Abstract
In proteomics, protein identifications are reported and stored using an unstable reference system: protein identifiers. These proprietary identifiers are created individually by every protein database and can change or may even be deleted over time.
To estimate the effect of the searched protein sequence database on the long-term storage of proteomics data we analyzed the changes of reported protein identifiers from all public experiments in the Proteomics Identifications (PRIDE) database by November 2010. To map the submitted protein identifier to a currently active entry, two distinct approaches were used. The first approach used the Protein Identifier Cross Referencing (PICR) service at the EBI, which maps protein identifiers based on 100% sequence identity. The second one (called logical mapping algorithm) accessed the source databases and retrieved the current status of the reported identifier.
Our analysis showed the differences between the main protein databases (International Protein Index (IPI), UniProt Knowledgebase (UniProtKB), National Center for Biotechnological Information nr database (NCBI nr), and Ensembl) in respect to identifier stability. For example, whereas 20% of submitted IPI entries were deleted after two years, virtually all UniProtKB entries remained either active or replaced. Furthermore, the two mapping algorithms produced markedly different results. For example, the PICR service reported 10% more IPI entries deleted compared with the logical mapping algorithm. We found several cases where experiments contained more than 10% deleted identifiers already at the time of publication. We also assessed the proportion of peptide identifications in these data sets that still fitted the originally identified protein sequences. Finally, we performed the same overall analysis on all records from IPI, Ensembl, and UniProtKB: two releases per year were used, from 2005. This analysis showed for the first time the true effect of changing protein identifiers on proteomics data. Based on these findings, UniProtKB seems the best database for applications that rely on the long-term storage of proteomics data.
Proteomics data is produced in constantly growing quantities. Like in other high-throughput approaches, one of the major challenges for a proteomics laboratory is the storage and management of huge amounts of information. Therefore, Laboratory Information Management Systems (LIMS) (1–3) have been developed and are heavily used as in-house data repositories to store the performed experiments for years to come. In addition, by means of standardized data formats, the new publication guidelines from scientific journals, and the requirements related to public data availability of some funding agencies, an increasing amount of proteomics data is being submitted to public proteomics repositories. Experiments are then stored in resources like the PRoteomics IDEntifications database (PRIDE)1 (4), PeptideAtlas (5), or Tranche (6).
Storing digital data for a potentially indefinitely long period of time invariably raises the big question of how long we will be able to read the data. A prominent example of lost data happened when the NASA discovered that they could no longer read their data from the first two manned moon missions (7). They simply no longer possessed a working model of the tape reader required to read the produced magnetic tapes. Proteomics does not require highly specialized hardware to store its data. Nevertheless, there still is a considerable risk that some of the produced data might be lost in the future because protein identifications are reported and stored using an unstable reference system: protein identifiers.
In Mass Spectrometry (MS) based experiments, the most common approach relies on the use of search engines to match sequences to mass spectra through a comparison of recorded peptide fragmentation spectra with theoretical spectra derived from a protein sequence database (8). The potentially identified proteins are then reported using the searched database's proprietary identifiers. These identifiers are unstable and can change or may even be deleted over time. The latter happens if, for instance, hypothetical proteins are removed when gene prediction algorithms are updated or new biological evidence is created.
The four main comprehensive protein databases used for proteomics experiments are the International Protein Index (IPI) (9), the UniProt Knowledgebase (UniProtKB) (10), Ensembl (11), and NCBI's nonredundant (nr) database (12). Because each database has a different focus, the databases can vary in terms of completeness, degree of redundancy, and quality of annotations. IPI is a nonredundant protein database built from different source databases. Its main characteristic is that it clusters the entries from the different source databases, which are believed to represent the same protein. The clusters are created by combining the results of sequence similarity comparisons with information derived from pre-existing cross-references (9). Thus, IPI provides a good balance between the degree of redundant records and its completeness. There are different IPI databases for different species such as human, mouse, rat, zebrafish, Arabidopsis, cow, and chicken. IPI will be discontinued in September 2011.
UniProtKB is a component of the UniProt suite of databases and actually consists of two databases: UniProtKB/Swiss-Prot and UniProtKB/TrEMBL. UniProtKB/Swiss-Prot is a high quality manually annotated protein knowledgebase whereas UniProtKB/TrEMBL holds computationally analyzed records enriched with automatic annotation and classification (10). Both databases use a shared space of protein identifiers, and identifiers from both databases are often mixed in experiments. Therefore, these two databases will not be distinguished in this paper. The major strengths of UniProtKB are the quality of its records and its minimal degree of redundancy.
The NCBI nr database compiles all protein sequences available from the following databases: “GenBank” translations, the Protein Data Bank (PDB) (13), UniProtKB/Swiss-Prot, PIR, and PRF (see http://www.ncbi.nlm.nih.gov/blast/blast_databases.shtml). The NCBI assigns “GenInfo” (gi) numbers to every sequence processed. Thus, experiments using NCBI nr or reporting their identifications using gi numbers are reported as “NCBI gi” in this paper. Records in the NCBI nr database possess a high level of redundancy. However, it is widely used for studies involving nonmodel organisms because it has a better representation of these species.
Finally, Ensembl is a genomics centric resource that integrates the information for a comprehensive set of mainly vertebrate genomes and provides automatic annotations derived from genome sequences. Ensembl produces protein sequence sets for each organism directly derived from the gene predictions (11). Ensembl's major strength is the easy connection between proteomics and biological knowledge as it directly links proteins to genes and transcripts. In PRIDE, these four are by far the most popular searched databases, together with other databases specific to certain model organisms like The Arabidopsis Information Resource (TAIR) (14).
When protein identifications have been generated from different databases, making results comparable can be troublesome (15). This is because of the existence of heterogeneous and changing identifiers referring to the same protein in different resources. To overcome this common problem in proteomics, the Protein Identifier Cross Referencing (PICR) service was launched in 2007 at the EBI (16). PICR uses the archive database of UniProt (UniParc) (10) as a data warehouse to offer protein cross-references based on 100% sequence identity from over 70 distinct source databases loaded into UniParc.
The impact of the selected searched protein sequence database on the sensitivity, specificity and speed of the search has been described before. For instance, it was shown that more inclusive bigger protein databases will take longer to search and may result in more false-positive identifications as well as reduced statistical significance (17). In this study we investigate the influence of the searched protein database on the long-term storage of proteomics data, estimating the rate of changing protein identifiers. Data coming from all public experiments from the PRIDE repository were processed using two distinct protein identifier mapping algorithms. Furthermore, the same analysis was performed on the complete database releases. Up to our knowledge, this is the first study that has investigated this aspect of the proteomics data generation workflow.
EXPERIMENTAL PROCEDURES
To analyze the effect of changing protein identifiers on the storage of real data, all public experiments available in the PRIDE database were used as a test data set. At the time of performing this study (November 2010), there were a total of 8500 public experiments containing 2,075,324 protein identifications (see Supplementary Table S1). The whole data set can be accessed via the PRIDE BioMart interface (http://www.ebi.ac.uk/pride/prideMart.do).
Database Release Date Curation
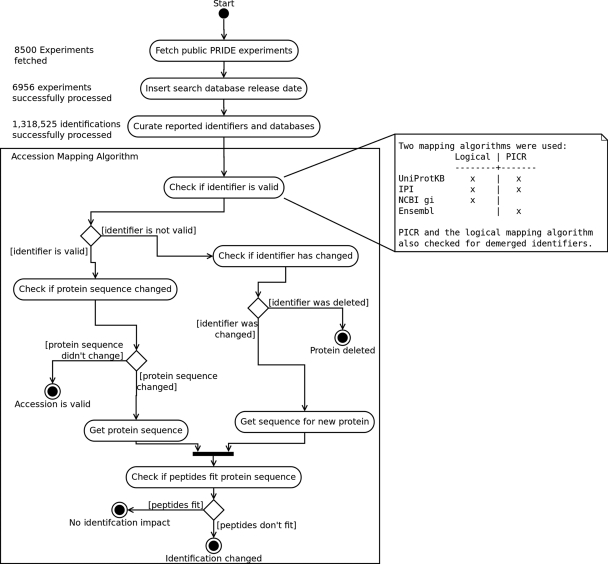
The workflow followed can be seen in Fig. 1. As a first step, the used search database and its version were manually curated. If the used database and/or database version was not available in the PRIDE submission metadata, the corresponding publication was downloaded and the information extracted from there. In case the PRIDE entry did not contain a link to a publication, the submitted contact's details were used as input for a PubMed search to retrieve a possible publication. In total, 6956 experiments containing a total of 1,402,837 protein identifications could be mapped successfully to specific searched database release dates (see Table I).
Fig. 1.
Flowchart representing the protein mapping algorithm(s). All public experiments available in PRIDE were used as initial test data set. This data set was then manually curated before the protein identifier mappings were performed. The box represents the protein mapping algorithms and depicts the different steps of the mapping process.
Table I. Searched database release date curation.
| Status | Number of Experiments |
|---|---|
| Successfully mapped | 6956 |
| No database version given in publication | 1349 |
| No publication found | 102 |
| Data not public | 15 |
| Publication not accessible | 2 |
| Custom database used | 76 |
As a next step, the reported protein identifiers and used searched databases were curated. This had to be done at the identification level as several experiments reported protein identifications using multiple protein identifier systems. Out of the 1,402,837 protein identifications mapped to a database release date, 1,307,505 identifications were successfully processed (see Table II). This set of identifications formed the basis for the here presented study. The group of “exotic” databases contained either custom built databases (for instance, “fgcz_6239_20050304.fasta,” “MSDB,” or “wormpep150-phg-orf-ecoli-rand.fasta”) or databases used in too few PRIDE experiments to be thoroughly evaluated (for instance, Flybase, Saccharomyces Genome Database or the EMBL Nucleotide Sequence Database). Invalid protein identifiers were defined as identifiers that did not confer to the format expected by the submitted database (for instance “BIOCTM_gi_88041_pir__A31994” as a UniProtKB accession number or “CZB00000043” as an IPI identifier).
Table II. Protein identifier and searched database mapping.
| Status | Number of Identifications |
|---|---|
| Successfully Mapped | 1,307,505 |
| Decoy Database Entries | 594 |
| Exotic Search Database | 93,334 |
| Invalid Protein Identifiers | 960 |
Protein Identifier Mapping Algorithms
To map reported protein identifiers to active identifiers, two different approaches were used (Fig. 1). The first mapping approach was performed using the PICR service at the EBI (http://www.ebi.ac.uk/Tools/picr) (16). As previously described, PICR maps protein identifiers based on 100% sequence identity. Thus, PICR ensures that a mapped identifier refers to the same protein sequence as the original one. Unfortunately, NCBI gi numbers could not be processed using this approach as PICR relates these entries to their source database at the NCBI and provides no data on the changes of the NCBI gi numbers themselves.
The second mapping approach (termed “logical mapping” algorithm) was meant to resemble the way a user tries to get a current protein entry. Source databases were accessed and the current status of the reported identifier retrieved. In addition to changes to the protein identifier, the current protein sequences were downloaded—either for the still active entry or for the new one. Ensembl entries were not supported by this approach as Ensembl is purely gene based and thus provides very limited information on protein identifier changes. Reference Sequence (RefSeq) database (18) identifiers were processed like NCBI gi numbers and are reported as part of the NCBI gi numbers. The main reason for this decision was that several studies using RefSeq reported NCBI gi numbers. This then left to few “true” RefSeq identifiers to get significant results.
Both mapping approaches returned four possible states: entry is active, entry was deleted, entry was replaced and entry was demerged (for instance, into distinct identifiers for every species).
Impact of Changing Protein Sequences on Peptide Identifications
For both mapping algorithms, we checked whether the originally identified peptide sequences still fitted the reported identified protein's sequence. As mentioned above, the logical mapping algorithm downloaded the protein sequence of the currently active entry. This sequence was then used to check whether the reported peptides still fitted this sequence. In addition, when available, the search engine scores (Mascot, Sequest, SpectrumMill, OMSSA, X!Tandem, and PeptideProphet) for the peptide spectrum matches were retrieved from the PRIDE database when the underlying protein sequence changed. These scores were then used to investigate whether peptides that no longer fitted the protein sequence showed a different score distribution compared with peptides that still fitted the sequence. The score distributions were analyzed using R (package stats version 2.12). Significant differences between the average score of peptides fitting and not fitting the protein sequence were defined as a Student's t test's p < 0.01.
The PICR based mapping algorithm followed exactly the same criteria as are applied to protein identifications submitted to the PRIDE database. To make submitted data to PRIDE more usable PRIDE automatically maps submitted protein identifiers to several other protein databases using PICR on a regular basis. PRIDE only reports an active mapped identification if all the identified peptides still match the reported protein sequence. In addition, the “PRIDE PICR” mapping process retrieves the current protein sequence. Thereby, protein sequence changes that do not lead to protein identifier changes are already taken into consideration and can lead to “missing” mappings even though the protein identifiers remained unchanged. It is important to highlight that the “PRIDE PICR” mappings are, by design, very pessimistic and use direct string matching. Although isobaric amino acids are taken into account, in cases of ambiguous/unknown amino acids, these peptides would not be correctly mapped to the protein sequence.
Rate of Change of Complete Protein Databases
We also analyzed the rate of change of protein identifiers from complete releases of IPI, UniProtKB, and Ensembl. Unfortunately, this analysis could not be done for the NCBI nr database because it is built on a daily basis and previous versions are not available. To calculate the rate of change, at least two complete releases per year of the respective databases (starting from 2005 - IPI version 3.01, UniProtKB version 2005_02, Ensembl release 33) were used. Again, both algorithms (logical mappings and PICR) were used, except for Ensembl, where it was only possible to use PICR for the reasons explained above.
For the logical mappings, each complete release was compared with a recent version of each respective resource (April 2011 - IPI version 3.82, UniProtKB version 2011_04). To generate the UniProtKB database, the respective UniProtKB/SwissProt and UniProtKB/TrEMBL identifiers were merged. As the complete UniProtKB/TrEMBL database was too big to perform the analysis, we only investigated the human and mouse specific identifiers and database builds.
RESULTS
Protein Identifier Mappings
For the presented analysis, an initial pool of 8500 PRIDE experiments was used containing 2,075,324 protein identifications. As explained in Experimental Procedures a prefiltering step of the data present in PRIDE was performed. In the end, 1,307,505 identifications coming from 6956 PRIDE experiments formed the basis for the present study. In this data set, IPI was by far the most commonly used database for performing searches comprising 59.5% of all valid identifications reported (see Table III). The second one was UniProtKB (20.1%) followed by TAIR (10.6%), Ensembl (5.6%), and NCBI nr (4.2%). Even though 10.6% of all processed identifications came from TAIR, it was not included in the detailed analysis as all these identifiers came from only two TAIR releases (see supplemental Fig. S1). Thus, there were not enough time points to perform a detailed analysis.
Table III. Mapped protein identifiers per searched database and mapping algorithm. “PRIDE PICR Mapped Ident.” refers to the mapping service as it is performed in the PRIDE database (see main text).
| Search Database | PICR Service Mapped Ident. | Logical Mapped Ident. | PRIDE PICR Mapped Ident. | Total Number of Ident. | |||
|---|---|---|---|---|---|---|---|
| ENSEMBL | 73,559 | (100%) | – | 67,057 | (91.2%) | 73,559 | |
| IPI | 777,848 | (100%) | 777,848 | (100%) | 609,345 | (78.3%) | 777,848 |
| NCBI gi | – | 54,225 | (97.8%) | – | 55,423 | ||
| TAIR | 137,958 | (100%) | – | 114,410 | (82.9%) | 137,958 | |
| UniProtKB | 253,658 | (96.6%) | 253,658 | (96.6%) | 211,111 | (80.4%) | 262,717 |
As previously stated, two different protein mapping approaches were used: the PICR service and an algorithm based on logical database mappings. Both approaches successfully mapped virtually all proteins from supported databases (see Table III). PICR successfully mapped 100% of Ensembl, IPI, and TAIR protein identifiers. The logical mappings achieved the same for IPI. There was a small percentage (about 3%) of UniProtKB protein identifiers that could not be mapped using the two approaches. This was caused by the existence of protein identifiers that most probably came from other databases but were reported to be UniProtKB entries. In addition, ∼2% of NCBI gi protein identifiers could not be mapped using the logical mapping algorithm. In all of these cases nucleotide instead of protein identifiers were submitted and thus caused the protein identifier mapping to fail.
As explained in Experimental Procedures, both approaches returned four different possible states for the mapped protein identifier: entry is active, entry was deleted, entry was replaced and entry was demerged (see Table IV). Overall, UniProtKB was the database with the highest percentage of active entries (over 90% in both mapping algorithms). UniProtKB was furthermore the resource with the fewest entries deleted (around 1%) and changed (between 4 and 5%). UniProtKB was the only database to contain demerged entries (around 0.5%). It is important to mention that the results presented in Table IV are based on all the protein identifications processed. Thus, large data submissions using one database release alter the overall results for the respective database.
Table IV. Total number of identification for every status and database per mapping algorithm.
| Database | Active |
Deleted |
Changed |
Demerged |
||||
|---|---|---|---|---|---|---|---|---|
| Logical | PICR | Logical | PICR | Logical | PICR | Logical | PICR | |
| ENSEMBL | – | 49,597 | – | 12,695 | – | 8,060 | – | – |
| (67.4%) | (17.3%) | (11.0%) | ||||||
| IPI | 605,376 | 508,926 | 78,344 | 233,787 | 94,128 | 35,135 | – | – |
| (77.8%) | (65.4%) | (10.1%) | (30.1%) | (12.1%) | (4.5%) | |||
| NCBI gi | 41,945 | – | 4,512 | – | 7,768 | – | – | – |
| (77.4%) | (8.3%) | (14.3%) | ||||||
| UniProtKB | 238,418 | 236,613 | 3,149 | 2,333 | 10,427 | 13,400 | 1,664 | 1,312 |
| (94.0%) | (93.3%) | (1.2%) | (0.9%) | (4.1%) | (5.3%) | (0.7%) | (0.5%) | |
IPI contained the lowest fraction of active entries (from 65.2 to 77.8% for the logical and PICR mapping algorithms, respectively) and the highest proportion of deleted entries (10.1 and 30.0%, respectively). The number of processed identifications using NCBI gi numbers was considerably smaller compared with UniProtKB and IPI. As previously described, NCBI gi numbers were just processed with the logical mapping approach. Only 75.0% of processed NCBI gi entries were active whereas 8.1% were deleted and 14.0% changed. Ensembl protein identifiers were only processed using the PICR based mapping algorithm and 67.4% of submitted entries using Ensembl were still active, 17.3% deleted and 10.1% changed.
Change of Protein Identifiers Over Time
The processed protein identifiers were used to analyze the stability of protein identifiers from the different searched databases in the test data set over time. Out of the five databases processed using the PICR service mapping algorithm, only IPI and UniProtKB contained sufficient entries to allow a detailed interpretation (see supplemental Fig. S1). As described above, the logical mapping algorithm processed IPI, UniProtKB, and NCBI gi entries. The number of entries processed per species and database are shown in supplemental Fig. S1. Other databases were not supported, as the test data set did not contain sufficient entries for a detailed analysis.
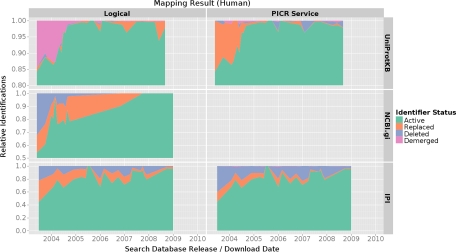
The analysis of protein identifier changes over time revealed distinct differences between the investigated protein databases. Fig. 2 shows the relative number of active, changed, deleted, and demerged entries per database and database release date for all species.
Fig. 2.
The combined protein identifier mapping result for all species. Some of the outliers were caused by very small submissions only consisting of a limited number of identifications. The number of identifications available from certain database release dates was normalized. Thus, the graph represents the proportion of active, replaced, deleted, and demerged identifications for specific release dates and databases. Different scales were used for the different searched databases to provide a more detailed view on the data.
UniProtKB
As stated before, UniProtKB showed to be the most stable database over time. For instance, in experiments reporting UniProtKB entries from releases before 2005, over 85% of entries were still active compared with 45 and 55% from IPI and NCBI gi, respectively. More importantly, apart from three data outliers (see below) virtually all UniProtKB entries were either active or could still be mapped to an alternative active entry (see Fig. 2 and Fig. 3).
Fig. 3.
The combined protein identifier mapping result for human data only. For a detailed description see Fig. 2. Different scales were used for the different searched databases to provide a more detailed view on the data as in Fig. 2.
Both mapping algorithms produced comparable results for UniProtKB entries, apart from the three outliers mentioned below. Identifications reported as demerged using the logical mapping algorithm are included in the portion of changed identifiers in the PICR mapping results. The three outliers were caused by submissions from less well characterized species: in March 2004 (all dates refer to the used searched database's release, not the submission date) a submission on rat (PRIDE experiment accessions 99–107, 3866–7954 (19)), in January 2005 a submission on chicken (PRIDE experiment accessions 1621–1626 (20)), and in August 2009 a submission on Drosophila (PRIDE accession number 8170). As expected, these outliers disappeared when only human submissions were considered (see Fig. 3).
NCBI gi Identifiers
As mentioned before, NCBI gi numbers were only mapped using the logical mapping algorithm. The analysis on all species contained several outliers (see Fig. 2). The low fraction of active entries in October 2004 was caused by a rather big submission on chicken data (PRIDE accession numbers 1654–59). The observed peak in June 2005 was caused by a submission on mouse containing only one mappable identification (PRIDE accession number 2381 (21), see supplemental Fig. S3). The last drop observed in October 2008 was caused by one large submission on Indian rice (PRIDE accessions 10726–10740 (22), see supplemental Fig. S2).
Irrespective of these outliers, NCBI gi numbers are less stable than UniProtKB entries as can be seen in the analysis on human data alone (see Fig. 3). Only 55% of NCBI entries from mid 2003 were still active with 32% deleted completely. The portion of deleted entries decreased considerably until 2006. In submissions using 4-year-old or younger database versions, only a few percent of identifiers were deleted. The portion of changed identifiers varied considerably across the different species. For human data, the portion of replaced identifiers decreased linearly from 15% in January 2005 to 0% in October 2007. From that date onwards, human NCBI gi numbers in PRIDE experiments were stable.
2.3. IPI
The results retrieved for IPI using the two mapping algorithms differed considerably (see Fig. 2). As mentioned before, IPI identifiers showed to be significantly less stable than UniProtKB or NCBI gi identifiers. Only 46% of logically mapped identifiers from mid 2003 were still active. This improved to about 75% active entries in mid 2004. Experiments reporting IPI entries continued to contain at least 5% of deleted identifiers until mid 2008. The results retrieved processing all identifications and the results from human identifications were similar (see Fig. 2 and Fig. 3).
The PICR service returned about 10% more deleted identifiers for IPI releases before 2005 compared with the logical mapping algorithm. From then onwards, the number of deleted identifiers became comparable between the two mapping approaches. PICR mapped identifiers contained one significant outlier. This large increase of deleted identifiers in August 2007 was caused by a very large submission on zebrafish (PRIDE accession numbers 3386–3532 (23), see supplemental Fig. S1).
The detailed analysis of two projects using the IPI database confirmed the above mentioned results: the data from the Human Proteome Organization (HUPO) Plasma Proteome Project (PPP) (PRIDE accession numbers 4–98 (24)) and the data from the HUPO Brain Proteome Project (BPP) (PRIDE accession numbers 1669–1750 (25)) (see supplemental Fig. S4). The HUPO PPP using an IPI release from July 2003 contained only 44.7% active identifiers. The HPO BPP data using an IPI release from April 2005 had 83.5% active identifiers for both the mouse and the human data.
Impact of Changing Protein Sequences on Peptide Identifications
In addition, we investigated the impact of changed protein sequences on the original peptide identifications using two distinct approaches. As previously described, all sequences of protein identifications processed using the logical mapping algorithm were downloaded. The identified peptides were then checked whether they were still part of the protein's sequence. Out of the 996,107 identifications that had not been deleted (“active entries”) 72,037 identifications (7.2%) contained at least one peptide that no longer fitted the protein sequence (see Table V). Identifications reported using NCBI gi numbers contained a significantly higher portion of proteins with not fitting peptides (20.5%). This was caused by the fact that the investigated NCBI gi entries were dominated by less characterized species (see supplemental Fig. S1): 42.6% and 15.5% of active identifications from chicken and Indian rice contained nonfitting peptides, respectively.
Table V. Portion of peptides fitting the protein sequence for active entries.
| Database | Active Entries | Missing Peptide Mapping | |
|---|---|---|---|
| UniProtKB | 248,404 | 18,899 | (7.6%) |
| NCBI gi | 49,680 | 10,162 | (20.5%) |
| IPI | 698,023 | 42,976 | (6.2%) |
As a second approach to investigate the percentage of nonfitting peptides we used the method currently applied in the PRIDE database to create and maintain protein identifier mappings. This method uses the PICR service but only accepts a returned identifier if all the identified peptides fit the returned protein sequence. This approach is more stringent than using logical mappings and thus seems most suited to be used for data repositories like PRIDE. This approach returned 10–20% more deleted identifiers compared with the results obtained with the PICR mapping algorithm (see Table III and supplemental Fig. S5). Two additional outliers were observed: an increase of deleted identifiers in March 2004 caused by two submissions on rat (PRIDE accession numbers 99–107 and 3866–7963(19)) and an increase of active entries in May 2005 caused by only one mapped identifier on human (PRIDE accession number 1647).
Peptide Score Distribution of Nonfitting Peptides
We were able to retrieve the search engine peptide scores for 55.7% of peptides fitting and 15.6% of peptides not fitting the protein sequence (see Table VI and supplemental Fig. S6). For this analysis only peptides where the underlying protein sequence changed were considered. The fraction of peptides for which we were able to retrieve the search engine scores was significantly lower for peptides that did not fit the protein sequence. Many of the nonfitting peptides belong to old data sets that were submitted before the PRIDE Converter (26) submission tool was introduced. Through PRIDE Converter the presence of peptide scores in data submissions became the general rule.
Table VI. Number of peptides and average peptide scores (± standard deviation) of peptides fitting and not-fitting the protein sequence per search engine. Numbers refer to peptides where the search engine score was retrieved successfully.
| Mascot | Peptide Prophet | Sequest | SpectrumMill | X!Tandem | |
|---|---|---|---|---|---|
| No. Fitting peptides | 1380884 | 38008 | 21827 | 47860 | 95231 |
| No. Nonfitting pep. | 48440 | 4031 | 21536 | 498 | 24168 |
| Av. score fitting peptides | 40.63 ± 19.80 | 7.87 ± 11.56 | 2.40 ± 1.28 | 13.34 ± 3.24 | 21.22 ± 8.81 |
| Av. score Nonfitting peptides | 40.78 ± 22.36 | 2.35 ± 6.06 | 1.66 ± 0.52 | 11.66 ± 3.75 | 13.31 ± 5.18 |
The average peptide score of peptides no longer fitting the protein sequence was significantly lower than the average peptide score of peptides still fitting the protein sequence for all search engines but Mascot (Student's t test, p < 0.01, see supplemental Fig. S6). Mascot scores showed no significant difference between peptides that fitted the protein sequence and peptides that no longer fitted the sequence (p = 0.1473, see Table VI).
Rate of Change of Complete Protein Databases
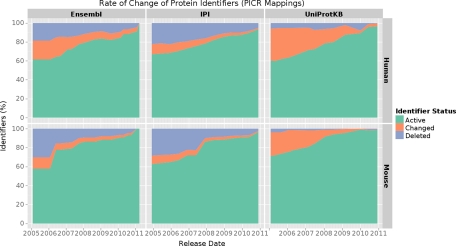
As an independent analysis we investigated the rate of change in time of complete releases of IPI, UniProtKB and Ensembl (see Experimental Procedures). The overall rate of change of the species specific builds of IPI and UniProtKB is comparable to the rate of change found in the respective identifiers from PRIDE (the PRIDE centric analysis did not contain sufficient data points for a detailed analysis on Ensembl, see Fig. 3, Fig. 4, and supplemental Fig. S3).
Fig. 4.
Rate of change of identifiers in complete releases of UniProtKB, IPI, and Ensembl for human and mouse (PICR mappings). Two releases per year were considered from 2005. The UniProtKB database contains the species specific identifiers from UniProtKB/SwissProt and UniProtKB/TrEMBL.
There were no significant differences between the results retrieved using the PICR mapping algorithm and the logical one (see Fig. 4 and supplemental Fig. S7). Consistent with the results presented above, IPI again showed to be less stable than UniProtKB with more than twice as many identifiers deleted at any point in time. As expected, identifiers from the two investigated species changed at different rates. In IPI identifiers from mouse were slightly less stable than identifiers from human (28.5% deleted mouse identifiers compared with 22.5% deleted human identifiers at the beginning of 2004). This changed at the end of 2007 when suddenly mouse identifiers became more stable than human ones (13.5% deleted human identifiers compared with 9.0% deleted mouse identifiers June 2008). Surprisingly, in UniProtKB mouse identifiers were always more stable than human identifiers. Although, for example, about 10% of human identifiers were deleted from the January release in 2010 virtually no mouse identifiers changed.
DISCUSSION
In this study, we analyzed the influence of the searched database on the long-term storage of proteomics data. We found distinct differences in the stability of protein identifiers between the investigated protein databases: UniProtKB, the only database that contains a high proportion of manually curated records (UniProtKB/Swiss-Prot), proved to be significantly more stable than IPI and NCBI gi numbers. As mentioned above UniProtKB/Swiss-Prot and UniProtKB/TrEMBL were treated as one unique searched database in this study (UniProtKB).
Given the fact that IPI was by far the most commonly used database in PRIDE submissions, the stability of IPI identifiers seems especially problematic. For example, already 10% to 20% (depending on the mapping algorithm) of the reported protein identifications were deleted after only two years. A concerning and, at the same time, surprising point is that several of these investigated experiments were published in 2010 and thus contained a considerable amount of basically outdated or invalid data already at the time of publication—data that is published and immediately perished (2, 22, 27). One such example would be a study done by Gammulla et al. in Indian rice (22) (PRIDE accessions 10726–10740) using the NCBI nr database from August 2008. This study contained 18.8% deleted identifiers when it was published in August 2010 (see above). This effect of changing protein identifiers on published data increased considerably over time. For instance, another study (28) published in May 2009, already contained 33.0% of deleted protein identifiers at the time of investigation (November 2010, PRIDE accessions 3706–3714). A representative example of a much older project is the data from the HUPO PPP project: at the time of the here presented study data from the HUPO PPP only contained 44.7% active identifiers (see above). These results are not caused by any misconduct of the respective authors but reflect the instability of certain protein databases for specific species.
The primary focus of this study was the quantification of changing protein identifiers from different databases over time. The results presented in Fig. 2, Fig. 3, and Fig. 4 do not fully reflect the true impact of changing protein databases on the long-term storage of proteomics data. To correctly approximate this effect the consequences of changing protein sequences on the actual peptide identifications need to be taken into consideration. Based on our results when investigating the fraction of peptides no longer fitting the current protein sequences we expect that the fraction of deleted identifiers is actually twice as high. Comparing the distribution of peptide scores between fitting and nonfitting peptides, we found that the scores of the still fitting ones were statistically significantly different (higher) than the scores of the nonfitting ones (except for Mascot). It thus seems probable that the fraction of nonfitting peptides contains more false-positive identifications than the fraction of fitting peptides. However, without carefully considering how the protein inference (29) was done in each particular case it is impossible to reach further conclusions.
Deleted identifiers are the worst but not the only problem caused by changes in protein databases. Cases where protein identifiers from UniProtKB are demerged into several new identifiers may also alter the original significance of the data. UniProtKB/Swiss-Prot has historically “merged” 100% identical protein sequences from different genes in the same species into one single record. However, UniProt recently started to demerge entries containing multiple individual genes coding for 100% identical protein sequences into individual UniProtKB/Swiss-Prot entries containing a single gene (see UniProt release 2010_09 notes, http://www.uniprot.org/news/2010/08/10/release). This development might cause significant problems when comparing old and more recent data. For example protein P05209, identified in several PRIDE experiments, was demerged and currently maps to 13 different identifiers. For human, mouse, and rat there are even two different mappings for each species. Another problematic example is protein P59641, identified in an experiment performed on human (PRIDE accession number 1645). Currently, P59641 maps to four UniProtKB/Swiss-Prot entries but none of which is human. The time when UniProtKB identifiers were demerged into entries for every species can clearly be seen in Fig. 2. The majority of these cases could be resolved based on the investigated species and thus have only a limited negative effect on the stored data.
After studying the stability of the protein identifiers stored in PRIDE, we decided to compare these findings with the total rate of change of the underlying protein databases. For this analysis we used at least two releases of UniProtKB, IPI, and Ensembl per year since 2005 (human and mouse only). The overall rate of change was comparable to the one found when only the identifiers reported in PRIDE experiments for UniProtKB and IPI were taken into account (see Fig. 3, Fig. 4, and supplemental Fig. S3). The different identifier stability of IPI and UniProtKB as well as the different stability of mouse and human identifiers was also reflected in the analysis of the complete database builds. A possible reason for the higher instability of IPI is the varying quality (based on the improvements of genome annotation) of the source databases' records that are used to create the “clustered” IPI protein entries: UniProt, Ensembl, RefSeq, TAIR, H-inv (30), and Vega (31).
Surprisingly, human identifiers were significantly less stable than mouse identifiers in UniProtKB even though human is considered the “more stable” species. This might be caused by a stronger curation effort put into human than in mouse data. Nevertheless, the constant number of about 6% deleted human identifiers compared with about 2% deleted mouse identifiers until the middle of 2010 is striking.
The instability of protein identifiers is not only a problem for published data but can furthermore cause unforeseen problems in long-term projects as, for example, clinical studies. In these cases, samples are generally collected over several years but often need to be processed immediately. If the raw data from these experiments is not reprocessed before the final overall data interpretation, invalid results may be retrieved. In such cases, the observed variation in results caused by changing protein databases will be considerably higher than the here reported numbers as we could only assess the loss of data. When reprocessing MS data the changes in protein databases do not only cause a potential loss of data but will also result in new findings. If this effect is not taken into consideration long-term studies might produce invalid results.
The instability of certain protein databases reported in this study does not only influence the storage of “pure” proteomics data. Other biological resources such as Reactome (32), that process and curate proteomics data from publications need to find ways to handle these changes of protein sequence databases. The two protein identifier mapping algorithms produced significantly different results. Although the PICR mapping algorithm seems more stringent it sometimes reported twice as many IPI identifiers deleted compared with the logical mapping algorithm when mapping protein identifiers retrieved from PRIDE (see Fig. 2). This difference was not observed when mapping whole database releases (see Fig. 4 and supplemental Fig. S7). These results clearly suggest that it is imperative to carefully pick the used protein identifier mapping algorithm for specific applications and thoroughly test its effect. The logical mapping approach seems more suited for applications that are focused on extracting biological knowledge from proteomics data. Especially in manually curated databases like UniProtKB/Swiss-Prot, protein identifier changes are curated according to the biological meaning. Thus, the logical mapping approach seems most likely to maintain the biological significance. A striking example of the differences between the two mapping algorithms can be found when looking at the results from less characterized species. The PICR service, for instance, reported 50% of UniProtKB identifiers deleted from the submission of chicken data in January 2005 (see above) compared with little more than 10% reported deleted by the logical mappings. A similar example is the submission of data on zebrafish in August 2007 (see above) where the PICR service reported virtually all of the identifiers to be deleted compared with 20% based on the logical mappings. Nevertheless, PICR's approach seems to be more stringent and thus better suited for data repositories like PRIDE.
In this study we could show that changing protein identifiers are a risk for the long-term storage of proteomics data as well as the evaluation of long-term proteomics studies. There is a significant difference between the different protein databases concerning identifier stability. Based on the here presented findings UniProtKB seems the best database for applications that rely on the long-term storage of proteomics data. Nevertheless, there are several applications were UniProtKB cannot be used. This is the case when, for example, investigated species are not present in UniProtKB. It is therefore imperative to take the effect of changing protein identifiers into consideration when performing proteomics experiments and evaluating proteomics data. The results from the two protein identifier mapping algorithms used in this study differed considerably. These differences have to be taken into consideration when choosing a protein database and mapping algorithm for a specific task to prevent the misinterpretation of proteomics data.
Acknowledgments
We thank María Martín, Claire O'Donovan, and Rolf Apweiler, for their input in the manuscript.
Footnotes
* J. G. is supported by the Wellcome Trust [grant number WT085949MA]. R. G. C. is supported by EU FP7 grant SLING [grant number 226073]. C. G. is supported by the Austrian Science Fund, FWF [grant number L 670-B13 (to C. G.)]. J. A. V. is supported by the EU FP7 grants LipidomicNet [grant number 202272] and ProteomeXchange [grant number 260558].
 This article contains supplemental Fig. S1 to S7 and Table S1.
This article contains supplemental Fig. S1 to S7 and Table S1.
1 The abbreviations used are:
- BPP
- (HUPO) Brain Proteome Project
- gi
- GenInfo (number)
- HUPO
- Human Proteome Organisation
- IPI
- International Protein Index
- LIMS
- Laboratory Information Management System
- NCBI
- National Center for Biotechnological Information
- nr
- nonredundant
- PICR
- Protein Identifier Cross Referencing
- PPP
- (HUPO) Plasma Proteome Project
- PRIDE
- PRoteomics IDEntifications (database)
- RefSeq
- Reference Sequence (database)
- TAIR
- The Arabidopsis Information Resource
- UniParc
- UniProtKB Archive
- UniProtKB
- UniProt Knowledgebase.
REFERENCES
- 1. Häkkinen J., Vincic G., Månsson O., Wårell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J. Proteome Res. 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 2. Ubaida, Mohien C., Hartler J., Breitwieser F., Rix U., Remsing, Rix L., Winter G. E., Thallinger G. G., Bennett K. L., Superti-Furga G., Trajanoski Z., Colinge J. (2010) MASPECTRAS 2: An integration and analysis platform for proteomic data. Proteomics 10, 2719–2722 [DOI] [PubMed] [Google Scholar]
- 3. Helsens K., Colaert N., Barsnes H., Muth T., Flikka K., Staes A., Timmerman E., Wortelkamp S., Sickmann A., Vandekerckhove J., Gevaert K., Martens L. (2010) ms lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 4. Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith B. E., Hill J. A., Gjukich M. A., Andrews P. C. (2011) Tranche distributed repository and ProteomeCommons.org. Methods Mol. Biol. 696, 123–145 [DOI] [PubMed] [Google Scholar]
- 7. Laursen L. (2009) Apollo scientist dusts off ‘lost’ lunar data. Nature, published online 24. April 2009 (http://dx.doi.org/2010.1038/news.2009.2397)
- 8. Sadygov R. G., Cociorva D., Yates J. R., 3rd (2004) Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat. Methods 1, 195–202 [DOI] [PubMed] [Google Scholar]
- 9. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 10. The UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S., Gordon L., Hendrix M., Hourlier T., Johnson N., Kähäri A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Larsson P., Longden I., McLaren W., Overduin B., Pritchard B., Riat H. S., Rios D., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Spudich G., Tang Y. A., Trevanion S., Vandrovcova J., Vilella A. J., White S., Wilder S. P., Zadissa A., Zamora J., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernández-Suarez X. M., Herrero J., Hubbard T. J., Parker A., Proctor G., Vogel J., Searle S. M. (2011) Ensembl 2011. Nucleic Acids Res. 39, D800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., Geer L. Y., Kapustin Y., Khovayko O., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Ostell J., Miller V., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Sirotkin K., Souvorov A., Starchenko G., Tatusov R. L., Tatusova T. A., Wagner L., Yaschenko E. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35, D5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rose P. W., Beran B., Bi C., Bluhm W. F., Dimitropoulos D., Goodsell D. S., Prlíc A., Quesada M., Quinn G. B., Westbrook J. D., Young J., Yukich B., Zardecki C., Berman H. M., Bourne P. E. (2011) The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 39, D392–D401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weems D., Miller N., Garcia-Hernandez M., Huala E., Rhee S. Y. (2004) Design, implementation and maintenance of a model organism database for Arabidopsis thaliana. Comp. Funct. Genomics 5, 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bell A. W., Deutsch E. W., Au C. E., Kearney R. E., Beavis R., Sechi S., Nilsson T., Bergeron J. J. (2009) A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods 6, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Côté R. G., Jones P., Martens L., Kerrien S., Reisinger F., Lin Q., Leinonen R., Apweiler R., Hermjakob H. (2007) The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics 8, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards N. J. (2011) Protein identification from tandem mass spectra by database searching. Methods Mol. Biol. 694, 119–138 [DOI] [PubMed] [Google Scholar]
- 18. Pruitt K. D., Tatusova T., Klimke W., Maglott D. R. (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 37, D32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., Bergeron J. J. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 20. McCarthy F. M., Burgess S. C., van den Berg B. H., Koter M. D., Pharr G. T. (2005) Differential detergent fractionation for non-electrophoretic eukaryote cell proteomics. J Proteome Res 4, 316–324 [DOI] [PubMed] [Google Scholar]
- 21. Lam L., Arthur J., Semsarian C. (2007) Proteome map of the normal murine ventricular myocardium. Proteomics 7, 3629–3633 [DOI] [PubMed] [Google Scholar]
- 22. Gammulla C. G., Pascovici D., Atwell B. J., Haynes P. A. (2010) Differential metabolic response of cultured rice (Oryza sativa) cells exposed to high- and low-temperature stress. Proteomics 10, 3001–3019 [DOI] [PubMed] [Google Scholar]
- 23. Lemeer S., Pinkse M. W., Mohammed S., van Breukelen B., den Hertog J., Slijper M., Heck A. J. (2008) Online automated in vivo zebrafish phosphoproteomics: from large-scale analysis down to a single embryo. J. Proteome Res. 7, 1555–1564 [DOI] [PubMed] [Google Scholar]
- 24. Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. (2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core data set of 3020 proteins and a publicly-available database. Proteomics 5, 3226–3245 [DOI] [PubMed] [Google Scholar]
- 25. Hamacher M., Apweiler R., Arnold G., Becker A., Blüggel M., Carrette O., Colvis C., Dunn M. J., Frohlich T., Fountoulakis M., van Hall A., Herberg F., Ji J., Ji J., Kretzschmar H., Lewczuk P., Lubec G., Marcus K., Martens L., Palacios Bustamante N., Park Y. M., Pennington S. R., Robben J., Stühler K., Reidegeld K. A., Riederer P., Rossier J., Sanchez J. C., Schrader M., Stephan C., Tagle D., Thiele H., Wang J., Wiltfang J., Yoo J. S., Zhang C., Klose J., Meyer H. E. (2006) HUPO Brain Proteome Project: summary of the pilot phase and introduction of a comprehensive data reprocessing strategy. Proteomics 6, 4890–4898 [DOI] [PubMed] [Google Scholar]
- 26. Barsnes H., Vizcaíno J. A., Eidhammer I., Martens L. (2009) PRIDE Converter: making proteomics data-sharing easy. Nat. Biotechnol. 27, 598–599 [DOI] [PubMed] [Google Scholar]
- 27. Liang C. R., Tan S., Tan H. T., Lin Q., Lim T. K., Liu Y., Yeoh K. G., So J., Chung M. C. (2010) Proteomic analysis of human gastric juice: A shotgun approach. Proteomics 10, 3928–3931 [DOI] [PubMed] [Google Scholar]
- 28. Aye T. T., Mohammed S., van den Toorn H. W., van Veen T. A., van der Heyden M. A., Scholten A., Heck A. J. (2009) Selectivity in enrichment of cAMP-dependent protein kinase regulatory subunits type I and type II and their interactors using modified cAMP affinity resins. Mol. Cell Proteomics 8, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 30. Yamasaki C., Murakami K., Fujii Y., Sato Y., Harada E., Takeda J., Taniya T., Sakate R., Kikugawa S., Shimada M., Tanino M., Koyanagi K. O., Barrero R. A., Gough C., Chun H. W., Habara T., Hanaoka H., Hayakawa Y., Hilton P. B., Kaneko Y., Kanno M., Kawahara Y., Kawamura T., Matsuya A., Nagata N., Nishikata K., Noda A. O., Nurimoto S., Saichi N., Sakai H., Sanbonmatsu R., Shiba R., Suzuki M., Takabayashi K., Takahashi A., Tamura T., Tanaka M., Tanaka S., Todokoro F., Yamaguchi K., Yamamoto N., Okido T., Mashima J., Hashizume A., Jin L., Lee K. B., Lin Y. C., Nozaki A., Sakai K., Tada M., Miyazaki S., Makino T., Ohyanagi H., Osato N., Tanaka N., Suzuki Y., Ikeo K., Saitou N., Sugawara H., O'Donovan C., Kulikova T., Whitfield E., Halligan B., Shimoyama M., Twigger S., Yura K., Kimura K., Yasuda T., Nishikawa T., Akiyama Y., Motono C., Mukai Y., Nagasaki H., Suwa M., Horton P., Kikuno R., Ohara O., Lancet D., Eveno E., Graudens E., Imbeaud S., Debily M. A., Hayashizaki Y., Amid C., Han M., Osanger A., Endo T., Thomas M. A., Hirakawa M., Makalowski W., Nakao M., Kim N. S., Yoo H. S., De, Souza S. J., Bonaldo, Mde F., Niimura Y., Kuryshev V., Schupp I., Wiemann S., Bellgard M., Shionyu M., Jia L., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Zhang Q., Go M., Minoshima S., Ohtsubo M., Hanada K., Tonellato P., Isogai T., Zhang J., Lenhard B., Kim S., Chen Z., Hinz U., Estreicher A., Nakai K., Makalowska I., Hide W., Tiffin N., Wilming L., Chakraborty R., Soares M. B., Chiusano M. L., Suzuki Y., Auffray C., Yamaguchi-Kabata Y., Itoh T., Hishiki T., Fukuchi S., Nishikawa K., Sugano S., Nomura N., Tateno Y., Imanishi T., Gojobori T. (2008) The H-Invitational Database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res. 36, D793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilming L. G., Gilbert J. G., Howe K., Trevanion S., Hubbard T., Harrow J. L. (2008) The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 36, D753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Croft D., O'Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., Jupe S., Kataskaya I., Mahajan S., May B., Ndegwa N., Schmidt E., Shamovsky V., Yung C., Birney E., Hermjakob H., D'Eustachio P., Stein L. (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]