Abstract
Skeletal muscle is a key tissue site of insulin resistance in type 2 diabetes. Human myotubes are primary skeletal muscle cells displaying both morphological and biochemical characteristics of mature skeletal muscle and the diabetic phenotype is conserved in myotubes derived from subjects with type 2 diabetes. Several abnormalities have been identified in skeletal muscle from type 2 diabetic subjects, however, the exact molecular mechanisms leading to the diabetic phenotype has still not been found. Here we present a large-scale study in which we combine a quantitative proteomic discovery strategy using isobaric peptide tags for relative and absolute quantification (iTRAQ) and a label-free study with a targeted quantitative proteomic approach using selected reaction monitoring to identify, quantify, and validate changes in protein abundance among human myotubes obtained from nondiabetic lean, nondiabetic obese, and type 2 diabetic subjects, respectively. Using an optimized protein precipitation protocol, a total of 2832 unique proteins were identified and quantified using the iTRAQ strategy.
Despite a clear diabetic phenotype in diabetic myotubes, the majority of the proteins identified in this study did not exhibit significant abundance changes across the patient groups. Proteins from all major pathways known to be important in type 2 diabetic subjects were well-characterized in this study. This included pathways like the trichloroacetic acid (TCA) cycle, lipid oxidation, oxidative phosphorylation, the glycolytic pathway, and glycogen metabolism from which all but two enzymes were found in the present study. None of these enzymes were found to be regulated at the level of protein expression or degradation supporting the hypothesis that these pathways are regulated at the level of post-translational modification. Twelve proteins were, however, differentially expressed among the three different groups. Thirty-six proteins were chosen for further analysis and validation using selected reaction monitoring based on the regulation identified in the iTRAQ discovery study. The abundance of adenosine deaminase was considerably down-regulated in diabetic myotubes and as the protein binds propyl dipeptidase (DPP-IV), we speculate whether the reduced binding of adenosine deaminase to DPP-IV may contribute to the diabetic phenotype in vivo by leading to a higher level of free DPP-IV to bind and inactivate the anti-diabetic hormones, glucagon-like peptide-1 and glucose-dependent insulintropic polypeptide.
The constant increase in obesity in industrialized countries resulting from a sedentary lifestyle and overly rich nutrition has resulted in an elevation in the number of people diagnosed with diabetes worldwide (1–4). In the United States alone, 23.6 million people have been estimated to have diabetes of which 17.9 million have been diagnosed, 90% of them with type 2 diabetes (T2D)1 (5, 6). Furthermore, increased prevalence in the United States has lead the American Centers for Disease Control and Prevention to publically classify the disease as an epidemic. The worldwide picture shows the same trends and today T2D is seen as one of the main threats to human health in the Western world (7). T2D is a complex disorder characterized by impaired insulin secretion from β-cells and insulin resistance in the major metabolic tissues such as skeletal muscle, liver, and fat cells (8, 9). Skeletal muscle is a key tissue site of insulin resistance, with evidence of decreased insulin-stimulated glucose uptake and storage as glycogen. Although several abnormalities have been identified in skeletal muscle from T2D subjects, the exact molecular mechanisms leading to insulin resistance have still not been established (8, 10).
For the last two decades, measurements of energy stores and metabolites, morphological studies, assays of enzyme activity and phosphorylation, as well as gene and protein expression have been carried out in skeletal muscle biopsies to study the molecular mechanisms underpinning insulin resistance (11). Changes in both mRNA and protein abundance in skeletal muscle have been linked to a large number of metabolic disorders, including insulin resistance (12, 13). Most studies of changes in muscle structure and metabolism under different physiological and pathophysiological conditions have, however, only focused on a few genes or proteins (12). In addition, most of these studies have been performed on individual patient samples in which the risk of patient variability is very high. Furthermore, the level of mRNA may not precisely reflect the exact protein abundance because of post-transcriptional control, alternative splicing and different lifetime, and no information on post-translational modifications (PTMs) is obtained from such analysis (14, 15). Moreover, it is still uncertain whether the pathophysiological mechanisms leading to the diabetic phenotype e.g. insulin resistance will be found at the level of protein abundance or PTM. To obtain a better understanding of the molecular mechanisms leading to such disorders it is necessary to perform large-scale quantitative studies. Strategies such as proteomics and gene expression profiling on skeletal muscle biopsies have recently been implemented to obtain a more global overview of the human skeletal muscle proteome. The focus however, has mainly been on mitochondrial proteins and only a few quantitative studies have been published.
Cultured human muscle cells (myotubes) are a valuable tool for the investigation of T2D (16–21). Human myotubes are primary skeletal muscle cells displaying the morphological, metabolic, and biochemical properties of adult skeletal muscle (16, 22). Furthermore, they express the diabetic phenotype when established from subjects with T2D (16, 21) and are as such a unique model system to distinguish between genetic and environmental factors in the etiology of insulin resistance and T2D. We and others have reported several potential intrinsic defects in myotubes established from subjects with T2D. These include reduced insulin-mediated glucose uptake, glucose oxidation, and glycogen synthesis (18, 21). In addition, lipid oxidation and trichloroacetic acid (TCA) flux was decreased (23, 24) and very recently, it was found that the ATP synthesis rate during ATP utilization was reduced in isolated mitochondria from diabetic myotubes (25). The use of primary cell lines is challenged by the fact that they can only be cultured for a few passages to preserve their primary phenotype. This precludes the use of metabolic labeling of proteins such as N14/15-labeling (26) and stable isotope labeling by amino acids in cell culture (SILAC) (27, 28) for quantitation as a certain number of passages are needed to obtain full incorporation. Instead, alternative stable isotopic labeling methods such as isobaric peptide tags for relative and absolute quantification (iTRAQ) are useful for labeling of primary cells. Using this method, the labels are added at the peptide level and therefore no demands are placed on the number of cell culture passages. In addition, this also facilitates the implementation of the strategy onto muscle biopsies (29).
In the present study, we investigated whether the basal proteome in diabetic myotubes varies from that of nondiabetic controls. We therefore combined a quantitative proteomic approach using iTRAQ labeling with a label-free study to provide independent data sets for the discovery phase. Labeling with iTRAQ in combination with sample prefractionation and nano liquid chromatography-tandem MS (LC-MS/MS) for identification and quantitation of the basal proteome of myotubes established from obese, diabetic subjects compared with that of nondiabetic lean and nondiabetic obese subjects revealed almost 3000 proteins. Furthermore, when performing quantitative proteomics to investigate a biological hypothesis, it is important to validate any quantitative findings. To strengthen the data, all proteins that were identified as differentially abundant in the iTRAQ discovery study were subsequently validated in a targeted study using selected reaction monitoring (SRM) (30). The SRM strategy is highly sensitive making it possible to detect even precursor ions present in low copy number in a complex sample (31). Thirty-six proteins were analyzed using SRM and the abundances found in the iTRAQ data were validated. Twelve of these proteins were found to be differentially expressed between the three different groups.
MATERIALS AND METHODS
Human Myotubes
Ten lean, ten obese control subjects, and ten obese T2D patients participated in the study (see Table I). Their clinical characteristics have previously been published (32, 33). Fasting plasma glucose, serum insulin, and HbA1c levels were significantly higher in the diabetic group compared with both the lean and obese controls. Fasting HDL cholesterol concentrations were lower in T2D patient compared with lean controls. The obese controls showed higher fasting serum insulin levels compared with lean controls. The glucose infusion rates during the steady state of the hyperinsulinaemic euglycemic clamp period were significantly lower in T2D subjects compared with both lean and obese control subjects, and glucose infusion rates was significantly lower in obese controls compared with lean control subjects. Muscle biopsies were obtained from the vastus lateralis muscle by needle biopsy ad modum Bergstrøm (34) under local anesthesia. Diabetic subjects were treated either with diet alone or in combination with sulfonylurea, metformin, or insulin, withdrawn 1 week before the study. The subjects suffered from no diabetic complications except for simplex retinopathy. The control subjects had normal glucose tolerance and no family history of diabetes. All subjects gave written, informed consent, and the local ethics committee of Funen and Vejle County approved the study. The setup enabled us to specifically study proteome changes in diabetic and obese subjects (diabetic and nondiabetic obese subjects), which could provide insight into regulation occurring at a prestate of T2D. To identify overall differences and trends among the type 2 diabetic myotubes and controls, myotubes from each group were pooled thereby avoiding variations among subjects.
Table I. This table shows the physiological and biochemical details of the 30 subjects32. We use muscle biopsies from a total of 30 subjects (10 subjects from each category). Data are means ± S.E. *Significantly different from the lean controls (p < 0.05), #significantly different from the lean and obese controls (p < 0.05).
| Control, lean | Control, obese | T2D | |
|---|---|---|---|
| n | 10 | 10 | 10 |
| Age (years) | 51 ± 1 | 49 ± 1 | 50 ± 1 |
| Weight (kg) | 71.6 ± 3.0 | 105.5 ± 6.4* | 102.2 ± 4.1* |
| BMI (kg/m2) | 24.2 ± 0.5 | 33.7 ± 1.4* | 33.5 ± 1.1* |
| Fasting plasma glucose (mm) | 5.7 ± 0.1 | 5.7 ± 0.2 | 10.0 ± 0.7# |
| Fasting serum insulin (pm) | 24.3 ± 5.7 | 52.7 ± 5.0* | 94.6 ± 10.1# |
| Glucose infusion rate (mg/min) | 383.3 ± 20.4 | 257.9 ± 28.3* | 117.8 ± 18.6# |
| HbA1c (%) | 5.5 ± 0.1 | 5.4 ± 0.1 | 7.7 ± 0.5# |
| Fasting total cholesterol (mm) | 5.29 ± 0.22 | 5.43 ± 0.41 | 5.42 ± 0.37 |
| Fasting LDL cholesterol (mm) | 2.94 ± 0.22 | 3.33 ± 0.33 | 3.20 ± 0.27 |
| Fasting HDL cholesterol (mm) | 1.85 ± 0.15 | 1.48 ± 0.15 | 1.36 ± 0.03* |
| Fasting plasma triglyceride (mm) | 1.12 ± 0.16 | 1.35 ± 0.18 | 1.93 ± 0.40 |
Cell Culture
Cell cultures were established as previously described (22, 35, 36). In brief, muscle tissue was minced, washed, and dissociated for 60 min by three treatments with 0.05% trypsin-EDTA. The harvested cells were pooled and fetal calf serum (FCS) was added to stop trypsination. The cells obtained were seeded for up-scaling on ECM-gel coated dishes after 30 min of preplating. Cell cultures were established in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1.25 μg/ml amphotericin B. After 24 h, cell debris and nonadherent cells were removed by change of growth medium to Dulbecco's modified Eagle's medium supplemented with 2% FCS, 2% Ultroser G, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1.25 μg/ml amphotericin B. Cells were subcultured twice before final seeding (4–6 weeks). At 75% confluence, the growth medium was replaced by basal medium (Dulbecco's modified Eagle's medium supplemented with 2% FCS, 50 U/ml penicillin, 50 μg/ml streptomycin, 1.25 μg/ml amphotericin B, and 25 pmol/l insulin) to induce differentiation (37). The cells were cultured in humidified 5% CO2 ATM at 37 °C, and medium was changed every 2–3 days. Satellite cell were differentiated into myotubes in passage four and myotubes were harvested by cell lysis day 4 after induction of differentiation (22). Myotubes established from diabetic and control subjects were visually inspected under phase-contrast microscopy, and did not differ in appearance.
Cell Lysis
The media was removed from the cells and 1.5 ml lysis buffer (0.2% SDS, 50 mm Tris-Base, 1 mm MgCl2, 60°C) was added. Complete protease inhibitor mixture (Roche) and Benzonase (Merck, 750 units.) was subsequently added to the lysate to avoid inactivation of these at 60 °C. The cell lysates were transferred to 15-ml Falcon tubes. The remaining cell debris were scraped off using a rubber policeman and pooled with the cell lysate. Cell lysates from 10 T2D subjects, 10 nondiabetic lean, or 10 nondiabetic obese subjects were pooled, respectively and the lysates were snap-frozen in liquid nitrogen.
Test of Different Protocols for Protein Precipitation
After thawing the myotube lysates, the lysates were sonicated (on ice three times 20 s with intervals, output 40% using a probe sonicator). Five different protocols for protein precipitation were tested in four replicates. In all 20 experiments, 133 μl cell lysate was used to test each protocol as 133 μl was the maximum volume to be used in the chloroform/methanol protocol when performing the precipitation in a 1.5 ml Eppendorf tube.
Protein Precipitation Using Acetone
One hundred thirty-three microliters cell lysate was transferred to a fresh 1.5-ml Eppendorf tube. One milliliter ice-cold acetone was added. The sample was vortexed for 1 min and incubated overnight at –20 °C. After incubation, proteins were precipitated by centrifugation at 15,000 rpm at 4 °C for 30 min. The protein pellet was subsequently washed three times using 1 ml ice-cold acetone. As lyophilization of the protein pellet will often form a hard rubber-like protein pellet that is very difficult to resolubilize, the precipitated protein was left in the fume hood at room temperature overnight to evaporate remaining acetone.
Protein Precipitation Using Chloroform/Methanol
One hundred thirty-three microliter of cell lysate was transferred to a fresh Eppendorf tube and 532 μl ice-cold methanol was added. The lysate was vortexed for 1 min after which 133 μl ice-cold chloroform was added. The lysate was vortexed for 1 min and 400 μl ice-kold ddH2O was added. The lysate was again vortexed and then centrifuged at 15000 rpm at 4 °C for 2 min. The upper layer containing methanol, water, and dissolved salts was carefully removed without disturbing the white protein layer in the middle. The lower layer contained chloroform. Six hundred microliters of ice-cold methanol was added and the sample was vortexed for 1 min. Proteins were precipitated by centrifugation at 15000 rpm at 4 °C for 2 min. The protein pellet was subsequently washed three times using 1 ml ice-cold methanol. The precipitated protein was left in the fume hood at room temperature overnight to evaporate remaining methanol.
Protein Precipitation Using Trichloroacetic Acid, Acetone, and β-Mercaptoethanol
133 μl cell lysate was transferred was transferred to a fresh Eppendorf tube. 1400 μl ice-cold 10% trichloroacetic acid/∼90% acetone/0.07% β-mercaptoethanol was added. The sample was vortexed for 1 min and incubated for 1 h at - 20 °C. After incubation, proteins were precipitated by centrifugation at 15000 x rpm at 4 °C for 15 min. The protein pellet was washed three times using 1 ml ice-cold 10% trichloroacetic acid/90% acetone/0.07% β-mercaptoethanol and the sample was incubated for 1 h at - 20 °C between each washing step. The precipitated protein was subsequently left in the fumehood at room temperature overnight to evaporate remaining trichloroacetic acid/acetone/β-mercaptoethanol.
Protein Precipitation Using Ethanol/Acetone
One hundred thirty-three microliters of cell lysate was transferred to a fresh Eppendorf tube. To this 665 μl ice-cold absolute ethanol was added. The lysate was vortexed for 1 min after which 665 μl ice-cold acetone was added. The sample was vortexed and incubated overnight at −20 °C. After incubation, proteins were precipitated by centrifugation at 15000 rpm at 4 °C for 30 min. The protein pellet was subsequently washed three times using ice-cold 40% ethanol, 40% acetone, and 20% ddH2O. The precipitated protein was left in the fume hood at room temperature overnight to evaporate remaining ethanol/acetone.
Protein Precipitation Using Trichloroacetic Acid
One hundred thirty-three microliter of cell lysate was transferred to a fresh Eppendorf tube. To this 20% trifluoroacetic acid (TFA) was added and the sample was vortexed for 1 min. The sample was left on ice for 30 min to allow protein precipitation. After incubation, proteins were centrifuged at 15000 rpm at 4 °C for 15 min. The protein pellet was subsequently washed three times using 1 ml ice-cold acetone to remove trichloroacetic acid. The precipitated protein was left in the fume hood at room temperature overnight to evaporate remaining acetone.
The dry weight of the precipitated matter from all 20 precipitations was measures (supplemental Table 1). Each protein pellet was then dissolved using 20 μl 6 m urea and 2 M thiourea and incubated at –80°C for 30 min. The frozen samples were subsequently sonicated for 5 min in water bath to improve solubilization. After this 140 μl H2O was added to dilute urea (as subsequent iTRAQ labeling in the biological study was not compatible with NH4HCO3 and >1 M urea). Two microliters from each precipitation was measured for the amount of protein using the QubitTM quantitation platform (Invitrogen) (supplemental Table 1). From each sample 50 μl were transferred to fresh Eppendorf tubes and lyophilized in the speedvac to reduce the volume after which 15 μl SDS sample buffer was added and the samples were run on two gels along with a protein marker (supplemental Fig. 1).
Protein Precipitation Using Ethanol and Acetone for Myotube Proteomics
After thawing, the myotube lysates were sonicated (on ice three times 20 s with intervals, output 40%). One milliliter lysate was transferred to a fresh Eppendorf tube. Four ml ice-cold absolute ethanol was added. The lysate was thoroughly vortexed after which 4 ml ice-cold acetone was added. The sample was vortexed and incubated overnight at −20 °C. Proteins were precipitated by centrifugation at 15000 rpm at 4 °C for 30 min. The protein pellet was subsequently washed three times using ice-cold 40% ethanol and 40% acetone/20% ddH2O.
Reduction and Alkylation
The pellet was resuspended in 40 μl 8 m Urea (6 m urea (Sigma), 2 m thiourea (Merck, Darmstadt, Germany) and 10 mm dithiotreitol was added. The sample was incubated at room temperature for 35 min. Finally, 20 mm iodoacetamide was added followed by incubation for 35 min at room temperature in the dark.
Digestion with Lysyl Endopeptidase and Trypsin
The reduced and alkylated sample was incubated at room temperature for 3 h with 1 μg lysyl endopeptidase (Wako, Richmond, VA) per 50 μg protein. The lysyl endopeptidase-digested sample was diluted with iTRAQ dissolution buffer (Applied Biosystems, Foster City, CA) to make a five times dilution, because trypsin does not work well at high concentrations of urea. One μg chemically modified trypsin (Promega, Charbonnières, France) was added per 50 μg protein and the sample was incubated at room temperature for 18 h.
Amino Acid Analysis
Four microlliters from each sample was transferred to fresh 500-ml Eppendorf tubes. The samples were lyophilized and the tubes transferred to hydrolysis glass vials (22-ml screw-lid glass with mini-inert valves, Pierce) containing 400 μl hydrolysis buffer (6N HCl, 0.1% phenol, 0.1% thioglycolic acid) and 4–5 samples per vial. Oxygen was removed by argon after which the glasses were evacuated to less than 5 mBar. The vials were incubated at 110 °C overnight (18–20 h) and subsequently lyophilized and analyzed using a Biochrom 31 amino acid analyzer run as recommended by the manufacturer using the Sodium Accelerated Buffer System.
iTRAQ 4plex Labeling
A 100 μg peptide sample derived from the T2D myotube protein solution was transferred to fresh 500-μl Eppendorf tubes and labeled with iTRAQ 114 according to the company protocol (http://www3.appliedbiosystems.com/cms/groups/psm_support/documents/generaldocuments/cms_047330.pdf). A 100-μg sample of peptide from the lean myotube protein was labeled with iTRAQ 115 and the obese myotube peptides were labeled with iTRAQ 116.
Determination of iTRAQ Mixing Ratio
To determine the correct mixing ratio for the iTRAQ labeled samples, 1 μl of each sample was pooled into 20 μl 0.1% TFA. The pooled sample was subsequently desalted and concentrated on a 2 mm long reversed-phase micro-column. The micro-column was prepared using a GELoader tip as described previously (38) and sample loaded onto the reversed-phase micro-column using gentle air pressure (38). The micro-column was washed using 20 μl 0.1% TFA and the peptides were subsequently eluted directly onto a matrix-assisted laser desorption ionization (MALDI)-MS target using 0.5 μl α-cyano-4-hydroxycinnamic acid. The sample was analyzed by MALDI MS/MS using a Bruker Ultraflex MALDI-TOF/TOF instrument. All spectra were obtained in positive reflector mode. Mass spectrometric data analysis was performed using the Bruker Daltonics flexAnalysis 2.4 Software. The three labeled samples were subsequently mixed 1:1:1.
Strong Cation Exchange (SCX)
The mixed iTRAQ labeled sample was diluted 10 times in SCX loading buffer (30% acetonitrile, 1% acetic acid) and pH was adjusted to 2.7. The sample was loaded slowly onto an equilibrated SCX column (SCX cartridge from Applied Biosystems). The SCX column was washed with 1 ml SCX loading buffer and the peptides were subsequently eluted using increasing concentrations of KCl (200 μl of 50 mm, 100 mm, 150 mm, 200 mm, 250 mm, 300 mm, 350 mm, 400 mm, 500 mm, and 1 m KCl buffer). All eluents were subsequently lyophilized.
Desalting the Sample Using R3 Columns before Mass Spectrometry
POROS Oligo R3 Reversed Phase resin (Applied Biosystems) was dissolved in 100% acetonitrile (HPLC grade, Fisher Scientific). The R3 beads were packed in a P100 tip by stamping out a small plug of C18 material from a 3 m EmporeTM C18 extraction disk using an HPLC syringe needle, and placing this plug in the constricted end of P100 tips (39) to gain a micro-column of 1 cm. The R3 micro-column was washed with 50 μl 100% acetonitrile and equilibrated with 50 μl 0.1% TFA. Each acidified sample was loaded onto a R3 column of 1 cm. The flow-through was loaded onto another R3 column. The R3 micro-columns were washed with 50 μl 0.1% TFA and the peptides were eluted from the columns using 50 μl 50% acetonitrile and 0.1% TFA. After lyophilization the peptides were resuspended in 5% formic acid and the amount to be run on nanoLC-MS was tested on MALDI MS.
Nano-liquid Chromatography Tandem Mass Spectrometry (nanoLC-MS/MS)
iTRAQ Study
The iTRAQ labeled peptides from the SCX fractions were loaded onto an EASY nanoLC system (Proxeon, Denmark). They were loaded directly onto a 20 cm 100 μm inner diameter, 360 μm outer diameter, ReproSil - Pur C18 AQ 3 μm (Dr. Maisch, Ammerbuch-Entringen, Germany) reversed phase capillary column. The peptides were eluted using a gradient from 100% phase A (0.1% formic acid) to 40% phase B (0.1% formic acid, 90% MeCN) over 80–90 min at 200 nL/min directly into a LTQ-Orbitrap XL or Velos mass spectrometer (Thermo Scientific, San Jose, CA). The LTQ-Orbitrap XL was operated in a data-independent mode automatically switching among MS, MS/MS (collision-induced dissociation (CID) or electron transfer dissociation (ETD)), and Higher-energy Collision Dissociation (HCD) using a threshold of 50,000 for ion selection. The LTQ-Orbitrap Velos was operated in a data-independent mode automatically switching among MS (Resolution of 30,000), MS/MS (Resolution of 7500), and HCD using a threshold of 10,000 for ion selection.
For each MS scan on the LTQ-Orbitrap XL, the two most abundant precursor ions were selected for fragmentation using firstly CID or ETD MS/MS (CID: normalized collision energy 35; activation time 30 ms; ETD: target value of 200000 anions; activation time 100 ms) to facilitate peptide identification and then subsequently HCD (normalized collision energy 55; activation time 30 ms) to obtain reliable high abundant iTRAQ quantitative data (40). For each MS scan on the LTQ-Orbitrap Velos, the three most abundant precursor ions were selected for fragmentation using firstly MS/MS (normalized collision energy 35; activation time 30 ms) to facilitate peptide identification and then subsequently HCD (normalized collision energy 50; activation time 30 ms) to obtain reliable high abundant iTRAQ quantitative data (40). Raw data were viewed in Xcalibur v2.1 (Thermo Scientific).
Label-free (HCD only) Study
The nonlabeled peptides from the SCX fractions were loaded onto an EASY nanoLC system (Proxeon, Denmark). They were loaded directly onto a 20 cm 100 μm inner diameter, 360 μm outer diameter, ReproSil - Pur C18 AQ 3 μm (Dr. Maisch, Ammerbuch-Entringen, Germany) reversed phase capillary column. The peptides were eluted using a gradient from 100% phase A (0.1% formic acid) to 40% phase B (0.1% formic acid, 90% MeCN) over 120 min at 250 nL/min directly into a LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose CA). The LTQ-Orbitrap Velos was operated in a data-independent mode automatically switching between MS (Resolution of 30,000) and HCD using a threshold of 10,000 for ion selection. For each MS scan, the seven most abundant precursor ions were selected for fragmentation (normalized collision energy 45; activation time 5 ms). Raw data were viewed in Xcalibur v2.1 (Thermo Scientific).
Data Processing and Protein Sequence Database Searching
The data were processed using Proteome Discoverer, beta-version 1.1.0.228. The files were subsequently submitted to an in-house MASCOT server (version 2.2.05) (Matrix Science Ltd., London, U.K.) for database searching through the Proteome Discoverer program. The data were searched against the Swissprot human sequence database (Sprot 2010 (517100 sequences; 182146551 residues), Taxonomy: Homo sapiens (human) (20367 sequences) as the study was performed on human myotubes. The search was performed choosing trypsin as specific enzyme. A maximum of 2 missed cleavages were allowed. Carbamidomethyl (C) was chosen as fixed modification. As variable modifications, iTRAQ 4plex (K), iTRAQ 4plex (N-term) and Oxidation (M) were chosen. The data were searched with a peptide mass tolerance of 10 ppm and a fragment mass tolerance of 0.8 Da (CID/ETD) or 0.1 Da (HCD). Finally, the data were searched against the human decoy database to calculate a false discovery rate (FDR) of 1 and 5% using the Proteome Discoverer program. Protein grouping was performed in the Proteome Discoverer program.
Evaluation of iTRAQ Data
For the initial identification, peptides with a false discovery rate (FDR) of ≤5% and a Mascot ion score of at least 20 were accepted. For quantitation, only proteins identified by at least two peptides and with FDR of ≤1% and mascot peptide ion scores of at least 20 were considered. A Mascot score of 20 was accepted as the quantiatative finding would subsequently be validated using SRM. In addition, only peptides containing N-terminal iTRAQ labeling or peptides containing iTRAQ labeled lysine residue in combination with another N-terminal modification were used for quantitation. The iTRAQ data were normalized on protein median using the Proteome Discoverer software as the majority of the proteins with valid iTRAQ information were not differentially expressed (supplemental Fig. 2). The data set was divided into 18 different bins depending on their ion intensities. For each bin the mean value, standard deviation and 95% confidence was calculated and based on these values we decided to use the value 1.3 for the entire data set.
Selected Reaction Monitoring (SRM)
For each protein to be validated using SRM, proteotypic peptides (41 ), and matching SRM transitions were selected based on a label-free discovery experiment using HCD MS/MS on a mixture of all three biological replicates. Test runs were performed to establish the retention time window for each peptide ion. A minimum of four transitions, mainly y-ions, was chosen for each peptide. When possible, three peptides were used per protein and all SRM analyses were run in five replicates. Twenty-five μg peptide derived from the T2D, lean and obese myotube protein samples, respectively, were desalted as described above. On lyophilization the peptides were resuspended in 25 μl 5% formic acid to gain a concentration of 1 μg/μl. 5 μg peptides were used for each SRM run. The peptides were traped on a 2 cm precolumn (100 μm inner diameter, 360 μm outer diameter, ReproSil - Pur C18 AQ 5 μm (Dr. Maisch, Ammerbuch-Entringen, Germany)) and separated using a 15 cm analytical column (100 μm inner diameter, 360 μm outer diameter, ReproSil - Pur C18 AQ 3 μm) reversed phase capillary column. The peptides were eluted using a gradient from 0% to 34% phase B (0.1% formic acid, 90% MeCN) over 100 min at 250 nL/min directly into a triple quadrupole mass spectrometer (TSQ Vantage, Thermo Scientific, San Jose CA). The TSQ Vantage was operated in a nano-electrospray mode. For ionization, 2300 V of spray voltage and 200 °C capillary temperature were used. The selectivity for Q1 was set to 0.7 and Q3 at 0.7 Da (FWHM). The collision gas pressure of Q2 was set at 1.5 mTorr argon. The collision energy was calculated by Pinpoint 1.0 (Thermo Scientific). The instrument acquisition parameters, including collision energy and intensity were generated by using Pinpoint 1.0 software (Thermo Scientific). The cycle time of 3 s was used as default for the 777 transitions used for targeting 158 peptides. The complete transition list can be found in the supplemental Table 2. All raw files were processed by Pinpoint 1.0. The targeted peptides were verified by manual interpretation. For each targeted peptide that fulfilled the verification criteria, the software computed the integrated peak areas of all transitions for quantitation and calculated %CVs for the five replicates.
Bioinformatic Analysis
Venn diagrams were prepared using the Venny program (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The Protein Center software was used to investigate the cellular localization of the proteins identified using Gene Ontology (GO) annotations as well as to study which KEGG pathways were over-represented in the dataset (ThermoFisher, www.proxeon.com). The Ingenuity® pathway analysis (IPA) software was used for biological pathway analyses (Ingenuity® Systems, www.ingenuity.com).
RESULTS AND DISCUSSION
Strategy
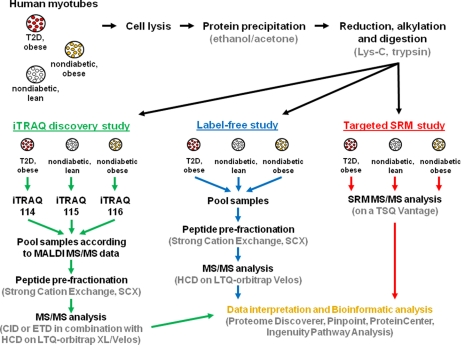
In the present study, we investigated whether there is a change in the basal protein abundance in primary diabetic muscle cells (myotubes) when compared with that of nondiabetic lean and nondiabetic obese controls. This is important in order to obtain a more global overview of the skeletal muscle cell proteome, i.e. to investigate whether the mechanisms leading to the diabetic phenotype e.g. insulin resistance, are at the level of protein expression. The strategies used in this study are illustrated in Fig. 1. The human myotubes were harvested at passage four to keep them as close to the in vivo system as possible. At this stage, the myotubes from 10 subjects were pooled within each group to avoid patient variability in this discovery study. As for in vivo samples, primary cell lines are often limited by the amount of material available. It is therefore crucial to minimize any sample loss during sample preparation. To obtain the best protein precipitation, five different protocols were tested in four replicates (supplemental Fig. S1 and Table 1). The experiments showed that protocols using ethanol and acetone (v/v: 40%/40%) or 100% acetone gave the best protein precipitation from human myotubes. Based on protein measurements using the QubitTM both methods gave ∼100 μg protein from 133 μl cell lysate. However, the dry weight of the precipitated matter from the four replicates isolated by 100% acetone precipitation had an average weight of 1.16 mg whereas ethanol and acetone precipitation only resulted in an average weight of 0.65 mg (supplemental Table 1). It therefore seems that precipitation using acetone alone coprecipitated approximately twice as much nonprotein matter than the ethanol/acetone protocol. Another protocol using chloroform/methanol gave a less efficient protein precipitation on these cells whereas protocols using trichloroacetic acid/acetone/β-mercaptoethanol or just trichloroacetic acid gave poor protein precipitations. This was mainly because of the fact that the precipitated protein pellets were difficult to resolubilize, which also resulted in poor reproducibility (supplemental Fig. 1). The results illustrated the importance of optimizing protein precipitation protocols on the individual samples to be analyzed and based on these results the ethanol and acetone method was used in this study. Proteins harvested from the myotubes (total cell lysates) were reduced, alkylated and subsequently digested using Lys-C and trypsin. The resulting peptides were then analyzed in three different set-ups: (1) an iTRAQ discovery study to identify proteins differentially expressed among the three groups, (2) a label-free qualitative study to obtain HCD MS/MS information on peptides chosen for subsequent validation in; (3) a targeted SRM study. In the iTRAQ discovery study, peptides from diabetic myotubes were labeled using iTRAQ 114, peptides from nondiabetic, lean myotubes were labeled using iTRAQ 115, and peptides from nondiabetic, obese myotubes were labeled using iTRAQ 116. The labeled peptides were subsequently mixed in a 1:1:1 ratio, as determined by MALDI MS/MS analysis. The peptide sample was then fractionated using SCX chromatography to reduce sample complexity before mass spectrometric analysis. The resulting ten peptide fractions were analyzed using nanoLC-electrospray ionization (ESI)-MS/MS. CID or ETD MS/MS was applied for obtaining peptide sequence information, whereas HCD MS/MS was used to obtain quantitative information from the iTRAQ ions in the lower mass area. CID and ETD provide complementary fragmentation and by using both strategies on different replicates we are able to increase the number of peptide identifications; however CID and ETD are both performed in the linear ion trap (LTQ), which has poor resolution in the lower mass region. Neither approach is therefore suitable for the quantitation of iTRAQ ions meaning that analytical space needed to be devoted to HCD scans to provide high resolution information on the relative iTRAQ reporters. By analyzing the sample using a combination of CID or ETD with HCD, we are able to obtain the best conditions for both peptide sequencing and iTRAQ quantitation. A limitation to this strategy, however, is the considerable amount of filling time needed to get good ion statistics on the HCD data. This gives a large reduction in the number of peptide identifications compared with experiments using only CID or ETD for fragmentation. This is, however, needed to obtain strong quantitative data. The experiments in this study were performed as four sample replicates that were analyzed using either CID/HCD on a LC-ESI-LTQ-Orbitrap XL (replicate 1 and 2), ETD/HCD on a LC-ESI-LTQ-Orbitrap XL (replicate 3), or CID/HCD on a LC-ESI-LTQ-Orbitrap Velos (replicate 4). By this we expected to strengthen the data set by increasing the overall number of peptide and protein identifications as well as by obtaining overlapping information at both peptide and protein level among the different replicates.
Fig. 1.
The figure illustrates the strategies used in this study. On cell lysis, proteins are isolated, reduced, alkylated, and digested. The resulting peptides are then used for three different setups; (1) an iTRAQ discovery study in which peptides and thereby proteins that are differentially expressed among the three groups are identified, (2) a label-free study to identify the peptides best suited for MS/MS analysis from each of the proteins found to be regulated by iTRAQ, and (3) a targeted selected reaction monitoring (SRM) study that is set up based on the information found in the two prior studies to validate quantitative findings from the iTRAQ discovery study.
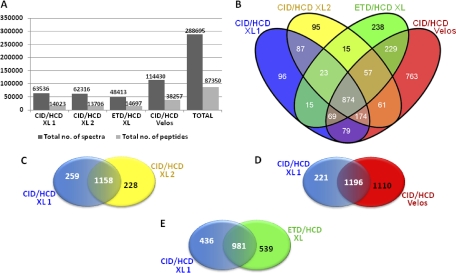
In replicate 1 and 2 (CID/HCD on XL) 63,536 and 62,316 spectra were recorded, respectively (Fig. 2A). This resulted in the identification of 14,023 and 13,706 peptides, respectively. Fewer spectra (48,413) were recorded for replicate 3 (ETD/HCD on XL) as would be expected because ETD takes longer because of the filling-time required for the anions, leaving time for fewer MS/MS. However, more peptides were identified in replicate 3 (14,697). This may be a direct result of labeling the peptides using iTRAQ. Recently, we found that the derivatization of peptides with isobaric tags such as iTRAQ and tandem mass tags (42) resulted in increased charging in electrospray ionization, reducing peptide identification efficiency using CID (43). As ETD performs better on multiply-charged peptides, increased charging would therefore be expected to improve ETD analysis (44). 114,430 spectra were recorded in replicate 4 (CID/HCD on the LTQ Orbitrap Velos) as a result of the high scan speed and sensitivity of the instrument (45). This led to the assignment of 38,257 peptides. In total, 87,350 peptides were identified across all four experiments of which 12,328 were nonredundant peptides from 3147 proteins. This resulted in 2832 unique proteins after protein grouping (supplemental Table 3). The Venn diagram in Fig. 2B illustrates the distribution of the 2832 proteins identified. Because of the complexity of the sample, only 874 proteins were identified in all four replicates whereas 1197 proteins were identified in at least three of the replicates and 1192 proteins were only identified in one replicate. By performing the study in four replicates we were able to increase the coverage of the proteome but as a total cell lysate is highly complex, additional prefractionation on both protein and peptide level and longer LC gradients would undoubtedly aid in overcoming the sampling effects seen here. Running the experiment in two replicates on a LTQ-Orbitrap XL allowed the identification of ∼250 additional proteins with the majority of the protein identifications shared between the two replicates (Fig. 2C). As discussed above, replicates performed on an LTQ-Orbitrap Velos resulted in a considerable increase in the number of protein identifications, and the use of ETD/HCD gave rise to a slight increase in the number of proteins identifications most likely because of iTRAQ super-charging assisting ETD fragmentation (Fig. 2D, 2E).
Fig. 2.
A, The table shows the total number of spectra recorded from each sample as well as the total number of peptides assigned by database searching. B, A total of 2832 unique proteins were identified in this study. This Venn diagram illustrates the distribution of the proteins among the four different experiments. The number of proteins unique as well as shared among different experimental setups. C, CID/HCD versus CID/HCD on a LTQ-orbitrap XL, D, CID/HCD on a LTQ-orbitrap XL versus CID/HCD on a LTQ-orbitrap Velos, and E, CID/HCD versus ETD/HCD on a LTQ-orbitrap XL.
Computational and Bioinformatic Analysis
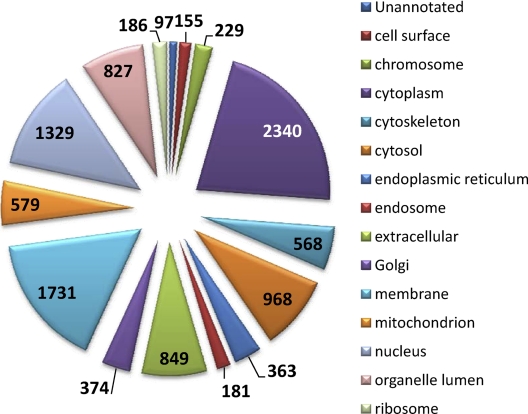
The proteins identified in this study were analyzed to obtain an overview of their functional roles in a biological context. The 2832 proteins were examined with respect to cellular localization using GO annotation analysis in ProteinCenter (Fig. 3). As proteins often belong to several cellular categories, the total percentage will be higher than 100. ProteinCenter revealed that 2340 (82.6%) proteins were assigned to the cytoplasm, 1731 (61.1%) proteins were assigned to the membrane, and 1329 (46.9%) proteins were assigned to the nucleus. From the 2,832 proteins, 579 (20.4%) were assigned to the mitochondria. Muscle tissue is rich in mitochondria and previous proteomic studies of muscle tissue also reveal a high number of mitochondrial proteins (12).
Fig. 3.
The 2832 unique proteins were examined with respect to cellular localization using GO annotation analysis in ProteinCenter (www.proxeon.com).
Quantitative Proteomic Analysis of the Diabetic and Nondiabetic Control Myotubes
For the quantitative analysis, only peptides containing complete labeling (i.e. N-terminal iTRAQ labeling or iTRAQ labeled lysine residue in combination with another N-terminal modification) were considered, which accounted for 99% of the assigned peptide spectra. In addition, only proteins identified with at least two peptides with a minimum Mascot score of 20 and a FDR ≤ 1% were considered. The standard deviation and 95% confidence were calculated for all iTRAQ data manually (data not shown) and based on these data a fold-change of at least 1.3 was accepted for a protein being significantly regulated.
Despite a clear diabetic phenotype observed in myotubes established from diabetic subjects, the majority of the proteins identified in this study proved not to be differentially expressed between the diabetic myotubes and the two nondiabetic controls (supplemental Fig. 2). Also, none of the 579 mitochondrial proteins identified were found to be differentially abundant. Microarray studies of muscle biopsies have revealed several changes in gene expression when comparing skeletal muscle cells from diabetic and nondiabetic subjects (46). Sixty-one of the proteins found to be differentially abundant in human muscle biopsies by Sreekumar and coworkers (46) were in fact identified as not regulated at protein level in this study. In addition, other microarray studies of human myotubes showed no significant changes at the gene expression level (47). A plausible explanation could be that the proteins found to be differentially abundant in human muscle biopsies may be highly influenced by factors such as stress, physical activity, aging and fiber type composition. Human myotubes are unaffected by environmental factors and as such offer a unique model system for studying the genetic influence of the T2D phenotype.
All enzymes from the TCA cycle were identified in this study (including two proteins based on single peptide identifications with a FDR ≤ 5%). None of the enzymes showed any differential abundance in diabetic myotubes when compared with the two nondiabetic controls (supplemental Table 8). A previous study revealed a reduction in citrate synthase activity in diabetic myotybes when compared with nondiabetic myotubes (32). As this enzyme was identified from a strong set of iTRAQ data (90 peptides, 34 of which were nonredundant) in this study, it illustrates that the enzyme activity is not regulated through protein expression nor protein degradation. Additional proteins from other pathways known to be highly important in T2D were also well-characterized in this study. All but two enzymes from lipid oxidation, oxidative phosphorylation, the glycolytic pathway and glycogen metabolism were identified in this study and these pathways were also not found to be regulated at the level of protein expression or degradation (supplemental Table 8). Insulin resistance in skeletal muscle in vivo has been linked to reduced lipid oxidation. Studies in human myotubes indicated that the reduction in lipid oxidation observed in diabetic skeletal muscle in vivo may be of genetic origin as the reduction was also observed in cultured myotubes (20, 24). As all but one enzyme from the fatty acid transport and oxidation were identified in this study and none of the enzymes identified in diabetic myotubes showed a significant change in abundance when compared with the two nondiabetic controls, we expect that the regulation of lipid oxidation is performed at PTM level.
A well-described defect in T2D is reduced glycogen synthesis in skeletal muscle. This pathway regulates the disposal of glucose in skeletal muscle and is in many ways controlled by the insulin-sensitive enzyme glycogen synthase (GS). Myotubes established from patients with T2D, precultured under normophysiological conditions, express an impaired insulin-stimulated GS activation indicating that it is in part a genetic defect (21). As high plasma glucose and/or insulin levels are often observed in obese and diabetic subjects it has been suggested that these metabolic abnormalities might play an important role in the development of acquired defects in insulin action (17). Henry and coworkers concluded that exposure of human skeletal muscle cells to high insulin induces severe insulin resistance through multiply acquired posttranslational defects as no changes were found at the level of GS mRNA or protein expression (17). Also in the present study no differential abundance was observed for GS when compared with the two nondiabetic controls confirming that regulation should be found at PTM level. Glycogen synthase kinase-3 is a serine and threonine protein kinase, which on phosphorylation of GS, inhibits its actions. Studies in myotubes, however, have shown that the failure of insulin to decrease GS phosphorylation and thereby increase its activity was not based on changes in Glycogen synthase kinase-3 expression or activity (19). Instead suggesting that the primary defect in GS involved abnormal activity of another protein kinase or phosphatase (19).
In addition, it has been reported that insulin-mediated glucose oxidation is reduced in myotubes established from human diabetic subjects (18), however, all enzymes from the glycolytic pathway were identified in this study and none of the enzymes showed any differential abundance between diabetic and nondiabetic myotubes.
Overall, this strongly supports the hypothesis that regulation of these pathways may be performed at the level of PTM. Nevertheless, some studies have found the abundance of certain proteins to be differentially regulated in diabetes. However, only a low number of proteins have been identified as regulated. In this study, 12 proteins were found to be differentially abundant among the three groups. Table II lists the 12 proteins found to be differentially abundant along with their iTRAQ ratios. Some of the structural proteins identified in this study like myosin-2, myosin-3, myosin-8 (all identified by both common as well as isoform-specific peptides) and Tenascin were identified as up-regulated in diabetic myotubes and might reflect intrinsic alteration of diabetic myotubes. Sreekumar et al. compared gene expression in skeletal muscle samples from nondiabetic control subjects and type 2 diabetic subjects before and after intensive insulin therapy and identified 85 differentially expressed genes (46). The expression of 74 genes normalized after insulin therapy whereas the expression of 11 genes remained unaffected by the insulin therapy. Six of these proteins were identified with a down-regulated expression in the diabetic muscle (myosin heavy chain isoform I, troponin I, troponin C, tropomyosin, and calmodulin type I). This regulation might be because of other external factors such as physical activity or because of intrinsic alteration of muscle tissue in type 2 diabetes. Moreover, the composition of skeletal muscle fiber has been found to be altered in both type 2 diabetic patients (48) and their nondiabetic first degree relatives (49), with a greater proportion of type IIb (nonoxidative) compared with type I (oxidative) muscle fibers. At present it is still unknown whether altered myosin isoform expression is important for the development of muscle fiber distribution and development of insulin resistance. However, an up-regulation of structural proteins is likely a secondary effect resulting from the diabetic phenotype and not the direct cause leading to insulin resistance and T2D.
Table II. The 12 proteins found to be differentially abundant are listed in this table. The accession number for each protein is listed along with the number of unique peptides assigned to each protein. The overall protein ratios identified by iTRAQ are indicated for each protein. These values are based on the total peptide information obtained for the individual proteins. *2 were isoform-specific. **51 were only assigned to Myosin-3, most were isoform-specific. ***9 were only assigned to Myosin-8, 2 were isoform-specific.
| Protein name | Acc. No. | Unique peptides | Average (Lean/T2D) | Std dev. (Lean/T2D) | Average (Obese/T2D) | Std dev. (Obese/T2D) |
|---|---|---|---|---|---|---|
| Adenosine deaminase | P00813 | 3 | 1.45 | 0.46 | 1.63 | 0.48 |
| Alcohol dehydrogenase 1B | P00325 | 13 | 1.68 | 0.38 | 2.46 | 0.76 |
| Apolipoprotein A-I | P02647 | 2 | 1.32 | 0.02 | 1.27 | 0.10 |
| Eukaryotic initiation factor 4A-I | P60842 | 15 | 0.72 | 0.15 | 0.79 | 0.13 |
| Fatty acid-binding protein | P05413 | 6 | 0.75 | 0.12 | 0.93 | 0.07 |
| Heat shock protein beta-6 | O14558 | 7 | 1.31 | 0.23 | 1.54 | 0.28 |
| Hemoglobin subunit alpha | P69905 | 5 | 1.55 | 0.47 | 1.26 | 0.19 |
| Macrophage-capping protein | P40121 | 3 | 1.30 | 0.05 | 1.15 | 0.02 |
| Myosin-2 | Q9UKX2 | 12* | 0.84 | 0.28 | 0.95 | 0.35 |
| Myosin-3 | P11055 | 80** | 0.76 | 0.14 | 0.77 | 0.16 |
| Myosin-8 | P13535 | 37*** | 0.76 | 0.13 | 0.72 | 0.15 |
| Tenascin | P24821 | 13 | 0.78 | 0.18 | 0.60 | 0.21 |
In the present study, proteins like adenosine deaminase, alcohol dehydrogenase 1, apolipoprotein A-I, and heat shock protein beta-6 were all found to be down-regulated in diabetic myotubes when compared with nondiabetic controls. Pathway analysis was performed on the regulated proteins using IPA and ProteinCenter. However, no common pathway or link among the regulated proteins could be established. Fatty acid-binding protein was identified as slightly down-regulated in myotubes established from obese subjects (both obese diabetic and nondiabetic) whereas hemoglobin subunit alpha was up-regulated in obese myotubes. Hemoglobin is an iron-containing oxygen-transport protein in the red blood cells and their progenitors. The up-regulation of the subunit alpha identified in this study is likely an artifact resulting from hemoglobin adsorption during the biopsy process, and insufficient dilution at the early passages during the establishment of the satellite cell cultures. Peptides originating from the β and δ subunits were also identified with a slight up-regulation (below the threshold). However, peptides identified from all three subunits also contained sites of possible PTM which could explain the variation among the ratios of the different subunits.
Another 24 proteins were found not to be significantly different among the three groups but contained at least one peptide showing abundance differences. These regulated peptides were all ambiguous in the way that they had either been identified from spectra with poor ion statistics or with a high standard deviation for the iTRAQ ratios, which made them less trustworthy. In addition, several of these peptides carried amino acids that could be targeted by various PTMs, which could affect the quantitative results. Furthermore, several regulated peptides only displayed minor differences in regulation among the three groups. Supplementary Table S4 and S5 lists the iTRAQ data for the 36 proteins analyzed by SRM (12 regulated and 24 nonregulated proteins, respectively). The average ratio and standard deviation for each peptide is listed. Overall, the consistency in iTRAQ ratio measurements of peptides from the same protein was good; however, many peptides had sites of possible PTMs which could be a plausible explanation for any variation in the ratios detected. For this reason an average ratio (and standard deviation) was calculated for each protein as a mean value from the peptide values. Peptides carrying oxidized methionine residues were not included in the calculations as a differential abundance of the modification would shift the protein ratio.
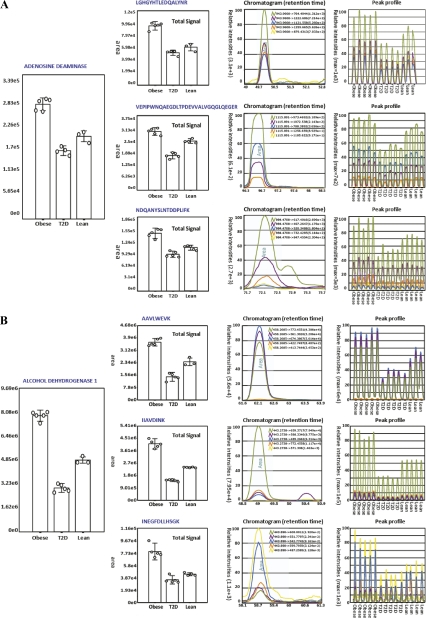
To validate the results from the 12 differentially abundant proteins and to confirm whether the abundance of the additional 24 proteins described above was altered, we designed a targeted SRM experiment (Fig. 1). To obtain data for making transitions for each peptide in SRM, a label-free analysis of the myotube peptides was performed using HCD MS/MS only. This study enabled us to identify proteotypic peptides (41) and the matching SRM transitions. A minimum of four transitions, mainly y-ions, was chosen for each protein and when possible, three peptides were used per protein. In addition, all SRM analyses were run in five replicates. The SRM data were subsequently normalized based on data from three housekeeping proteins because these were known to be present in a 1:1:1 ratio. In addition, equal amounts of a tryptic digest of β-lactoglobulin had been spiked into each sample before the SRM analysis and upon normalization this protein also proved to be present in a 1:1.1 ratio (data not shown). A total of 777 transitions were used for targeting 158 peptides. The complete transition list can be found in the supplemental Table 2. For each targeted peptide that fulfilled the verification criteria, the software computed the integrated peak areas of all transitions for quantification and calculated %CVs for the five replicates. Fig. 4 gives two examples of protein quantitation using SRM. The figure illustrates the SRM data of adenosine deaminase and alcohol dehydrogenase 1 showing the overall abundance of the proteins in the different myotube groups as well as the overall peptide abundance based on all transitions from each peptide. The figure clearly illustrates how both proteins are down-regulated in diabetic myotubes.
Fig. 4.
This figure shows two examples of protein quantitation using SRM: A, Quantitation of adenosine deaminase; The total signal from three peptides selected for SRM analysis illustrating the overall abundance of the protein. Standard deviations are indicated. The overall peptide abundance based on all transition from each peptide is illustrated in “Peptide ratio.” The “Total Signal” is an average of the five technical replicates in each group. The “Chromatogram” shows the peak profile of the different transitions from one of the 12 runs. The “Peak profile” displays the peak profile for all 12 runs and each group is indicated. B, Quantitation of alcohol dehydrogenase 1; The total signal from three peptides selected for SRM analysis including standard deviations. The “Peptide ratio,” “Total Signal,” and “Chromatogram”.
The SRM data proved to be very consistent with the iTRAQ data. Supplemental Table 6 lists the SRM data for the 12 regulated proteins selected for SRM analysis, whereas supplemental Table 7 lists the SRM data for the 24 nonregulated proteins selected for SRM analysis. For each peptide analyzed by SRM the average ratio and standard deviation were calculated based on the transitions for each peptide. The overall protein ratio is listed as a mean of these values. In addition, when available the iTRAQ data for the same peptides (and proteins) are listed along with the average ratio and standard deviations based on the data in supplemental Tables 4 and 5.
The 24 proteins found not to be differentially regulated in the iTRAQ study, though still having peptides displaying differences in regulation, were confirmed as nonregulated using SRM. For 11 proteins, the differentially abundant peptides were amenable for SRM (17 peptides). For the remaining 13 proteins SRM had to be performed on other peptides than the ones identified in the iTRAQ discovery study. The regulations observed for the 12 regulated proteins using iTRAQ were also confirmed by SRM. The exact fold regulation was not always confirmed by SRM, but in many cases the regulation was highly similar to that identified by iTRAQ and in all cases a down- or up-regulation found by iTRAQ was confirmed by SRM. Two proteins, Eukaryotic initiation factor 4A-I and Macrophage-capping protein were identified by iTRAQ as having a slight regulation. SRM confirmed this, but with values below the threshold for acceptable regulation.
Because of the low number of differentially abundant proteins, we were unable to generate functional clusters associated with T2D or obesity. Several of the individual proteins identified in this study, however, do provide functional insight into the biological processes occurring in these samples. For example, adenosine deaminase was identified as 1.45-fold down-regulated in diabetic myotubes when compared with the nondiabetic, lean control using iTRAQ (confirmed as 1.35-fold down-regulated by SRM). The protein was found as 1.63-fold down-regulated when compared with the nondiabetic, obese control using iTRAQ (confirmed as 1.76-fold down-regulated by SRM as illustrated in Fig. 4A). In 1991, Bell and coworkers identified a connection between adenosine deaminase and Maturity-onset diabetes of the young (MODY) (50). In a study of the largest and best-studied Maturity-onset diabetes of the young pedigree, the RW family (51, 52), they found that a DNA polymorphism in the adenosine deaminase gene (ADA) cosegregated with Maturity-onset diabetes of the young. The majority of the diabetic subjects from the RW family were characterized by having reduced or delayed insulin secretory response to glucose. A few years before this, Leighton et al. had shown that changes in the level of adenosine in skeletal muscle affected insulin sensitivity independent of fiber type composition of the muscle (53). They showed that the presence of adenosine deaminase in the medium of soleus, extensor digitorum longus, and hemi-diaphragm muscles of rats resulted in increased insulin sensitivity (53). Another interesting point is that propyl dipeptidase (DPP-IV) is known to bind to adenosine deaminase. Homodimeric DPP-IV is a membrane-bound exoprotease anchored to the plasma membrane of endothelia of almost all organs (54). It also exists as a soluble circulating form in body fluids such as blood plasma and cerebrospinal fluid having both adenosine deaminase-binding and enzymatic activity (55, 56). DPP-IV is involved in the proteolytic cleavage of insulin-sensing incretin hormones such as glucagon-like peptide 1 (GLP-1) and glucose-dependent insulintropic polypeptide (GIP). By proteolytic cleavage, DPP-IV negatively regulates the in vivo concentrations of these hormones. Incretin hormones have potent insulin-secretory activity and are as such antidiabetic. As DPP-IV inactivates GLP-1 and GIP, inhibitors of DPP-IV have been found useful in the treatment of type 2 diabetes and other diseases (54, 57–63).
We observed a reduced abundance of adenosine deaminase in myotubes established from T2D subjects when compared with nondiabetic controls. This decrease would lead to a higher concentration of adenosine, which would contribute to decreased insulin sensitivity in the diabetic myotubes. However, the exact molecular role of adenosine deaminase in diabetes has not yet been established and we speculate whether the diabetic phenotype in vivo is also in part a result of the down-regulation of adenosine deaminase leading to a higher number of DPP-IV molecules free to bind and inactivate GLP-1 and GIP besides from the increase in the adenosine concentration.
Another protein found in this study to be down-regulated in diabetic myotubes is apolipoprotein A-I. This protein was identified from 3 peptides with good ion scores in the iTRAQ experiments that all indicated a down-regulation in T2D (1.3-fold). Only one of these peptides could be successfully validated using SRM. SRM confirmed a slight decrease in the protein abundance in diabetic myotubes, however, the down-regulation was only 1.1-fold which was not considered to be significant. However, because the SRM data were weak compared with the iTRAQ data, we considered the regulation of this protein to be possible. Though, further experiments are needed to validate a regulation. A down-regulation of this protein in diabetic myotubes would in fact correlate with studies showing that Apo A-I expression is reduced both in plasma samples from T2D humans (64) as well as in animal models of diabetes and cell cultures treated with high concentrations of glucose (65).
Alcohol dehydrogenase 1 was also found to be down-regulated in T2D myotubes. It was identified with a 1.68-fold down-regulation in diabetic myotubes when compared with the nondiabetic, lean control using iTRAQ (confirmed as 1.61-fold down-regulated by SRM) and a 2.46-fold down-regulation when compared with the nondiabetic, obese control using iTRAQ (confirmed as 2.56-fold down-regulated by SRM). The protein was identified and quantitated from 13 nonredundant iTRAQ-labeled peptides of which almost all were found in multiple runs (a total of 57 peptides). The iTRAQ-labeled peptides had been assigned to alcohol dehydrogenase 1B, however, the peptides were in fact common between alcohol dehydrogenase 1A, 1B, and 1C. A protein alignment was performed using ClustalW to find the peptides that were sequence specific to the three isoforms and SRM was subsequently performed on these and other selected peptides. SRM confirmed the down-regulation found by iTRAQ (based on three different SRM peptides), however, limited information was found on the isoform-specific peptides simply because of the fact that not all peptides are ionized and analyzed equally well by MS. It was therefore not possible to conclude whether one, two or all three isoforms were regulated. Fig. 4B illustrates the SRM data of alcohol dehydrogenase 1 illustrating the results obtained by SRM.
CONCLUSIONS
To our knowledge, no large-scale quantitative proteomic analysis had been performed on human myotubes to elucidate protein expression profiles between diabetic subjects and nondiabetic lean and nondiabetic obese subjects, respectively. In the present study, a label-based discovery study using iTRAQ was employed to identify and quantify changes in protein expression in human myotubes established from diabetic obese subjects versus nondiabetic subjects. All proteins identified as regulated were subsequently validated using a targeted label-free approach (SRM) and there was a good correlation between the iTRAQ data and the SRM data. The majority of the proteins identified in this study proved not to be differentially expressed between the diabetic myotubes and the two nondiabetic controls. Proteins from all major pathways known to be important in T2D were well-characterized in this study. All but two enzymes from the TCA cycle, lipid oxidation, oxidative phosphorylation, the glycolytic pathway, and glycogen metabolism were identified in the present study and none of these enzymes were found to be differentially abundant in myotubes established from diabetic subjects when compared with nondiabetic controls. These findings support the hypothesis that these specific pathways are not regulated at the level of protein expression or protein degradation but instead through some degree of PTM. Twelve proteins were, however, differentially abundant between the three different groups and it would be interesting for future studies to perform more targeted experiments in which the individual patient samples would be quantitatively screened using SRM to determine the level of these proteins in each patient. Adenosine deaminase was clearly down-regulated in myotubes established from T2D subjects. As DPP-IV, which is known to bind and inactivate the antidiabetic hormones, GLP-1 and GIP, also binds to adenosine deaminase, we speculate whether the diabetic phenotype in vivo is in part a result of the down-regulation of adenosine deaminase leading to a higher number of DPP-IV molecules free to bind GLP-1 and GIP as well as the increase in the adenosine concentration. Because only minor changes were found in protein expression, the pathophysiological mechanisms leading to the diabetic phenotype may also be associated with a change in the level of PTMs of proteins or in molecular mechanisms derived from the action of insulin in these cells. Future studies investigating the effects of insulin stimulation on myotubes may therefore provide additional insight into the mechanisms leading to the diabetic phenotype.
Acknowledgments
We wish to acknowledge Irene Lynfort for her assistance in cell culturing and Kristina Egede Budtz for her assistance in the amino acid analysis. Dr. Arkadiusz Nawrocki is acknowledged for his assistance during the initial data processing and Associate Professor Stuart J. Cordwell for his critical appraisal of the manuscript.
Footnotes
* This work was supported by The Research Foundation of Odense University Hospital, the Danish medical Research Council, and the Novo Nordisk Foundation.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S8.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S8.
1 The abbreviations used are:
- T2D
- type 2 diabetes
- Acc
- accession number
- CID
- collision-induced dissociation
- DPP-IV
- propyl dipeptidase
- EC
- enzyme commission number
- ESI
- electrospray
- ETD
- electron transfer dissociation
- FDR
- false discovery rate
- GIP
- glucose-dependent insulintropic polypeptide
- GLP-1
- glucagon-like peptide 1
- GS
- glycogen synthase
- HCD
- higher-energy collision dissociation
- IPA
- ingenuity® pathway analysis
- iTRAQ
- isobaric peptide tags for relative and absolute quantification
- LC
- liquid chromatography
- LTQ
- linear ion trap
- MALDI
- matrix-assisted laser desorption/ionization
- MS/MS
- tandem mass spectrometry
- PQD
- pulsed q collision induced dissociation
- SCX
- strong cation exchange chromatography
- SILAC
- stable isotope labeling by amino acids in cell culture
- SRM
- selected reaction monitoring
- TCA
- citric acid cycle
- FWHM
- Full Width at Half Maximum
- KEGG
- kyo to encyclopedia of Genes and Genomes.
REFERENCES
- 1. Amos A. F., McCarty D. J., Zimmet P. (1997) The rising global burden of diabetes and its complications: estimates and projections to the year 2010. Diabet. Med. 14 Suppl 5, S1–85 [PubMed] [Google Scholar]
- 2. King H., Aubert R. E., Herman W. H. (1998) Global burden of diabetes, 1995–2025: prevalence, numerical estimates, and projections. Diabetes Care 21, 1414–1431 [DOI] [PubMed] [Google Scholar]
- 3. Drewnowski A. (2009) Obesity, diets, and social inequalities. Nutr. Rev. 67 Suppl 1, S36–9 [DOI] [PubMed] [Google Scholar]
- 4. Zimmet P., Alberti K. G., Shaw J. (2001) Global and societal implications of the diabetes epidemic. Nature 414, 782–787 [DOI] [PubMed] [Google Scholar]
- 5. Inzucchi S., Sherman R. (2005) The prevention of type 2 diabetes mellitus. Endocrinol. Meab. Clin. North Am. 34, 199–219 [DOI] [PubMed] [Google Scholar]
- 6. American Diabetes Association, Total Prevalence of Diabetes and Pre-diabetes. http://www.diabetes.org/diabetes-statistics/prevalence.jsp 2008
- 7. Zimmet P. (2000) Globalization, coca-colonization and the chronic disease epidemic: can the Doomsday scenario be averted? J. Intern. Med. 247, 301–310 [DOI] [PubMed] [Google Scholar]
- 8. Beck-Nielsen H. (1998) Mechanisms of insulin resistance in non-oxidative glucose metabolism: the role of glycogen synthase. J. Basic Clin. Physiol. Pharmacol. 9, 255–279 [DOI] [PubMed] [Google Scholar]
- 9. Beck-Nielsen H., Groop L. C. (1994) Metabolic and genetic characterization of prediabetic states. Sequence of events leading to non-insulin-dependent diabetes mellitus. J. Clin. Invest. 94, 1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beck-Nielsen H., Vaag A., Poulsen P., Gaster M. (2003) Metabolic and genetic influence on glucose metabolism in type 2 diabetic subjects–experiences from relatives and twin studies. Best Pract. Res. Clin. Endocrinol. Metab. 17, 445–467 [DOI] [PubMed] [Google Scholar]
- 11. Højlund K., Gaster M., Beck-Nielsen H. (2007) Tissue Biopsies in Diabetes Research. Clinical Diabetes Research: Methods and Techniques, ISBN 9780470017289, John Wiley and Son [Google Scholar]
- 12. Yi Z., Bowen B. P., Hwang H., Jenkinson C. P., Coletta D. K., Lefort N., Bajaj M., Kashyap S., Berria R., De Filippis E. A., Mandarino L. J. (2008) Global relationship between the proteome and transcriptome of human skeletal muscle. J. Proteome Res. 7, 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Højlund K., Yi Z., Hwang H., Bowen B., Lefort N., Flynn C. R., Langlais P., Weintraub S. T., Mandarino L. J. (2008) Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell Proteomics 7, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anderson L., Seilhamer J. (1997) A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18, 533–537 [DOI] [PubMed] [Google Scholar]
- 15. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henry R. R., Abrams L., Nikoulina S., Ciaraldi T. P. (1995) Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes 44, 936–946 [DOI] [PubMed] [Google Scholar]
- 17. Henry R. R., Ciaraldi T. P., Mudaliar S., Abrams L., Nikoulina S. E. (1996) Acquired defects of glycogen synthase activity in cultured human skeletal muscle cells: influence of high glucose and insulin levels. Diabetes 45, 400–407 [DOI] [PubMed] [Google Scholar]
- 18. Gaster M., Beck-Nielsen H. (2004) The reduced insulin-mediated glucose oxidation in skeletal muscle from type 2 diabetic subjects may be of genetic origin–evidence from cultured myotubes. Biochim. Biophys. Acta 1690, 85–91 [DOI] [PubMed] [Google Scholar]
- 19. Gaster M., Brüsgaard K., Handberg A., Højlund K., Wojtaszewski J. F., Beck-Nielsen H. (2004) The primary defect in glycogen synthase activity is not based on increased glycogen synthase kinase-3alpha activity in diabetic myotubes. Biochem. Biophys. Res. Commun. 319, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 20. Gaster M., Rustan A. C., Aas V., Beck-Nielsen H. (2004) Reduced lipid oxidation in skeletal muscle from type 2 diabetic subjects may be of genetic origin: evidence from cultured myotubes. Diabetes 53, 542–548 [DOI] [PubMed] [Google Scholar]
- 21. Gaster M., Petersen I., Højlund K., Poulsen P., Beck-Nielsen H. (2002) The diabetic phenotype is conserved in myotubes established from diabetic subjects: evidence for primary defects in glucose transport and glycogen synthase activity. Diabetes 51, 921–927 [DOI] [PubMed] [Google Scholar]
- 22. Gaster M., Kristensen S. R., Beck-Nielsen H., Schrøder H. D. (2001) A cellular model system of differentiated human myotubes. APMIS 109, 735–744 [DOI] [PubMed] [Google Scholar]
- 23. Gaster M. (2007) Reduced TCA -flux in diabetic myotubes: A governing influence on the diabetic phenotype ? Biochem. Biophys. Res. Commun. 1772, 755–765 [DOI] [PubMed] [Google Scholar]
- 24. Gaster M. (2009) Reduced lipid oxidation in human myotubes established from obese and type 2 diabetic subjects. Biochem. Biophys. Res. Commun. 382, 766–770 [DOI] [PubMed] [Google Scholar]
- 25. Minet A. D., Gaster M. (2010) ATP synthesis is impaired in isolated mitochondria from myotubes established from type 2 diabetic subjects. Biochem. Biophys. Res. Commun. 402, 70–74 [DOI] [PubMed] [Google Scholar]
- 26. Oda Y., Huang K., Cross F. R., Cowburn D., Chait B. T. (1999) Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 96, 6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 28. Zhu H., Pan S., Gu S., Bradbury E. M., Chen X. (2002) Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun. Mass Spectrom. 16, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 29. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 30. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ortenblad N., Mogensen M., Petersen I., Højlund K., Levin K., Sahlin K., Beck-Nielsen H., Gaster M. (2005) Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: evidence for an intrinsic oxidative enzyme defect. Biochim. Biophys. Acta 1741, 206–214 [DOI] [PubMed] [Google Scholar]
- 33. Gaster M., Beck-Nielsen H. (2006) Triacylglycerol accumulation is not primarily affected in myotubes established from type 2 diabetic subjects. Biochim. Biophys. Acta 1761, 100–110 [DOI] [PubMed] [Google Scholar]
- 34. Bergstrom J. (1975) Percutaneous needle biopsy of skeletal muscle in physiological and clinical research. Scand. J. Clin. Lab. Invest. 35, 609–616 [PubMed] [Google Scholar]
- 35. Gaster M., Beck-Nielsen H., Schrøder H. D. (2001) Proliferation conditions for human satellite cells. The fractional content of satellite cells. APMIS 109, 726–734 [DOI] [PubMed] [Google Scholar]
- 36. Gaster M., Schrøder H. D., Handberg A., Beck-Nielsen H. (2001) The basal kinetic parameters of glycogen synthase in human myotube cultures are not affected by chronic high insulin exposure. Biochim. Biophys. Acta 1537, 211–221 [DOI] [PubMed] [Google Scholar]
- 37. Gaster M. (2009) The dynamic of lipid oxidation in human myotubes. Biochim. Biophys. Acta 1791, 17–24 [DOI] [PubMed] [Google Scholar]
- 38. Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116 [DOI] [PubMed] [Google Scholar]
- 39. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC - A phosphoproteomic strategy for the rapid separation of mono-phosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 40. Rewitz K. F., Larsen M. R., Lobner-Olesen A., Rybczynski R., O'Connor M. B., Gilbert L. I. (2009) A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem. Mol. Biol. 39, 475–483 [DOI] [PubMed] [Google Scholar]
- 41. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 42. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 43. Thingholm T. E., Palmisano G., Kjeldsen F., Larsen M. R. (2010) Undesirable Charge-Enhancement of Isobaric Tagged Phosphopeptides Leads to Reduced Identification Efficiency. J. Proteome Res 9, 4045–4052 [DOI] [PubMed] [Google Scholar]
- 44. Kjeldsen F., Giessing A. M., Ingrell C. R., Jensen O. N. (2007) Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal. Chem. 79, 9243–9252 [DOI] [PubMed] [Google Scholar]
- 45. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sreekumar R., Halvatsiotis P., Schimke J. C., Nair K. S. (2002) Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51, 1913–1920 [DOI] [PubMed] [Google Scholar]
- 47. Frederiksen C. M., Højlund K., Hansen L., Oakeley E. J., Hemmings B., Abdallah B. M., Brusgaard K., Beck-Nielsen H., Gaster M. (2008) Transcriptional profiling of myotubes from patients with type 2 diabetes: no evidence for a primary defect in oxidative phosphorylation genes. Diabetologia 51, 2068–2077 [DOI] [PubMed] [Google Scholar]
- 48. Gaster M., Staehr P., Beck-Nielsen H., Schrøder H. D., Handberg A. (2001) GLUT4 is reduced in slow muscle fibers of type 2 diabetic patients: is insulin resistance in type 2 diabetes a slow, type 1 fiber disease? Diabetes 50, 1324–1329 [DOI] [PubMed] [Google Scholar]
- 49. Nyholm B., Qu Z., Kaal A., Pedersen S. B., Gravholt C. H., Andersen J. L., Saltin B., Schmitz O. (1997) Evidence of an increased number of type IIb muscle fibers in insulin-resistant first-degree relatives of patients with NIDDM. Diabetes 46, 1822–1828 [DOI] [PubMed] [Google Scholar]
- 50. Bell G. I., Xiang K. S., Newman M. V., Wu S. H., Wright L. G., Fajans S. S., Spielman R. S., Cox N. J. (1991) Gene for non-insulin-dependent diabetes mellitus (maturity-onset diabetes of the young subtype) is linked to DNA polymorphism on human chromosome 20q. Proc. Natl. Acad. Sci. U.S.A. 88, 1484–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fajans S. S. (1989) Maturity-onset diabetes of the young (MODY). Diabetes Metab. Rev. 5, 579–606 [DOI] [PubMed] [Google Scholar]
- 52. Fajans S. S. (1990) Scope and heterogeneous nature of MODY. Diabetes Care 13, 49–64 [DOI] [PubMed] [Google Scholar]
- 53. Leighton B., Lozeman F. J., Vlachonikolis I. G., Challiss R. A., Pitcher J. A., Newsholme E. A. (1988) Effects of adenosine deaminase on the sensitivity of glucose transport, glycolysis and glycogen synthesis to insulin in muscles of the rat. Int. J. Biochem. 20, 23–27 [DOI] [PubMed] [Google Scholar]
- 54. Green B. D., Flatt P. R., Bailey C. J. (2006) Dipeptidyl peptidase IV (DPP IV) inhibitors: A newly emerging drug class for the treatment of type 2 diabetes. Diab. Vasc. Dis. Res. 3, 159–165 [DOI] [PubMed] [Google Scholar]
- 55. Gorrell M. D., Gysbers V., McCaughan G. W. (2001) CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand J. Immunol. 54, 249–264 [DOI] [PubMed] [Google Scholar]
- 56. Drucker D. J. (2003) Therapeutic potential of dipeptidyl peptidase IV inhibitors for the treatment of type 2 diabetes. Expert Opin. Investig. Drugs 12, 87–100 [DOI] [PubMed] [Google Scholar]
- 57. Ahn J. H., Shin M. S., Jun M. A., Jung S. H., Kang S. K., Kim K. R., Rhee S. D., Kang N. S., Kim S. Y., Sohn S. K., Kim S. G., Jin M. S., Lee J. O., Cheon H. G., Kim S. S. (2007) Synthesis, biological evaluation and structural determination of beta-aminoacyl-containing cyclic hydrazine derivatives as dipeptidyl peptidase IV (DPP-IV) inhibitors. Bioorg. Med. Chem. Lett. 17, 2622–2628 [DOI] [PubMed] [Google Scholar]
- 58. Augustyns K., Bal G., Thonus G., Belyaev A., Zhang X. M., Bollaert W., Lambeir A. M., Durinx C., Goossens F., Haemers A. (1999) The unique properties of dipeptidyl-peptidase IV (DPP IV / CD26) and the therapeutic potential of DPP IV inhibitors. Curr. Med. Chem. 6, 311–327 [PubMed] [Google Scholar]
- 59. Augustyns K., Van der Veken P., Senten K., Haemers A. (2005) The therapeutic potential of inhibitors of dipeptidyl peptidase IV (DPP IV) and related proline-specific dipeptidyl aminopeptidases. Curr. Med. Chem. 12, 971–998 [DOI] [PubMed] [Google Scholar]
- 60. Ferraris D., Belyakov S., Li W., Oliver E., Ko Y. S., Calvin D., Lautar S., Thomas B., Rojas C. (2007) Azetidine-based inhibitors of dipeptidyl peptidase IV (DPP IV). Curr. Top Med. Chem. 7, 597–608 [DOI] [PubMed] [Google Scholar]
- 61. Ferraris D., Ko Y. S., Calvin D., Chiou T., Lautar S., Thomas B., Wozniak K., Rojas C., Kalish V., Belyakov S. (2004) Ketopyrrolidines and ketoazetidines as potent dipeptidyl peptidase IV (DPP IV) inhibitors. Bioorg. Med. Chem. Lett. 14, 5579–5583 [DOI] [PubMed] [Google Scholar]
- 62. Kowalchick J. E., Leiting B., Pryor K. D., Marsilio F., Wu J. K., He H., Lyons K. A., Eiermann G. J., Petrov A., Scapin G., Patel R. A., Thornberry N. A., Weber A. E., Kim D. (2007) Design, synthesis, and biological evaluation of triazolopiperazine-based beta-amino amides as potent, orally active dipeptidyl peptidase IV (DPP-4) inhibitors. Bioorg. Med. Chem. Lett. 17, 5934–5939 [DOI] [PubMed] [Google Scholar]
- 63. Green B. D., Irwin N., Duffy N. A., Gault V. A., O'harte F, P., Flatt P. R. (2006) Inhibition of dipeptidyl peptidase-IV activity by metformin enhances the antidiabetic effects of glucagon-like peptide-1. Eur. J. Pharmacol. 547, 192–199 [DOI] [PubMed] [Google Scholar]
- 64. Liu X., Feng Q., Chen Y., Zuo J., Gupta N., Chang Y., Fang F. (2009) Proteomics-based identification of differentially-expressed proteins including galectin-1 in the blood plasma of type 2 diabetic patients. J. Proteome Res. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 65. Mooradian A. D., Haas M. J., Wong N. C. (2004) Transcriptional control of apolipoprotein A-I gene expression in diabetes. Diabetes 53, 513–520 [DOI] [PubMed] [Google Scholar]