Abstract
A membrane cell for hydrogen and deuterium exchange on-line with mass spectrometry has been developed to monitor protein-protein interactions and protein conformations. It consists of two channels separated by a semipermeable membrane, where one channel carries the protein sample and the other deuterium oxide. The membrane allows transfer of deuterium oxide into the sample flow. The labeling time is controlled via the flow rate in the sample channel. This cell was validated against three models commonly used in hydrogen-deuterium exchange mass spectrometry: monitoring of folded and unfolded states in a protein, mapping the protein secondary structure at the peptide level, and detection of protein and antibody interactions. The system avoids the conventionally used sample dilution and handling, allowing for potential automation.
Hydrogen/deuterium exchange (HDX)1 measured by mass spectrometry (HDX-MS) is a powerful tool to probe the structure and dynamics of proteins in solution (1–4). It is based on the principle that amide hydrogens of the protein backbone can be exchanged for deuterons when the protein is exposed to a deuterated solvent. The rate of exchange is influenced by the extent to which the amide hydrogens are involved in hydrogen bonding, thus reflecting binding interactions, secondary structure elements, and solvent accessibility. Monitoring the exchange rates in individual segments of the backbone can give information on local conformations (1), as well as on effects of altered states, such as in ligand binding (2) or aggregation.
Conventional, manual approaches require coordinated pipeting involving both the labeling and quenching steps followed by freezing of the protein sample. The protein is then labeled through dilution in deuterated buffer, which is usually produced by lyophilization of pH-adjusted phosphate- or Tris-buffers, followed by resuspension in deuterium oxide. After different incubation times, the labeling reaction is stopped by addition of cold acidic quenching solution (pH 2.4 and ∼4 °C), and snap-freezing in liquid nitrogen. Before mass spectrometry analysis, each sample is rapidly thawed and immediately injected into a cooled high performance liquid chromatography (HPLC) system using an ice-cold syringe. The HPLC system usually combines on-line pepsin digestion with desalting and separation of the proteolytic peptides by reverse phase chromatography, directly followed by electrospray ionization (ESI) MS analysis (1). There are several shortcomings limiting this application of classical HDX-MS. The extensive sample handling necessary to produce the labeled protein provides in itself a source of error, particularly with short incubation times and manual pipeting. Therefore, automation of the labeling procedure would be an advantage.
To avoid extensive manual pipeting and to ensure accurate timing and reproducibility a dual pipeting robot has been developed (5).
Previously, it has been shown that extremely rapid deuterium labeling of proteins can be carried out by mixing the protein sample with deuterated buffer in a continuous flow setup and injection of the mixture directly into an ESI-MS instrument, in which protein solution is mixed with deuterated solvent using a T-connector (4). Because of the constant sample flow, this technique can be combined with electron capture dissociation of the labeled protein to achieve HDX profiles with single-residue resolution (6). However, this approach involves dilution of the sample solution in deuterated buffer, whereas variation of the labeling time requires changing of the labeling capillary to a different length and/or diameter.
Here, we describe a method for dilution-free on-line deuterium labeling of proteins using an HDX cell with two flow channels separated by an ion-selective membrane (7). The layout of this cell is illustrated in Fig. 1. One flow channel carries the sample, the other the deuterium oxide. Deuterium oxide is delivered to the sample channel via the ion-selective membrane to yield dilution-free on-line labeling of the protein sample. Labeling times can be controlled by variation of the sample flow rate, and the deuterium content in the sample channel by the flow rate of the deuterium oxide.
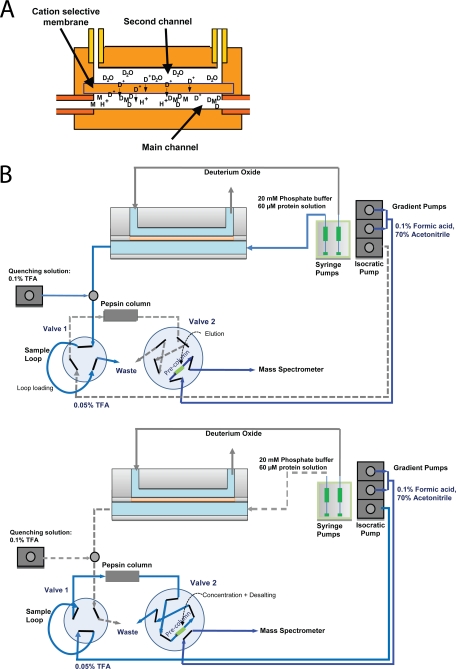
Fig. 1.
Schematic representation of the HDX cell and the automated HDX setup. A, The schematic illustration shows the sample (main) and deuterium oxide (second) channel, which are separated by an ion-selective membrane. B, Layout of the automated HDX cell system in deuterate/elute (top panel) and digest/desalt (bottom panel) positions (see Experimental Methods).
An earlier version of a cell developed for on-line pH scanning in liquid chromatography (LC)-MS has been previously used by us (8). Here, we demonstrate that the current HDX cell in combination with ESI-MS can be applied to monitor protein folding at the global and peptide level and to detect protein-protein interactions. Taken together, we present an automatable setup for HDX-MS that can be applied to investigate protein structure dynamics.
EXPERIMENTAL PROCEDURES
The cell has two channels separated by a cation-selective membrane like that used in electrocapture cells (9) (Fig. 1A). It is made from two identical PEEK blocks each having a drilled rectangular groove of 101 μm depth, 381 μm width, and 17 mm length. Inlet and outlet channels are drilled perpendicularly from the grooves and equipped with 10–32 female threaded connector ports. The fluidic cell is assembled by locating a 125 μm thick Nafion membrane (Ion Power Inc., New Castle, DE) between the two PEEK blocks and sealing it using four screws. The volume of each fluidic channel is 654 nL.
To probe the effect of deuterium oxide (D2O) flow rate on the deuterium ion content in the sample channel, the setup can be operated with or without addition of quenching solution (30% acetonitrile and 0.1% formic acid, pH 2.4) (supplemental Fig. S1). For this control, 2 μm [Glu1]-fibrinopeptide B in 20 mm ammonium acetate, pH 7.5, was infused into the mass spectrometer via the HDX cell at a constant flow rate of 0.5 μl/min using a syringe pump (Harvard Apparatus, Holliston, MA), and quenching solution was added at a flow rate of 2 μl/min, while the D2O flow rate was varied between 0.1 and 10 μl/min.
To estimate the D2O content in the sample channel under quenching conditions, 2 μm solutions of [Glu1]-fibrinopeptide B containing 20 mm ammonium acetate and 0–90% D2O were prepared and incubated for 5 min. Exchange was quenched by addition of four volumes of 30% acetonitrile containing 0.1% formic acid, pH 2.4, and the solutions were directly infused into the mass spectrometer.
Horse myoglobin was purchased from Ultra Scientific (North Kingstown, RI). For folding studies, the HDX cell was coupled to the inlet of an ESI-MS TOF mass spectrometer (supplemental Fig. S1 B). Myoglobin was introduced into the system using a syringe pump at a concentration of 10 μm in 100% H2O or 65% H2O with 35% acetonitrile at flow rates between 0.25 and 3.33 μl/min. D2O flow rates were kept at double the sample flow rates.
The N-terminal domain of Major Ampullate Spidroin 1 (MaSp1 NT) was purified as described (10). For HDX-MS analysis of MaSp1 NT, the HDX cell was connected to the 5 μl sample loop of a cooled HPLC system via a T-connector delivering 0.1% trifluoroacetic acid, pH 2, as quenching solution (Fig. 1B). 60 μm MaSp1 NT in 20 mm phosphate buffer, pH 7, was infused into the sample loop via the HDX cell at flow rates between 0.25 and 2 μl/min using a syringe pump. The D2O flow rate was kept at 10 times the sample flow rate, and quenching solution was added at the same flow rate as the sample. At each flow rate, the system was allowed a 60 min delay for stabilization of the flow and to ensure complete filling of the sample loop prior to injection.
On-line digestion and spectra acquisition were as described (11). Briefly, protein samples were digested online in a Porozyme Immobilized Pepsin Cartridge (Applied Biosystems, Foster City, CA) operated at 17 μl/min in 0.05% trifluoroacetic acid, pH 2. Peptic peptides were desalted using a Waters (Milford, MA) Symmetry C18 trap column and eluted in a single step with 70% acetonitrile containing 0.1% formic acid, pH 2.4, at a flow rate of 17 μl/min. Digestion and desalting were carried out in a single step for 10 min. Samples were delivered to the mass spectrometer through a tapered tip emitter with an opening of 50 μm (New Objective, Milford, MA) coupled to the HPLC via a T-connector.
To correct for back exchange, a solution of 60 μm MaSp1 NT in 80% D2O containing 20 mm phosphate buffer, pH 7, was heated for 2 h at 50 °C. The deuterated sample was quenched and loaded into the sample loop of the HPLC system with a syringe pump at a flow rate of 0.5 μl/min. Peptic peptide identification and calculations of deuterium incorporation were carried out as described (11). For reverse-phase purification of on-line labeled myoglobin, the setup was used without the pepsin column with a desalting time of 4 min.
Spectra (except for those from interleukin studies, next paragraph) were acquired in the positive-ion mode with a Waters Ultima API mass spectrometer equipped with a Z-spray source. The source temperature was 80 °C, the capillary voltage 1.7 kV and the cone and RF lens 1 potentials were 100 and 38 V, respectively. The mass spectrometer was operated in single-reflector mode to achieve a resolution of 10,000 (full width half maximum definition). The mass scale was calibrated using [Glu1]-fibrinopeptide. Scans were acquired for 5 min at a rate of one scan per 2 s between 200 and 2000 m/z. Data were analyzed using the MassLynx software package (Waters).
Interleukin 1β (IL-1β) and a Fab fragment were obtained from UCB S.A. (Brussels, Belgium). For HDX cell interaction studies, 20 μl of 230 μm IL-1β, mixed with 2.2 μl of 2 mm Fab fragment or 2.2 μl ammonium acetate, pH 8, were incubated for 1 h at 22 °C. Aliquots were diluted ten times in 10 mm ammonium acetate, pH 8, before HDX-MS analysis. Deuteration was carried out at a flow rate of 0.8 μl/min, quenching by addition of 4% formic acid via a T-connector at a flow rate of 10 μl/min, and the mixture directly infused into the mass spectrometer (supplemental Fig. S1 B). Average mass ± standard deviation of deuterated IL-1β in the presence and absence of Fab fragment was determined from the deconvoluted ESI-MS spectra of six repeats. Spectra were acquired on a Perkin Elmer TOF mass spectrometer equipped with an electrospray source that operates at ground potential (Biomotif AB, Sweden) and analyzed using Aviator 2.0 Software (Analytica of Branford, Branford, CT).
RESULTS
On-line Deuteration of a Short Peptide Using the HDX Cell
To test the feasibility of on-line deuteration for HDX-MS, we carried out on-line labeling of [Glu1]-fibrinopeptide B, a 14-residue peptide with no known secondary structure elements. Consequently, the degree of labeling of [Glu1]-fibrinopeptide B should be solely dependent on the deuterium content during the labeling reaction.
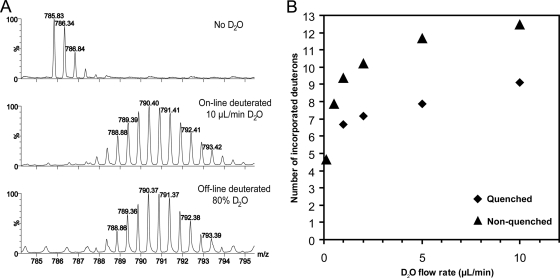
To investigate the effect of D2O flow rate on HDX cell deuteration efficiency, [Glu1]-fibrinopeptide B was infused at a constant flow rate of 0.5 μl/min whereas that in the channel carrying deuterium oxide was varied between 0.1 and 10 μl/min. Under both quenching and nonquenching conditions, we observed an increase in [Glu1]-fibrinopeptide B deuteration with increasing D2O flow rate (Fig. 2). To estimate the concentration of deuterium in the sample channel at different D2O flow rates, we compared on-line deuteration of [Glu1]-fibrinopeptide B with off-line deuteration in solutions containing 10–90% D2O and found that at a D2O flow rate of 10 μl/min, the deuterium content in the sample channel reaches ∼80% (supplemental Fig. S2).
Fig. 2.
Deuteration of [Glu1]-fibrinopeptide B using the HDX cell. A, On-line and off-line deuteration of [Glu1]-fibrinopeptide B. On-line deuteration of [Glu1]-fibrinopeptide B at a flow rate of 0.5 μl/min and a D2O flow rate of 10 μl/min (middle panel) yields the same deuteration level as conventional off-line deuteration in 80% D2O (lower panel), indicating that isotope delivery via the membrane is sufficient for deuteration of a short peptide. B, Dependence of [Glu1]-fibrinopeptide B deuteration on D2O flow rate. A 2 μm peptide solution was infused into the mass spectrometer through the HDX cell at 0.5 μl/min with or without subsequent quenching (30% acetonitrile and 0.1% formic acid at 2 μl/min). The flow rate of D2O was varied between 0.1 and 10 μl/min. With increasing D2O flow rate, the deuteration level is observed, indicating that the deuterium content in the sample channel is correlated with the D2O flow rate.
HDX Cell Deuteration Distinguishes Between Folded and Unfolded Proteins
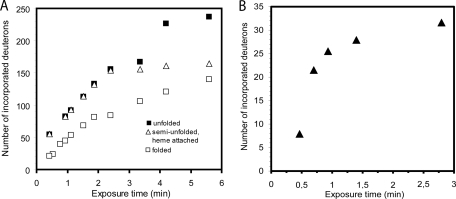
To investigate whether monitoring of flow rate dependent deuterium labeling can be facilitated by the HDX cell, we tested on-line deuteration of recombinant myoglobin using direct infusion from the HDX cell into the ESI source of the mass spectrometer. The myoglobin solution was pumped using flow rates between 0.25 and 3.33 μl/min. This flow rate interval corresponds to between 5 min 8 s and 25 s of exposure time in the HDX cell (using the volume of the cell, 654 nL, plus the volume of the PEEK tubing leading to the T-connector used for quenching, 744 nL, as an approximation). We observed that the incorporation of deuterium into the protein increased from 21 to 140 deuterons per molecule when the flow rate was lowered from 3.33 to 0.25 μl/min (Fig. 3A and supplemental Fig. S3A). To investigate whether the observed mass increase could be modified by changing the folding state of the protein, we tested on-line deuteration of myoglobin in the presence of 35% acetonitrile, which is known to cause unfolding and partial loss of the heme group (12). We then found incorporation of 54 to 237 deuterons for this denaturated myoglobin at the same flow rates as those used for the nondenaturated protein, corresponding to a 60–70% increase in deuteration on denaturation. Interestingly, an intermediate increase in deuteration was observed for the semiunfolded myoglobin population that retained its heme group, ranging from 55 to 163 deuterons (Fig. 3A). Taken together, these data demonstrate that the HDX cell can deuterate proteins in a fold-dependent manner, and that the degree of deuterium labeling can be controlled through the flow rate in the HDX cell.
Fig. 3.
Deuterium incorporation into folded and unfolded myoglobin using the HDX cell. Exposure times in the cell were calculated using the volumes of the sample channel (652 nL) and the connecting PEEK tubing (744 nL) and the respective sample flow rates. The same diagrams with μl/min instead of exposure time can be found as supplemental Fig. S3. A, Myoglobin exhibits a flow-rate dependent increase in deuterium incorporation. The presence of 35% acetonitrile significantly increases the deuterium incorporation in unfolded myoglobin. Holo-myoglobin (that retained its heme group) under these conditions represents a partially unfolded population with intermediate deuterium incorporation, indicating that protein labeling using the HDX cell can distinguish between different folding states. B, Flow-rate dependent deuterium incorporation can be detected after reverse-phase purification of HDX cell-labeled folded myoglobin, indicating that backbone amides can be successfully labeled using the HDX cell.
However, even at low flow rates the number of incorporated deuterons exceeds the number of backbone amides, indicating contributions from side-chain deuteration. To determine the extent of backbone labeling reached by the HDX cell at given exposure times, we coupled the HDX cell to the injection loop of a cooled HPLC setup equipped with a C18 desalting column used for conventional HDX-MS. The protein was deuterated in the HDX cell at flow rates between 3.0 and 0.5 μl/min and subjected to 4 min desalting under quenching conditions before ESI-MS analysis. Because inclusion of a desalting step is known to cause a significant amount of back-exchange, a lower degree of deuterium incorporation can be expected (13). It was found that at 0.5 μl/min, which corresponds to about 2 min 47 s of exposure time, myoglobin retains 30.6 deuterons (Fig. 3B), corresponding to 36% of the deuteration observed in the nondenaturated protein.
HDX Cell Deuteration Identifies Secondary Structure Elements
Analysis of deuterium incorporation rates is usually carried out using peptic peptides of a deuterium-labeled protein. Therefore, we compared the HDX cell labeling of MaSp1 NT from Euprosthenops australis in the HDX-MS HPLC setup with that in conventional HDX-MS analysis, which was previously characterized in our laboratory (11). MaSp1 NT has a five helix-bundle structure (14), in which peptic peptides from all five helices were found to exhibit a slower backbone amide exchange rate than their neighboring non-helical segments when analyzed by conventional HDX-MS (11). To combine the HDX cell with a standard HPLC system, the cell was coupled to the sample loop of a cooled HPLC system via a T-connector for delivery of quenching solution. The loop is loaded with the deuterated and quenched protein sample, which allowed us to use standard on-line pepsin digestion and desalting (11).
With HDX cell deuteration, significantly lower labeling was detected compared with conventional off-line HDX-MS of MaSp1 NT. For the maximum deuteration control (incubated for 2 h at 50 °C in 80% D2O and quenched and infused into the HPLC), a total deuterium incorporation of 30.79 deuterons was achieved, which is significantly lower than that achieved by resuspending freeze-dried MaSp1 NT in 99.9% D2O followed by snap-freezing in liquid nitrogen (94.2 deuterons) (11). Similar differences were observed when comparing the on-line deuteration at 0.25 μl/min, corresponding to a labeling time of 2 min 36 s (15.4 deuterons) with that achieved by 1 and 5 min off-line labeling (50 and 64 deuterons, respectively).
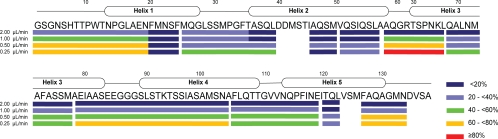
Analysis of the degree of deuteration of the peptic peptides from the HDX cell-labeled MaSp1 NT reveals that peptides covering helical segments have lower deuterium incorporation than fragments from nonhelical segments (Fig. 4 and supplemental Fig. S4). This indicates that the observed backbone deuteration accurately reflects secondary structure elements. Hence, HDX cell deuteration can identify their location in a manner similar to that of conventional HDX-MS (11).
Fig. 4.
Deuterium labeling using the HDX cell accurately reflects secondary structure elements of MaSp1 NT. Incorporation of deuterium at 0.25, 0.5, 1.0, and 2.0 μl/min is indicated by color coded underlining of the amino acid sequence. Each line corresponds to an observed peptic fragment. The corresponding deuteration levels in percent are given on the lower right. The positions of the five α-helices and residue numbers are indicated (14). The resulting deuteration pattern indicates the location of secondary structure elements in a manner similar to that observed with off-line HDX-MS (11), proving that HDX cell deuteration is secondary structure-dependent.
Interleukin-Antibody Interactions Detected Using the HDX Cell
To assess the ability of HDX cell-based deuteration to probe protein-protein interactions in a protein-antibody system, we tested whether pre-incubation with a specific Fab fragment could decrease the deuterium incorporation in recombinant interleukin 1β (IL-1β). Because the excess of undeuterated quenching solution causes rapid isotope equilibration, side-chain deuteration should not be detectable under these conditions (15). We found that deuteration at 1 μl/min with subsequent quenching produces a deuterium incorporation of 74.8 ± 0.3 deuterons for IL-1β alone and 69.8 ± 0.5 for the IL-1β/Fab fragment complex. Hence, the HDX cell is a useful tool for probing protein-protein interactions.
DISCUSSION
We present a system for on-line HDX-MS that alleviates two problems of conventional deuterium labeling procedures: the need for manual pipeting and sample dilution. Using a membrane-driven deuteration principle, we are able to carry out on-line protein labeling to detect protein folded states, secondary structure elements, and protein-protein interactions.
During initial evaluation studies we found that the degree of labeling, which for an unstructured peptide is directly proportional to the D2O concentration, was increased with increasing D2O flow rate when the sample flow rate was kept constant until a maximal deuteration level was obtained (Fig. 2). This is not surprising because the D2O diffusion across the membrane is a gradient-driven process, and a more complete exchange in the sample channel can be achieved against a larger volume of deuterium oxide. Comparison of the on-line deuteration with the manual “in-tube” deuteration showed a maximal D2O concentration of ∼80% (supplemental Fig. S2).
In addition to the expected deuteration differences between apo- and holo-myoglobin, an intermediate form of unfolded holo-myoglobin was observed (Fig. 3). This form shows a higher degree of unstructured elements than the holo-myoglobin, but less than the apo-myoglobin. At shorter exposure times, the unfolded holo-myoglobin shows labeling kinetics similar to apo-myoglobin. This suggests that both structures share similar unstructured elements, but that the yet attached heme group in the unfolded holo-myoglobin retains some of the structured elements of the holo-myoglobin. Such an intermediate unfolding event has been suggested also in other experiments (12).
Because our myoglobin studies involve major structural changes, we thought that an antigen/antibody experimental model may form a better test to evaluate the present system. Notably, our results then showed that the presence of a specific antibody fragment reduced the global deuteration of IL-1β, which supports the conclusion that backbone (and side-chain) accessibility governs polypeptide deuteration by the HDX cell.
A further step to validate the use of the HDX cell, the device was connected to a standard dual-valve HDX system. In this system, the protein sample is deuterated online, digested, desalted, and analyzed by ESI-MS. This setup was used to test whether the HDX-cell deuteration accurately reflects secondary structure elements at the peptide level. For this, we performed online deuteration of MaSp1 NT followed by standard pepsin digestion and desalting, as previously characterized in our laboratory using conventional off-line HDX-MS. Loading the HPLC sample loop from the current HDX cell setup results in hold-up times and therefore increases the back exchange even under quenching conditions. This leads to lower overall labeling in on-line than in off-line deuteration. However, the relative deuteration allows for the identification of slow and fast exchanging fragments (Fig. 4 and supplemental Fig. S4). Therefore, the location of secondary structure elements can be accurately traced with the HDX cell system, and the results are consistent with those obtained from conventional HDX setups (11).
A wide variety of semipermeable membranes is commercially available, which have different ion selectivity and diffusion efficiency. Many future possibilities therefore exist regarding general labeling in different buffers and at varying salt concentrations by selecting the appropriate membrane and buffer system. Because high salt concentrations and most standard buffers with Tris or phosphate are unsuitable for direct infusion into a mass spectrometer, a trapping and desalting system can be necessary. In this study, we demonstrate the use of the HDX cell in combination with such a system by on-line deuteration of MaSp1 in 20 mm phosphate buffer and observe no adverse effects. We further show that the [Glu1]-fibrinopeptide B labeling experiment described here can serve as a simple test for deuterium labeling efficiency when a different membrane or buffer system is used.
In summary, we show that the HDX cell is a suitable tool for determination of protein folding states, protein-protein interactions, and secondary structure elements by on-line, pipet- and dilution-free HDX mass spectrometry. The system allows separate adjustments of deuterium content and labeling time by control of the flow rates in the deuterium oxide and the sample channels, respectively. The system is automatable, e.g. in combination with an autosampler, which should make it suitable for use in high-throughput assays. Combinations of the HDX cell with existing on-line HDX systems such as rapid quench-flow and top-down sequencing (4, 6) would represent further new developments in HDX-MS.
Footnotes
* This work was supported by grants from the Swedish Research Council (03X-3532 and 13X-10371), Karolinska Institutet (PhD grant to M. L.), Biomotif AB, Solna, Sweden (to J. A.-W.), and the Knut and Alice Wallenberg Foundation (to H. J.).
 This article contains supplemental Figs. S1 to S4.
This article contains supplemental Figs. S1 to S4.
Conflict of interest statement: J. A.-W. and H. J. are shareholders of Biomotif AB, Solna, Sweden.
1 The abbreviations used are:
- HDX
- Hydrogen/deuterium exchange
- HDX-MS
- HDX with mass spectrometry
- ESI
- electrospray ionization
- LC
- liquid chromatography.
REFERENCES
- 1. Zhang Z., Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jørgensen T. J., Gårdsvoll H., Danø K., Roepstorff P., Ploug M. (2004) Dynamics of urokinase receptor interaction with Peptide antagonists studied by amide hydrogen exchange and mass spectrometry. Biochemistry 43, 15044–15057 [DOI] [PubMed] [Google Scholar]
- 3. Kheterpal I., Zhou S., Cook K. D., Wetzel R. (2000) Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc. Natl. Acad. Sci. U.S.A. 97, 13597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konermann L., Collings B. A., Douglas D. J. (1997) Cytochrome c folding kinetics studied by time-resolved electrospray ionization mass spectrometry. Biochemistry 36, 5554–5559 [DOI] [PubMed] [Google Scholar]
- 5. Chalmers M. J., Busby S. A., Pascal B. D., He Y., Hendrickson C. L., Marshall A. G., Griffin P. R. (2006) Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 6. Pan J., Han J., Borchers C. H., Konermann L. (2009) Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J. Am. Chem. Soc. 131, 12801–8 [DOI] [PubMed] [Google Scholar]
- 7. Astorga-Wells J., Whitehouse C., Jörnvall H. (2009) Membrane-assisted sample preparation for online ESI-MS analysis of biomolecules, ASMS Abstract 1917. Mol. Cell Proteomics 8, 14–15 [Google Scholar]
- 8. Jörnvall H., Lindahl E., Astorga-Wells J., Lind J., Holmlund A., Melles E., Alvelius G., Nerelius C., Mäler L., Johansson J. (2010) Oligomerization and insulin interactions of proinsulin C-peptide: Threefold relationships to properties of insulin. Biochem. Biophys. Res. Commun. 391, 1561–1566 [DOI] [PubMed] [Google Scholar]
- 9. Astorga-Wells J., Vollmer S., Bergman T., Jörnvall H. (2005) Microfluidic systems and proteomics: applications of the electrocapture technology to protein and peptide analysis. Anal. Biochem. 345, 10–17 [DOI] [PubMed] [Google Scholar]
- 10. Hedhammar M., Rising A., Grip S., Martinez A. S., Nordling K., Casals C., Stark M., Johansson J. (2008) Structural properties of recombinant nonrepetitive and repetitive parts of major ampullate spidroin 1 from Euprosthenops australis: implications for fiber formation. Biochemistry 47, 3407–3417 [DOI] [PubMed] [Google Scholar]
- 11. Landreh M., Askarieh G., Nordling K., Hedhammar M., Rising A., Astorga-Wells J., Alvelius G., Casals C., Knight S. D., Johansson J., Jörnvall H., Bergman T. (2010) A pH-dependent Dimer Lock in Spider Silk Protein. J. Mol. Biol. 404, 328–336 [DOI] [PubMed] [Google Scholar]
- 12. Lin X., Zhao W., Wang X. (2010) Characterization of conformational changes and noncovalent complexes of myoglobin by electrospray ionization mass spectrometry, circular dichroism and fluorescence spectroscopy. J. Mass Spectrom. 45, 618–626 [DOI] [PubMed] [Google Scholar]
- 13. Zhang H. M., Bou-Assaf G. M., Emmett M. R., Marshall A. G. (2009) Fast reversed-phase liquid chromatography to reduce back exchange and increase throughput in H/D exchange monitored by FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 20, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Askarieh G., Hedhammar M., Nordling K., Saenz A., Casals C., Rising A., Johansson J., Knight S. D. (2010) Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465, 236–238 [DOI] [PubMed] [Google Scholar]
- 15. Wales T. E., Engen J. R. (2006) Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]