Abstract
The mitochondrial respiratory chain is comprised of four different protein complexes (I–IV), which are responsible for electron transport and generation of proton gradient in the mitochondrial intermembrane space. This proton gradient is then used by FoF1-ATP synthase (complex V) to produce ATP by oxidative phosphorylation. In this study, the respiratory complexes I, II, and III were affinity purified from Trypanosoma brucei procyclic form cells and their composition was determined by mass spectrometry. The results along with those that we previously reported for complexes IV and V showed that the respiratome of Trypanosoma is divergent because many of its proteins are unique to this group of organisms. The studies also identified two mitochondrial subunit proteins of respiratory complex IV that are encoded by edited RNAs. Proteomics data from analyses of complexes purified using numerous tagged component proteins in each of the five complexes were used to generate the first predicted protein-protein interaction network of the Trypanosoma brucei respiratory chain. These results provide the first comprehensive insight into the unique composition of the respiratory complexes in Trypanosoma brucei, an early diverged eukaryotic pathogen.
Mitochondria are dynamic organelles essential for cellular life, death, and differentiation of virtually every eukaryotic cell. They house systems for energy production through oxidative phosphorylation, synthesis of key metabolites, and iron-sulfur cluster assembly. The oxidative phoshorylation system of eukaryotic mitochondria comprises five major complexes located in the mitochondrial (mt)1 inner membrane, and often abbreviated as mt complexes I–V. The redox energy of the substrates NADH and succinate is first converted into an electrochemical proton potential across the inner mt membrane by respiratory complexes I (NADH:ubiquinone reductase), II (SDH, succinate:ubiquinone reductase), III (bc1, ubiquinone:cytochrome c reductase), and IV (cytochrome c oxidase). The electrochemical proton potential is then used by complex V (FoF1-ATP synthase) to synthesize ATP from ADP and inorganic phosphate, a mechanism that has essentially remained unchanged from bacteria to human (1). However, parasitic organisms have exploited unique energy metabolic pathways by adapting to their natural host habitats (2). Indeed, the respiratory systems of parasites typically show greater diversity in electron transfer pathways than those of their host, and Trypanosoma brucei is no exception to this rule (3).
T. brucei, the causative agent of human African trypanosomiasis (HAT), or sleeping sickness, is a blood-borne pathogenic parasite transmitted by tsetse flies. It has a complex life cycle that alternates between the bloodstream forms (BF) in the mammalian host and several stages in the insect vector starting with the procyclic form (PF) in the midgut. During T. brucei differentiation between the distinct life-cycle stages, the mitochondrion undergoes morphological and functional changes, and the parasite switches its energy metabolism from amino acid to glucose oxidation (4). BF cells, which live in sugar-rich environment, use energy metabolism predominantly through the glycolytic pathway (5). They contain no cytochrome-mediated respiratory chain and they possess a unique electron transport chain in the mitochondria, the glycerol-3-phosphate dehydrogenase and the salicyl hydroxamic acid (SHAM)-sensitive alternative oxidase, which is known as the trypanosome alternative oxidase (TAO) (6). Despite the absence of complete cytochrome-containing complexes III and IV in BF trypanosomes, a mt membrane potential is maintained and involves the hydrolytic activity of the FoF1-ATP synthase complex (7). Conversely, PF cells are dependent on the cytochrome-containing respiratory chain and ATP generated by conventional function of the FoF1-ATP synthase complex for their energy production (8, 9). The branched electron-transport chain contains four complexes that donate electrons to the ubiquinone pool, two NADH:ubiquinone oxidoreductases (complex I and a rotenone-insensitive enzyme), complex II, and glycerol-3-phosphate dehydrogenase. Reduced ubiquinol can be reoxidized by the transfer of electron to either the TAO, which does not translocate protons, or to the cytochrome-containing complexes III and IV that produce a proton motive force by translocation of protons and thus create essential membrane potential (10).
Although the T. brucei genome has been sequenced (11), little information is available on the subunit composition of mt complexes I–V based on similarity searches. However, some respiratory complexes have been partially characterized in other trypanosomatids such as Crithidia fasciculata, T. cruzi, and Leishmania tarentolae (12–15). In recent studies, we have determined the protein composition of complexes IV and V, and part of complex I purified from mitochondria of T. brucei PF cells (8, 16, 17, 25). These analyses revealed the uniqueness of respiratory complexes in trypanosomes, where large numbers of component proteins have no homologs outside of the Kinetoplastida.
In this study, we focus on the comprehensive characterization of all respiratory complexes in T. brucei, collectively termed the respiratome. We report the composition of complexes II and III from PF cells, and extend the characterization of complex I by identifying additional protein constituents. This included the identification of two subunits of the respiratory complex IV, both encoded by mt edited RNAs. We also present a predicted protein-protein interaction network of the respiratome, which was generated using proteomics data collected from numerous tagged proteins in each of the complexes I–V. Our results provide a comprehensive insight into the unique composition of the respiratory complexes in one of the life-cycle stages of T. brucei.
EXPERIMENTAL PROCEDURES
Cell Growth
T. brucei PF cells IsTat 1.7a were grown in vitro at 27 °C in SDM-79 media containing hemin (7.5 mg/ml) and 10% (v/v) fetal bovine serum to a density of 1–2 × 107 cells/ml. PF T. brucei strain 29.13 (18), which contains integrated genes for T7 polymerase and the tetracycline repressor, was grown in the presence of G418 (15 μg/ml) and hygromycin (25 μg/ml). The cells were harvested by centrifugation at 6000 × g for 10 min at 4 °C.
Tandem Affinity Purification (TAP)-tagged Cell Lines
To create constructs for the inducible expression of TAP tagged proteins in T. brucei, the open reading framess of interest were PCR amplified from the genomic DNA of T. brucei strain Lister 427. A detailed list of all tagged proteins and primers used to amplify the selected open reading frames is provided in the supplemental material. The PCR products were cloned into pGEM-T easy vector, digested with BamHI or BglII and HindIII enzymes, and ligated into the pLEW79-MHT vector (19, 20). The plasmids were linearized with NotI enzyme, transfected into PF T. brucei 29.13 cell line, phleomycin-resistant clones were selected, and checked for tetracycline-regulated expression. The transgenic PF cell lines expressing a TAP-tagged protein were supplemented with 2.5 μg/ml phleomycin.
Sample Preparation for Native Electrophoresis
Mt vesicles were isolated from IsTat 1.7a cells by hypotonic lysis followed by density gradient flotation in 20–35% linear Percoll gradients (21). The enriched vesicles were solubilized by adding n-dodecyl-β-d-maltoside corresponding to n-dodecyl-β-D-maltoside/protein ratio of 2 (g/g) and incubated on ice for 30 min. Insoluble material was removed by centrifugation at full speed in a microcentrifuge for 30 min at 4 °C.
Blue Native Electrophoresis (BNE) and High-resolution Clear Native Electrophoresis (hrCNE)
For BNE, 100 μg of mt lysate was supplemented with a 5% (w/v) stock solution of Coomassie Blue G-250 in 500 mm 6-aminohexanoic acid and analyzed on a 3–12% acrylamide gradient gel (8) using 50 mm Tricine, 15 mm Bis-Tris, pH 7.0, 0.02% Coomassie Blue G-250 as cathode buffer, and 50 mm Bis-Tris, pH 7.0 as anode buffer. For hrCNE (22), 100 μg of mt lysate was supplemented with 50% glycerol, 0.1% Ponceau S, and analyzed on a 4–13% acrylamide gradient gel using 50 mm Tricine, 7.5 mm imidazole, pH 7.0, 0.05% deoxycholate as cathode buffer, and 25 mm imidazole, pH 7.0 as anode buffer. All the native gels were run at 4οC at 100 V until the dye marker approached the gel front.
In-gel Catalytic Activity Assays
Following electrophoresis, complex I–V bands were visualized by activity staining as previously described (22). Briefly, to visualize complex I activity, the gel was incubated in 100 mm Tris-HCl, 0.1 mg/ml NADH, and 2.5 mg/ml nitrotetrazolium blue, pH 7.4. To visualize complex II activity, the gel was incubated in 50 mm phosphate buffer, 20 mm sodium succinate, 0.2 mm phenazine methasulfate, 2.5 mg/ml nitrotetrazolium blue, pH 7.4. For complex III, the gel was incubated in 50 mm phosphate buffer, pH 7.2 and 0.05% 3,3′-diaminobenzidine tetrahydrochloride; and the specificity of the reaction was demonstrated using 2 mm antimycin A. For complex IV, the gel was incubated in 50 mm phosphate buffer, pH 7.2, 0.05% 3,3′-diaminobenzidine tetrahydrochloride and 50 μm horse heart cytochrome c. Reactions were carried out at room temperature by slow agitation overnight and stopped by fixing the gels in 45% methanol and 10% acetic acid. Complex V was visualized as previously reported (8).
Native Western Blot Analysis for Complex I and III
Following electrophoresis, the native gels were transferred onto an Immobilon membrane overnight at 20 V at 4οC. The membranes were then blocked with Odyssey® Blocking Buffer, probed with mAb52 (1:10), which recognize a native epitope of subcomplex Iα (16) or with rabbit polyclonal antibodies against T. brucei apocytochrome c1 (apoc1) (1:1000) (23) to detect complex III. IRDye680 conjugated goat anti-rabbit (LiCor 926–32221) and IRDye800 conjugated goat anti-mouse (Rockland 610–132-121) were used as secondary antibodies at 1:15,000. Protein signal was detected by Odyssey™ Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) using one-color fluorescence detection at 700 or 800 nm and analyzed with Odyssey version 3.0 software.
Tandem Affinity Purification of Tagged Complexes
TAP-tagged proteins were expressed by induction with tetracycline (100 ng/ml of culture) for 48 h and 500 ml of cells were harvested by centrifugation at a density of ∼2 × 107 cells/ml. Tagged complexes were purified from cell lysates by two methods as previously described (8, 16). Briefly, in method 1 (M1) the harvested cells were lysed by 1% Triton-X 100 and the tagged complexes were isolated by IgG affinity chromatography. The bound complexes were eluted by TEV protease cleavage and fractionated on 10–30% glycerol gradients by centrifugation for 5 h at 38,000 rpm at 4 °C in Beckman Ultracentrifuge (SW40 Ti swinging bucket rotor). The sedimentation profiles of the tagged complexes were monitored by Western blot analyses using anti-His6 mAb. Peak reactive fractions were pooled and further purified by calmodulin affinity chromatography. In method 2, the tagged complexes were purified from cells lysed with 0.25% Nonidet P-40, cleared by low speed centrifugation and the supernatant was further treated with 1.25% Nonidet P-40 and cleared by high speed centrifugation. The tagged complexes were isolated by sequential IgG and calmodulin affinity columns.
Immunofluorescence Assay
Subcellular localizations of the expressed tagged proteins within the cell were determined by immunofluorescent assay using anti c-myc antibody (Sigma) as described (16). Colocalization analysis was performed using mAb 78 against mt heat shock protein 70 (16) coupled with Texas® Red-X conjugated secondary antibody (Invitrogen).
Protein Identification
We prepared and analyzed the samples by gel-based and gel-free approaches as described previously by us (16, 24, 25). Briefly, proteins in gel pieces were reduced with 10 mm dithiotreitol at 56 °C for 30 min, and alkylated with 55 mm iodoacetamide at room temperature for 45 min, and digested with sequencing-grade modified trypsin (Promega, Madison, WI) at 37 °C overnight; and the resulting peptides were extracted. The TAP tag-purified protein samples were precipitated with 6 volumes of acetone, and the proteins were denatured with 8 m urea, 1 mm dithiotreitol; diluted 1:4, and digested in-solution with trypsin. The resulting peptides were purified using C18 beads (Magnetic Dynabeads RPC18, Invitrogen). The peptides from gel pieces or complex mixtures were fractionated by nanoflow liquid chromatography using a 10-cm-long × 75-μm-inner diameter C18 capillary column and analyzed on line by electrospray ionization tandem mass spectrometry using a linear trap quadrupole mass spectrometer (Thermo Electron). The bound peptides were eluted from the C18 column at a flow rate of 200 nl/min with a 45-min linear gradient of 5–40% acetonitrile in 0.4% acetic acid followed by a 5-min linear gradient of 40–80% acetonitrile in 0.4% acetic acid.
Xcalibur 1.4 SR1 version software was used to collect MS data, and the mass range for the MS scan was m/z 400–1400. The tandem MS (MS/MS) data of the five most intense ions were collected sequentially following each MS scan using the dynamic exclusion parameter, where a specific ion was sequenced only twice and was excluded from the list for 45 s. The MS data were analyzed against T. brucei v4.0 predicted protein sequence database (11), which contains 9211 protein entries and additionally 18 predicted protein sequence from mt encoded edited and unedited RNAs, and bovine serum albumin, immunoglobulin heavy and light chains and keratin sequences. The peak list generation and search against database were carried out using SEQUEST module of Bioworks 3.1, cluster version SR1 (parameters: molecular weight range, 400–3500; mass tolerance, 1.0 Da; group scan, 25; minimum ion count, 15; no enzyme was specified during the search; no fixed modification was set for any of the amino acids, but differential modification for “M” and “C” were set at 15.994 and 57.000 Da, respectively). The output from SEQUEST search was filtered and compiled using PeptideProphet and ProteinProphet programs (26, 27). Our error rate of protein assignment is less than 1% as determined by comparing the proteomic data with six-frame translated T. brucei genomic sequence database that included 271,892 polypeptide entries STOP codon to-STOP codon (25). The data set presented here includes only the doubly tryptic peptides that have a minimum peptide identification probability of 0.9 and have a minimum SEQUEST X correlation value of 1.5 for +1 ions, 1.8 for +2 ions, and 2.5 for +3 ions. We excluded any peptide containing more than one missed trypsin cleavage site in the sequence. Proteins containing these peptides and with minimum identification probability of 0.9 were considered positive. All except one protein presented here was identified with two or more peptide matches and with a protein identification probability of ≥0.95. They all are unique proteins. For the protein identified with only one peptide match the MS/MS spectra and relevant scores are provided in supplemental Table S2 and supplemental Fig. S1.
Sequence Analysis
The probable functions of the proteins were assigned based on GeneDB annotation and for proteins with unknown function possible motifs and/or domains were searched in InterPro (http://www.ebi.ac.uk/Tools/InterProScan/), Pfam (http://pfam.sanger.ac.uk/) and NCBI CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases. The protein transmembrane topology was predicted using the TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM).
Prediction of Protein-Protein Interactions
The composition of each complex was assessed by tagging multiple proteins individually as described above and by performing multiple MS analyses of each sample. The .out files (SEQUEST output) were compiled and filtered using the DTASelect program (28) to identify proteins in each analysis. Proteins identified with at least two tryptic peptides and with minimum peptide and protein identification probability of ≥0.9 were included in the list except for known non-mt proteins and highly abundant mt protein that are often seen in various TAP-tag experiments (17). The .raw files were converted to mzXML format and the Census program (29) was used to determine the spectral count of each of the peptides. For each individual protein, the number of spectra per peptide was calculated (sum of spectral count of all peptides respective to the protein/number of unique peptides). The relative ratio of spectra per peptide to bait protein (tagged protein) was calculated by dividing spectra per peptide value of each individual protein identified in the TAP-tag experiment by the spectra per peptide value of tagged protein. We used the Cytoscape software (http://www.cytoscape.org) to predict protein-protein interaction networks where a target protein was linked to bait protein if it had relative ratio of ≥1.0. A value of ≥1.25 was considered as high-confidence interactions.
RESULTS
In-gel Activity Staining of T. brucei mt Respiratory Complexes
Native electrophoresis and subsequent in-gel catalytic activity assays were used for visualization of respiratory chain complexes I–V (Figs. 1A–C). MS analyses were carried out on gel slices to assess the presence of the component protein(s) of complexes in the respective stained gel bands. Numerous proteins were identified in each gel slice (supplemental Table S1), some of which included known components of respiratory complexes. The large majority of the proteins that were detected are not part of the respective respiratory complexes, which likely reflects the complexity of the mt lysate samples and comigration of large complexes in native gels. However, some of these proteins, especially those with unknown functions, may be associated with their respective respiratory complexes.
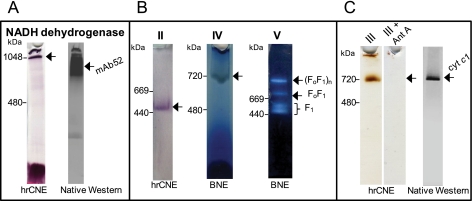
Fig. 1.
In-gel activity staining of T. brucei mt respiratory complexes separated by hrCNE and BNE. Mt lysate was fractionated by hrCNE and BNE PAGE, and stained for (A) NADH dehydrogenase activity. The hrCNE gel was subsequently blotted and probed with mAb52, which recognizes a native epitope of the oxidoreductase complex (16). (B) Activity of complexes II, IV and V, and (C) activity of complex III in the absence and presence of antimycin A (Ant A). The hrCNE gel was also blotted and probed with polyclonal antibodies specific to the apoc1 subunit of complex III. Arrows mark the major stained band in each case. The sizes of high molecular weight standards are indicated on the left.
NADH dehydrogenase activity revealed two stained bands (Fig. 1A). One band was in the well of the gel, suggesting that some material was not solubilized, very high molecular weight complexes and/or highly hydrophobic membrane-bound protein complexes that did not enter the gradient separation gel. The other band was observed at an apparent molecular weight of 1 MDa. MS analyses of the gel slice containing the ∼1 MDa NADH dehydrogenase activity band predominantly identified Tb11.01.1740, Tb11.47.0004, Tb11.01.3550, and Tb11.01.8470 (supplemental Table S1), four proteins described as components of the 2-oxoglutarate dehydrogenase complex (OGDC) (16). Although the theoretical molecular mass of the bovine complex I was reported as 980 kDa (30), only ten proteins out of the 30 nuclear-encoded subunits that were either predicted (31) or identified as part of the oxidoreductase complex (16, 25) were identified in this gel slice (supplemental Table S1, band 2). However, it was previously reported that T. brucei complex I has a mass of ∼600 kDa (9, 32), thus we analyzed the entire lane of the native gel by MS including the well (supplemental Table S1, bands 1–10). MS analyses of the ∼ 1 MDa to ∼ 700 kDa region identified a total of 20 proteins out of the 30 subunits assigned to complex I (supplemental Table S1, bands 3 and 4). Western analysis using mAb52, which recognizes a native epitope in the oxidoreductase complex that corresponds to subcomplex Iα ((16) and this work) recognized the same region of the gel (Fig. 1A). These results suggest that NADH dehydrogenase activity staining reported earlier as complex I activity (9, 32) may not only be specific for complex I in PF cells, but it is also likely because of OGDC.
The complex II in-gel activity assay resembled the NADH dehydrogenase activity assay except that succinate rather than NADH was used as electron donor (Fig. 1B). MS analyses of the ∼500 kDa reactive gel band identified Tb09.160.4380, Tb927.8.6580, and Tb927.8.3380, the three known core components of T. brucei SDH complex (supplemental Table S1), indicating that the stained band corresponds to T. brucei complex II. Specific in-gel complex IV staining revealed a band of an apparent molecular mass of ∼ 700 kDa (Fig. 1B), and MS analyses identified 15 known subunits of complex IV (supplemental Table S1 and (17)). Complex V was stained by an ATP hydrolysis assay that can identify holo-FoF1 complex and catalytically active F1 moiety as previously described in our study of T. brucei ATP synthase complex (Fig. 1B and (8)).
For the in-gel staining of complex III, we applied the method described previously for bovine heart complex III, in which 3,3′-diaminobenzidine tetrahydrochloride is used to stain complex III heme specifically (22). MS analyses of the gel slice containing the ∼720 kDa activity band (Fig. 1C) identified two known subunits of complex III; Tb927.8.1890 and Tb09.211.4700 (supplemental Table S1). The specificity of the catalytic activity was further demonstrated by incubating the mt lysates with 2 mm antimycin A, a specific inhibitor of complex III before hrCNE electrophoresis (Fig. 1C). Upon inhibition with antimycin A, the ∼720 kDa activity band was no longer visible, and only residual activities remained visible in the well, thus supporting that the stained band corresponds to complex III. Furthermore, native Western analysis with antibodies against apoc1 (33), a core component of complex III, confirmed the size of the native complex III (Fig. 1C).
In summary, we visualized under native conditions the T. brucei mt respiratory complexes II–V by activity staining using the characteristic enzyme activities associated with each complex. The primary observed NADH dehydrogenase activity is most likely because of OGDC, however, T. brucei complex I was detected using a specific monoclonal antibody.
Identification of Mitochondrial-encoded Subunits of Respiratory Chain Complexes
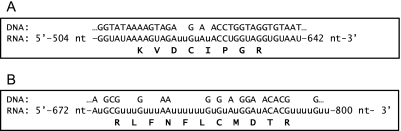
The mt genome of T. brucei, like that of most other organisms, encodes subunits of the respiratory complexes, such as apocytochrome b (complex III), cytochrome c oxidase subunits I (COI), II (COII), and III (COIII), and subunit A6 of ATP synthase (34). To date, apocytochrome b and COI from Leishmania tarentolae are the only proteins identified that are translated from edited mRNAs (35–37), which are the processed RNA products of kinetoplastid mt DNA (kDNA). Liquid chromatography (LC)-MS/MS analyses of the ∼1 MDa to ∼700 kDa region (Fig. 1A) identified three unique peptides that match to the COII subunit of complex IV (Fig. 2 and supplemental Table S1) and COIII, another subunit of complex IV was identified by a single tryptic peptide in the gel slice containing the ∼720 kDa activity band (Fig. 1C) (Fig. 2, supplemental Table S2 and supplemental Fig. S1). Fig. 2 shows the amino acid sequence of a tryptic peptide that matches the edited region of COII (Fig. 2A), and another peptide sequence that matches an edited region of COIII (Fig. 2B). Hence, peptides from two mt encoded-protein subunits of complex IV and their edited mRNAs were detected.
Fig. 2.
Identification of the mitochondrial encoded subunits COII and COIII. A portion of the gene (DNA), edited mRNA (RNA) and amino acid sequence of the peptides identified by MS that match an edited region of (A) COII and of (B) COIII from T. brucei. The uridylate residues inserted by RNA editing are shown in lowercase. nt, nucleotide.
Tagged Respiratory Complexes
The data obtained by MS analyses from the gel slices as described above did not allow us to specifically determine the composition of complexes I–III. Therefore, we purified these complexes from T. brucei PF cells by tandem affinity purification (TAP) using two previously published methods (M1 and M2) (8, 16), and analyzed the protein composition by LC-MS/MS. The mt localization of almost all tagged proteins was confirmed by immunofluorescence analysis using anti-myc Ab (anti-tag), indicating that the tags as well as over-expression did not alter the mt localization (supplemental Fig. S2).
NADH Dehydrogenase Complex and Subunit Composition
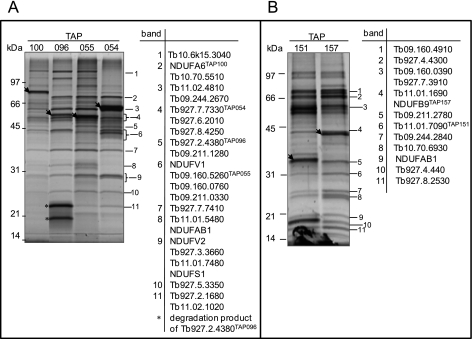
Complex I is the largest and least understood complex of the respiratory chain. Mammalian complex I consists of ∼46 different subunits that are assembled into a ∼1 MDa structure. The subunit composition and general topology of complex I has been defined through its dissection into three subcomplexes (Iα + λ, Iβ, and Iγ) (38). The T. brucei genome database contains seven mt encoded and 12 nuclear encoded genes with homology to eukaryotic subunits (Table I). To purify complex I from T. brucei, Tb10.61.1790 (NDUFA6), Tb11.01.0640 (NDUFA13), Tb10.05.0070 (NDUFA9), Tb11.47.0017 (NDUFS7), Tb09.244.4620 (NDUFA9), and Tb10.70.3150 (NDUFA5) subunits of the subcomplex Iα + γ as well as Tb11.01.7460 (NDUFB9), a subunit of the subcomplex Iβ were tagged resulting in generation of TAP100, TAP205, TAP204, TAP206, TAP081, TAP203, and TAP157 cell lines, respectively (Table I). Unfortunately, we were not able to tag any subunits of subcomplex Iγ because it contains mt encoded subunits and methodology for expression of these subunits is not yet available. Purification of intact complex I was not observed potentially because of its instability during lysis and purification. MS analyses of the TAP100, TAP081, TAP203, and TAP157 purified complexes yielded two separate sets of proteins, of which many were annotated as hypothetical (supplemental Table S3). TAP204, TAP205, and TAP206 were not incorporated into the complex I because only the tagged proteins were identified by MS analyses (supplemental Table S3). Given that these proteins are predicted to be subunits of complex I (25, 31), most likely the tag interfered with the folding of the tagged protein or with its integration into the complex. To assess the validity of the association of the hypothetical [novel] proteins identified by MS analyses, three additional proteins associated with subcomplexes Iα + λ Tb927.2.4380 (TAP096), Tb09.160.5260 (TAP055 (16)), Tb927.7.7330 (TAP054), and one protein associated with subcomplex Iβ Tb11.01.7090 (TAP151) were also TAP-tagged. The SDS-PAGE of the subcomplexes Iα and Iβ (Figs. 3A and B) showed similar protein profiles for their respective subcomplexes, which differed mainly with respect to the abundance and position of the tagged bait as detected by Western blot analysis (data not shown). All of the most prominent gel bands in subcomplex Iα+λ were individually analyzed by MS and the respective proteins were identified (Fig. 3A and supplemental Table S3). Twenty four proteins were identified in this subcomplex. The most prominent bands in TAP151 purifications were also analyzed by MS and twelve proteins were identified in this subcomplex (Fig. 3B). These twelve proteins were also identified in TAP157 purification with the exception of NDUFAB1 and Tb09.160.0390 (supplemental Table S3). In addition, five other proteins were identified in at least two samples of the subcomplex Iα+λ (Tb11.01.8630, Tb10.70.3150, Tb09.244.2620, Tb927.1.730, Tb927.6.1410) and three more proteins (Tb11.01.7460, Tb11.02.2070, Tb927.8.5560) in subcomplex β when the SDS-PAGE step was omitted and samples were directly submitted to trypsin cleavage and LC-MS/MS analysis (supplemental Table S3).These proteins are most likely associated with the Iα+λ and β sub-complexes although their affinity of association and relative concentration (stoichiometry) may be lower compared with the proteins identified by gel-band analysis. These results suggest that subcomplexes Iα + γ and Iβ were purified, each subcomplex contains at least 32 and 15 subunits, respectively and they had no overlapping composition except for NDUFAB1 (Tb927.3.860) subunit, which was identified in both subcomplexes. All twelve nuclear-encoded proteins that have similarity to the eukaryotic complex I subunits as predicted by genome and proteome analysis (25, 31) have been identified in our TAP-tag complex purifications. However, we were not able to identify the seven mt encoded subunits ND1, ND3, ND4, ND5, ND7, ND8, and ND9 most likely because of their high hydrophobicity and properties such as few tryptic cleavage sites and resulting cysteine rich peptides or not present in PFs (see discussion). Analysis of peptide sequences generated in silico indicated only six peptides has high probability of being detected by the MS approach taken in this study (data not shown). Also we were not able to identify in our purifications the two proteins that share identity with subunits of plant complex I ((31) and Table I), suggesting that these proteins may not be part of T. brucei complex I. Of all 46 proteins identified in purified complex I (subcomplexes Iα+λ and Iβ combined), 24 are currently annotated as hypothetical in the GeneDB database. Several of the latter possess domains/motifs that are indicative of possible function(s) in oxidoreductase processes (Table I). All together, we found that the T. brucei complex I is purified as two subcomplexes, and composed of at least 46 subunits (Table I).
Table I. Trypanosoma brucei complex I subunits.
√ indicates that only proteins identified in at least two experiments and by at least two unique tryptic peptides are shown. ND, protein has not been detected by MS. TAPProteins used as baits.
| Protein_Idd | Genomef/Proteome analysisg | Complex I Subcomplex Iαh | Complex I Subcomplex Iβi | Domains/Motifse | Homology with human/bovine proteins |
|---|---|---|---|---|---|
| Nuclear encoded | |||||
| Tb10.05.0070TAP204 | √ | √ | ND | NADH-ubiquinone oxidoreductase complex I subunit | NDUFA9/39-kDa |
| Tb10.389.1140 | √ | √ | ND | NADH-ubiquinone oxidoreductase, 75kDa 2Fe-2S ferredoxin-like | NDUFS1/75-kDa |
| Tb10.61.1790TAP100 | √ | √ | ND | NADH-ubiquinone oxidoreductase, subunit B14 | NDUFA6/B14 |
| Tb11.01.0640TAP205 | √ | √ | ND | NADH-ubiquinone oxidoreductase, subunit B16.6 | NDUFA13/B16.6 |
| Tb11.01.7460TAP157 | √ | ND | √ | NADH-ubiquinone oxidoreductase, subunit NI2M | NDUFB9/B22 |
| Tb11.01.8630 | √ | √ | ND | NADH-ubiquinone oxidoreductase, subunit B8 | NDUFA2/B8 |
| Tb11.47.0017TAP206 | √ | √ | ND | NADH-ubiquinone oxidoreductase, subunit NdhK | NDUFS7/PSST |
| Tb927.3.860 | √ | √ | √ | Acyl carrier protein | NDUFAB1/SDAP |
| Tb927.5.450 | √ | √ | ND | NADH-ubiquinone oxidoreductase, 51 kDa | NDUFV1/51-kDa |
| Tb927.7.6350 | √ | √ | ND | NADH-ubiquinone oxidoreductase, 24 kDa | NDUFV2/24-kDa |
| Tb09.160.0760 | √ | √ | ND | Alpha/beta-Hydrolases | |
| aTb09.160.5260TAP055 | √ | √ | ND | 2-enoyl thioester reductase | Trans-2-enoyl-CoA reductase |
| aTb927.7.7410 | √ | √ | ND | 2-enoyl thioester reductase | Trans-2-enoyl-CoA reductase |
| Tb09.211.0330 | √ | √ | ND | Heat shock protein DnaJ | DnaJ homolog |
| Tb09.244.2620TAP081 | √ | √ | ND | NADB_Rossman super family, NDUFA9_like_SDR_a | |
| Tb09.244.2670 | √ | √ | ND | ||
| Tb10.6k15.3040 | √ | √ | ND | Vacuolar H+-ATPase V1 sector, subunit C | |
| Tb10.70.3150TAP203 | √ | √ | ND | NADH-ubiquinone oxidoreductase, 13 kDa-B subunit | NDUFA5/B13 |
| Tb10.70.5510 | √ | √ | ND | Adrenodoxin reductase, putative; ferredoxin | Adrenodoxin oxidoreductase; ferredoxin |
| Tb11.01.5480 | √ | √ | ND | ||
| bTb11.01.7480 | √ | √ | ND | Manganese and iron superoxide dismutase | Superoxide dismutase |
| bTb927.5.3350 | √ | √ | ND | Manganese and iron superoxide dismutase | Superoxide dismutase |
| Tb11.02.4810 | √ | √ | ND | Phosphoenolpyruvate carboxykinase | |
| Tb927.1.730 | √ | √ | ND | NB-ARC domain | |
| Tb927.2.4380TAP096 | √ | √ | ND | ||
| Tb927.3.3660 | √ | √ | ND | ||
| Tb927.6.1410 | √ | √ | ND | ||
| cTb927.6.2010 | √ | √ | ND | Acyl-CoA synthetase-like | Acyl-CoA synthetase |
| cTb11.02.2070 | √ | ND | √ | Acyl-CoA synthetase-like | Acyl-CoA synthetase |
| Tb927.7.7330TAP054 | √ | √ | ND | ||
| Tb927.8.4250 | √ | √ | ND | Tetratricopeptide repeat | Tetratricopeptide repeat |
| Tb11.02.1020 | √ | √ | ND | ||
| Tb09.160.0390g | √ | ND | √ | Flavoprotein monooxygenase | Kynurenine 3-monooxygenase |
| Tb09.160.4910 | √ | ND | √ | Flavoprotein monooxygenase | Kynurenine 3-monooxygenase |
| Tb09.211.1280 | √ | √ | ND | ||
| Tb09.211.2780 | ND | ND | √ | ||
| Tb09.244.2840 | √ | ND | √ | ||
| Tb10.70.6930 | √ | ND | √ | ||
| Tb11.01.1690 | √ | ND | √ | ||
| Tb11.01.7090TAP151 | √ | ND | √ | Complex1 LYR | LYR motif |
| Tb927.2.1680 | √ | √ | ND | Cyclophilin-type peptidyl-prolyl cis-trans isomerase | Peptidyl-prolyl cis-trans isomerase |
| Tb927.4.4300 | √ | ND | √ | SET domain | |
| Tb927.4.440 | √ | ND | √ | ||
| Tb927.7.3910 | √ | ND | √ | P-loop containing nucleoside triphosphate hydrolases | |
| Tb927.8.2530 | √ | ND | √ | Cytochrome C oxidase copper chaperone | |
| Tb927.8.5560 | √ | ND | √ | S-adenosyl-l-methionine-dependent methyltransferases | Methyltransferase |
| Mitochondrial encoded | |||||
| M94286 | ND | ND | ND | 36 kDa subunit 1 | ND1/Chain1 |
| AAA20887 | ND | ND | ND | 18 kDa CR5/G5 cryptogene | ND3/Chain3 |
| AAB59224 | ND | ND | ND | 52 kDa subunit 4 | ND4/Chain4 |
| P04540 | ND | ND | ND | 71 kDa subunit 5 | ND5/Chain5 |
| P21301 | ND | ND | ND | 45 kDa 4Fe-FS protein | ND7/49kDa |
| gi 552291 gb AAA91499.1 | ND | ND | ND | 17 kDa subunit 8 | ND8/TYKY |
| gi 162166 gb AAA03749.1 | ND | ND | ND | 14 kDa subunit 9 | ND9/30kDa |
| Plant origin | |||||
| Tb11.02.5070 | √ | ND | ND | 33 kDa plant 2 | |
| Tb927.4.4980 | ND | ND | ND | 18 kDa plant 6 |
a, b, and c Protein pairs with sequence homology.
d GeneDB accession number (http://www.genedb.org) or GeneBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) for the mitochondrially encoded polypeptides.
e All proteins have orthologs in T. cruzi and L. major.
f [31].
g [25] and native gel analysis (this study).
h Proteins identified in pull-downs with mAb 52, 63, 69 [16] and/or TAP054, TAP055 [16],TAP081,TAP096, TAP100, TAP203, TAP204, TAP205 and TAP206 affinity purification.
i Proteins identified in TAP151 and TAP157 affinity purification.
Fig. 3.
TAP tag purified T. brucei NADH:ubiquinone dehydrogenase complex. A, TAP100, TAP096, TAP055 and TAP054 complexes were purified and separated on a 10–14.5% polyacrylamide Tris-glycine gel and stained with Sypro Ruby. The listed proteins were identified in the bands shown with corresponding numbers. The sizes of the protein marker are indicated. The position of tagged proteins is indicated by arrowheads. B, TAP151 and TAP157 complexes were purified and separated on a 10–14.5% and labeled as in A.
SDH Complex and Subunit Composition
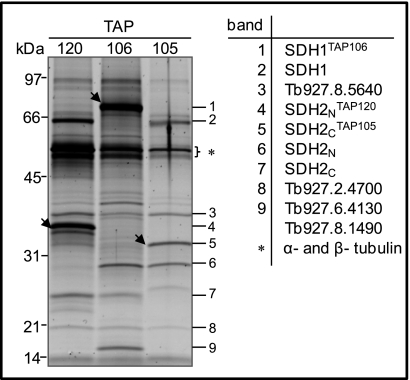
The large flavoprotein subunit Tb927.8.6580 (SDH1) and the smaller iron-sulfur subunit that is heterodimeric in Trypanosomatids (39) Tb927.8.3380, SDH2N; Tb09.160.4380, SDH2C were tagged resulting in generation of TAP106, TAP120, and TAP105 cell lines (Table II). All visible gel bands (Fig. 4) were individually analyzed by MS and the respective proteins SDH1, SDH2N, SDH2C, and four hypothetical proteins Tb927.8.5640, Tb927.2.4700, Tb927.6.4130, and Tb927.8.1490, were identified in all three tagged complexes with at least two peptides (Fig. 4 and supplemental Table S4). In addition, two other proteins (Tb927.7.3590 and Tb10.70.4500) were identified in all three purifications when the SDS-PAGE step was omitted (supplemental Table S4).
Table II. Trypanosoma brucei complex II subunits. Only proteins identified in at least two experiments and by at least two unique tryptic peptides are shown. TAPProteins used as baits.
| Protein_IDa | Homolog/Domainb | Shotgun approachc | TAP Tag | Ortholog in T. cruzid |
|---|---|---|---|---|
| Tb927.8.6580TAP106 | SDH1 | √ | √ | SDH1 |
| Tb09.160.4380TAP105 | SDH2C | √ | √ | SDH2C |
| Tb927.8.3380TAP120 | SDH2N | √ | √ | SDH2N |
| Tb927.2.4700 | √ | √ | SDH8 | |
| Tb927.8.5640 | √ | √ | SDH6 | |
| Tb10.70.4500 | √ | √ | SDH9 | |
| Tb927.6.4130 | √ | √ | SDH3 | |
| Tb927.7.3590 | √ | √ | ||
| Tb927.8.1490 | DUF1674 (domain of unknown function) | √ | √ |
a GeneDB accession number (http://www.genedb.org).
b All proteins have orthologs in L. major.
c [25] and native gel analysis (this study).
d [15].
Fig. 4.
TAP tag purified T. brucei SDH Complex. Sypro Ruby-stained SDS-PAGE protein profile of complexes, which were purified using TAP120, TAP106 and TAP105. The proteins identified by MS analysis of the corresponding band are shown and the position of tagged proteins is indicated by arrowheads. The asterisk indicates α- and β- tubulin.
In mammalian and bacterial mitochondria, SDH1 and SDH2 are anchored to the inner mt membrane by two small hydrophobic subunits, SDH3 and SDH4, which are required for electron transfer and ubiquinone reduction. These two subunits were not identified in the genome of the T. brucei by BLAST search suggesting that they may be highly diverged in sequence. However, two hypothetical proteins (Tb927.2.4700 and Tb927.7.3590) out of the nine proteins that were found associated with complex II possess predicted transmembrane domains and are similar in size to SDH3 and SDH4 (supplemental Table S4). This implies that they may play a role in electron transfer and perhaps function similar to SDH3 and SDH4 subunits, however direct comparison with these proteins did not reveal any predicted structural or motif similarity. In summary, we assigned nine proteins with high confidence to T. brucei SDH complex (Table II). This suggests that the complexity of the T. brucei complex is similar to that in T. cruzi, which is comprised of six hydrophobic and six hydrophilic subunits (15).
bc1 Complex and Subunit Composition
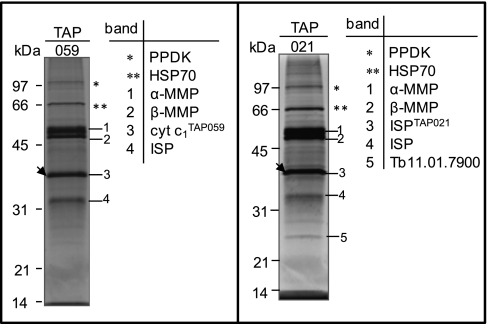
In trypanosomatids, as in other eukaryotes, the bc1 complex contains three subunits with active redox centers: the mt encoded cytochrome b, and the two nuclear encoded proteins, Tb927.8.1890 (cytochrome c1) and the Rieske iron-sulfur protein (Tb09.211.4700, ISP). The latter two were tagged resulting in generation of TAP059 and TAP021 cell lines (Table III). Because the members of the mt processing peptidase family have been shown to be core subunits of complex III, as in yeast and mammals (40), Tb11.02.1480 the T. brucei homolog of the α-MMP was also tagged (TAP066). All visible gel bands (Fig. 5 and supplemental Table S5) were individually analyzed by MS and the respective proteins cytochrome c1, ISP, α-MMP and Tb927.5.1060 (β-MMP) were identified in both purifications. Surprisingly, cytochrome c1 was not detected by MS analysis in the purified TAP021 samples although, detected when tagged itself (TAP059). Perhaps the TAP-tag interferes with the binding of cytochrome c1 with ISP protein. The additional ISP band in TAP021 (Fig. 5) is probably a product of proteolysis of the tagged protein because two bands were also detected by Western blot analysis (data not shown). Tb11.01.7900 has been identified in gel band analysis in TAP021 and TAP066 purifications as well as in TAP059 purification when the SDS-PAGE step was omitted (Fig. 5 and supplemental Table S5). In addition, two other annotated hypothetical proteins (Tb11.01.8225 and Tb10.70.2970) were identified only in TAP066 purification (data not shown and supplemental Table S5). Although Tb10.70.2970 has been identified only in one TAP purification, we assigned it to complex III based on its ubiquinol-cytochrome c reductase domain and its 28% sequence identity to the Saccharomyces cerevisiae Qcr7p subunit, which has a role in the assembly of bc1 complex (41). All together we were able to assign six proteins to the T. brucei complex III with high confidence (Table III).
Table III. Trypanosoma brucei complex III subunits. Only proteins identified in at least two experiments and by at least two unique tryptic peptides are shown. TAPProteins used as baits.
| Protein_IDa | Homolog/Domainb | Shotgun approachc | TAP Tag |
|---|---|---|---|
| Nuclear encoded | |||
| Tb09.211.4700TAP021 | ISP | √ | √ |
| Tb927.8.1890TAP059 | Cytochrome c1 | √ | √ |
| Tb11.02.1480TAP066 | α-MMP | √ | √ |
| Tb927.5.1060 | β-MMP | √ | √ |
| Tb10.70.2970d | Cytochrome bd ubiquinol oxidase, 14kDa subunit | √ | √ |
| Tb11.01.7900 | √ | √ | |
| Mitochondrial encoded | |||
| AAA32115 | apocytochrome b | ND | ND |
a GeneDB accession number (http://www.genedb.org) or GeneBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) for the mitochondrially encoded polypeptide.
b All proteins have orthologs in L. major and in T. cruzi.
c [25] and native gel analysis (this study).
d Only identified in TAP066.
Fig. 5.
TAP tag purified T. brucei cytochrome bc1 complex. SYPRO Ruby-stained SDS-PAGE gels of TAP059 and TAP021 purifications. The proteins identified by MS analysis of the corresponding bands are indicated. The position of tagged proteins is indicated by arrowheads. PPDK, pyruvate phosphate dikinase; HSP70, heat shock 70 kDa protein.
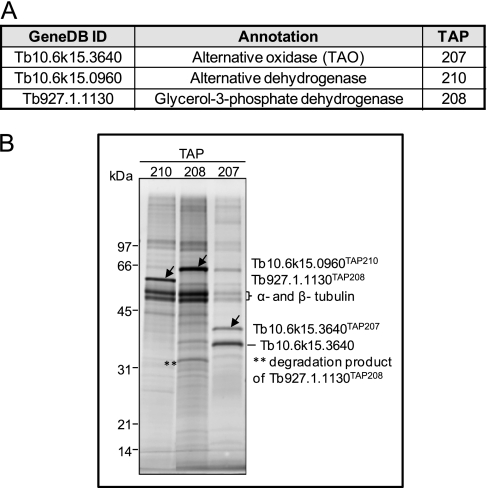
Besides the respiratory complexes described above, T. brucei mitochondrion contains a branched electron transport chain that is composed of the mt FAD-linked glycerol-3-phosphate dehydrogenase (Tb927.1.1130) and alternative oxidase TAO (Tb10.6k15.3640) (Fig. 6A). Moreover, an alternative rotenone-insensitive NADH dehydrogenase (Tb10.6k15.0960), which also occurs in plants and fungi, was characterized in T. brucei cells (42). All three enzymes are predicted to work as single proteins; however, these predictions have not been experimentally verified. Thus, all three proteins were tagged resulting in TAP208, TAP207, and TAP210 cell lines respectively, and TAP-purified samples were then subjected to SDS-PAGE fractionation and to LC-MS/MS analyses (Fig. 6B). SyproRuby-stained gel and MS data showed that alternative dehydrogenase and glycerol-3-phosphate dehydrogenase were purified as single protein since except typical contamination arising from α- and β-tubulin, only the bait protein was identified by MS. In TAP207 two protein bands were apparent on SDS-PAGE gel (Fig. 6B), the higher one corresponding to the tagged version of the TAO and the second one representing the endogenous protein, suggesting that this enzyme exists as a dimer or multimer.
Fig. 6.
TAP tag purified T. brucei glycerol-3-phosphate dehydrogenase, alternative dehydrogenase and alternative oxidase. A, Annotated and tagged enzymes. B, Sypro Ruby staining of TAP-tag purified proteins. The arrowheads indicate the tagged proteins that were identified by MS analysis. The sizes of the protein marker are indicated on left.
Protein-Protein Interaction Map
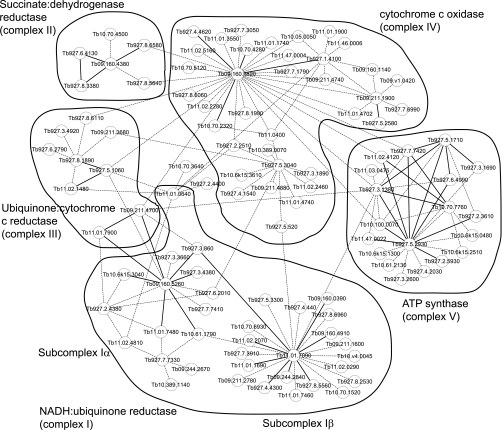
The acquired MS data from TAP-tag purified multiprotein complexes was used to predict a protein-protein interaction network of the respiratome (Fig. 7). The bold lines in Fig. 7 indicate high confidence predicted direct-interactions between the indicated proteins, and dotted lines indicate additional proteins pulled-down by the bait (tagged protein). As described above, complex I exists as multiple subunits, and results from the interaction map suggest Tb927.3.860 (NDUFAB1) interacts with Tb11.01.7090 and connects subcomplex Iα and Iβ. Six additional proteins (Tb10.70.1520, Tb11.02.0290, Tb10.v4.0045, Tb09.211.1600, Tb927.8.6960, and Tb927.5.3300) with unknown function were identified in complex I map that did not match our criteria of being identified in two separate TAP-tag experiments (Fig. 7), thus they are not included as bona-fide components of complex I in Table I. However, these are possible components of complex I or other complex(es) that may associate with complex I. Similarly, a number of proteins that did not match our criteria were identified in complex III, IV, and V map (Fig. 7). We also identified two components of complex III (Tb09.211.4700 and Tb11.01.7900) in purified subcomplex Iα. Similarly, four components of complex V (Tb927.7.7420/7430, α; Tb927.3.1380, β; Tb10.100.0070, γ; and Tb11.02.4120) were identified in complex IV preparation, and a potential new component of complex V Tb927.2.5930 was identified by this analysis as we included additional TAP-tag data than published earlier (8). We also found “MIX” protein (Tb927.5.3040) associated with components of complex IV, which we previously reported (17). These results identify the possible linking partners between the five respiratory complexes. However, the interactions have not yet been identified from genuine reciprocal interactions between the different subunits, thus further experimental evidence is required for validating these predictions especially as spurious association of proteins or highly abundant mt proteins may result in erroneous prediction. Overall, aggregate data may be useful for identifying component compositions of multi-protein complexes, where data from multiple TAP-tags and multiple MS runs are available.
Fig. 7.
Predicted protein-protein interaction network of the T. brucei respiratome. This Cytoscape graphical representation shows probable interactions within each respiratory complexes I-V and within the respiratome. The bold line indicates high confidence potential direct-interaction between the indicated proteins, and dotted line indicates additional proteins that were pulled-down by the bait.
DISCUSSION
The composition of the respiratory complexes was explored using native gel electrophoresis combined with in-gel activity staining, Western blot analysis and TAP-tag complex purification coupled with MS analysis. Respiratory complexes I, II, and III were purified using multiple tagged subunits and protein assignments were made based on the results from several purifications. In addition, the aggregate spectral counts of identified peptides from the TAP-tag studies were used to generate a preliminary protein-protein interaction map of the respiratory complexes. The interactions are consistent with the known structure of the respiratory complexes, e.g. complex I organized in subcomplexes, association of “MIX” protein with complex IV (17).The interaction map is consistent with our finding that RNAi knockdown of Tb10.70.7760 in complex V, resulted in the loss of the FoF1 complex interactions that contains the interactor, but the F1 moiety was retained (8). Also, we report the first evidence of MS identification of mt proteins encoded by edited RNAs in T. brucei. Two tryptic peptides match edited region of COII and COIII, indicating that translated products from fully edited mRNA were detected. The results suggest that electrophoresis under native conditions might avoid the loss of these highly hydrophobic subunits, which have a tendency to aggregate and precipitate when in SDS (13, 33).
Successful activity staining of the respiratory complex II and III helped to estimate the native size of these complexes which range from ∼450 to ∼720 kDa. Complex II is an integral membrane protein complex involved in both the Krebs cycle and the respiratory electron transport chain. The total molecular mass of SDH complex is calculated to be ∼220 kDa if it contained equimolar amounts of subunits. However, the activity band detected in hrCNE gel runs at ∼450 kDa which may suggest that complex II exists as a homodimer. It is also possible that the complex is a monomer but migrates at a higher size in native gel because of the effect of salt, detergent etc. Nevertheless dimeric complex II has been reported for T. cruzi (15). Among the 12 subunits that compose T. cruzi complex II, our TAP tag analyses did not identify the ortholog of SDH4, SDH5 and SDH11, whereas we identified two other subunits of T. brucei complex II that are not present in the T. cruzi complex (Table II). Moreover, we identified the ortholog of SDH7 and SDH10 but they did not match our criteria of being identified in two separate TAP-tag experiments and/or by more than one peptide (supplemental Table S4). The homologs of the membrane bound subunits SDH3 and SDH4 are difficult to identify with conventional BLAST programs due to their highly diverged sequence. However, their putative homologs were identified in T. cruzi purified complex II based on the presence of quinone/heme binding motifs (15). The ortholog of T. cruzi SDH3 was identified in our preparation, but it appears that this protein (Tb927.6.4130) does not have a predicted transmembrane domain in T. brucei (supplemental Table S4). The supramolecular organization of T. brucei complex II resembles the T. cruzi complex, but is divergent from the S. cerevisiae and mammalian SDH complex structure, indicating that trypanosomatid SDH complex contains additional non-catalytic subunits as it was previously suggested for plant complex II (43).
In S. cerevisiae, mutation in any one of the four genes leads to the loss of complex II function and the inability of S. cerevisiae to grow by respiration (reviewed in (44)). In contrast, T. brucei RNAi knockdown of the core subunits of complex II resulted in a complete loss of oxidative phosphorylation in response to succinate, but were conditionally lethal only in the absence of glucose (45, 46). Our functional analysis of the core subunits by RNAi showed that the complex II is not essential for the growth of T. brucei cells even in the absence of glucose (supplemental Fig. S3 and data not shown). These results suggest that Trypanosoma complex II is not the main entrance for electrons into the respiratory chain in PF cells.
bc1 complex (complex III) is typically a structurally dimeric protein complex. In yeast and in bovine mitochondria, it exists in association with one and two copies of complex IV (47, 48). The total mass of the six proteins identified in this study is 237 kDa (supplemental Table S5), but it migrates at ∼720 kDa indicating that T. brucei complex III could similarly form a multimer. The same size band (∼720kDa) is observed for cytochrome c oxidase activity, which has a predicted monomer size of ∼360 kDa (17). Hence, the stained band may correspond to the bc 1-cytochrome c oxidase heteromultimeric-supercomplex as described in yeast (48). A bc1-cytochrome c oxidase supercomplex is also suggested by MS analyses of the ∼720 kDa activity band that revealed five core components of complex IV (supplemental Table 1). Nevertheless, this possible association does not appear to be essential for the structural integrity of either complex because repression of components of either complex that result in the loss of one complex do not result in the loss of the other complex (9).
Purified complexes III from Crithidia fasciculate (13, 33) and L. tarentolae (14) consist of ten and eleven subunits, respectively. Whereas only six subunits were found in the complex purified from T. brucei. This suggests a differential composition of complex III among trypanosomatids, or nonspecific association of other proteins with Leishmania and Crithidia complexes. It is also possible that the protein components identified in TAP purified complex III in this analysis are the more stably associated proteins. Other proteins that correspond to those seen in the other species may not be stably associated with the complex, but may have similar functions in vivo.
Although hrCNE combined with NADH:NTB reductase was successfully applied for the detection of bovine complex I (22), we found that the T. brucei NADH dehydrogenase activity stained band corresponded primarily to OGDC. In T. brucei, dihydrolipoyl dehydrogenase, one of the four subunits of OGDC is shared between OGDC and the pyruvate dehydrogenase complex (16). Its activity was detected using the same approach in mammalian mitochondria, implying that this assay is not specific for complex I activity (49). Furthermore, using a nongradient BNE method, both complex I and dihydrolipoyl dehydrogenase protein were detected simultaneously on the same gel (50), suggesting that the gel-based method used to separate mt respiratory complexes could be critical. Moreover, Trypanosoma subcomplex Iα and OGDC cosediment in glycerol gradients (16), implying that the native size of both complexes might be very similar and their respective activities may overlap in native gel. However, MS analysis revealed the presence of OGDC rather than T. brucei complex I in the NADH activity band, suggesting that the activity of complex I cannot be readily detected in this system. These results may imply that complex I has a limited activity and role in NADH reoxidation in culture-grown PF cells. This assumption is further supported by a novel finding that complex I is not essential for the growth of PF cells (51). Thus it is plausible to hypothesize that most of the mitochondrially generated NADH is regenerated in PFs by an alternative dehydrogenase which is also localized in the inner membrane (52). In addition, it has been reported that deletions in the ND4, ND5, and ND7 genes that encode for complex I mt subunits occurring in natural mutants of T. cruzi have no consequence to the mt redox state, indicating that complex I can have limited function in NADH oxidation in this species (53).
In general, complex I is composed of 14 subunits in prokaryotes and up to 46 subunits in eukaryotes. Yet, only 19 subunits of Trypanosoma complex I were found to have some degree of homology to these subunits based on homology search (25, 31). Out of these 19 subunits, all 12 nuclear-encoded ones were identified in our purified complexes, however none of the seven mt encoded subunits were detected by MS possibly due to the characteristics of these proteins, making their peptide identification by routine mass spectrometry analysis difficult. Moreover, some of these subunits may be present at very low abundance/absent in PF as they are preferentially edited in BF cells (54).
Previous purification of the hydrophilic subcomplex Iα+λ of complex I suggested that T. brucei complex I is a unique multifunctional complex in trypanosomes that is involved not only in respiration but also in fatty acid synthesis (16). Our high confidence interaction map (Fig. 7) shows Tb11.02.2070, a long-chain-fatty acid-CoA ligase protein to be associated with subcomplex Iβ. In addition, the acyl carrier protein (NDUFAB1) that links subcomplex Iα with Iβ has homology to two acyl carrier proteins (ACPM1 and ACPM2) of Yarrowia lipolytica, which play roles in mitochondrial fatty acid metabolism as well as complex I assembly (55). Two flavoprotein monooxygenases were also identified as subunits of complex I. The members of the flavoprotein monooxygenases are usually involved in the microbial degradation of aromatic compounds, in biosynthesis of ubiquinone, and in detoxification of drugs and other xenobiotics (56). Thus, presence of such proteins reinforce that the complex I in trypanosomes is multifunctional.
Overall, this study characterized the composition of respiratory complexes I, II and III, identified not only the predicted subunits but also novel complex-associated proteins. These results expand our knowledge of the unique mt respiratory machinery in these early diverged parasitic protozoa. We also created a protein-protein interaction network of the respiratome. Although, we have not validated our prediction network, it provides a framework and candidates for downstream analyses.
Acknowledgments
We thank Yuko Ogata for help with mass spectrometry; Steve L. Hajduk (University of Georgia, Athens) for the apoc1 antibody.
Footnotes
* This work was supported by National Institutes of Health grant AI065935 to KS. AZ received support from grant 204/09/P563 from the Grant Agency of the Czech Republic. Research was conducted using equipment made possible by support from the Economic Development Administration - US Department of Commerce and the M.J. Murdock Charitable Trust.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S5.
1 The abbreviations used are:
- mt
- mitochondrial
- BNE
- Blue Native Electrophoresis
- hrCNE
- high resolution Clear Native Electrophoresis
- mAb
- monoclonal antibody
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- OGDC
- 2-oxoglutarate dehydrogenase complex
- PF
- procyclic form
- TAP
- tandem affinity purification
- T. brucei
- Trypanosoma brucei.
REFERENCES
- 1. von Ballmoos C., Cook G. M., Dimroth P. (2008) Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 37, 43–64 [DOI] [PubMed] [Google Scholar]
- 2. Tielens A. G., Rotte C., van Hellemond J. J., Martin W. (2002) Mitochondria as we don't know them. Trends Biochem. Sci. 27, 564–572 [DOI] [PubMed] [Google Scholar]
- 3. Tielens A. G., van Hellemond J. J. (2009) Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 25, 482–490 [DOI] [PubMed] [Google Scholar]
- 4. Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 41, 105–114 [DOI] [PubMed] [Google Scholar]
- 5. Bienen E. J., Maturi R. K., Pollakis G., Clarkson A. B., Jr. (1993) Non-cytochrome mediated mitochondrial ATP production in bloodstream form Trypanosoma brucei brucei. Eur. J. Biochem. 216, 75–80 [DOI] [PubMed] [Google Scholar]
- 6. Chaudhuri M., Ott R. D., Hill G. C. (2006) Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 22, 484–491 [DOI] [PubMed] [Google Scholar]
- 7. Schnaufer A., Clark-Walker G. D., Steinberg A. G., Stuart K. (2005) The F(1)-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 24, 4029–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziková A., Schnaufer A., Dalley R. A., Panigrahi A. K., Stuart K. D. (2009) The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 5, e1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horváth A., Horakova E., Dunajciková P., Verner Z., Pravdová E., Slapetová I., Cuninková L., Lukes J. (2005) Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol. Microbiol. 58, 116–130 [DOI] [PubMed] [Google Scholar]
- 10. Besteiro S., Barrett M. P., Rivière L., Bringaud F. (2005) Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 21, 185–191 [DOI] [PubMed] [Google Scholar]
- 11. Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Lennard N. J., Renauld H., Bartholomeu D., Caler E., Hamlin N., Haas B., Böhme U., Harris B. R., Hannick L., Barrell B., Donelson J., Hall N., Fraser C. M., Melville S. E., El-Sayed N., Shallom J., Aslett M., Hou L., Atkin B., Barron A. J., Bringaud F., Brooks K., Cherevach I., Chillingworth T., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R., Doggett J., Djikeng A., Feldblyum T., Fraser A., Goodhead I., Hance Z., Harper A. D., Hauser H., Hostetler J., Jagels K., Johnson D., Johnson J., Jones C., Kerhornou A., Koo H., Larke N., Larkin C., Leech V., Line A., MacLeod A., Mooney P., Moule S., Mungall K., Norbertczak H., Ormond D., Pai G., Peterson J., Quail M. A., Rajandream M. A., Reitter C., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A., Van Aken S., Walker D., Wanless D., White B., White O., Whitehead S., Wortman J., Barry J. D., Fairlamb A. H., Field M. C., Gull K., Landfear S., Marcello L., Martin D. M., Opperdoes F., Ullu E., Whickstead B., Alsmark C., Arrowsmith C., Carrington M., Embley T. M., Ivens A., Lord A., Morgan G. M., Peacock C. S., Rabbinowitsch E., Salzberg S., Wang S., Woodward J., Adams M. D. (2005) The genome of the African trypanosome, Trypanosoma brucei. Science 309, 416–422 [DOI] [PubMed] [Google Scholar]
- 12. Speijer D., Muijsers A. O., Dekker H., de, Haan A., Breek C. K., Albracht S. P., Benne R. (1996) Purification and characterization of cytochrome c oxidase from the insect trypanosomatid Crithidia fasciculata. Mol. Biochem. Parasitol. 79, 47–59 [DOI] [PubMed] [Google Scholar]
- 13. Speijer D., Breek C. K., Muijsers A. O., Hartog A. F., Berden J. A., Albracht S. P., Samyn B., Van Beeumen J., Benne R. (1997) Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol. Biochem. Parasitol. 85, 171–186 [DOI] [PubMed] [Google Scholar]
- 14. Horváth A., Berry E. A., Huang L. S., Maslov D. A. (2000) Leishmania tarentolae: a parallel isolation of cytochrome bc(1) and cytochrome c oxidase. Exp. Parasitol. 96, 160–167 [DOI] [PubMed] [Google Scholar]
- 15. Morales J., Mogi T., Mineki S., Takashima E., Mineki R., Hirawake H., Sakamoto K., Omura S., Kita K. (2009) Novel mitochondrial complex II isolated from Trypanosoma cruzi is composed of 12 peptides including a heterodimeric Ip subunit. J. Biol. Chem. 284, 7255–7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Panigrahi A. K., Ziková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. D. (2008) Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics 7, 534–545 [DOI] [PubMed] [Google Scholar]
- 17. Ziková A., Panigrahi A. K., Uboldi A. D., Dalley R. A., Handman E., Stuart K. (2008) Structural and functional association of Trypanosoma brucei MIX protein with cytochrome c oxidase complex. Eukaryot Cell 7, 1994–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wirtz E., Leal S., Ochatt C., Cross G. A. M. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 19. Jensen B. C., Kifer C. T., Brekken D. L., Randall A. C., Wang Q., Drees B. L., Parsons M. (2007) Characterization of protein kinase CK2 from Trypanosoma brucei. Mol. Biochem. Parasitol. 151, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panigrahi A. K., Schnaufer A., Ernst N. L., Wang B., Carmean N., Salavati R., Stuart K. (2003) Identification of novel components of Trypanosoma brucei editosomes. RNA. 9, 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Panigrahi A. K., Schnaufer A., Stuart K. D. (2007) Isolation and Compositional Analysis of Trypanosomatid Editosomes, In: Gott J. M. (ed), Methods in Enzymology, Vol 424 Ed., pp. 3–24, Elsevier Inc; [DOI] [PubMed] [Google Scholar]
- 22. Wittig I., Karas M., Schägger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell Proteomics. 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 23. Priest J. W., Hajduk S. L. (2003) Trypanosoma brucei cytochrome c1 is imported into mitochondria along an unusual pathway. J. Biol. Chem. 278, 15084–15094 [DOI] [PubMed] [Google Scholar]
- 24. Ziková A., Panigrahi A. K., Dalley R. A., Acestor N., Anupama A., Ogata Y., Myler P. J., Stuart K. (2008) Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell Proteomics 7, 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panigrahi A. K., Ogata Y., Ziková A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. (2009) A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 27. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 28. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome. Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park S. K., Venable J. D., Xu T., Yates J. R., 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. (2003) Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2, 117–126 [DOI] [PubMed] [Google Scholar]
- 31. Opperdoes F. R., Michels P. A. (2008) Complex I of Trypanosomatidae: does it exist? Trends Parasitol. 24, 310–317 [DOI] [PubMed] [Google Scholar]
- 32. Fang J., Wang Y., Beattie D. S. (2001) Isolation and characterization of complex I, rotenone-sensitive NADH: ubiquinone oxidoreductase, from the procyclic forms of Trypanosoma brucei. Eur. J. Biochem. 268, 3075–3082 [DOI] [PubMed] [Google Scholar]
- 33. Priest J. W., Hajduk S. L. (1992) Cytochrome c reductase purified from Crithidia fasciculata contains an atypical cytochrome c1. J. Biol. Chem. 267, 20188–20195 [PubMed] [Google Scholar]
- 34. Stuart K., Feagin J. E. (1992) Mitochondrial DNA of kinetoplastids. Int. Rev. Cytol. 141, 65–88 [DOI] [PubMed] [Google Scholar]
- 35. Horváth A., Berry E. A., Maslov D. A. (2000) Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 287, 1639–1640 [DOI] [PubMed] [Google Scholar]
- 36. Horvath A., Kingan T. G., Maslov D. A. (2000) Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. Evidence for translation of unedited mRNA in the kinetoplast. J. Biol. Chem. 275, 17160–17165 [DOI] [PubMed] [Google Scholar]
- 37. Hashimi H., Benkovicová V., Cermaková P., Lai D. H., Horváth A., Lukes J. (2010) The assembly of F(1)F(O)-ATP synthase is disrupted upon interference of RNA editing in Trypanosoma brucei. Int. J Parasitol. 40, 45–54 [DOI] [PubMed] [Google Scholar]
- 38. Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M. (2009) Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793, 78–88 [DOI] [PubMed] [Google Scholar]
- 39. Gawryluk R. M., Gray M. W. A split and rearranged nuclear gene encoding the iron-sulfur subunit of mitochondrial succinate dehydrogenase in Euglenozoa. BMC Res. Notes. 2: 16:16, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwata S., Lee J. W., Okada K., Lee J. K., Iwata M., Rasmussen B., Link T. A., Ramaswamy S., Jap B. K. (1998) Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71 [DOI] [PubMed] [Google Scholar]
- 41. Hemrika W., De, Jong M., Berden J. A., Grivell L. A. (1994) The C-terminus of the 14-kDa subunit of ubiquinol-cytochrome-c oxidoreductase of the yeast Saccharomyces cerevisiae is involved in the assembly of a functional enzyme. Eur. J Biochem. 220, 569–576 [DOI] [PubMed] [Google Scholar]
- 42. Fang J., Beattie D. S. (2002) Novel FMN-containing rotenone-insensitive NADH dehydrogenase from Trypanosoma brucei mitochondria: isolation and characterization. Biochemistry 41, 3065–3072 [DOI] [PubMed] [Google Scholar]
- 43. Millar A. H., Eubel H., Jänsch L., Kruft V., Heazlewood J. L., Braun H. P. (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 56, 77–90 [DOI] [PubMed] [Google Scholar]
- 44. Lemire B. D., Oyedotun K. S. (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim. Biophys. Acta 1553, 102–116 [DOI] [PubMed] [Google Scholar]
- 45. Bochud-Allemann N., Schneider A. (2002) Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849–32854 [DOI] [PubMed] [Google Scholar]
- 46. Coustou V., Biran M., Breton M., Guegan F., Rivière L., Plazolles N., Nolan D., Barrett M. P., Franconi J. M., Bringaud F. (2008) Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J. Biol. Chem. 283, 16342–16354 [DOI] [PubMed] [Google Scholar]
- 47. Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cruciat C. M., Brunner S., Baumann F., Neupert W., Stuart R. A. (2000) The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275, 18093–18098 [DOI] [PubMed] [Google Scholar]
- 49. Yan L. J., Yang S. H., Shu H., Prokai L., Forster M. J. (2007) Histochemical staining and quantification of dihydrolipoamide dehydrogenase diaphorase activity using blue native PAGE. Electrophoresis 28, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 50. Yan L. J., Forster M. J. (2009) Resolving mitochondrial protein complexes using nongradient blue native polyacrylamide gel electrophoresis. Anal. Biochem. 389, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verner Z., Cermakova P., Skodova I., Kriegova E., Horvath A., Lukes J. (2010) Complex I (NADH:ubiquinone oxidoreductase) is active in but non-essential for procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. doi:10.1016/j.molbiopara.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 52. Fang J., Beattie D. S. (2003) Alternative oxidase present in procyclic Trypanosoma brucei may act to lower the mitochondrial production of superoxide. Arch. Biochem. Biophys. 414, 294–302 [DOI] [PubMed] [Google Scholar]
- 53. Carranza J. C., Kowaltowski A. J., Mendonca M. A., de Oliveira T. C., Gadelha F. R., Zingales B. (2009) Mitochondrial bioenergetics and redox state are unaltered in Trypanosoma cruzi isolates with compromised mitochondrial complex I subunit genes. J. Bioenerg. Biomembr. 41, 299–308 [DOI] [PubMed] [Google Scholar]
- 54. Corell R. A., Myler P. J., Stuart K. (1994) Trypanosoma brucei mitochondrial gene CR4 encodes an extensively edited mRNA with completely edited sequence only in the bloodstream forms. Mol. Biochem. Parasitol. 64, 65–74 [DOI] [PubMed] [Google Scholar]
- 55. Dobrynin K., Abdrakhmanova A., Richers S., Hunte C., Kerscher S., Brandt U. (2010) Characterization of two different acyl carrier proteins in complex I from Yarrowia lipolytica. Biochim. Biophys Acta 1797, 152–159 [DOI] [PubMed] [Google Scholar]
- 56. van Berkel W. J., Kamerbeek N. M., Fraaije M. W. (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689 [DOI] [PubMed] [Google Scholar]