (44-fold) and appears to be de novo synthesized after onset of stress (Fig. 4AP). It is a very large protein of 3606 or 4935 amino acids (according to gene models estExt_fgenesh2_pg.C_440074 or au5.g5194_t1, respectively). The protein contains ankyrin repeats, a p-loop containing NTPase and an IQ-calmodulin binding region. It exhibits some similarity with other conserved large proteins of unknown function harboring a similar domain architecture. The second protein with ID 148088 increased 1.6-fold during heat stress and also is a huge protein of 2942 or 3530 amino acids (according to gene models Chlre2_kg.scaffold_20000230 or au5.g4118_t1, respectively). The protein contains repeats found in chlamydial polymorphic membrane proteins and accordingly contains several predicted transmembrane domains. Moreover, it contains a pectin lyase fold.
The seven proteins of bin 32 decreasing during heat stress are: a putative spermine synthase (SPS1; by 21%); a conserved expressed protein with DUF1499 (CPLD48; by 36%); an iron-sulfur domain protein (ID 190138; by 22%); mannitol dehydrogenase (ID 190503; by 24%); a putative protein required for cell differentiation (RCD2; by 26%); a putative programmed cell death protein six interacting protein (ID 194623; by 20%); and a MoxR-like chaperone putatively involved in metal cluster assembly (ID 179100; by 54%) (Fig. 4AP).
For 115 of the 1116 quantified proteins no functional annotation could be made (Table I, bin 33). Among these, seven proteins declined during heat stress (Fig. 4AQ). Although levels of six of the seven proteins declined by 15–44%, levels of protein with ID 178937 were unaffected during the first 30 min of stress, but appeared to decline to zero at later time points. Two of the seven declining proteins are predicted to be very large (3547 amino acids for IDs 144849 and 3528 amino acids for ID 178937). These and 27 other quantified proteins may represent fusions of several gene models (supplemental Table S1).
Proteins Whose Levels Change During Heat Stress can be Assigned to Five Clusters According to Their Kinetic Behavior
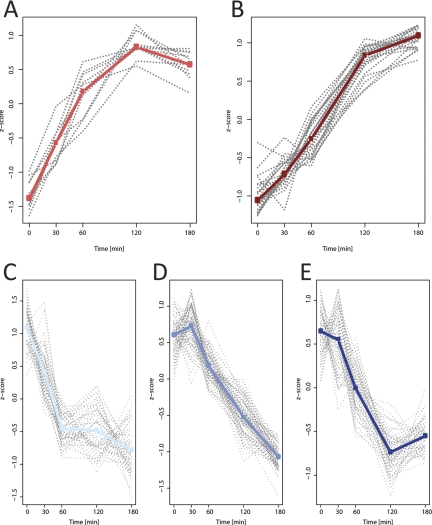
The function of an uncharacterized protein may be inferred if it reacts to changes in environmental conditions with kinetics similar to proteins with known functions. To allow for such analyses, we assigned 230 of the 244 proteins, whose levels were found to change during heat stress, to five different clusters (Fig. 5 and Table I). Missing time points in kinetics of 14 of these 244 proteins led to their exclusion from this analysis. The percentual occupation of clusters within the 47 functional bins is shown in supplemental Fig. S1.
Fig. 5.
Clustering of the kinetic behavior of 230 Chlamydomonas proteins found to be differentially regulated by heat stress. A, UP fast (11 proteins). B, UP delayed (24 proteins). C, DOWN fast (51 proteins). D, DOWN delayed (85 proteins). E, DOWN delayed, into steady state (59 proteins). Information on which protein belongs to which cluster is compiled in Table I. The medians of all proteins within a cluster are shown in bold.
Positive Correlations Between Transcript and Protein Levels are Observed for 19 Out of 24 Selected Target Genes
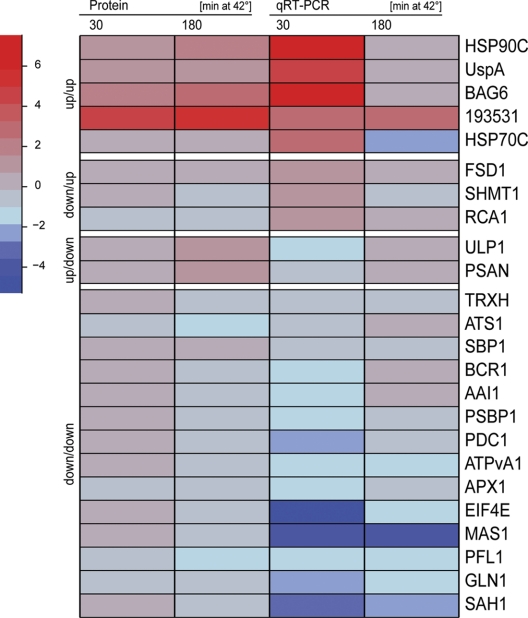
We wondered to which extent the heat stress-induced increase or decrease in the content of a given protein correlated with its transcript levels. To address this question we used qRT-PCR and measured transcript levels of seven (17) genes encoding proteins whose levels appeared to increase (decrease) during heat stress. Based on the direction of change in transcript levels 30 min after heat shock and that in protein levels 180 min after heat shock (transcripts change fast, proteins more slowly), we observed positive correlations between transcript and protein levels for 19 of the 24 chosen targets (Fig. 6 and supplemental Table S6). Negative correlations were observed only for five targets: whereas protein levels of PSAN and uncharacterized lumenal polypeptide 1 (ULP1) appeared to continuously increase during heat stress (Figs. 4O and 4Q), levels of their transcripts decreased 30 min after onset of heat stress, but recovered again after 180 min at 42 °C (Fig. 6 and supplemental Table S6). In contrast, while levels of Fe-superoxide dismutase 1 (FSD1), Rubisco activase (RCA1), and serine hydroxymethyltransferase 1 (SHMT1) decreased during heat stress (Figs. 4T and 4AH), their transcript levels increased. Elevated transcript levels were detected for RCA1 30 and 180 min after onset of heat stress, but for FSD1 and SHMT1 only at the 30-min time point (Fig. 6 and supplemental Table S6).
Fig. 6.
Heat map representation of protein and transcript abundance during heat stress. Relative changes in the abundance of transcript levels of 24 selected target genes in Chlamydomonas cells exposed to heat stress for 30 and 180 min were determined by qRT-PCR using the relative 2−ΔΔCt method. Transcripts of the CBLP2 house-keeping gene were used as internal standard. Values are the average of two biological replicates, each measured in triplicate (all values are compiled in supplemental Table S6). For each of the 24 targets the relative changes in protein levels at the two time points, as determined by quantitative shotgun proteomics, are also shown (values are from supplemental Table 3). Fold changes are log2 transformed.
Heat Stress Results in Immediate, but Reversible Arrest of Cell Division
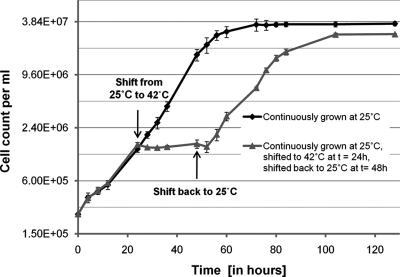
The quantitative protein data revealed that enzymes involved in several different anabolic reactions, like photosynthetic carbon assimilation, acetate metabolism/glyoxylate cycle, N-metabolism, amino acid biosynthesis, protein biosynthesis and subunits of cytosolic ribosomes decrease during heat stress (Table I; Fig. 4). This indicated that Chlamydomonas may not sustain full growth under heat stress conditions. To get an estimate for growth, we measured cell division rates of Chlamydomonas cells at 25 °C and after shifting them to 42 °C (Fig. 7). Although cells doubled with a generation time of ∼8.3 h at 25 °C, cell division ceased immediately after the shift to 42 °C. Although the proteomics data indicated that impaired growth is an acclimation process, we could not rule out that it is caused by the irreversible damage of essential cellular functions. To address this question we continued to monitor division rates of cells that have been incubated at 42 °C for 24 h and were shifted back to 25 °C. Despite the back-shift to 25 °C cell numbers remained constant for several hours after which cells suddenly resumed dividing at the same rate as before exposure to heat stress. Hence, cell cycle arrest of Chlamydomonas cells shifted from 25 °C to 42 °C is indeed an acclimation process. Interestingly, cells that have experienced heat stress for 24 h did not reach the same final cell density as unstressed cells did. We suppose that cells convert part of the acetate within the medium into energy to maintain protein homeostasis during stress, which nonstressed cells can invest into biomass production.
Fig. 7.
Growth curves of unstressed and heat stressed Chlamydomonas cells. Chlamydomonas cells were grown at 25 °C to a cell density of ∼5 × 106 cells/ml and diluted 20-fold in two flasks with fresh TAP medium. Both cultures were incubated at 25 °C and cell densities were measured in triplicate during 130 h after dilution. One culture was kept continuously at 25 °C (black line), the other was incubated in a 42 °C water bath after t = 24 h (gray line) for 24 h and shifted back to 25 °C at t = 48 h. Error bars represent standard deviations from three independent growth experiments.
DISCUSSION
Comparison of Two-dimensional Gel-MS/MS with Quantitative Shotgun Proteomics
In almost all proteomic studies performed to date to dissect the plant heat stress response, protein extracts from stressed and control plants were displayed by two-dimensional-PAGE and proteins in differentially stained spots were identified by mass spectrometry (4–7, 9–15). Advantages of this approach are: (1) information on pI and MW is gained; (2) MS is done only on those proteins that appear to be differentially expressed; (3) if a protein is identified in multiple spots conclusions on extent and potential nature of protein modifications or degradation products may be derived. However, this approach also bears many disadvantages: (1) proteins shifted out of a bulk spot through modifications may be misinterpreted as being downregulated and proteins shifting into a bulk spot by loss of modifications may be misinterpreted as being up-regulated; (2) differential expression may be attributed to the wrong protein, if several proteins are present in the same spot; (3) low abundant proteins, very small or very large proteins, or proteins with extreme basic or acidic pI will not be displayed; (4) tryptic peptides may be only partially recovered from gels (66); (5) the immense work load associated with gel-based approaches precludes extensive time course analyses with multiple biological replicates.
In a previous study, the heat stress response in Arabidopsis was studied by a new approach (8). Here, four plant populations whose proteomes were labeled to different percentages with 15N and heat stressed for different time periods were mixed, separated by SDS-PAGE, and after digestion peptides were analyzed by nanoLC-MS/MS. From which sample peptides originated was determined from their relative shift toward higher masses. This method allowed for the identification and quantification of 267 proteins in a quadruplex sample. However, only four to five time points in a time course can be measured as multiplexing is limited to few 15N dilutions and mass analyzers with very high resolution power (like FT-ICR) are required to separate the isotope envelopes.
In this study we report a novel approach to study the plant heat stress response and used Chlamydomonas reinhardtii to establish it. We generated a uniform 15N-labeled reference proteome which is extracted together with the proteome to be studied from samples of heat stress time courses (Fig. 1). Extracted proteins are digested and peptides analyzed in shotgun mode by nanoLC-MS/MS. Changes in the abundance of proteins in the time course are determined from the relative change of ratios of light to heavy peptides.
The advantages of quantitative shotgun proteomics versus two-dimensional gel-MS/MS approaches are: (1) sample processing is very fast (sampling and processing to the peptide level for an entire time course with three biological replicates may be performed within a week), leaving most of the work to the mass spectrometer and computers for data evaluation and is thus ideal for high-throughput approaches; (2) the early mixing of labeled and unlabeled proteomes maintains the ratio of light to heavy peptides constant despite losses during processing; (3) systematic under- or overestimation of peptide ion intensities are irrelevant, as we measure the change of ratios over time. The pooling of 15N-labeled samples taken during the time course ensures that the uniform standard always contains a certain amount of each protein, even if that protein is absent at some time points; (4) also slight changes in protein abundance are picked up, as peptide identification and quantification is untargeted; (5) the shotgun approach avoids problems with peptide extraction from gels.
However, the shotgun approach also bears some disadvantages: (1) although it is fast, the large complexity limits quantification to the most abundant cellular proteins (Fig. 2B). Additional off-gel fractionation steps e.g. by isoelectric focusing (67, 68) or MudPIT (69) would increase coverage, however, at the expense of increased measuring time and cost; (2) information on pI, MW and degradation products is lost; (3) if a protein is multiply modified under the applied physiological condition, it would be interpreted as declining (if quantification is based on too few peptides). Similarly, if quantities of precursors or splicing isoforms vary under the applied condition, peptides derived from transit peptides or from spliced regions may result in erroneous quantifications. Nevertheless, the ease of the shotgun approach, the large numbers of identified peptides and proteins and the high accuracy of quantification (Fig. 3) clearly indicate that it is the method of choice for quantitative proteomics.
An essential part of the quantitative shotgun approach presented in this study is our in-house developed IOMIQS framework. IOMIQS wraps up several published, sophisticated tools for peptide identification (Mascot, OMSSA, X!Tandem and Sequest) and quantification (XPRESS) and provides interfaces for data exchange between these tools for batch job processing of large raw data sets from quantitative proteomics based on 15N metabolic labeling. 15N-labeling is the labeling method of choice for plant models (24, 28), which as autotrophic organisms are generally capable of synthesizing all amino acids and therefore make SILAC approaches difficult (31). We will provide IOMIQS as a web-based tool in close future (Manuscript in preparation) and expect that it will be of considerable interest especially to the plant community.
Quantitative Shotgun Proteomics Using a Uniform 15N Standard Provides New Insights Into the Stress Response of Chlamydomonas
Regarding the heat stress response of Chlamydomonas (and plant systems in general), do we learn something new by the quantitative shotgun proteomics approach employed here? Assuming that not all of the ∼15,000 genes in Chlamydomonas (16) are constitutively expressed, we estimate that the 1116 proteins quantified in this study represent the 10–20% most abundant proteins of the whole Chlamydomonas proteome. These 1116 proteins cover comprehensively the enzymes involved in the important metabolic pathways of the plant cell and therefore allow deep insights into how these pathways are affected by heat stress (Fig. 8). In fact, we found that levels of 244 of these 1116 proteins significantly changed during heat stress, indicating major responses of the cell to heat stress at the protein level. That these changes are indeed part of an acclimation process and not the prelude of cell death is supported by the observation that Chlamydomonas cells remained fully viable even after experiencing heat stress for 24 h (Fig. 7).
Fig. 8.
Schematic overview of the quantitative shotgun proteomics results. Shown is a crude outline of the biochemical pathways and cellular processes in which most of the 1116 quantified proteins participate. Boxes next to each pathway or process represent the 47 functional bins, bin numbers as listed in Table I are in gray letters. Bins with underlined numbers and names are significantly enriched in proteins changing in response to heat stress with respect to the total protein population. White boxes are proteins that do not change during heat stress or whose changes are not significant. Red boxes represent proteins increasing, blue boxes proteins decreasing during heat stress. The color intensity indicates the magnitude of change.
Surprisingly, of the 244 proteins significantly changing during heat stress levels of only 38 increased. Twenty-five of these 38 proteins are (co-)chaperones of the Hsp100, Hsp90, Hsp70, Hsp60 and sHsp families (70). The bias of the shotgun approach towards abundant proteins indicates that these 25 (co-)chaperones are the major players required for re-establishing protein homeostasis during heat stress (supplemental Table S4). A new steady state appears to be established within the first 2–3 h after onset of stress, as accumulation of chaperones reached a plateau (Hsp100s and Hsp70s) or was close to it (Hsp90s, Hsp60s and sHsps) (Figs. 4A–4E). The most dramatic increases in protein concentrations were detected for (co-)chaperones in cytosol and chloroplast, while increases for chaperones in ER and mitochondria were only moderate or could not be significantly detected (supplemental Table S4). This suggests either that heat sensitive structures are predominantly in nucleocytosol and chloroplast, or that protein (re-)folding capacities in ER and mitochondria are high already under nonstress conditions.
Only levels of cytosolic CLPB1/HSP101 and of the three cytosolic/chloroplast sHsps increased by 13 to 26-fold during stress, suggesting that these are the only chaperones practically synthesized de novo during heat stress and hence appear to play roles exclusively under stress. sHsps are known to be strongly induced by heat shock and to bind partially denatured proteins, thereby preventing irreversible protein aggregation during stress (71). The presence of sHsps in protein aggregates facilitates the access of ATP-dependent chaperones ClpB and Hsp70/Hsp40, which catalyze the refolding of the complexed, denatured proteins back to the native state (72). Accordingly, a plant sHsp has been shown to facilitate protein refolding by Hsp70 in vitro (73) and Arabidopsis HSP101 (the ortholog of Chlamydomonas CLPB1) was shown to be essential for thermotolerance (74). Levels of the other 21 chaperones increased by less than sevenfold indicating that in addition to restoring protein homeostasis under heat stress they also play roles in housekeeping functions. These comprise Hsp90s, Hsp70s, organellar Hsp60s, and the two organelle-targeted ClpBs (CLPB3/4), which in Arabidopsis were reported also to play roles in plastid development (75). The importance of the ClpBs for thermotolerance appears to be underscored by the observation that they all clustered with the group of proteins that react fast to heat stress, while most of the other chaperones clustered with the group of less rapidly up-regulated proteins (Fig. 5; supplemental Fig. S1; Table I). Whether the other 8 proteins in the cluster of fast-responding proteins also are essential for thermotolerance needs to be demonstrated.
Novel Chlamydomonas Heat Stress Inducible Proteins
Two of the 25 (co-)chaperones increasing during heat stress turned out to be putative cochaperones of cytosolic HSP70A (Fig. 4D; supplemental Table S4). The first was annotated as MOT53, which resulted from comparative genomics in order to identify gene products present only in organisms having motile (MOT) cilia (16). In fact, MOT53 is an armadillo-fold containing protein homologous to HspBP1, a protein interacting with cytosolic Hsp70s and acting as a nucleotide exchange factor (76). The second with protein ID 150437 was also strongly up-regulated at the transcript level (Fig. 6 and supplemental Table S6). This protein was not annotated and contains an IQ calmodulin binding motif immediately followed by a BAG domain. This domain organization is also found in Arabidopsis BAG6 (Bcl-2 associated athanogene), one of 7 BAG genes in the Arabidopsis genome (77). The Arabidopsis BAG6 gene was shown to be heat shock inducible and regulated by the HsfA2 transcription factor (77, 78). Apparently, BAG6 and HSP70 upon heat shock reciprocally influence each other's accumulation (79). In yeast and plants, overexpression of Arabidopsis BAG6 induced cell death phenotypes consistent with programmed cell death (80).
In addition to the 25 (co-)chaperones, levels of 13 other proteins were found to increase during heat stress. These are FKB62, USPA, PSAN, LHCA3, LHCSR3, ULP1, THI4, CRT2, AKC2, two DNA-binding proteins, and two proteins of unknown function. In the following, we will briefly discuss potential functions of these novel heat shock inducible proteins.
FKB62 is the only one of 10 quantified immunophilins/cyclophilins whose levels significantly increase during heat stress (by 11.5-fold). FKB62 is a close homolog of Arabidopsis FKBP62/65 (ROF1/2) (59), which were shown to be heat stress inducible, to interact with cytosolic Hsp90 via their TPR domains and to mediate long term acquired thermotolerance (81, 82). Levels of the six other members of the immunophilin and cyclophilin family significantly declined during stress, suggesting that they play roles in housekeeping functions. Accordingly, a function of CYN38 in assembly and maintenance of the PS II supercomplex has been reported recently (83).
Nonannotated protein with ID 154979 was also up-regulated at the transcript level (Fig. 6 and supplemental Table S6) and turned out to be homologous to bacterial UspA. E. coli UspA was shown to be induced by a large variety of stress conditions, like nutrient starvation, heat, oxidants, metals, uncouplers, or ethanol (84). By an unknown mechanism UspA enhances the rate of cell survival during prolonged exposure to such conditions, suggesting that it asserts a general “stress endurance” activity. The heat stress-inducible USPA described here is conserved in the green lineage, but is not orthologous to the ethylene-inducible OsUsp1 described previously (85). Hence, the function of USPA in plants is unknown.
We found PSAN and LHCA3, two components of photosystem (PS) I, to significantly increase during heat stress. PSAN, an extrinsic lumenal protein, was shown to mediate efficient electron transfer between plastocyanin and P700+ (86). In contrast, PS II components LHCBM1, LHCBM3, PSBO, and PSBP1 significantly decreased. PSBO and PSBP1 are part of the oxygen evolving complex known to be sensitive to heat stress (87). These results suggest that heat stress leads to reduced activity of PS II and increased activity of PS I, presumably resulting in increased cyclic electron flow at the expense of linear electron flow. This hypothesis is in line with the observed modest, but significant reduction in levels of ferredoxin-NADP reductase (FNR), which transfers electrons from linear electron transport to NADPH. In fact, increased cyclic electron flow as a consequence of mild heat stress was recently demonstrated in Arabidopsis by spectrometry (88). In this regard it is tempting to speculate that heat stress induced uncharacterized lumenal polypeptide ULP1 might be involved in improving PS I activity, while unknown thylakoid membrane protein TEF8 might be functionally associated with PS II. It is important to note that we observed a negative correlation between transcript and protein levels for PSAN and ULP1 in that transcripts declined, while proteins appeared to increase (Figs. 4O, 4Q, and 6). As quantification of both proteins was based only on single peptides (supplemental Table S1), we can offer 3 possible explanations for this discrepancy: (1) the accumulation of both proteins is regulated at the level of translation initiation or stability. (2) The quantified peptides (or residues close to the tryptic cleavage site) are modified under non-stress conditions and the modification is removed during heat stress. (3) The extractability of both proteins from the thylakoid lumen is increased during heat stress. We are currently raising antisera against both proteins to distinguish between these possibilities.
The slight, but significant increase in levels of the LHCSR3 protein during heat stress was verified by immunoblotting and might point to a requirement for the dissipation of light energy under heat stress conditions to protect the photosynthetic apparatus (89).
The heat stress induced THI4a protein, a homolog of plant THI1, is involved in the biosynthesis of the thiazol moiety of thiamine pyrophosphate. Interestingly, results from yeast show that yeast thi4 mutants are sensitive to (heat stress-induced) oxidative damage of mitochondrial DNA despite the presence of thiamine in the medium (90, 91). These findings point to a role of THI4 in stress protection in addition to its activity in thiamine biosynthesis.
As judged from its C-terminal ‘HDEL’ ER retention motif, Calreticulin 2 (CRT2) is an ER lumenal protein. Calreticulins are involved in regulation of intracellular Ca2+ homoeostasis and ER Ca2+ capacity, thus explaining their involvement in many biological processes (92). Calreticulins are also involved in the folding of newly synthesized proteins and glycoproteins in the ER, presumably explaining why we found levels of CRT2 to slightly increase during heat stress.
AKC2 is a member of the ABC1 family of Ser/Thr kinases, with 17 putative family members in Arabidopsis (93) and at least seven in Chlamydomonas. AKC2 is a GreenCut protein, which are proteins present in green plant/algal lineages, but not in organisms that do not perform photosynthesis (16). Note that AKC2 is not orthologous to the previously described chloroplast-targeted ABC1 family member AtOSA1, which was suggested to be involved in the cadmium and oxidative stress responses (93).
Interestingly, two non-annotated DNA-binding proteins were found to be heat shock inducible: the first protein with ID 24073 is homologous to TIP49 (TATA-box binding protein interacting protein 49), which possesses ATP-dependent DNA helicase activity. Only previously it was demonstrated that human TIP49 and TIP48 via their ATPase activities play a major role in catalyzing H2A acetylation-induced H2A.Z exchange (94). The histone H2A variant H2A.Z is preferentially localized to specific sites within promoter regions and might establish unique promoter architectures for transcription regulation, both positive and negative. The second heat stress induced putative DNA-binding protein contains part of a domain present in bacterial structural maintenance of chromosome (SMC) proteins, which bind DNA and act in organizing and segregating chromosomes for partition (95). Close homologs of the Chlamydomonas SMC-like protein are found only in Volvox and Micromonas.
The two very large (>2900 amino acids) heat stress-induced proteins with unknown function appear not to be fusions of several gene models, as conserved proteins with similar domain architecture exist in other organisms. Protein with ID 193531 was up-regulated ∼44-fold during heat stress, which is even more than observed for CLPB1 and the small HSPs, therefore indicating that this protein is synthesized de novo during heat stress. Also transcript levels of this protein increased during heat stress and, remarkably, stayed high also 180 min after the onset of stress, while transcript levels of the other heat stress-inducible genes had declined by then (Fig. 6 and supplemental Table S6). Unfortunately, we have no hints to the function of this interesting novel heat shock protein.
Enzymes Participating in Many Anabolic Reactions are Down-regulated During Heat Stress
We observed that Chlamydomonas cells immediately ceased to divide after onset of heat stress. After prolonged exposure to 42 °C cells required a lag-phase of several hours before they resumed dividing at the same rates as before stress (Fig. 7). Assuming that the arrest of cell division comes along with a growth arrest the 206 declining proteins appear to be actively degraded rather than diluted out by growth. Protein degradation may enable scavenging of amino acids for the synthesis of chaperones and other proteins required for stress acclimation. The finding that none of 53 quantified proteases and peptidases significantly increased during stress suggests that protein degradation can be accomplished by the degradation capacity present already under nonstress conditions. The reasons for why proteins are actively degraded during stress may be twofold: first, thermosensitive proteins may unfold, aggregate and become degraded because they cannot be folded to the native state at the elevated temperature. Likely examples for such proteins are Fe-superoxide dismutase (FSD1), Rubisco activase (RCA1) and serine hydroxymethyltransferase (SHMT1) as their transcript levels increased—at least 30 min after onset of stress—while protein levels declined (Fig. 6 and supplemental Table S6). By increasing transcription, the cell might attempt to counteract the loss of function of thermosensitive proteins. In fact, Rubisco activase was previously reported to be heat sensitive (96). Second, proteins dispensable during heat stress may be selectively degraded, either by stopping their de novo synthesis, while normal turnover continues, and/or by tagging them for degradation. That this scenario is true for most of the Chlamydomonas proteins found to decline during heat stress is indicated by the observation that in 14 out of 17 cases investigated a decline in protein levels correlated with a decline in transcript levels (Fig. 6 and supplemental Table S6).
Looking at the kinetics of decline (Fig. 5, supplemental Fig. S1 and Table I) unfortunately does not help distinguishing between a selected down-regulation of proteins involved in specific processes as part of an acclimation response and protein degradation necessitated by thermolability. However, this distinction can be made by Gene Ontology (GO) enrichment, which allows estimating whether levels of proteins of the same functional category are significantly altered when compared with proteins of other functional categories (52). As expected, significant GO enrichment for proteins increasing during heat stress was observed only for the bins containing members of the five major chaperone families (Table I). For proteins decreasing during heat stress, significant GO enrichment was observed for the following bins: Proteins of photosynthetic and photorespiratory carbon metabolism, acetate metabolism/glyoxylate cycle, N-metabolism, C1-metabolism and cytosolic ribosomes (SSU and LSU). This suggests that specifically proteins are down-regulated under heat stress that direct carbon flux from CO2 or from acetate present in the medium to protein biosynthesis (Fig. 8). This also correlates with the observed rapid cell division arrest of cells exposed to heat stress (Fig. 7). A reduced rate of protein biosynthesis may directly circumvent problems with de novo folding of nascent chains during heat stress and cell cycle arrest may prevent temperature sensitive processes like DNA replication or chromosome segregation to run out of control. Based on their kinetics of down-regulation during heat stress it might even be possible to distinguish between proteins required for maintaining essential cellular functions during heat stress and those being more dispensable: the former are expected to be present in clusters C and E (entering a steady-state after 3 h at 42 °C), while the latter are expected in cluster D, which comprises proteins that continue to decline even after 3 h of heat stress (Fig. 5).
In fact, heat shock or feeding of yeast cells with the proline analog azetidine 2-carboxylic acid was shown to lead to a block in the G1/S transition and to a reduced expression of ribosomal protein genes (97, 98). Cell cycle arrest appears to be mediated by a heat shock factor dependent reduction of cyclin 1/2 levels. A similar mechanism appears to be responsible for cell cycle arrest also in mammalian cells (99). Hence, cell cycle arrest and reduced protein synthesis appear to be highly conserved responses to heat stress in eukaryotic cells.
Can Results from Chlamydomonas be Compared with Results from Higher Plants?
In our literature search we found 11 studies that addressed the consequences of heat stress on proteome composition in diverse plant models (4–14) (for a review see Neilson et al. (3)). In Table II we have compiled all proteins that were reported to be differentially expressed during heat stress in at least two studies, including our own. Regarding proteins up-regulated during heat stress, a broad consensus between earlier studies and ours was reached for members of the five major chaperone families, for the thiamine biosynthesis protein and for FKBP65 orthologs (the latter were shown to be up-regulated by heat stress also in noncomprehensive studies (81, 82)). Consensus on up-regulated proteins was also reached for cyclophilin A-2, ATP synthase CF1α, glycine dehydrogenase, glutathione-S-transferase, GTP-binding nuclear protein Ran, and granule-bound glycogen starch synthase. However, for neither of them we could detect significant changes. Broad consensus on proteins down-regulated by heat stress between several studies including ours was reached for ferredoxin-NADP(H) oxidoreductase, Rubisco, methionine synthase, adenosylhomocysteinase, and S-adenosylmethionine synthetase. For 12 more proteins reported in earlier studies to be downregulated during heat stress, down-regulation was confirmed for nine by our study.
Table II. Compilation of proteomics results from heat stress experiments performed with other plant species focusing on consensus and contradictory expression profiles. From the study of Ferreira et al. (4) only changes after 6 h of heat stress were considered. (*) Protein data from this study. The asterisk is underlined and in boldface when the protein data positively correlated with qRT-PCR data.
| Protein name | Functional category | References up | References down |
|---|---|---|---|
| sHsps | Small heat shock proteins | (5–8, 12–14), (*) | |
| Chloroplast chaperonins | HSP60s | (4, 5, 11), (*) | |
| Hsp70s | HSP70 and co-chaperones | (5, 6, 8, 9, 11), (*) | |
| Hsp90s and cochaperones | HSP90 and co-chaperones | (4, 6–8, 10, 11), (*) | |
| ClpB3/4, Hsp101 | HSP100 | (5, 8), (*) | |
| Peptidyl-prolyl isomerase AtFKBP65/ROF2 | FKBPs/Cyclophilins | (8), (*) | |
| Cyclophilin A-2 | - “ - | (9, 10) | |
| ATP synthase CF1α | Other proteins associated with light reactions | (4, 5) | |
| Glycine dehydrogenase | Proteins of photosynthetic and photorespiratory carbon metabolism | (5, 11) | |
| Glutathione-S-transferase | Redox regulation/Oxidative stress response | (5, 8, 10) | |
| Thiamine biosynthesis protein | Vitamin biosynthesis | (4, 5, 11), (*) | |
| GTP-binding nuclear protein Ran | Signal transduction | (4, 7, 9, 10) | |
| Granule-bound glycogen starch synthase | Starch synthesis & degardation | (6, 7) | |
| PS I-N | Proteins associated with PS I | (*) | (9) |
| Oxygen-evolving enhancer protein 2 | Proteins associated with PS II | (9) | (12), (*) |
| ATP synthase CF1β | Other proteins associated with light reactions | (11) | (11) (9) |
| Ribulose biphosphate carboxylase/oxygenase activase | Proteins of photosynthetic and photorespiratory carbon metabolism | (7, 11) | (4, 9), (*) |
| Phosphoribulokinase | - “ - | (11) | (5, 9), (*) |
| Glyceraldehyde 3-phosphate dehydrogenase | - “ - | (4, 10, 14) | (9, 11) |
| Chloroplast phosphoglycerate kinase | - “ - | (11) | (9) |
| Triosephosphate isomerase | - “ - | (11) | (12), (*) |
| Transketolase | - “ - | (5, 11) | (4), (*) |
| Chloroplast seduheptulose-1,7-bisphosphatase | - “ - | (11) | (9), (*) |
| NADP-malic enzyme | - “ - | (8) | (4) |
| Ketol-acid reductoisomerase, chloroplast precursor | Amino acid metabolism | (4) | (*) |
| Eucaryotic translation initiation factor 4E | Protein biosynthesis (not ribosome) | (7) | (*) |
| Eucaryotic translation initiation factor 5A | - “ - | (14) | (12) |
| Elongation factor TU | - “ - | (5, 7) | (*) |
| Dehydroascorbate reductase | Redox regulation/Oxidative stress response | (5) | (12) |
| Thioredoxin h | - “ - | (4, 5) | (*) |
| Superoxide dismutase | - “ - | (10) | (*) |
| Ascorbate peroxidase | - “ - | (11) | (*) |
| Pyruvate dehydrogenase, SU E1α | TCA | (5) | (*) |
| Nucleoside diphosphate kinase | Nucleotide metabolism | (5) | (10) |
| Adenosine diphosphate glucose pyrophosphatase | Sugar nucleotide metabolism | (9) | (12) |
| Cytosolic GAPDH | Glycolysis and gluconeogenesis | (9) | (9, 10) |
| Vacuolar ATP synthase | Pumps and transporters | (6, 7) | (*) |
| Glycine-rich RNA-binding protein (Grp1A) | RNA processing/binding | (14) | (*) |
| Plastidic ATP sulphurylase (APS) | S-assimilation | (4, 10) | (*)a |
| Biotin carboxylase | Glycerolipid metabolism | (4) | (*)a |
| Mitochondrial ATP synthase beta | Mitochondrial electron transport chain and ATP synthase | (4) | (5, 7, 11) |
| Mannitol dehydrogenase | Miscellaneous | (14) | (*) |
| Oxygen evolving complex protein 1 (PSBO) | Proteins associated with PS II | (9), (*) | |
| Ferredoxin-NADP(H) oxidoreductase | Other proteins associated with light reactions | (5, 10, 11), (*) | |
| RBCS2 | Proteins of photosynthetic and photorespiratory carbon metabolism | (5, 9) | |
| RbcL | - “ - | (9), (*) | |
| Fructose-bisphosphate aldolase | - “ - | (10), (*) | |
| Serine hydroxymethyltransferase | - “ - | (10), (*) | |
| Glycine/serine hydroxymethyltransferase | - “ - | (9), (*) | |
| Cytosolic glutamine synthetase | N-metabolism | (10), (*) | |
| Methionine synthase (METE) | Amino acid metabolism | (9, 10), (*) | |
| O-Acetylserine (thiol) lyase | - “ - | (4), (*) | |
| FtsH-like protein | Proteases/Peptidases | (4, 9) | |
| 2-phosphoglycerate dehydratase | Glycolysis and gluconeogenesis | (9), (*) | |
| Cytosolic Pyruvate kinase | - “ - | (10), (*) | |
| Malate dehydrogenase (MDH) | TCA | (9, 10) | |
| Cytoplasmic aconitate hydratase | Acetate metabolism /Glyoxylate cycle | (10), (*) | |
| Adenosylhomocysteinase | C1-metabolism | (4, 10), (*) | |
| S-adenosylmethionine synth(et)ase | - “ - | (7, 10, 11), (*) | |
| Glucose-1-phosphate adenyltransferase | Sugar nucleotide metabolism | (6, 7) |
a Two isoforms were detected in our study of which only one decreased significantly.
However, contradictory results were obtained for as many as 29 proteins participating in diverse cellular functions (Table II). Thirteen of these 29 proteins were in contradiction only with our study, 6 were in contradiction with our and earlier studies, and for 10 proteins contradictions were only between earlier studies. In the latter case the respective proteins were not identified in our study, they did not change, or changes were not significant. From this comparison we conclude that the up-regulation of chaperones, FKBP65 orthologs, and thiamine biosynthesis protein and the down-regulation of ferredoxin-NADP(H) oxidoreductase, Rubisco, methionine synthase, adenosylhomocysteinase, and S-adenosylmethionine synthetase are conserved in the green lineage. We can envision two not mutually exclusive explanations for this rather modest consensus of 12 proteomics studies: first, differential expression of several proteins may be species-dependent, e.g. because variations in amino acid sequence had consequences on thermostability of individual proteins, or because chaperoning capacities are higher in more thermotolerant species. Second, as 10 of the 11 earlier studies were based on two-dimensional-gel MS/MS approaches, differential expression of many proteins may have been misinterpreted due to the shortcomings of this approach outlined above. The latter interpretation is supported by the following observations: In a recent study, the proteins phosphoglycerate kinase, transketolase, phosphoribulokinase, sedoheptulose-1,7-bisphosphatase, and glycine dehydrogenase by a two-dimensional-gel MS/MS approach were found to increase during heat stress, while their transcripts declined (11). In contrast, we could demonstrate a positive correlation between levels of transcripts and proteins for 10 of the 19 cases where our and earlier studies were in conflict regarding down-regulated proteins (Fig. 6, Table II and supplemental Table S6) (only two cases gave negative correlations (PSAN and FSD1), the seven others were not tested).
Outlook
We are convinced that quantitative shotgun proteomics using a uniform 15N-labeled standard over all time points in time-course analyses is the method of choice for analyzing proteome dynamics in plant models. In particular, when the IOMIQS framework is available as a web-based platform to the plant community to allow for automatized peptide identification by four search engines, quantification, and data deposit in a database. Integration of these proteomics data with data from transcriptomics, metabolomics, and lipidomics analyses will allow for comprehensive systems biology approaches on plant models.
Regarding the heat stress response in plants we are now able to raise new questions: What are the roles of the many new proteins found to be up-regulated during heat stress, like USPA, BAG6, ULP1, AKC2, the two proteins involved in chromatin remodeling and the two large proteins of unknown function? As this study focused only on soluble proteins and covered membrane proteins like the LHCs only by chance it will certainly be a future challenge to follow membrane proteome dynamics during heat stress.
Acknowledgments
We thank Waltraud Schulze for fruitful discussions, Kaiyao Huang and Joel Rosenbaum (Yale University, New Haven, CT) for the antiserum against HSP70A, Francis-André Wollman (IBPC, Paris, France) for the antiserum against CF1β, and Daniela Strenkert for help with the primer design for qRT-PCR.
Footnotes
* This work was supported by the Max Planck Society and grants from the Deutsche Forschungsgemeinschaft (Schr 617/5–1) and the Bundesministerium für Bildung und Forschung (Systems Biology Initiative FORSYS, project GoFORSYS).
 This article contains supplemental Fig. S1 and Tables S1 to S6.
This article contains supplemental Fig. S1 and Tables S1 to S6.
Abbreviations for Chlamydomonas proteins can be found at http://www.chlamy.org/nomenclature.html
1 The abbreviations used are:
- TAP
- Tris-acetate-phosphate
- ABC
- ATP-binding cassette
- ANOVA
- analysis of variance
- BAG6
- Bcl-2 associated athanogene 6
- BCA
- bicinchoninic acid
- BCL2
- B-cell lymphoma 2
- DUF
- domain of unknown function
- ECL
- enhanced chemiluminescence
- ER
- endoplasmic reticulum
- FKBP
- FK506 binding protein
- GO
- gene ontology
- HSF
- heat shock factor
- Hsp
- heat shock protein
- HspBP1
- heat shock protein 70 binding protein 1
- ID
- identity
- IOMIQS
- integration of mass spectrometry identification and quantification software
- LSU
- large subunit
- mgf
- Mascot generic format
- MOT
- motile flagella
- MS
- mass spectrometry
- MSSQL
- Microsoft structured query language
- MudPIT
- multidimensional protein identification technology
- MW
- molecular weight
- OMSSA
- open mass spectrometry search algorithm
- OPP
- oxidative pentose phosphate pathway
- pI
- isoelectric point
- ppm
- parts per million
- PS
- photosystem
- qRT-PCR
- quantitative real-time PCR
- SILAC
- stable isotope labeling by amino acids in cell culture
- SMC
- structural maintenance of chromosome
- SNF
- sucrose non-fermenting
- SPM
- spectra-to-peptide mapping
- SSU
- small subunit
- TCA
- tricarboxylic acid cycle
- TEF
- thylakoid enriched fraction
- TIP
- TATA-box binding protein interacting protein
- TPR
- tetratricopeptide repeat
- ULP1
- uncharacterized lumenal polypeptide 1
- USPA
- universal stress protein A
- XML
- extensible markup language.
REFERENCES
- 1. Barnabas B., Jager K., Feher A. (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant, Cell Environment 31, 11–38 [DOI] [PubMed] [Google Scholar]
- 2. González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S. S., Grossman A. R. (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22, 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neilson K. A., Gammulla C. G., Mirzaei M., Imin N., Haynes P. A. (2010) Proteomic analysis of temperature stress in plants. Proteomics 10, 828–845 [DOI] [PubMed] [Google Scholar]
- 4. Ferreira S., Hjernø K., Larsen M., Wingsle G., Larsen P., Fey S., Roepstorff P., Salomé Pais M. (2006) Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann. Bot. 98, 361–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee D. G., Ahsan N., Lee S. H., Kang K. Y., Bahk J. D., Lee I. J., Lee B. H. (2007) A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 7, 3369–3383 [DOI] [PubMed] [Google Scholar]
- 6. Majoul T., Bancel E., Triboi E., Ben Hamida J., Branlard G. (2003) Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from total endosperm. Proteomics 3, 175–183 [DOI] [PubMed] [Google Scholar]
- 7. Majoul T., Bancel E., Triboi E., Ben Hamida J., Branlard G. (2004) Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from non-prolamins fraction. Proteomics 4, 505–513 [DOI] [PubMed] [Google Scholar]
- 8. Palmblad M., Mills D. J., Bindschedler L. V. (2008) Heat-shock response in Arabidopsis thaliana explored by multiplexed quantitative proteomics using differential metabolic labeling. J. Proteome Res. 7, 780–785 [DOI] [PubMed] [Google Scholar]
- 9. Xu C., Huang B. (2010) Differential proteomic response to heat stress in thermal Agrostis scabra and heat-sensitive Agrostis stolonifera. Physiol. Plant [DOI] [PubMed] [Google Scholar]
- 10. Xu C., Huang B. (2008) Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 59, 4183–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scafaro A. P., Haynes P. A., Atwell B. J. (2010) Physiological and molecular changes in Oryza meridionalis Ng., a heat-tolerant species of wild rice. J. Exp. Bot. 61, 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Süle A., Vanrobaeys F., Hajós G., Van Beeumen J., Devreese B. (2004) Proteomic analysis of small heat shock protein isoforms in barley shoots. Phytochemistry 65, 1853–1863 [DOI] [PubMed] [Google Scholar]
- 13. Skylas D. J., Cordwell S. J., Hains P. G., R., L. M. (2002) Heat shock of wheat during grain filling: proteins associated with heat tolerance. J. Cereal Sci. 35, 175–188 [Google Scholar]
- 14. Valcu C. M., Lalanne C., Plomion C., Schlink K. (2008) Heat induced changes in protein expression profiles of Norway spruce (Picea abies) ecotypes from different elevations. Proteomics 8, 4287–4302 [DOI] [PubMed] [Google Scholar]
- 15. Koussevitzky S., Suzuki N., Huntington S., Armijo L., Sha W., Cortes D., Shulaev V., Mittler R. (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 283, 34197–34203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernandez E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schroda M. (2004) The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 82, 221–240 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka Y., Nishiyama Y., Murata N. (2000) Acclimation of the photosynthetic machinery to high temperature in Chlamydomonas reinhardtii requires synthesis de novo of proteins encoded by the nuclear and chloroplast genomes. Plant Physiol. 124, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Gromoff E. D., Treier U., Beck C. F. (1989) Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell Biol. 9, 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kloppstech K., Meyer G., Schuster G., Ohad I. (1985) Synthesis, transport and localization of a nuclear coded 22-kd heat-shock protein in the chloroplast membranes of peas and Chlamydomonas reinhardi. EMBO J. 4, 1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schulz-Raffelt M., Lodha M., Schroda M. (2007) Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas Plant J. 52, 286–295 [DOI] [PubMed] [Google Scholar]
- 22. Nover L., Bharti K., Döring P., Mishra S. K., Ganguli A., Scharf K. D. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Analyt. Bioanalyt. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 24. Nelson C. J., Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 [DOI] [PubMed] [Google Scholar]
- 25. Washburn M. P., Ulaszek R., Deciu C., Schieltz D. M., Yates J. R., 3rd (2002) Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74, 1650–1657 [DOI] [PubMed] [Google Scholar]
- 26. Wu C. C., MacCoss M. J., Howell K. E., Matthews D. E., Yates J. R., 3rd (2004) Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76, 4951–4959 [DOI] [PubMed] [Google Scholar]
- 27. Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 28. Engelsberger W. R., Erban A., Kopka J., Schulze W. X. (2006) Metabolic labeling of plant cell cultures with K(15)NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 30. Allmer J., Naumann B., Markert C., Zhang M., Hippler M. (2006) Mass spectrometric genomic data mining: Novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics 6, 6207–6220 [DOI] [PubMed] [Google Scholar]
- 31. Heide H., Nordhues A., Drepper F., Nick S., Schulz-Raffelt M., Haehnel W., Schroda M. (2009) Application of quantitative immunoprecipitation combined with knockdown and cross-linking to Chlamydomonas reveals the presence of vesicle-inducing protein in plastids 1 in a common complex with chloroplast HSP90C. Proteomics 9, 3079–3089 [DOI] [PubMed] [Google Scholar]
- 32. Terashima M., Specht M., Naumann B., Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 34. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 35. Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 36. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schroda M., Vallon O., Wollman F. A., Beck C. F. (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harris E. H. (2008) The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, 2nd Ed., Elsevier/Academic Press, San Diego, CA [Google Scholar]
- 39. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Willmund F., Schroda M. (2005) HEAT SHOCK PROTEIN 90C is a bona fide Hsp90 that interacts with plastidic HSP70B in Chlamydomonas reinhardtii. Plant Physiol. 138, 2310–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lemaire C., Wollman F. A. (1989) The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constitutive subunits. J. Biol. Chem. 264, 10228–10234 [PubMed] [Google Scholar]
- 42. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 43. Eng J., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Mass Spectrom 111, 976–989 [DOI] [PubMed] [Google Scholar]
- 44. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 45. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 46. Han D. K., Eng J., Zhou H., Aebersold R. (2001) Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19, 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 48. Goldschmidt-Clermont M., Rahire M. (1986) Sequence, evolution and differential expression of the two genes encoding variant small subunits of ribulose bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. J. Mol. Biol. 191, 421–432 [DOI] [PubMed] [Google Scholar]
- 49. Schroda M., Vallon O., Whitelegge J. P., Beck C. F., Wollman F. A. (2001) The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13, 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 [Google Scholar]
- 51. Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 52. Rivals I., Personnaz L., Taing L., Potier M. C. (2007) Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23, 401–407 [DOI] [PubMed] [Google Scholar]
- 53. Ward J. H. (1963) Hierarchical grouping to optimize an objective function. J. Amer. Statist. Assoc. 236–244 [Google Scholar]
- 54. Dagda R. K., Sultana T., Lyons-Weiler J. (2010) Evaluation of the Consensus of Four Peptide Identification Algorithms for Tandem Mass Spectrometry Based Proteomics. J. Proteomics Bioinformat. 3, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Drzymalla C., Schroda M., Beck C. F. (1996) Light-inducible gene HSP70B encodes a chloroplast-localized heat shock protein in Chlamydomonas reinhardtii. Plant Mol. Biol. 31, 1185–1194 [DOI] [PubMed] [Google Scholar]
- 56. Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brackley K. I., Grantham J. (2009) Activities of the chaperonin containing TCP-1 (CCT): implications for cell cycle progression and cytoskeletal organisation. Cell Stress Chaperones 14, 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vainberg I. E., Lewis S. A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H. L., Cowan N. J. (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93, 863–873 [DOI] [PubMed] [Google Scholar]
- 59. Vallon O. (2005) Chlamydomonas immunophilins and parvulins: survey and critical assessment of gene models. Eukaryot. Cell 4, 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Atteia A., Adrait A., Brugiere S., Tardif M., van Lis R., Deusch O., Dagan T., Kuhn L., Gontero B., Martin W., Garin J., Joyard J., Rolland N. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evolution 26, 1533–1548 [DOI] [PubMed] [Google Scholar]
- 61. Iliev D., Voytsekh O., Schmidt E. M., Fiedler M., Nykytenko A., Mittag M. (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 142, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schwarz C., Elles I., Kortmann J., Piotrowski M., Nickelsen J. (2007) Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19, 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yohn C. B., Cohen A., Rosch C., Kuchka M. R., Mayfield S. P. (1998) Translation of the chloroplast psbA mRNA requires the nuclear-encoded poly(A)-binding protein, RB47. J. Cell Biol. 142, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beck R., Rawet M., Ravet M., Wieland F. T., Cassel D. (2009) The COPI system: molecular mechanisms and function. FEBS Lett. 583, 2701–2709 [DOI] [PubMed] [Google Scholar]
- 65. Fowkes M. E., Mitchell D. R. (1998) The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell 9, 2337–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Havlis J., Shevchenko A. (2004) Absolute quantification of proteins in solutions and in polyacrylamide gels by mass spectrometry. Anal. Chem. 76, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 67. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 69. Chen E. I., Hewel J., Felding-Habermann B., Yates J. R., 3rd (2006) Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT). Mol. Cell Proteomics 5, 53–56 [DOI] [PubMed] [Google Scholar]
- 70. Schroda M., Vallon O. (2008) Chaperones and Proteases. In: The Chlamydomonas Sourcebook, Second Edition, Elsevier/Academic Press [Google Scholar]
- 71. Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 72. Mogk A., Schlieker C., Friedrich K. L., Schönfeld H. J., Vierling E., Bukau B. (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042 [DOI] [PubMed] [Google Scholar]
- 73. Lee G. J., Vierling E. (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 122, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Queitsch C., Hong S. W., Vierling E., Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee U., Rioflorido I., Hong S. W., Larkindale J., Waters E. R., Vierling E. (2007) The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 49, 115–127 [DOI] [PubMed] [Google Scholar]
- 76. Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 77. Doukhanina E. V., Chen S., van der Zalm E., Godzik A., Reed J., Dickman M. B. (2006) Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281, 18793–18801 [DOI] [PubMed] [Google Scholar]
- 78. Nishizawa-Yokoi A., Yoshida E., Yabuta Y., Shigeoka S. (2009) Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. Biosci. Biotechnol. Biochem. 73, 890–895 [DOI] [PubMed] [Google Scholar]
- 79. Corduan A., Lecomte S., Martin C., Michel D., Desmots F. (2009) Sequential interplay between BAG6 and HSP70 upon heat shock. Cell Mol. Life Sci. 66, 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kang C. H., Jung W. Y., Kang Y. H., Kim J. Y., Kim D. G., Jeong J. C., Baek D. W., Jin J. B., Lee J. Y., Kim M. O., Chung W. S., Mengiste T., Koiwa H., Kwak S. S., Bahk J. D., Lee S. Y., Nam J. S., Yun D. J., Cho M. J. (2006) AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differentiation 13, 84–95 [DOI] [PubMed] [Google Scholar]
- 81. Meiri D., Breiman A. (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 59, 387–399 [DOI] [PubMed] [Google Scholar]
- 82. Meiri D., Tazat K., Cohen-Peer R., Farchi-Pisanty O., Aviezer-Hagai K., Avni A., Breiman A. (2010) Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 72, 191–203 [DOI] [PubMed] [Google Scholar]
- 83. Fu A., He Z., Cho H. S., Lima A., Buchanan B. B., Luan S. (2007) A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kvint K., Nachin L., Diez A., Nystrom T. (2003) The bacterial universal stress protein: function and regulation. Current Opinion Microbiol. 6, 140–145 [DOI] [PubMed] [Google Scholar]
- 85. Sauter M., Rzewuski G., Marwedel T., Lorbiecke R. (2002) The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331 [DOI] [PubMed] [Google Scholar]
- 86. Haldrup A., Naver H., Scheller H. V. (1999) The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 17, 689–698 [DOI] [PubMed] [Google Scholar]
- 87. Allakhverdiev S. I., Kreslavski V. D., Klimov V. V., Los D. A., Carpentier R., Mohanty P. (2008) Heat stress: an overview of molecular responses in photosynthesis. Photosynth. Res. 98, 541–550 [DOI] [PubMed] [Google Scholar]
- 88. Zhang R., Sharkey T. D. (2009) Photosynthetic electron transport and proton flux under moderate heat stress. Photosynth. Res. 100, 29–43 [DOI] [PubMed] [Google Scholar]
- 89. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 90. Medina-Silva R., Barros M. P., Galhardo R. S., Netto L. E., Colepicolo P., Menck C. F. (2006) Heat stress promotes mitochondrial instability and oxidative responses in yeast deficient in thiazole biosynthesis. Research Microbiol. 157, 275–281 [DOI] [PubMed] [Google Scholar]
- 91. Machado C. R., Praekelt U. M., de Oliveira R. C., Barbosa A. C., Byrne K. L., Meacock P. A., Menck C. F. (1997) Dual role for the yeast THI4 gene in thiamine biosynthesis and DNA damage tolerance. J. Mol. Biol. 273, 114–121 [DOI] [PubMed] [Google Scholar]
- 92. Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 93. Jasinski M., Sudre D., Schansker G., Schellenberg M., Constant S., Martinoia E., Bovet L. (2008) AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 147, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choi J., Heo K., An W. (2009) Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 37, 5993–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hirano T. (2005) SMC proteins and chromosome mechanics: from bacteria to humans. Phil. Trans. Roy. Soc. Lon. 360, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Crafts-Brandner S. J., Salvucci M. E. (2000) Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proc. Natl. Acad. Sci. U.S.A. 97, 13430–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Trotter E. W., Berenfeld L., Krause S. A., Petsko G. A., Gray J. V. (2001) Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 7313–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rowley A., Johnston G. C., Butler B., Werner-Washburne M., Singer R. A. (1993) Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 13, 1034–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kühl N. M., Rensing L. (2000) Heat shock effects on cell cycle progression. Cell Mol. Life Sci. 57, 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]