Abstract
Human blood plasma can be obtained relatively noninvasively and contains proteins from most, if not all, tissues of the body. Therefore, an extensive, quantitative catalog of plasma proteins is an important starting point for the discovery of disease biomarkers. In 2005, we showed that different proteomics measurements using different sample preparation and analysis techniques identify significantly different sets of proteins, and that a comprehensive plasma proteome can be compiled only by combining data from many different experiments. Applying advanced computational methods developed for the analysis and integration of very large and diverse data sets generated by tandem MS measurements of tryptic peptides, we have now compiled a high-confidence human plasma proteome reference set with well over twice the identified proteins of previous high-confidence sets. It includes a hierarchy of protein identifications at different levels of redundancy following a clearly defined scheme, which we propose as a standard that can be applied to any proteomics data set to facilitate cross-proteome analyses. Further, to aid in development of blood-based diagnostics using techniques such as selected reaction monitoring, we provide a rough estimate of protein concentrations using spectral counting. We identified 20,433 distinct peptides, from which we inferred a highly nonredundant set of 1929 protein sequences at a false discovery rate of 1%. We have made this resource available via PeptideAtlas, a large, multiorganism, publicly accessible compendium of peptides identified in tandem MS experiments conducted by laboratories around the world.
Blood plasma contains a combination of subproteomes derived from different tissues, and thus, it potentially provides a window into an individual's state of health. Therefore, a detailed analysis of the plasma proteome holds promise as a source of biomarkers that can be used for the diagnosis and staging of diseases, as well as for monitoring progression and response to therapy.
For many years, before the era of proteomics, the classic multivolume reference, The Plasma Proteins by Frank Putnam (1975–1989) (1), provided a foundation for studies of plasma proteins. In 2002, Anderson and Anderson (2) published a review of 289 plasma proteins studied by a wide variety of methods, and quantified primarily with immunoassays, providing an early plasma proteome reference set.
Subsequently, the widespread adoption of liquid chromatography-tandem MS (LC-MS/MS)1 techniques resulted in a rapid increase in plasma proteome-related data sets that needed to be similarly integrated to form a next-generation comprehensive human plasma proteome reference set. In 2002, the Human Proteome Organization (HUPO) launched Phase I of its Human Plasma Proteome Project (PPP) and provided reference specimens of serum and EDTA-, citrate-, and heparin-anticoagulated plasma to 55 laboratories. Eighteen laboratories contributed tandem MS findings and protein identifications, which were integrated by a collaborative process into a core data set of 3020 proteins from the International Protein Index (IPI) database (3) containing two or more identified peptides, plus filters for smaller, higher confidence lists (4, 5). A stringent re-analysis of the PPP data, including adjustment for multiple comparisons, yielded 889 proteins (6).
Meanwhile, in 2004, Anderson et al. (7) published a compilation of 1175 nonredundant plasma proteins reported in the 2002 literature review and in three published experimental data sets (8–10). Only 46 were reported in all four sources, suggesting variability in the proteins detected by different methods, high false positive rates because of insufficiently stringent identification criteria, and nonuniform methods for assigning protein identifications. Shen et al. (11) reported 800 to 1682 proteins from human plasma, depending on the proteolytic enzymes used and the criteria applied for identification; Omenn et al. (4) re-analyzed those raw spectra with HUPO PPP-I search parameters and matched only 213 to the PPP-I core data set. Chan et al. reported 1444 unique proteins in serum using a multidimensional peptide separation strategy (12), of which 1019 mapped to IPI and 257 to the PPP-I core data set. These previous efforts highlight the challenges associated with accurately determining the number of proteins inferred from large proteomic data sets, and with comparing the proteins identified in different data sets.
In 2005, we used a uniform method based on the Trans-Proteomic Pipeline (13) to create the first Human Plasma PeptideAtlas (14), containing 28 LC-MS/MS data sets and over 1.9 million spectra. Using a PeptideProphet (15) probability threshold of p > = 0.90, 6929 peptides were identified at a peptide false discovery rate (FDR) of 12%, as estimated by PeptideProphet's data model, mapping to about 960 distinct proteins. Comparison of protein identifiers with those from studies cited above showed quite limited overlap.
From the 2005 Human Plasma PeptideAtlas, as well as the PPP-I collaboration, we concluded that different proteomics experiments using different samples, depletion, fractionation, sample preparation, and analysis techniques identify significantly different sets of proteins. We decided that a comprehensive plasma proteome could be compiled only by combining data from many diverse, high-quality experiments, and strove to collect as much such data as possible. The resulting 2007 Human Plasma PeptideAtlas (unpublished), encompassing 53 LC-MS/MS data sets, identified 27,801 distinct peptides—four times the number in the 2005 Atlas—and 2738 proteins.
In 2008, Schenk et al. (16) published a high-confidence set of 697 nonimmunoglobulin human plasma proteins based on measuring a single pooled sample on two high-end MS instruments after depletion, prefractionation, and protease inhibition, with stringent validation methods. This highly nonredundant set of proteins likely contains fewer false-positives than any previous MS-derived plasma proteome reference set.
The goal of the present work was to compile a larger human plasma proteome reference set of similar high confidence by creating a new release of the Human Plasma PeptideAtlas incorporating more data than in 2007 and interpreting the data using more stringent criteria. We searched raw data sets submitted to PeptideAtlas and performed peptide validation using a uniform pipeline (Fig. 1), compiled several sets of corresponding protein identifications at different clearly defined levels of redundancy (Fig. 2), and, using a spectral counting technique, provided a rough estimate of concentrations for a highly nonredundant set of protein sequences to guide blood-based diagnostic efforts such as doping using stable isotope-labeled synthetic reference peptides for selected reaction monitoring (SRM) experiments (Fig. 3). The result is a plasma proteome reference set (Fig. 4) (supplemental Tables S3 and S6) containing 1929 highly nonredundant protein sequences at an estimated 1% FDR.
Fig. 1.
Left: Search, analysis, and validation steps for each LC-MS/MS experiment. Spectra were searched against a spectral library or sequence database. The resulting PSMs were then processed using the TPP, including a new component, iProphet, to improve discrimination (see text for details). Right: The PeptideAtlas build process. ProteinProphet combines PSMs passing the FDR threshold for all experiments to create lists of distinct peptides, protein identifications, and protein groups. These data, along with supporting information such as consensus spectra, genome mappings, and proteotypic peptides, comprise a PeptideAtlas build.
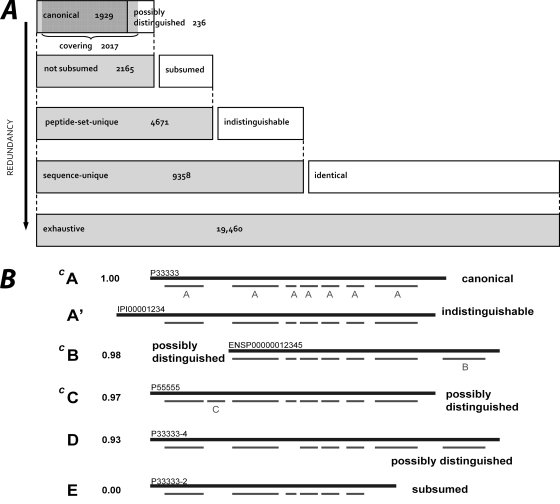
Fig. 2.
A, Six shaded bars (two of which overlap) represent sets of protein identifications at various levels of redundancy under the Cedar scheme. Tallies are for the Human Plasma PeptideAtlas. Beginning at bottom: ●Exhaustive set: contains any protein sequence in the atlas' combined protein sequence database (Swiss-Prot 2010–04 + IPI v3.71 + Ensembl v57.37) that includes at least one identified peptide. ●Sequence-unique set: exhaustive set with exact duplicates removed. ●Peptide-set-unique set: a subset of the sequence-unique set within which no two protein sequences include the exact same set of identified peptides. ●Not subsumed set: peptide-set-unique set with subsumed protein sequences removed (those for which the identified peptides form a proper subset of the identified peptides for another protein sequence). ●Canonical set: a subset of the not subsumed set within which no protein sequence includes more than 80% of the peptides of any other member of the set. Protein sequences that are not subsumed, but not canonical are called possibly distinguished, because each has a peptide set that is close, but not identical, to that of a canonical protein sequence. ●Covering set: a minimal set of protein sequences that can explain all of the identified peptides. B, Peptide-centric illustration of six protein sequences in a hypothetical ProteinProphet protein group, in order of descending ProteinProphet probability. Heavy lines represent protein chains (with invented identifiers); lighter lines represent observed peptides. Vertically aligned peptides are identical in sequence, and one instance of each is labeled with the letter of the highest probability protein to which it maps. A' is indistinguishable from A because it contains exactly the same set of observed peptides; both are equally likely to exist in the sample(s), but A is labeled canonical because its Swiss-Prot protein identifier is preferred. E is subsumed by A because its observed peptides form a subset of A's peptides; it is also subsumed by A', C, and D. Protein sequences B, C, and D are labeled possibly distinguished because the peptide set for each is slightly different from that of A. The three protein sequences with superscript C comprise the smallest subset of sequences sufficient to explain all the observed peptides in the group, and thus belong to the covering set.
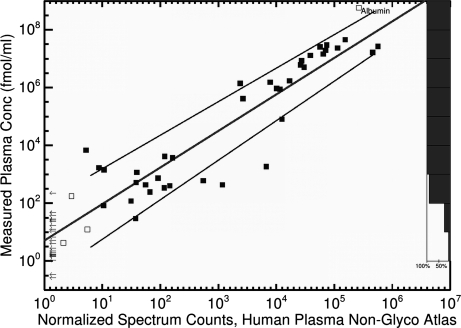
Fig. 3.
Plasma protein concentrations determined using immunoassay and antibody microarray analysis (40) versus normalized spectral counts from the Human Plasma Non-glyco PeptideAtlas, plotted on a log scale. Each small square represents a protein found in both sources. Hollow squares represent proteins that were excluded when drawing the trend line (either depleted (albumin) or fewer than four spectrum counts). The line segments above and below the trend line are fit to the standard deviation of the y axis values computed at intervals of 0.1 (log scale). The arrows on the left represent proteins with reported concentrations in (40) but no spectrum counts. The histogram at the right depicts an estimate of the completeness of the Human Plasma Non-glyco PeptideAtlas as a function of concentration, calculated as the number of points divided by the total number of points and arrows within each decade. See supplemental Fig. S2, for N-Glyco atlas.
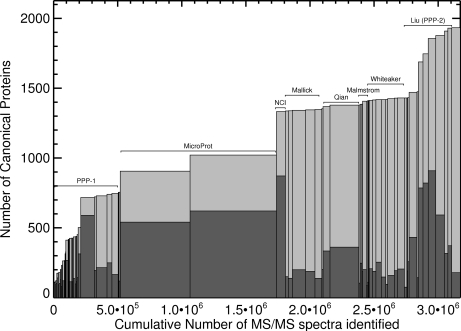
Fig. 4.
Proteins identified by each experiment. Each bar represents one of the 91 experiments, ordered as in supplemental Table S4. Height of dark bar = canonical protein sequences identified per experiment; total height (dark + light) = cumulative tally; width of bar = PSM count. See supplemental Fig. S5, for a similar graph of distinct peptides.
EXPERIMENTAL PROCEDURES
Searching of Data Sets
We collected raw spectra from 91 high-quality LC-MS/MS data sets ((4, 12, 17–21) and several unpublished; supplemental Table S4, Supplemental Data), including 44 from Phase I PPP experiments, 13 from PPP Phase II, the Chan data set, and several from corporate research laboratories. Data from both plasma and serum samples, a variety of sample preparation techniques (depleted/not depleted, various fractionation schemata, use of protease inhibitors, N-linked glycocapture enrichment (22)), and analysis on a variety of instruments were included. All samples were digested with trypsin. Each data set consisted of between one and 38,252 LC-MS/MS runs (median 22) for a total of 48,789 LC-MS/MS runs2. For analysis, we separated the data sets into two groups, glycocapture and nonglycocapture, and later combined the results.
The 69 data sets for the nonglycocapture samples were all selected from ion trap experiments because we wished to search them against an ion trap spectral library. Data were converted to mzXML (23) and searched with SpectraST version 4.0 (24) against a spectral library consisting of the NIST 3.0 human spectral library (261,777 consensus spectra) (25) plus one SpectraST-generated (26) decoy for each NIST spectrum. This library contains consensus spectra derived from actual identified spectra, some of which include missed cleavages and/or modifications. A precursor mass tolerance of 3.0 Th (thomson) was used. See supplemental Data for complete SpectraST parameters.
Analysis and Validation of Search Results
The search results for each experiment were processed using the Trans-Proteomic Pipeline (TPP) (13), as shown in Fig. 1, left (see supplemental Data for TPP parameters used). PeptideProphet (15) computed a probability for each peptide-spectrum match (PSM) for peptides of length seven or greater. iProphet (27) was applied to the PeptideProphet results to improve discrimination by modeling five additional properties of the data beyond those modeled by PeptideProphet, and adjusting peptide probabilities accordingly. The five models are number of sibling searches (rewards or penalizes identifications based on the output of multiple search engines, not applicable here), number of replicate spectra (models the assumption that precursor ions with multiple high probability identifications are more likely to be correct), number of sibling experiments (models the assumption that precursor ions observed in multiple experiments and matched to the same peptide sequence are more likely to be correct), number of sibling ions (rewards peptides identified by precursors with different charges), and number of sibling modifications (rewards peptides identified with different mass modifications).
RefreshParser mapped each PSM to a combined protein sequence database derived from Swiss-Prot 2010–04 including splice variants (28, 29), IPI v3.71, Ensembl v57.37 (30), and cRAP v1.0 (31). In many cases, the exact same protein sequence is included in the combined database multiple times because it is contained in multiple databases and/or because the Ensembl database includes many duplicates. Each PSM was mapped to all protein sequences containing the PSM's peptide sequence; in many cases this resulted in a PSM mapping to multiple protein sequences that are duplicates, splice variants, or paralogs.
For very large data sets, the FDR at the peptide level tends to be much larger than that at the PSM level, and, at the protein level, much larger still (32). Thus, in order to obtain a 1% decoy-estimated protein FDR for the final Human Plasma PeptideAtlas, a stringent PeptideProphet-estimated PSM FDR filter of 0.0002 (corresponding to probability cutoffs ranging from 0.9903 to 0.9998) was applied to each experiment.
ProteinProphet (33) was then run on each experiment, assigning to each distinct peptide the probability of its highest probability PSM, and further adjusting these probabilities using a number of sibling peptides model, which rewards peptides that map to proteins with many identified peptides.
The set of identified peptides for the HsSerum NCI Large Survey experiment (12) was found to contain many peptides that map to yeast but not human. Suspecting yeast contamination, we purged the peptide set for this experiment of all peptides that appear in the yeast genome.
Next, ProteinProphet was run again, this time combining the PSMs for all experiments, to assign probabilities to protein identifications and to group protein identifications with overlapping peptide sets. The PSMs passing threshold for all experiments and their corresponding distinct observed peptides and protein identifications were then compiled (Fig. 1, right) to form a Human Plasma Non-glyco PeptideAtlas build.
Classification of Protein Identifications
It is impossible to generate a definitive list of identified proteins because such a list depends on what is meant by “protein” and on what one considers sufficient evidence for the existence of a specific protein. Further, when the set of identified peptides mapping to the sequence of a protein is identical to, or a subset of, the set of peptides mapping to the sequence of another protein, it is quite possible that both proteins have been observed, but there is no way to determine this from the data.
To partially address this issue, we compiled several sets of protein identifications for this build at different levels of redundancy. For the purpose of this work, the redundancy of a set of protein identifications is the extent to which the set contains more sequences than necessary to reasonably explain all of the data. Note that redundancy is different from confidence and that the different redundancy levels do not correspond to different confidence levels. The four most useful protein sets are described below; the first two, exhaustive (most redundant) and canonical (least redundant), are used extensively throughout this report. All sets are summarized in Fig. 2, and a detailed explanation, including examples, is given in supplemental Data (Cedar). We have named this scheme Cedar to capture the somewhat tree-like pyramidal shape shown in Fig. 2A.
The exhaustive set includes any entry from the combined protein sequence database (Swiss-Prot 2010–04 + IPI v3.71 + Ensembl v57.37) to which any identified peptide maps. This highly redundant set includes multiple copies of identical sequences. To determine whether a protein corresponding to a particular identifier exists in the Human Plasma PeptideAtlas, one must check whether that identifier is in the exhaustive set. Assuming the identifier is in Swiss-Prot 2010–04, IPI v3.71, or Ensembl v57.37, its presence in the atlas' exhaustive set indicates that the protein sequence includes a peptide sequence in the atlas.
The canonical set is a highly nonredundant set of protein sequences explaining nearly all of the identified peptides and it serves as a proteome reference set. It includes the highest probability protein sequence from each ProteinProphet protein group, called the group representative. Swiss-Prot protein sequences are preferred for inclusion because of Swiss-Prot's comprehensive sequence documentation and curation, and because Swiss-Prot, a subset of Uniprot, is now considered to contain one entry for each currently known human protein coding gene (34), with a total of 20,251 entries in the 2010–10 release, of which 13,329 have evidence at the protein level [www.uniprot.org]. When a protein group includes protein sequences for which the peptide set has less than 80% overlap with the group representative, we label those sequences canonical as well (see supplemental Data (Cedar) for algorithm and justification for 80% threshold). The size of the canonical set is a conservative estimate of the number of distinct proteins observed. It is important to understand that the label canonical is with respect to a particular data collection; a protein sequence that is identified in two atlas builds may be labeled canonical in one collection and something else in another.
The possibly distinguished set includes protein sequences that have one or more peptides distinguishing it from all protein sequences in the canonical set, but with these peptides comprising fewer than 20% of the total number of identified peptides in each protein, making the case for independent existence less strong.
Finally, the covering set is a near-minimal set sufficient to explain all of the peptide identifications (see supplemental Data, (Cedar), for algorithm). This set consists of almost all of the canonical protein sequences plus some of the possibly distinguished protein sequences, and is usually somewhat larger than the canonical set. It is useful for assigning a “parent” protein identification to each identified peptide, as is necessary for estimating FDR using Mayu (32) or computing the empirical observability score described in Empirical Observability Score below.
See supplemental Data (Cedar) for settings to apply when using the PeptideAtlas web interface to obtain these protein sequence sets.
Analysis of N-linked Glycopeptide-enriched Samples
We then analyzed the 22 data sets from samples prepared using N-linked glycocapture enrichment. Our aim in including these samples was to detect low-abundance proteins, many of which are N-glycosylated. Sample preparation was as described in (35). Briefly, N-linked glycoproteins were conjugated to a solid support using hydrazide chemistry, proteins were digested with trypsin on the support, N-linked glycopeptides were optionally labeled with stable isotopes, and formerly N-linked glycosylated peptides were specifically released via peptide-N-glycosidase F (PNGase F) resulting in a N-linked glycopeptide-rich fraction, but with the glycans removed. Within this fraction, all asparagines (N) that had been glycosylated in the intact protein were now present as aspartic acid (D) residues. This fraction was analyzed via LC-MS/MS. We did not search against the NIST spectral library because it does not contain glycopeptide spectra; instead, data were searched with X!Tandem version 2009.10.01.1 (36) using a score plug-in implementing the COMET (k-score) function (13) against a target database consisting of IPI 3.54 (75,428 sequences) plus one decoy per target sequence generated by a random scrambling of each tryptic peptide in place. Peptides appearing in more than one target sequence were scrambled identically each time. The mass tolerance for precursor ions ranged from –2.1 to +4.1 Daltons. Modifications were allowed on cysteine (fixed, mass depending on modification used) and methionine (variable, oxidation). A maximum of two missed cleavages was allowed. A standard protocol (37) was employed so that D-[not P]-[S/T]-containing spectra could be matched against N-[not P]-[S/T]-containing database sequences. Briefly, we substituted the letter B for N in all N-glycosite motifs in the database (B commonly denotes “N or D” but in this context denotes “N presumed to be glycosylated”), then searched with the mass of B fixed to the mass of D, allowing B to behave as D during the search. Instances of B were then converted back to N in the search results. See supplemental Data for complete X!Tandem parameters. It is important to note that, whereas this computational protocol allows identification of peptides containing the (possibly de-amidated) N-glyco motif, it does not confirm whether the site was indeed glycosylated in the sample.
We then constructed a Human Plasma N-Glyco PeptideAtlas using the same methods as above, but with a PSM FDR threshold of 0.00002, yielding a protein-level FDR of 0.56%. We chose this threshold to achieve our goal of a 1% protein FDR after combining with the 0.86% FDR Non-glyco build described above. It was not practical to use identical FDRs for the component builds because even fine adjustments in the PSM FDR for a component build sometimes resulted in coarse changes in the protein FDR for the combined build.
Concentration Estimation
Spectral counting was applied to roughly estimate the absolute concentration of the group representative for each ProteinProphet protein group in each atlas. Spectral counting rests on the observation that the PSM count for a peptide correlates linearly with its molar concentration in the sample (38). We applied a simplification of the APEX method described by Lu and coworkers (39). For each protein sequence, i, identified in the Human Plasma Non-glyco PeptideAtlas, we begin with a ProteinProphet-adjusted count SCi of all PSMs that map to that protein sequence (ProteinProphet adjusts the actual PSM count downward according to the degeneracy of the peptide-protein mappings). SCi is then normalized by scaling it to the total number of available tryptic peptides. Specifically, we calculate a normalization factor, NFi, by dividing the number of tryptic peptides of length seven or more resulting from an in silico digestion, NTPi, by 25, which is very roughly the average number of tryptic peptides per protein sequence across the whole proteome, and then calculate the normalized spectrum count NSCi by dividing SCi by that factor:
 |
We calibrated the concentration scale to the published concentrations of individual proteins. In Fig. 3, we plot NSCi versus concentrations determined via immunoassay and antibody microarray in (40). In many cases, these concentrations reflect multiple isoforms and/or cross-reacting proteins.
Using the slope S and y-intercept K from this calibration plot, we then calculated an estimated concentration Ci for each group representative protein sequence with NSCi > = 4 (smaller counts have been found unreliable for this purpose (41)) in the Human Plasma Non-glyco PeptideAtlas:
Concentrations were converted to mass units (ng/ml) for storage in PeptideAtlas using molecular weights calculated from amino acid sequence.
The distance of the standard deviation curve from the trend line at the center of each decade on the x axis (between 100 and 101, between 101 and 102, etc.) was recorded as an uncertainty factor for the normalized PSM counts in that decade, ranging from less than 5× at high concentrations to 13× at low concentrations. See supplemental Table S2, for complete listing.
To estimate concentrations in the N-Glyco Plasma PeptideAtlas, we adjusted the technique to account for the N-linked glycopeptide enrichment. About half of the distinct peptides in this atlas contain the N-glycosite motif (N - [not P] - [S/T]), indicating a potential N-linked glycosylation site. Thus, in calculating the normalization factor NFi, we take into account both the total number of tryptic peptides in each protein sequence NTPi and the number of peptides containing the N-glycosite motif, NTGPi:
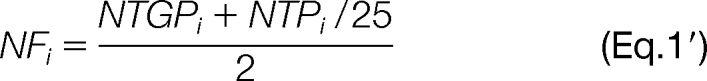 |
The calibration plot is shown in supplemental Fig. S2. Also shown is a plot correlating the estimated concentrations in the Non-glyco and N-Glyco atlases for protein sequences appearing in both (supplemental Fig. S3).
Construction of Combined PeptideAtlas Plasma Build
Finally, we combined the PSMs and peptides from these two atlases to form a Human Plasma PeptideAtlas build that includes results from all 91 plasma (or serum) experiments, both nonglycocapture and glycocapture. We ran ProteinProphet on the combined set of experiments and created protein identification sets as described above. Estimated concentrations from the Non-glyco atlas were used for protein sequences with values in both contributing atlases.
False Discovery Rate
Mayu, a software tool for estimating false discovery rates of protein identifications in large-scale data sets (32), was applied to each component atlas and to the combined atlas to estimate the protein-level FDR. Mayu implements a refinement of the common decoy-counting approach, improving accuracy by taking into consideration the size of the data set, the number of tryptic peptides in each protein, and proteome coverage.
Manual Validation of Single-PSM Protein Identifications
Three hundred fifty-seven of the 1999 canonical protein identifications that emerged after combining the Non-glyco and N-Glyco builds were supported by only a single PSM (supplemental Table S5). We manually validated these, judging a PSM positively for each of the following: identifications to b- or y-ions or neutral losses for nearly all of the tallest peaks in the spectrum, at least one series of four or more consecutive highly abundant fragment ions of the same type (b or y, preferably y) and charge state, highly abundant fragments corresponding to cleavage N-terminal to proline and C-terminal to aspartic acid (42), no missed tryptic cleavages, fragments observed above the noise level for at least 50% of the expected ions, internal positively charged amino acids to account for precursor charges above +2, and N-terminal acetylation only for peptides at N terminus of protein. We discarded 70 PSMs that failed to fulfill these criteria to the extent that, in our opinion, they had a greater than 10% chance of being incorrect identifications. So the user can view these 70 discarded identifications, they were not removed from the component (N-Glyco and Non-glyco) atlases.
Construction of Combined PeptideAtlas Plasma Build at 5% Protein FDR
We repeated the above atlas construction procedure to obtain a combined build with a protein FDR of ∼5%, as follows. We applied a PSM FDR of 0.001 to the non-glyco data and a PSM FDR of 0.0007 to the glyco data, obtaining in each case a build with a Mayu protein FDR of 4.8%. These were combined to yield a “Human Plasma FDR 5% PeptideAtlas” build (actual Mayu protein FDR is 4.6%). Single-PSM identifications were not manually validated, and all that passed our computational criteria were retained in this build.
Empirical Observability Score
For all peptides in each atlas, we calculated an empirical observability score (43), defined as the ratio of the number of samples in which a given peptide is observed divided by the number of samples in which the parent protein sequence is observed. For example, if peptide X is seen in five different samples and its parent protein sequence is observed in 10 samples, the empirical observability score is 0.5.
RESULTS
Size, Confidence, and Completeness of Proteome Reference Set
The 2010 Human Plasma PeptideAtlas, constructed from 91 LC-MS/MS data sets, contains 1929 canonical protein sequences with an estimated protein FDR of <0.98% (Fig. 4 and supplemental Table S3). As described under “Experimental Procedures,” the set of canonical protein sequences is a highly nonredundant protein sequence set with no protein sequence sharing more than 80% of its observed peptides with any other member of the set. This criterion may exclude closely related protein family members. The list of 1929 protein identifiers, along with estimated concentrations and number of supporting PSMs and distinct peptides, is given in supplemental Table S6.
Each canonical protein sequence in the Human Plasma PeptideAtlas is supported by between 1 and 521 distinct observed peptides (mean = 11, median = 3) and between 1 and 390,366 PSMs (mean = 1720, median = 10). Of the 1929 canonical protein sequences, 1642 are supported by more than one PSM, and 1313 are supported by more than one distinct peptide.
High Confidence Identifications
The previous Human Plasma PeptideAtlas contained 27,801 peptides mapping to 2738 nonredundant proteins (protein redundancy level corresponding roughly to that of the covering list for the 2010 atlas). The 2010 Human Plasma PeptideAtlas contains fewer identified peptides and protein sequences, but these fulfill much more stringent criteria. For lack of suitable methods, we could not accurately estimate the protein FDR of the 2007 build, but, because it was constructed using a very liberal PSM probability cutoff, its protein FDR is no doubt much higher than the 1% of the 2010 build. The high confidence level for the 2010 build, and the ability to estimate it, were accomplished by the inclusion of more data plus four methodological improvements:
1. Spectral Library Searching
Non-glyco query spectra were compared against consensus spectra derived from real spectra, rather than against theoretical spectra. This resulted in better discrimination between true and false identifications (24), giving a higher number of identifications at any given PSM FDR.
2. iProphet
A new component of the Trans-Proteomic Pipeline, iProphet (27), increased discrimination between true and false identifications in our atlas builds by modeling five additional properties of the data beyond those modeled by PeptideProphet (see Experimental Procedures).
3. PSM FDR Cutoff
For the 2007 build, we used a PSM probability cutoff of 0.9. Because experiments vary in the quality of their results, this uniform probability cutoff admitted a higher proportion of false PSMs for poor experiments than for high quality experiments. Therefore, here we instead used a PSM FDR threshold, adjusted to achieve a protein FDR of about 1% for the combined build. Corresponding probability cutoffs were one to three orders of magnitude more stringent than those for the 2007 build, admitting many fewer PSMs per experiment.
4. Decoy-estimated Protein FDR
By including decoys in our target database we were able to apply the recently developed tool Mayu to accurately estimate the protein FDR.
Single-PSM Protein Identifications
Three hundred fifty-seven single-PSM protein identifications passed our rigorous computational pipeline. This subpopulation has a Mayu decoy-estimated protein FDR of 3.4%. Because decoy analysis may under-estimate protein FDR (44) and because single-PSM protein identifications are especially in need of extra validation, we manually examined all 357 and discarded 70 that we believed had a greater than 10% chance of being false identifications (see details under “Experimental Procedures”). Assuming that the FDR decreased as a result, we state that the final protein FDR is <0.98%. Building a protein FDR 1% atlas excluding all single-PSM protein identifications would have included more multiple-PSM identifications, but fewer total protein identifications (see supplemental Data, Choice of Atlas Stringency Level, for analysis).
Estimated Concentrations
Although plasma protein concentration is dependent on the individual organism, its disease state, and its physiological status at time of sample collection, concentrations of relatively abundant proteins under relatively normal conditions generally do not vary more than an order of magnitude (45), and it is useful to have a rough estimate of normal protein concentration for purposes such as the spiking in of reference peptides for SRM or other targeted MS measurements. Spectral counting has been established as a reliable method for both relative (38, 41) and absolute (39, 46) quantification of proteins based on LC-MS/MS data. Comparison of raw spectral counts has previously been used for relative quantification between plasma samples (47). Here, following a simplification of the APEX method of Lu and coworkers (39), we obtain absolute quantification by normalizing spectral counts to adjust for the number of observable tryptic peptides per protein and by calibrating to previously measured protein concentrations.
The estimated concentrations are rough estimates and should not be mistaken as accurate quantitative values. Above 1 μg/ml, they are generally accurate within one to two orders of magnitude. Sixty-eight canonical proteins not used for spectral counting calibration appear in the Hortin et al. 2008 review of abundant plasma proteins (48); the estimated concentrations for 51% of these proteins are within a factor of 10 of the mean of the concentration range reported by Hortin et al., and 94% are within a factor of 100. Of course, there are considerable uncertainties about these previously published measurements as well, because of the nature of immunoassays and antibody specificities. Further, even a precise concentration measurement in a specific sample would not generate a general statement about plasma protein abundances because of the variation among individuals.
To the extent that these roughly estimated values are accurate, the very large amount of data contributes to the accuracy. Data heterogeneity may also add to accuracy by allowing averaging over many diverse samples. However, it may also detract because of the variety of instruments and settings used. Dynamic exclusion settings, for example, can be optimized to amplify the spectral counts of low abundance proteins relative to the counts for high abundance proteins (49); the mixing of results in PeptideAtlas from experiments with optimized and nonoptimized settings could reduce accuracy. Obviously, estimated concentrations are sensitive to the calibration values used; see supplemental Fig. S4, for illustration.
Concentration is estimated for the group representative for each protein group (as long as its ProteinProphet-adjusted PSM count is at least 4). This concentration must be considered to be shared among all protein sequences in the group, usually splice isoforms or paralogs. Some atlas data come from analysis of depleted samples; concentrations for depleted proteins (including those proteins that are inadvertently removed during the depletion process, see (50)) are underestimated. Plasma concentrations for cellular proteins can be elevated when there is nonphysiological breakage of blood cells during sample collection and preparation. The sum of the estimated concentrations for hemoglobin-α and -β, 71 μg/ml, is close to the 100–200 μg/ml measured in serum in (51), suggesting that such breakage was minimal.
The estimated concentrations based on spectral counting of the canonical protein sequences in the Human Plasma PeptideAtlas span 6.5 orders of magnitude, ranging from 1.6 × 106 ng/ml for serum albumin (P02768) down to 0.5 ng/ml for CEACAM1 (P13688, Carcinoembryonic antigen-related cell adhesion molecule 1). Serum albumin is known to be the most abundant protein in plasma with a normal range of 3.4–5.4 × 107 ng/ml (2, 52), but is underestimated in the atlas because of depletion.
N-linked Glycoproteome
Many proteins of medical interest, such as receptor extracellular portions, transport molecules, and hormones, are N-linked glycosylated. Ninety percent of the 485 canonical protein sequences in the Human Plasma N-Glyco PeptideAtlas contain the N-glycosite motif (N - [not P] - [S/T]) and are thus likely N-linked glycoproteins. However, we emphasize that our computational protocol does not confirm N-linked glycosylation for any particular protein and the N[115] notation does not indicate a confirmed deamidation site. See supplemental Data, Computational pipeline for N-Glyco atlas does not confirm glycosylation, for details. The employed glycocapture technique also purifies some non-glycosylated peptides, presumably through nonspecific binding to the base bead used (Table I).
Table I. Prevalence of N-glycosite motif in the component builds of the Human Plasma PeptideAtlas.
| Fraction of identifications containing N-glycosite motif |
||
|---|---|---|
| Non-glyco PeptideAtlas | N-Glyco PeptideAtlas | |
| Distinct peptides | 3.9% | 53% |
| Canonical proteins | 72% | 90% |
Eighty-six canonical protein sequences from the Human Plasma N-Glyco PeptideAtlas, all with estimated concentrations ≤ 25 ng/ml, are not found in the Human Plasma Non-glyco PeptideAtlas exhaustive set (supplemental Table S7). All but one of the 125 peptides mapping to these proteins has an N-glycosite motif. Because glycosylation hinders LC-MS/MS identification, it is highly unlikely that these peptides would be identified without the glycocapture protocol, which results in removal of glycan groups. Indeed, only four are present in the NIST 3.0 spectral library we used to search the non-glyco data.
Of the 86 proteins, 31% have no spectra in the NIST 3.0 library and thus could not have been identified by spectral searching. However, as explained in supplemental Data Completeness of spectral library searching, we expect very few additional canonical proteins would be identified were we to perform database searching on the non-glyco data. Therefore, we conclude that for nearly all of these 86 proteins, the reason they are missing from the Non-glyco atlas is because they are of low abundance in plasma.
Missed Cleavages; Semitryptic and Nontryptic Peptides
Both SpectraST and X!Tandem were set to allow matches to peptides with missed cleavages and/or peptides that were not fully tryptic; see Table II for tallies. Missed cleavages and nontryptic termini are usually penalized by ProteinProphet; penalties vary depending on the software's statistical modeling of each data set.
Table II. Summary of peptide and protein features. Fully-tryptic= both termini are either tryptic cleavage sites ([K/R][not-P]) or at the N- or C-terminus of a mapped protein. Some nontryptic termini can be explained by signal peptide cleavage or other natural post-translational cleavage. See text for further details.
| Human Plasma PeptideAtlas peptide and protein features | ||
|---|---|---|
| Distinct peptides (total 20,433) | ||
| contain missed cleavage ([K/R][not P]) | 6164 | 30% |
| of the 6164, matching Monigatti & Bernd pattern (53) | 4227 | 69% |
| not fully tryptic | 6180 | 30% |
| Canonical proteins (total 1929) | ||
| observed only in one or more trauma experiments | 455 | 24% |
| immunoglobulins | 124 | 6% |
| keratins | 36 | 2% |
Contribution of Trauma Experiments
Our intention was to catalog the proteins found in normal plasma; therefore, the 2010 Human Plasma PeptideAtlas almost exclusively includes experiments on samples originating from individuals with no known disease state or other unusual condition. Six included experiments, however, were performed on a pool of six severe trauma patients plus one healthy subject (20), and we found that 455, or 24%, of the canonical protein sequences in the Atlas were observed only in one or more of these experiments and not in any of the other 85, raising the question of whether these proteins are trauma-specific. The 455 are all low abundance with at most 145 PSMs per protein; we believe that most of these are difficult-to-detect proteins present in normal plasma, rather than trauma-specific proteins, because of the advanced technology employed in the experiments (depletion of the 12 most abundant plasma proteins; fractionation into cysteinyl and noncysteinyl peptides, glyco- and nonglycopeptide; separation of each fraction into 30 subfractions using strong cation exchange, analysis on a Thermo LTQ instrument), which yielded nearly twice the peptide identifications per experiment when compared with earlier experiments from the same lab (depletion of only six most abundant plasma proteins and, in some cases, a less advanced instrument (Thermo LCQ) employed) (54).
Keratins and Immunoglobulins
Some keratins are common contaminants in proteomic sample processing, and the immunoglobulins are a very large class of plasma proteins consisting of similar interchangeable subunits, so one may wish to omit these classes of protein sequences from a plasma proteome reference set. We estimated the number of canonical protein sequences that belong to these classes (Table II) by counting those identified as immunoglobulins or keratins in their descriptions, plus all those in the same protein group as such a sequence. We counted all keratins, even those that are internal cytokeratins and not skin contaminants. We did not count sequences annotated as immunoglobulin-like or immunoglobulin-related. Omitting these immunoglobulins and keratins leaves 1769 canonical protein sequences not belonging to these classes.
Evidence for Multiple Splice Isoforms and Single Nucleotide Polymorphisms
The human section of Swiss-Prot is curated to contain one entry per protein-coding gene, each with descriptions for known splice isoforms. There is only one Swiss-Prot entry for which two splice isoforms exist in the canonical set, and it is only this protein, mannan-binding lectin serine protease 1, which we confidently claim is present in more than one splice isoform in human plasma. Twelve additional Swiss-Prot alternative splice isoforms are noted as possibly distinguished; we are less confident that these are present as distinct isoforms because possibly distinguished protein sequences have only a small amount of peptide evidence distinguishing them from their canonical counterparts. Further, 131 canonical protein sequences come from the IPI or Ensembl databases, indicating that each includes at least one observed peptide that is not mappable to any Swiss-Prot entry. These might represent single nucleotide polymorphisms or sequence errors (see IPI00887739 in Complement C3 group in supplemental Fig. S1 for an example), or protein-coding genes or splice variants not described in Swiss-Prot.
DISCUSSION
Composition and Completeness of Proteome Reference Set
Our set of 1929 canonical protein sequences, by far the largest published so far at this confidence level, includes the highest concentration proteins as well as nearly complete coverage of the phosphoproteome described in (55) (details in Table III). Still, we believe it is far from a complete catalog of the human plasma proteome. First, our reference set and the MS-derived lists in Table III are all biased toward proteins that are readily detectable by MS techniques; proteins missing from one list are likely to be missing in the others, so coverage of the lists in Table III is not indicative of complete proteome coverage.
Table III. PeptideAtlas coverage of published protein lists. Percentages in column B are relative to numbers in column A.
| Plasma protein identification list | Total proteins | (A) In PeptideAtlas combined protein sequence databasea | (B) In Human Plasma PeptideAtlas exhaustive set (of those in column A) | |
|---|---|---|---|---|
| Hortin et al. highly abundant plasma proteins (48) | 150 | 126 | 126 | 100% |
| Kuzyk et al. moderate/high concentration plasma proteins reproducibly detectable by LC-MRM/MS analysis of whole tryptic digests; nearly all reported as potential disease markers (56) | 45 | 45 | 45 | 100% |
| Carrascal et al. LC-MS/MS derived plasma phosphoproteins with peptide identified by >1 search engine (55) | 44 | 44 | 43b | 98% |
| Schenk, et al. LC-MS/MS derived plasma proteins (non-immunoglobulin) (16) | 697 | 554 | 503 | 91% |
| Polanski and Anderson potential cancer biomarkers (57) | 1261 | 1054 | 326 | 31% |
a Swiss-Prot 2010–04 + IPI v3.71 + Ensembl v54.37.
b A single spectrum for the missing phosphoprotein was identified by our computational pipeline, but discarded during our manual validation process.
Other evidence suggests we are not close to full coverage of even the LC-MS/MS-observable proteome. Mayu analysis of the 5% protein FDR plasma atlas (see Experimental Procedures) shows that at least 410 correct identifications are excluded from the 1% protein FDR Human Plasma PeptideAtlas by its stringent FDR threshold. Fig. 4, showing the accumulation of canonical proteins as additional identified MS/MS spectra that were added to the Human Plasma PeptideAtlas, also suggests that we are not near complete coverage. The PPP-I data contributed about 38% of the total canonical proteins. Growth after PPP-1 was shallow, then jumped with the addition of experiments employing extensive depletion and fractionation and high mass accuracy instruments (19, 20). The curve will asymptotically approach the total number of proteins detectable with the techniques used, but is not yet nearing that limit.
In 2008, Schenk et al. published a plasma proteome reference set (16) of comparable confidence to ours (see supplemental Data, Comparison of confidence level with Schenk et al., for details). Of the 697 nonredundant, nonimmunoglobulin protein identifiers in (16), 51 are in our combined protein sequence database, but not in the Human Plasma PeptideAtlas exhaustive set, meaning that we identified no peptides for them (see supplemental Table S8). If the Schenk et al. data were added to the Human Plasma PeptideAtlas, most or all of these would appear in the resulting canonical list. This supports our conclusion that more data, preferably from different laboratories using different sample sources, depletion techniques, and preparation techniques, will continue to add significant numbers of high confidence protein sequences to the human plasma proteome.
Because we searched the nonglycocapture data against a spectral library and not against a sequence database, we only identified peptides that had been previously seen in LC-MS/MS experiments and included in the NIST spectral library. However, the NIST library is extremely comprehensive, including most of the human data in the PeptideAtlas (from plasma and many other sources), so nearly all human-derived spectra identifiable with a sequence search engine with standard parameters will be identified with our spectral library search. Very few, if any, canonical proteins would be added to the Atlas were we to incorporate sequence database search results (see supplemental Data, Completeness of spectra library searching for analysis).
Multi-tiered Protein Identifications: Alternatives for Comparison of Data Sets
As described under “Experimental Procedures,” we created our exhaustive identification set by mapping all identified peptides to a combined protein sequence database containing many sequences repeated identically or with only slight variations. Removing redundancy from such a set is always a problem in interpreting proteomics data, and no standard methods have been agreed upon.
In considering this issue, it is critical to understand that virtually no protein identification list for a given data set can be considered definitive. Once one eliminates exact duplicates, the process of removing redundancy necessarily involves choices that are somewhat arbitrary, as described in (58), and is at odds with the preservation of identifications consistent with the data. In most cases, a highly nonredundant list is necessarily a model or example list, each entry of which may represent several proteins that are as likely, or almost as likely, to exist in the sample. In particular, we emphasize that we do not claim to have definitive evidence for any of the specific isoforms in our canonical set; rather, we claim that, for each protein sequence in the set, there exists either that protein or a closely related one in at least one of the samples.
For some purposes, such as estimating the number of distinct proteins revealed by the data, a highly nonredundant protein identification set is desired. For other purposes, such as comparison with a nonredundant list for another proteome, filtering by molecular weight or pI, or selection of peptides for SRM experiment design, redundancy is desirable. As described under “Experimental Procedures,” we created several different protein sequence sets that could be used, alone or in combination, for different purposes.
Multitiered schemes are not novel and have been implemented in many proteomics studies. For example, the core data set for PPP-I contained 3020 protein sequences, but alternative threshold criteria were used to generate several other sets including a set of 889 protein sequences using very restrictive criteria with an adjustment for multiple hypothesis testing (6), roughly analogous to our canonical set, and an unintegrated set of 15,710 protein sequences based on only a single peptide, roughly analogous to the exhaustive set defined here.
With the current work, we make two contributions in this area. First, we present Cedar, a protein identification classification scheme based on the freely available ProteinProphet and applicable to any search results that can be converted to mzML (59) or mzXML (23). Protein identifications generated for different data sets using Cedar can be easily and meaningfully compared against each other. Although software is not yet available to automate Cedar, all steps except for the manual validation of single-PSM identifications are clearly defined and reproducible, and we propose Cedar as a standard for the community, including the HUPO Human Proteome Project.
Second, we assert that when evaluating the overlap between the protein identifications for two proteomics data sets, it is essential to map to the same sequence databases and to compare the highly nonredundant (Cedar's canonical) set for one against the maximally redundant (Cedar's exhaustive) set for the other. Otherwise, the overlap will be under-reported. For example, Schenk et al. reported that 242 of their 697 high confidence identifications were found on the HUPO high-confidence list. We compared their identifications against the exhaustive set for an atlas we built from most of the HUPO PPP-I data (see “Results”, Single-PSM Protein Identifications) and found an overlap of 362 identifications, which is 50% more.
Spectral Counting
Spectral counting has allowed us to provide rough estimates for protein concentrations in the Human Plasma PeptideAtlas down to 0.54 ng/ml, but even lower estimated concentrations are achievable. By including about 100 times as many PSMs as currently included, we could reach 7 × 10−3 ng/ml, the lowest concentration measured by antibody-based methods in (40). See supplemental Data, Completeness of spectral counting, for analysis.
Estimated concentrations in ng/ml, along with uncertainty factors, are now available in PeptideAtlas. Again, these are rough estimates and should not be mistaken as accurate quantitative values. Experimentally measured concentrations from (40), (57), and (Anderson, N. L. (2007) private communication) are provided as well. We plan to apply this same spectral counting method to atlas builds for other subproteomes such as human urine, mouse plasma, and various organ or cell type data sets that we acquire. Our goal is to develop a quantitative PeptideAtlas reflecting protein expression in multiple organs, cell types, and biofluids in health and disease.
Uses of the Human Plasma Peptide
Atlas Biomarker Discovery
Polanski and Anderson in 2006 (57) published a review of candidate cancer biomarkers listing 1261 proteins believed to be differentially expressed in patients with various cancers. Literature search revealed only 274 to be reported in plasma, but 326 appear in the Human Plasma PeptideAtlas exhaustive set (Table III), skewed toward lower concentrations. Those identified in (46) as “high priority” for biomarker development (about one-third of the 326) are listed in supplemental Table S9.
Experiment Design for Targeted Proteomics
When a protein is observed in a sample that is analyzed with LC MS/MS techniques, some of the protein's peptides are observed many times, whereas others are not observed at all, despite being in the observable mass range and otherwise having attributes consistent with MS analysis (60, 61). Several algorithms that attempt to predict observability based on sequence attributes have been put forward (39, 60, 61); these are heavily influenced by the data with which they are trained. As noted under “Experimental Procedures”, for all peptides in the Human Plasma PeptideAtlas, we calculated an empirical observability score that does not rely on prediction algorithms; however, it is highly dependent on MS data collection parameters, including dynamic exclusion settings, as in (49).
Because shotgun-style experiments of complex samples will always miss many proteins, especially low concentration proteins, a targeted approach in which the mass spectrometer selects only peptides contained within specific proteins of interest should be more successful, reproducible, and time efficient. Using the PeptideAtlas web interface, one can select peptides based on the empirical observability score and other attributes, such as number of observations, number of protein mappings, missed cleavages, semitryptic, or multiple genome locations, and present these as an inclusion list for the mass spectrometer.
PeptideAtlas includes several other features to support SRM experiment design. For peptides belonging to proteins not yet observed in PeptideAtlas, observability scores based on sequence attributes are calculated. When multiple spectra exist for the same precursor ion, they are combined to generate a consensus spectrum that can be visualized by the user. Transition lists can be generated automatically from these consensus spectra according to user-specified rules. For absolute protein abundance measurements, the estimated protein concentrations described above allow one to spike in synthetic reference peptides at concentrations similar to those expected in the sample. These features and others are described in (43). Finally, we and others are in the process of systematically generating reference fragment ion spectra from synthetic peptide libraries using the triple quadrupole instruments used for SRM measurements and we will make these publicly accessible as verified transition sets (63, 64).
CONCLUSION
PeptideAtlas is an integral part of the ProteomeXchange infrastructure for HUPO initiatives and other worldwide data submissions (figure published in (65)), together with the ProteomeCommons.org Tranche distributed file-sharing system (66) and the EBI PRIDE (67) database. PRIDE contains the investigators' original data sets; PeptideAtlas consolidates the raw data of individual studies into re-analyzed proteome reference sets. A significant aspect of PPP-II is the establishment of a standard method for the submission of data to the ProteomeXchange consortium. It is the policy of PPP-II that all published plasma data be submitted to Tranche or PRIDE, from which it will be stored in Tranche and incorporated into the PeptideAtlas.
The PeptideAtlas approach described here provides a framework for the continued analysis of human and other complex proteomes. Soon, MS/MS data interpretation based on translated genomes will be replaced by rich spectral libraries derived from both natural and synthetic peptide information, which outperform current database searching strategies. Already, there is a complete spectral database for the entire yeast proteome (64) and mouse and human are being completed (Deutsch et al., in preparation; Kusebauch et al., in preparation).
The 2010 Human Plasma PeptideAtlas, a comprehensive collection of high-confidence peptide and protein identifications, contains well over twice as many protein sequences as any previous collection at a similar confidence level. With estimated concentrations and a multitiered protein identification scheme, it is a useful resource for biomarker discovery and SRM experiment design. Peptide identifications, protein identifications, estimated concentrations, and raw data in mzXML (23) format are all offered freely to the public at www.PeptideAtlas.org.
Acknowledgments
We thank all those who contributed raw proteomics data to the Human Plasma PeptideAtlas, including Phillip A. Wilmarth and Larry L. David of Oregon Health and Science University; Wei-Jun Qian, Tao Liu and Richard Smith of Pacific Northwest National Laboratory; Jeffrey R. Whiteaker of the Fred Hutchinson Cancer Research Center, Manuel C. Peitsch of the University of Basel (formerly of Novartis), King C. Chan, David A. Lucas, and Thomas P. Conrads of the National Cancer Institute, and all participants in the PPP Phase I project (see (4)). Thanks to Rich Johnson for manual spectrum evaluation, to Leigh Anderson for concentration data, and to Leigh Anderson and Laura Beretta for critical review of the manuscript.
Footnotes
* This work has been funded in part by the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract No. N01-HV-28179 and grants R44HG004537, PM50 GMO7U547/Center for Systems Biology, NIH-NHGRI ARRA HG005805, NIH U54 DA021519, PB0ES017885, NIH GM087221, grant 31003A_130530 from the Swiss National Science Foundation, European Research Council (grant #ERC-2008-AdG 233226), and the Systems Biology Initiative of the Grand Duchy of Luxembourg.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S9.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S9.
2 This total includes two extraordinarily large experiments together comprising 45,160 runs.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography-tandem MS
- HUPO
- Human Proteome Organization
- PPP
- Plasma Proteome Project
- IPI
- International Protein Index
- TPP
- Trans-Proteomic Pipeline
- FDR
- false discovery rate
- PSM
- peptide-spectrum match
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Putnam F. W. ed. (1975–1989) The Plasma Proteins, 2nd Ed., Academic Press, New York [Google Scholar]
- 2. Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 3. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 4. Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. (2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core data set of 3020 proteins and a publicly-available database. Proteomics 5, 3226–3245 [DOI] [PubMed] [Google Scholar]
- 5. Omenn G. Editor (2006) Exploring the Human Plasma Proteome, Wiley-VCH, New York, NY [Google Scholar]
- 6. States D. J., Omenn G. S., Blackwell T. W., Fermin D., Eng J., Speicher D. W., Hanash S. M. (2006) Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat. Biotechnol. 24, 333–338 [DOI] [PubMed] [Google Scholar]
- 7. Anderson N. L., Polanski M., Pieper R., Gatlin T., Tirumalai R. S., Conrads T. P., Veenstra T. D., Adkins J. N., Pounds J. G., Fagan R., Lobley A. (2004) The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell Proteomics 3, 311–326 [DOI] [PubMed] [Google Scholar]
- 8. Pieper R., Gatlin C. L., Makusky A. J., Russo P. S., Schatz C. R., Miller S. S., Su Q., McGrath A. M., Estock M. A., Parmar P. P., Zhao M., Huang S. T., Zhou J., Wang F., Esquer-Blasco R., Anderson N. L., Taylor J., Steiner S. (2003) The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 3, 1345–1364 [DOI] [PubMed] [Google Scholar]
- 9. Adkins J. N., Varnum S. M., Auberry K. J., Moore R. J., Angell N. H., Smith R. D., Springer D. L., Pounds J. G. (2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell Proteomics 1, 947–955 [DOI] [PubMed] [Google Scholar]
- 10. Tirumalai R. S., Chan K. C., Prieto D. A., Issaq H. J., Conrads T. P., Veenstra T. D. (2003) Characterization of the low molecular weight human serum proteome. Mol. Cell Proteomics 2, 1096–1103 [DOI] [PubMed] [Google Scholar]
- 11. Shen Y., Jacobs J. M., Camp D. G., 2nd, Fang R., Moore R. J., Smith R. D., Xiao W., Davis R. W., Tompkins R. G. (2004) Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal. Chem. 76, 1134–1144 [DOI] [PubMed] [Google Scholar]
- 12. Chan K. C., Lucas D. A., Hise D. et al. (2004) Serum/Plasma Proteome. Clinical Proteomics 1, 101–225 [Google Scholar]
- 13. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deutsch E. W., Eng J. K., Zhang H., King N. L., Nesvizhskii A. I., Lin B., Lee H., Yi E. C., Ossola R., Aebersold R. (2005) Human Plasma PeptideAtlas. Proteomics 5, 3497–3500 [DOI] [PubMed] [Google Scholar]
- 15. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 16. Schenk S., Schoenhals G. J., de Souza G., Mann M. (2008) A high confidence, manually validated human blood plasma protein reference set. BMC Med. Genomics 1, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qian W. J., Monroe M. E., Liu T., Jacobs J. M., Anderson G. A., Shen Y., Moore R. J., Anderson D. J., Zhang R., Calvano S. E., Lowry S. F., Xiao W., Moldawer L. L., Davis R. W., Tompkins R. G., Camp D. G., 2nd, Smith R. D. (2005) Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell Proteomics 4, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whiteaker J. R., Zhang H., Eng J. K., Fang R., Piening B. D., Feng L. C., Lorentzen T. D., Schoenherr R. M., Keane J. F., Holzman T., Fitzgibbon M., Lin C., Zhang H., Cooke K., Liu T., Camp D. G., 2nd, Anderson L., Watts J., Smith R. D., McIntosh M. W., Paulovich A. G. (2007) Head-to-head comparison of serum fractionation techniques. J. Proteome Res. 6, 828–836 [DOI] [PubMed] [Google Scholar]
- 19. Liu T., Qian W. J., Gritsenko M. A., Camp D. G., 2nd, Monroe M. E., Moore R. J., Smith R. D. (2005) Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 4, 2070–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu T., Qian W. J., Gritsenko M. A., Xiao W., Moldawer L. L., Kaushal A., Monroe M. E., Varnum S. M., Moore R. J., Purvine S. O., Maier R. V., Davis R. W., Tompkins R. G., Camp D. G., 2nd, Smith R. D. (2006) High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell Proteomics 5, 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Armandola E. A. (2003) Proteome profiling in body fluids and in cancer cell signaling. Med. Gen. Med. 5, 18. [PubMed] [Google Scholar]
- 22. Zhang H., Yi E. C., Li X. J., Mallick P., Kelly-Spratt K. S., Masselon C. D., Camp D. G., 3rd, Smith R. D., Kemp C. J., Aebersold R. (2005) High Throughput Quantitative Analysis of Serum Proteins Using Glycopeptide Capture and Liquid Chromatography Mass Spectrometry. Mol. Cell. Proteomics 4, 144–155 [DOI] [PubMed] [Google Scholar]
- 23. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 24. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 25. NIST Peptide Mass Spectral Libraries, http://peptide.nist.gov. National Institute of Standards and Technology
- 26. Lam H., Deutsch E. W., Aebersold R. (2010) Artificial decoy spectral libraries for false discovery rate estimation in spectral library searching in proteomics. J. Proteome Res. 9, 605–610 [DOI] [PubMed] [Google Scholar]
- 27. Shteynberg D., Deutsch E., Lam H., Eng J., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (submitted) iProphet: Improved statistical validation of peptide identifications in shotgun proteomics. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boutet E., Lieberherr D., Tognolli M., Schneider M., Bairoch A. (2007) UniProtKB/Swiss-Prot. Methods Mol. Biol. 406, 89–112 [DOI] [PubMed] [Google Scholar]
- 29. Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., Gasteiger E., Martin M. J., Michoud K., O'Donovan C., Phan I., Pilbout S., Schneider M. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hubbard T. J., Aken B. L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. (2009) Ensembl 2009. Nucleic Acids Res. 37, D690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. cRAP, Common Repository of Adventitious Proteins; http://www.thegpm.org/cRAP
- 32. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 34. The Uniprot Consortium (2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 37, D169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 36. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 37. Atwood J. A., 3rd, Sahoo S. S., Alvarez-Manilla G., Weatherly D. B., Kolli K., Orlando R., York W. S. (2005) Simple modification of a protein database for mass spectral identification of N-linked glycopeptides. Rapid Commun. Mass. Spectrom. 19, 3002–3006 [DOI] [PubMed] [Google Scholar]
- 38. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 39. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 40. Haab B. B., Geierstanger B. H., Michailidis G., Vitzthum F., Forrester S., Okon R., Saviranta P., Brinker A., Sorette M., Perlee L., Suresh S., Drwal G., Adkins J. N., Omenn G. S. (2005) Immunoassay and antibody microarray analysis of the HUPO Plasma Proteome Project reference specimens: systematic variation between sample types and calibration of mass spectrometry data. Proteomics 5, 3278–3291 [DOI] [PubMed] [Google Scholar]
- 41. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 42. Kapp E. A., Schütz F., Reid G. E., Eddes J. S., Moritz R. L., O'Hair R. A., Speed T. P., Simpson R. J. (2003) Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal. Chem. 75, 6251–6264 [DOI] [PubMed] [Google Scholar]
- 43. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen Y., Tolić N., Hixson K. K., Purvine S. O., Pasa-Tolić L., Qian W. J., Adkins J. N., Moore R. J., Smith R. D. (2008) Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal. Chem. 80, 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Craig W. Y., Ledue T. B., Ritchie R. F. (2001) Plasma Proteins: Clinical Utility and Interpretation, Foundation for Blood Research, Scarborough, ME [Google Scholar]
- 46. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 47. Shen Z., Want E. J., Chen W., Keating W., Nussbaumer W., Moore R., Gentle T. M., Siuzdak G. (2006) Sepsis plasma protein profiling with immunodepletion, three-dimensional liquid chromatography tandem mass spectrometry, and spectrum counting. J. Proteome Res. 5, 3154–3160 [DOI] [PubMed] [Google Scholar]
- 48. Hortin G. L., Sviridov D., Anderson N. L. (2008) High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin. Chem. 54, 1608–1616 [DOI] [PubMed] [Google Scholar]
- 49. Zhang Y., Wen Z., Washburn M. P., Florens L. (2009) Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 81, 6317–6326 [DOI] [PubMed] [Google Scholar]
- 50. Gundry R. L., Fu Q., Jelinek C. A., Van Eyk J. E., Cotter R. J. (2007) Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clin. Appl. 1, 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Na N., Ouyang J., Taes Y. E., Delanghe J. R. (2005) Serum free hemoglobin concentrations in healthy individuals are related to haptoglobin type. Clin. Chem. 51, 1754–1755 [DOI] [PubMed] [Google Scholar]
- 52. The Merck Manual of Diagnosis and Therapy (2006) 18th Ed., Merck Research Laboratories, Whitehouse Station, NJ [Google Scholar]
- 53. Monigatti F., Berndt P. (2005) Algorithm for accurate similarity measurements of peptide mass fingerprints and its application. J. Am. Soc. Mass. Spectrom. 16, 13–21 [DOI] [PubMed] [Google Scholar]
- 54. Liu T., Qian W. J., Strittmatter E. F., Camp D. G., 2nd, Anderson G. A., Thrall B. D., Smith R. D. (2004) High-throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 76, 5345–5353 [DOI] [PubMed] [Google Scholar]
- 55. Carrascal M., Gay M., Ovelleiro D., Casas V., Gelpi E., Abian J. Characterization of the human plasma phosphoproteome using linear ion trap mass spectrometry and multiple search engines. J. Proteome Res. 9, 876–884 [DOI] [PubMed] [Google Scholar]
- 56. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Polanski M., Anderson N. L. (2007) A List of Candidate Cancer Biomarkers for Targeted Proteomics. Biomarker Insights 1, 1–48 [PMC free article] [PubMed] [Google Scholar]
- 58. Adamski M., Blackwell T., Menon R., Martens L., Hermjakob H., Taylor C., Omenn G. S., States D. J. (2005) Data management and preliminary data analysis in the pilot phase of the HUPO Plasma Proteome Project. Proteomics 5, 3246–3261 [DOI] [PubMed] [Google Scholar]
- 59. Deutsch E. (2008) mzML: a single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 60. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 61. Kuster B., Schirle M., Mallick P., Aebersold R. (2005) Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6, 577–583 [DOI] [PubMed] [Google Scholar]
- 62. Tang H., Arnold R. J., Alves P., Xun Z., Clemmer D. E., Novotny M. V., Reilly J. P., Radivojac P. (2006) A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics 22, e481–488 [DOI] [PubMed] [Google Scholar]
- 63. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 64. Picotti P., Lam H., Campbell D., Deutsch E., Roest H., Sun Z., Rinner O., Schmidt A., Shen Q., Frei A., Wollscheid B., Aebersold R. (submitted) A complete mass spectrometric reference map for the analysis of the yeast proteome. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Omenn G. S., Aebersold R., Paik Y. K. (2009) 7(th) HUPO World Congress of Proteomics: launching the second phase of the HUPOPlasma Proteome Project (PPP-2) 16–20 August 2008, Amsterdam, The Netherlands Proteomics 9, 4–6 [DOI] [PubMed] [Google Scholar]
- 66. Falkner J. A., Andrews P. C. (2007) Tranche: Secure Decentralized Data Storage for the Proteomics Community. J. Biomol. Tech. 18, 3 [Google Scholar]
- 67. Vizcaino J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hamilton J. A., Benson M. D. (2001) Transthyretin: a review from a structural perspective. Cell Mol. Life Sci. 58, 1491–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sato T., Susuki S., Suico M. A., Miyata M., Ando Y., Mizuguchi M., Takeuchi M., Dobashi M., Shuto T., Kai H. (2007) Endoplasmic reticulum quality control regulates the fate of transthyretin variants in the cell. EMBO J. 26, 2501–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]