Abstract
Phospholipid bilayer nanodiscs are model membrane systems that provide an environment where membrane proteins are highly stable and monodisperse without the use of detergents or liposomes. Nanodiscs consist of a discoidal phospholipid bilayer encircled by two copies of an amphipathic alpha helical membrane scaffold protein, which is modeled from apolipoprotein A-1. Hydrogen exchange mass spectrometry was used to probe the structure and dynamics of the scaffold protein in the presence and absence of lipid. On nanodisc self-assembly, the entire scaffold protein gained significant protection from exchange, consistent with a large, protein-wide, structural rearrangement. This protection was short-lived and the scaffold protein was highly deuterated within 2 h. Several regions of the scaffold protein, in both the lipid-free and lipid-associated states, displayed EX1 unfolding kinetics. The rapid deuteration of the scaffold protein and the presence of correlated unfolding events both indicate that nanodiscs are dynamic rather than rigid bodies in solution. This work provides a catalog of the expected scaffold protein peptic peptides in a nanodisc-hydrogen exchange mass spectrometry experiment and their deuterium uptake signatures, data that can be used as a benchmark to verify correct assembly and nanodisc structure. Such reference data will be useful control data for all hydrogen exchange mass spectrometry experiments involving nanodiscs in which transmembrane or lipid-associated proteins are the primary molecule(s) of interest.
Biophysical investigations of membrane proteins are notoriously difficult. A wide variety of methods have been developed to circumvent some of the difficulties, including strategies to keep membrane proteins stable and soluble in aqueous solution (1). Detergents are frequently used to increase solubility but, depending on the analysis method, may interfere with the assay or cause structural rearrangements not found in native lipid membranes (2–4). Membrane proteins can be incorporated into liposomes (5) and this has particular advantages for many protein systems, such as ion channels where compartmentalization of each side of the bilayer is required (6). Care must be taken, however, to control the size distribution of liposomes and the stoichiometry of protein-liposome mixtures. An alternative model membrane system, termed nanodiscs, was developed in 2002 (7).
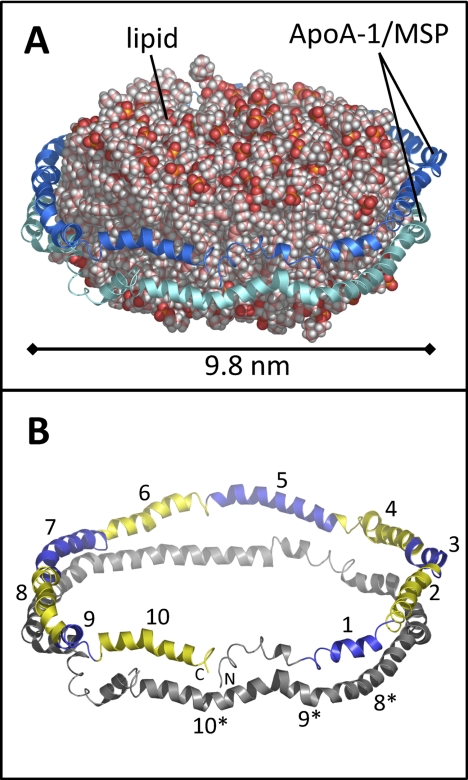
Nanodiscs provide an environment where membrane proteins are stable and compatible with aqueous environments in a native-like phospholipid bilayer. Nanodiscs (Fig. 1) are 8–15 nm diameter phospholipid bilayer discs encircled by two copies of an amphipathic helical protein called a membrane scaffold protein (MSP)1. The MSPs interact with the hydrophobic tails of the lipids around the circular edge of the disc, shielding the lipid from solvent. Nanodiscs self-assemble when detergent is slowly removed from a solubilized mixture of lipid and MSP (8, 9). Membrane protein incorporation into nanodiscs can be accomplished during the self-assembly process (10, 11). Many proteins, including numerous membrane bound cytochrome P450s [reviewed in: (12)], rhodopsin (13), anthrax toxin (14), and the human voltage dependent anion channel (15) have been successfully investigated while embedded in nanodiscs.
Fig. 1.
Model of a nanodisc, based on the discoidal double-belt apolipoprotein A-1 model (32). A, The phospholipid bilayer is encircled by two copies (blue and cyan) of apoA-1. In a nanodisc, the MSP takes the place of apoA-1. B, The model shown without lipid. The ten amphipathic helices in the upper copy of apoA-1/MSP are numbered and shown in alternating blue and yellow. Helices 8, 9, and 10 of the lower copy of apoA-1/MSP are numbered with asterisks.
A great strength of the nanodisc model membrane system is that nanodiscs make membrane proteins compatible with analytical methods that have primarily been limited to investigating soluble proteins [reviewed in: (16, 17)]. A wide variety of experimental techniques have been used to probe the function and structure of nanodisc-embedded membrane proteins. These techniques include, but are not limited to: surface plasmon resonance (18), solid-state NMR (19), solution NMR (15), various single-molecule fluorescence applications (20), electron paramagnetic resonance spectroscopy (21), and hydrogen exchange mass spectrometry (22).
The membrane scaffold protein used in many nanodiscs is an N-terminal truncation of apolipoprotein A-1 (apoA-1), which is the primary protein component of high-density lipoprotein (HDL) particles (23). Because MSP shares many properties with apoA-1, the structural characteristics of HDL particles outline much of the functional assembly of nanodiscs. Nascent HDL particles adopt a discoidal shape when they are cholesterol poor and the incorporation and subsequent esterification of cholesterol by lecithin:cholesterol acyltransferase causes the HDL particles to mature and assume a spherical shape with three apoA-1 molecules assuming what is believed to be a trefoil conformation (24) around the newly formed sphere (25). ApoA-1 itself contains an N-terminal globular domain of ∼43 residues followed by 10 amphipathic helices (two 11-residue helices and eight 22-residue helices), which have glycine or proline at either end. ApoA-1 is largely α-helical based on circular dichroism, displaying ∼50% helical content when lipid-free (26) and ∼80% helical content in HDL particles (27).
Although there is structural information for apoA-1 in the absence of lipids, structural details for the protein in the presence of lipids are lacking. The two crystal structures for lipid-free apoA-1 display similar (>80%) helical content whereas their tertiary structure is highly divergent. Full-length apoA-1 (28), PDB 2A01, contains an N-terminal four-helix bundle followed by two shorter C-terminal helices. An N-terminal truncation of apoA-1 (29), PDB 1AV1, adopts a discoidal shape with four copies of apoA-1. The structure of apoA-1 in the presence of lipid is much more challenging to obtain experimentally. To date, no high resolution structures for apoA-1 in discoidal HDL particles have been solved, although several models have been proposed. The models include the picket fence (30), helical hairpin (31), and double-belt (32) representation. Mounting computational and experimental evidence (33) supports the double-belt model of discoidal HDL structure [reviewed in (25, 34)] and it has become the dominant view. Chemical cross-linking and mass spectrometry have been used to validate the various discoidal HDL particle models and determine the spatial arrangement of the apoA-1 monomers in relation to each other (35–37). Recently, chemical cross-linking and mass spectrometry were applied to mature (spherical) HDL particles to discern the layout of apoA-1 (38). Hydrogen exchange (HX) mass spectrometry (MS) was previously applied to discoidal HDL to try to determine the spatial arrangement of apoA-1 in discoidal HDL particles (39). That work used tandem MS (MS/MS) by collision induced dissociation (CID) to attempt to measure deuterium at single residue resolution and resulted in a solar flares model for discoidal HDL. Localizing deuterium by means of CID has been shown to be nearly impossible because of deuterium scrambling in the gas phase; MS/MS of deuterated peptides requires much less internal excitation, such as afforded by electron transfer dissociation, to avoid scrambling (40, 41). The solar flares model derived from CID-assisted HX MS was later shown to be inaccurate (42) and the exact same data were reused (43) to support a different model, the proposed double superhelical model (44). This model has an elliptical cross section and is the first to deviate from the widely accepted discoidal shape; however the validity of this model remains controversial (45).
In terms of nanodiscs, as opposed to HDL particles, small-angle neutron and x-ray scattering (46), solid-state NMR spectroscopy (47), and molecular modeling (48) of nanodiscs generally support a double-belt model, similar to that of discoidal HDL particles. An example of the double-belt model structure of a nanodisc is shown in Fig. 1 where the two copies of apoA-1 encircle the lipid disc in an antiparallel manner and the two copies are rotated such that the maximum number of intermolecular salt-bridges are formed. When apoA-1 is wrapped around the perimeter of the lipid disc it is primarily helical in nature (the 10 amphipathic helices are highlighted in Fig. 1B). Because the conformation of lipid-free apoA-1 differs dramatically from that of lipid-associated apoA-1, it is reasonable to assume that lipid-free MSP also has a different conformation relative to lipid-associated MSP. To test this hypothesis, and to provide other vital data about the MSP, as described below, we examined the hydrogen exchange of lipid-free MSP and MSP in assembled nanodiscs.
We previously reported on HX MS methods to analyze the conformation of a transmembrane protein embedded in a nanodisc (22). In that work, a modified HX MS protocol was employed to enhance nanodisc disassembly and digestion as well as remove phospholipids prior to MS analysis. Although MSP was present in those experiments and deuterium exchange occurred in MSP, the MSP itself was not characterized. In the present work, both lipid-free MSP and MSP in fully assembled nanodiscs were studied and compared. The MSP [specifically MSP1D1, see (8)] was analyzed at a concentration of <5 μm where lipid-free MSP1D1 was monomeric and the nanodiscs were monodisperse. The results reveal a gross structural rearrangement and decreased deuteration of MSP1D1 upon lipid-association. However, MSP1D1 in nanodiscs remained dynamic and still became significantly deuterated even though the models indicate a high percentage of helical content and hydrophobic interactions with the lipids. EX1 kinetic signatures indicative of cooperative unfolding were observed in several regions of MSP in both the lipid-free and lipid-associated states. Our results provide important information for understanding MSP1D1 conformation in nanodiscs, demonstrate the feasibility of studying biologically relevant HDL particles by this method, offer essential control data that characterize and validate proper nanodisc assembly, and catalogue the peptides and results expected for the MSP1D1 background signals that will be present in any HX MS experiment involving nanodiscs to study peripheral or integral membrane proteins associated with nanodiscs.
EXPERIMENTAL PROCEDURES
Purification of Membrane Scaffold Protein (MSP1D1)
MSP1D1 expression and purification was carried out as previously described (8). In short, the MSP1D1 plasmid was obtained from Add Gene and expressed in Escherichia coli BL21 Codon Plus (DE3) cells. For purification, the protein was isolated by nickel affinity chromatography and purity was confirmed by polyacrylamide gel electrophoresis and electrospray mass spectrometry. Fractions containing MSP1D1 were pooled and dialyzed against 20 mm Tris/HCl pH 7.4, 0.1 m NaCl, 0.5 mm EDTA, and 0.02% NaN3. Protein concentration was determined by absorbance at 280 nm using ε = 21000 cm−1M−1.
Self-assembly of Nanodiscs
Nanodisc self-assembly was carried out as previously described (8). Purified MSP1D1 was added to a 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/deoxycholate solubilized mixture (2:1) to a ratio of 67:1 DOPC:MSP1D1 and incubated for one hour at 4 °C. After incubation, detergent was removed during a 2 h gentle rotation with damp Biobeads SM-2 (BioRad) at 4 °C. The disk preparations were purified by size exclusion chromatography on a Tosoh Biosciences TSKGEl BioAssist G3SWXL 7.8 × 300 mm (5 μm, 250 Å) column run with 50 mm Tris/HCl pH 7.0, 0.15 m NaCl, 0.02% NaN3 at 0.5 ml/minute. Fractions containing purified nanodiscs were isolated and concentrated by Millipore Microcon YM-30 centrifugal filters.
Hydrogen Exchange Mass Spectrometry
HX MS experiments were similar to those described previously (22). Both lipid-free and lipid-associated MSP1D1 were labeled with deuterium in an identical fashion to ensure that both the exchange reaction and quench conditions were the same for both conformational states. Continuous labeling exchange experiments were initiated by diluting 25 μl of sample solution (4 μm lipid-free MSP1D1 or 4 μm MSP1D1 in nanodiscs) 10-fold in 99% deuterium oxide buffer (50 mm Tris/HCl, 0.15 m NaCl, 0.02% NaN3, 2H2O, pD 7.0) at 21 °C. At predetermined time points (ranging from 1 s to 4 h) the labeling reaction was quenched to pH 2.5 by the addition of 0.68 μl of concentrated formic acid (using a 2-μl pipet with filter tip) and placed on ice. All labeling experiments were prepared by hand with pipets, including the short time points of 1 and 5 s (with practice, reproducible labeling and quenching can be accomplished with two manual pipets for a 1 s labeling time point). To retain as much of the deuterium label as possible, all solutions after the quenching step were maintained at pH 2.5 and 0–4 °C before analysis by UPLC/MS. Immediately after quenching, ice-cold sodium cholate solution (10 mm sodium cholate in H2O) was added to a final 25:1 cholate:DOPC molar ratio. POROS 20AL beads [6 μl as a 50% slurry in H2O, 0.08% TFA, pH 2.5, (Applied Biosystems, Foster City, CA)] containing covalently coupled porcine pepsin (49) were then added and the mixture was incubated on ice. After 4 mins of digestion, 3 mg of zirconium oxide coated silica particles (Supelco HybridSPE, #55261-U) were added to the digestion mixture to precipitate phospholipid. After 1 min of incubation with the ZrO2 particles the mixture was spin filtered (0.45 μm cellulose acetate filter, prechilled) at 4 °C for 1 min to remove the pepsin and ZrO2 beads. Flow through from the spin filtration was immediately injected into a Waters nanoAcquity UPLC with hydrogen exchange technology (50) in which the valves, sample loop, trap and analytical columns were all maintained at 0 °C. The regular online pepsin column normally used in this system was not present and was instead replaced by a union.
Upon injection, the peptic peptides were trapped on a pre-column (Waters VanGuard C18, 1.7 μm, 2.1 × 5 mm) (Milford, MA) and desalted with 0.05% formic acid in H2O, pH 2.5, 100 μl/minute for 5 min. Peptides were eluted from the trap to the analytical column (Waters XBridge C18, 1.7 μm, 1.0 mm × 100 mm). A second pre-column (identical to the above one) was placed before the analytical column to prevent any lipid that had not been precipitated from entering the analytical column. Peptides were separated with an 8–40% gradient of 0.05% formic acid in acetonitrile (pH 2.5) over 6 min at a flow rate of 40 μl/min. Throughout an entire experiment, the organic phase never exceeded 85% to ensure that any lipid captured on the trap or guard column did not elute into the mass spectrometer. After a complete HX MS time course analysis, the column and traps were regenerated by flowing 95% acetonitrile through the system. Note that the total amount of time with the modified protocol (22) is only 3 min longer than a typical off-line pepsin digestion protocol and that backexchange (∼30% in these experiments) is not significantly increased in the additional 3 min at quench conditions.
Mass spectral analyses were performed with a Waters QTof Premier equipped with a standard ESI source and Glu-Fibrinopeptide lockmass calibration as described previously (22). Peptic peptides were identified in undeuterated samples for both MSP1D1 in nanodiscs and lipid-free with Waters MSE (51) and Waters ProteinLynx Global Server 2.4 (PLGS) on the same UPLC/QTOF system used for the HX MS experiments. Peptic peptide maps for MSP1D1 with and without lipid are shown in supplementary Fig. S1.
To process the data, the amount of deuterium in each peptic peptide was determined by measuring the centroid of the isotopic distribution for each time point and subtracting the centroid of the isotopic distribution of the undeuterated version of each peptic peptide. The data were not corrected for back-exchange and as such, the plots show the relative deuterium level (50). HX MS analysis on this experimental system has been well characterized previously; the error of each data point generally does not exceed ±0.15 Da (52). For this work, differences in the relative deuterium level between two protein states which exceed 0.50 Da were considered to be significant. Deuterium uptake curves (see supplementary Fig. S2) were prepared using HX-Express (53) and the corresponding data were plotted on structural models with PyMOL (54). The presence or absence of EX1 unfolding kinetics was determined by visually inspecting the shape of the isotopic distribution for all peptides in addition to plotting the width of the isotopic distribution versus time. By plotting the width of the distribution over time, proteins and peptides that exhibit EX1 unfolding result in a distinct peak, with the peak maximum corresponding to the half-life of unfolding (55). The unfolding half-life of each peptide displaying EX1 kinetics was determined by an intersection method (56). Linear regression was performed on either side of the peak and the intersection point of the two lines was used as the half-life of unfolding. For peptides displaying EX1 kinetics, calculating deuterium uptake into both distributions was accomplished by fitting the spectrum with two Gaussian distributions such that their sum equaled the experimentally derived data using PeakFit Software (Systat Software Inc, San Jose, CA). The centroid values of these Gaussian distributions were used as the labeled masses for calculating relative deuterium levels.
RESULTS AND DISCUSSION
Hydrogen Exchange Mass Spectrometry
Hydrogen exchange mass spectrometry has been extensively reviewed (57–61). Briefly, as it applies to the study here, backbone amide hydrogens in proteins are constantly in flux with hydrogens in solution; this exchange is modulated by the presence of structure (i.e. hydrogen bonding) or solvent occlusion upon formation of higher order protein structure and protein complexes (62–64). By measuring the deuterium incorporation into proteins with mass spectrometry, structured regions can be distinguished from those with less structure. At the same time, information about protein motions in solution (protein dynamics) can be obtained because protein dynamics influence the rate of exchange. When comparing exchange into the same protein under two different conditions (in this case, with and without lipid) the location of changes in conformation can be determined (65, 66).
In the most widely accepted mechanism for hydrogen exchange under physiological conditions, the so called localized unfolding model (62), proteins undergo localized unfolding reactions that involve unfolding and refolding (breaking and reforming of hydrogen bonds and changes to solvent exposure of the backbone amide hydrogens). Most proteins follow an EX2 kinetic regime wherein the rate of the exchange reaction is much slower than the refolding reaction and the protein makes many visits to an exchange competent state eventually resulting in deuteration. EX1 kinetics, also known as cooperative or coordinated exchange, exists when the rate of protein refolding during localized motions is much slower than the rate of hydrogen exchange (55, 63, 67, 68). Multiple positions are simultaneously deuterated during EX1 kinetic events and the mass spectra are quite distinct from those in EX2 kinetics (67). Evidence for EX1 kinetics was found for the MSP in this study and this will be described in more detail below.
An investigation of lipid-free apoA-1 has previously been carried out by HX MS (69). The results showed protection from exchange in ∼50% of the sequence with much of the C terminus lacking protection or structure. One would expect, based on the 80% helical content in the models of apoA-1, that there would be more protection from exchange. This discrepancy between the HX data and crystal structure was rationalized by apoA-1 being monomeric (50 μg/ml, 2.5 μm) during the HX experiments whereas in the crystal structure work, it likely formed stabilized helices because of crystal packing. Greater helical content was also seen in EPR studies (70, 71) where similarly, because of experimental constraints, apoA-1 was analyzed at much higher concentrations (3 mg/ml, 100 μm). In the work presented here for lipid-free MSP1D1, which is similar but not identical to apoA-1, differing in the N terminus (for MSP1D1 a polyhistidine tag and cleavable linker replace residues 1–54 of apoA-1), we also maintained a low concentration (0.4 μm during deuterium labeling) to ensure a monomeric state of lipid-free MSP. Note that in assembled nanodiscs, there are two copies of MSP as a result of the assembly process and that these two copies remain in place as long as the nanodisc remains assembled.
Lipid-free MSP1D1 and MSP1D1 in the Nanodisc
Recall that there is no crystal structure for MSP1D1. All of our HX MS results for MSP1D1, in both lipid-free and lipid-associated forms, were modeled onto the structure and model for apoA-1. As MSP is derived from apoA-1, this is a logical choice for interpreting the data, although a few important differences exist between MSP1D1 and apoA-1, as described in the following sections.
Deuterium exchange into lipid-free MSP1D1 was monitored, in the first instance (see also below for additional time points), over the time course of 10 s to 4 h. The deuterium incorporation plots (See supplementary Fig. S2) show rapid rates of deuteration, consistent with a highly dynamic protein containing only limited regions of protection. A summary of the deuterium incorporation into lipid-free MSP1D1 is shown as relative percent deuteration plotted on the lipid-free full-length apoA-1 crystal structure (Fig. 2). In particular, we found the N terminus to be highly deuterated at the 10 s time point and nearly completely deuterated by 5 mins. The HX MS data partially correlate with MSP1D1 assuming a conformation similar to that known for lipid-free apoA-1, although it is impossible to tell what the structure of lipid-free MSP1D1 is from the HX MS data. Relative to the N terminus of apoA-1 in the crystal structure, the N terminus of MSP1D1 is shorter and contains a polyhistidine tag and linker rather than the full first helix of the four helix bundle found in the apoA-1 crystal structure (gray in Fig. 2). Given that this missing part of MSP1D1 would most likely form one helix of a four helix bundle, the overall stability of a putative four helix bundle may be significantly reduced. Indeed, the HX MS data for the MSP1D1 residues which comprise the likely second helix of the bundle indicated rapid deuteration consistent with a lack of structure. Only one region, encompassing residues 127–137, maintained 30–45% relative deuteration throughout the entire time course. Several regions (residues 72–82, 115–126 and 198–203) showed 15–30% relative deuteration (Fig. 2, blue) at the shortest time point but appeared to be very dynamic in nature as they acquired more deuterium over time and all three regions were maximally labeled (60–75% deuteration, orange) by the 30 min time point. The protected region encompassing residues 198–203 has been suggested to be both part of a “lipid sensitive conformational trigger” (71) and a site of potential oligomerization (69). The lipid sensitive conformational trigger was a region in MSP identified by EPR to change first when increasing amounts of lipid were added to the free protein. Several neighboring regions (residues 58–71, 138–146) showed 30–45% relative deuteration (Fig. 2, green) at the shortest time point and peptides from these regions were also dynamic in that they increased in deuterium content as labeling time increased. The “core” of lipid-free MSP1D1 appears to be bounded by residues 58 and 146 with an additional protected region near the C terminus (residues 198–203). However, there were two segments (residues 82–114, 127–137) within the “core” region which only showed 45–60% relative deuteration at the 10 s time point (Fig. 2, yellow). Overall, one would predict that a highly ordered four helix bundle with long alpha helices containing a large number of stabilizing hydrogen bonds would exchange much slower than what was observed. Based on circular dichroism measurements of MSP1D1 (see also below and supplementary Fig. S3) the helical content of lipid-free MSP1D1 (∼44%) was not consistent with the very ordered, highly helical (∼85%) four helix bundle structure in the crystal form of apoA-1. We conclude therefore, that if, in solution, MSP1D1 does assume a partial helical bundle similar to that shown in the lipid-free apoA-1 crystal structure, the bundle is not very stable and a great deal of protein flexibility exists.
Fig. 2.
Deuterium uptake of MSP1D1 plotted on the crystal structure of lipid-free apoA-1 (28), PDB 2A01, and the discoidal double-belt model of apoA-1 in nanodiscs (32). The color code shows the relative percent deuteration according to the scale at the bottom. N- and C termini are noted; in the nanodisc illustration (bottom) one pair of termini are asterisked to differentiate the individual copies of MSP1D1. The N-terminal region of the apoA-1 structure in these illustrations was not present in the MSP1D1 construct that was investigated and is therefore colored gray. For this figure, the N-terminal regions of MSP1D1 in the nanodisc model have been arbitrarily bent away from the double belt because they have been replaced by the histidine tag and linker in the MSP1D1 construct and most likely do not compliment the other copy of the scaffold protein.
In contrast, when MSP1D1 was part of a nanodisc, the entire protein, with the exception of the absolute N- and C- termini, showed much more <15% and 15–30% relative deuteration signatures (Fig. 2, bottom). After 10 s of deuterium labeling, there was <30% relative deuteration in the majority of the protein. Based on the current model for nanodiscs, the MSP structure is believed to be highly helical and consequently, the level of protection from exchange at the early time points is consistent with a protein in which ∼80% of the amide hydrogens are involved in the formation of the hydrogen bonds maintaining the helices. Circular dichroism of lipid-free and lipid-associated MSP1D1 confirms that there was a significant increase in α-helical content in MSP1D1 in nanodiscs relative to lipid-free MSP1D1 (supplementary Fig. S3). However, despite <30% relative deuteration at short labeling times, considerable exchange was observed at later labeling times. Much of MSP1D1 had >45% relative deuteration after 30 min of labeling and the majority of the protein reached the same level of deuteration as lipid-free MSP1D1 by the 2 h time point (see supplementary Fig. S2). The fact that MSP1D1 in nanodiscs reached nearly the same level of deuteration as did lipid-free MSP1D1 is evidence for a dynamic protein, implying that the structure of MSP in the nanodisc, and perhaps the nanodisc itself, is not a rigid body but is quite flexible, allowing for the breaking and reforming of hydrogen bonds along the entire protein backbone while it is wrapped around the lipid core.
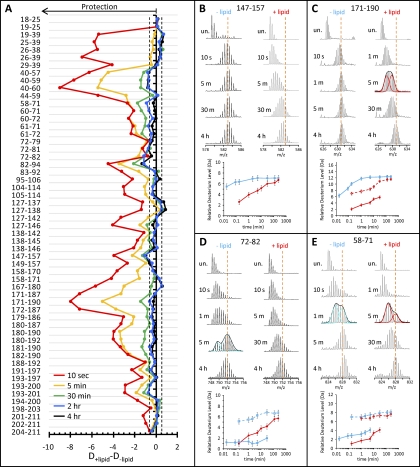
The most protected regions of MSP1D1 in the nanodisc (residues 72–83, 115–126 and 127–137) maintained <45% relative deuteration at the 30 min time point (Fig. 2, green) and correspond to regions in apoA-1 which were identified as lipid binding regions in early investigations (72–74). These regions are opposite each other in the double belt model and known to interact with one another via salt bridges. The HX MS results support the models that implicate these regions of MSP as having the greatest stability in the nanodisc. However, the region which is thought to exhibit the greatest lipid affinity (75), helices 9 and 10 (residues 177–211) in the nanodisc structure, did not show the same level of protection from exchange as the other two lipid binding regions. Although much of helix 9 had <15% relative deuteration (Fig. 2, purple at the shortest time point), indicative of its high affinity for lipid-association, peptides from this region were all very near or greater than 60% deuterated by the 30 min time point. In a native apoA-1 discoidal structure, the C-terminal helices of one copy of MSP1D1 are stabilized by salt bridges to the N-terminal helices of the other copy. As MSP1D1 was missing much of the first helix (replaced with the polyhistidine tag and linker), what was left of helix 1 was >75% deuterated by 30 min of labeling (Fig. 2). This level of disorder indicates that, most likely, salt bridges between the N terminus of the first copy of MSP1D1 and the C terminus of the second copy were not present in MSP1D1 in nanodiscs. The absence of these stabilizing interactions between the MSP1D1 copies may be the cause of the lack of protection from exchange seen in helices 9 and 10. The stark contrast between lipid-free MSP1D1 and MSP1D1 in nanodiscs can be clearly seen when the amounts of deuterium uptake for each condition are plotted together (supplementary Fig. S2) or by subtracting the relative deuterium level of the lipid-free form from the relative deuterium level of the lipid-associated form (Fig. 3A). The difference plot shows the magnitude of protection from exchange (larger negative numbers denote greater protection) when MSP1D1 is part of the nanodisc. Significant differences were seen throughout the MSP1D1 sequence. Two regions, spanning residues 26–59 and 171–178, showed the greatest increase in protection because of lipid association, especially at the short labeling time points. The second region with a large difference (residues 171–178) localizes to part of the two C-terminal helices in the lipid-free apoA-1 crystal structure. Similar to what was observed in HX MS of lipid-free apoA-1 (69), most of the C terminus of lipid-free MSP1D1 (including residues 171–178) was quickly deuterated. In nanodiscs, however, there was significant protection from exchange in this region at the earliest timepoints. The difference plot (Fig. 3A) again clearly shows that lipid-free MSP1D1 and MSP1D1 in nanodiscs have almost the same amount of deuterium in most regions by the 2 h time point.
Fig. 3.
Lipid dependent changes in MSP1D1 deuteration. A, Difference plot of relative deuterium incorporation into MSP1D1 in the presence of lipid minus relative deuterium incorporation into MSP1D1 in the absence of lipid. Negative numbers indicate lipid-induced protection. The residues in each peptic peptide are shown on the left. These results are the average of two independent experiments. The dashed lines indicate the boundary of significant differences (± 0.50 Da), as determined in (52). B–E, Mass spectra and deuterium uptake plots for selected peptides in both the lipid-free (blue) and lipid-associated (red) states; encompassing residues are noted at the top of each panel. Peptides which display EX1 kinetics can be distinguished by the presence of two isotopic distributions or a large increase in peak width. For peptides with EX1 unfolding, one time point was fitted with two Gaussian distributions. The center of these distributions was used to plot the deuterium uptake for both the low (solid line, filled symbols) and high (dashed line, empty symbols) mass populations. Error bars in the deuterium uptake curves have manually been set at ±0.50 Da as described in the text.
EX1 Kinetics in MSP1D1
As discussed above, hydrogen exchange is believed to proceed via exchange into partially or globally unfolded forms (62) and these protein fluctuations have two kinetic limits, EX2 and EX1 (63). These two kinetic limits can be distinguished in HX MS by the characteristic bimodal isotopic distributions of EX1 compared with the single distribution of EX2 (55, 67, 68). An example of the spectra produced as a result of EX2 kinetics is shown in Fig. 3B for both lipid-free and lipid-associated MSP1D1; three examples of EX1 kinetics can be seen in Fig. 3C–3E.
EX1 kinetics are much less common than EX2 kinetics, and false EX1 signatures must be ruled out. False EX1 signatures can be caused by several factors including aggregation and oligomerization of the protein before or during labeling, abnormal back exchange, and carryover between injections (76). Blank injections were run between samples to eliminate carryover and labeling reactions were carried out at 0.4 μm MSP1D1, which is well below the apoA-1 concentration (∼40 μm) where oligomerization was seen previously (69). Circular dichoism of MSP1D1 at ≤5 μm resulted in ∼44% helical content (supplementary Fig. S3) which is consistent with monomeric apoA-1 (69, 77). As a result of the careful controls, we are confident that the EX1 signatures are real and report of protein dynamics rather than experimental artifacts.
To properly characterize EX1 kinetics, including the half-life of unfolding (55), additional labeling time points outside of the initial time course (10 s–4 h) were needed. Three additional labeling time points for lipid-free MSP1D1 (1 s, 5 s, and 2 min) and two time points for MSP1D1 in nanodiscs (1 min and 10 min) were investigated. These additional time points were only processed to extract values for the half-life of EX1-related unfolding. Additional short timepoints (1 s, 5 s) were not acquired for MSP1D1 in nanodiscs as the degree of protection at 10 s in nearly all peptides was already <15% relative deuteration and we did not expect to gain any new information from such short labeling times.
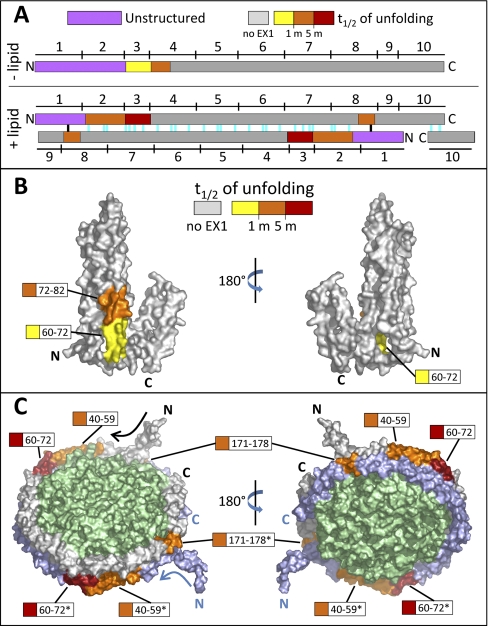
EX1 unfolding was identified by the appearance of bimodal isotope distributions (see Fig. 3C–3E) and localized to four distinct regions (Fig. 4A). In many cases, the regions of unfolding were shorter than the peptic peptides that were produced and sublocalization was accomplished with the use of overlapping peptic peptides (see supplemental Fig. S1). Two regions in MSP1D1, which showed EX1 signatures only when lipid-associated, (residues 40–59 and 171–178) had an EX1 unfolding half-life between 1 and 5 min. Fig. 3C shows the mass spectra of the peptide encompassing residues 171–190 at selected timepoints during the experiment. Peak width analysis of the spectra determined that this peptide displayed EX1 kinetics in the nanodisc form but not in lipid-free MSP1D1 (this can be seen in that the width of the isotope distribution changes to become wider in the lipid-associated form but remains of constant width in the lipid-free form). To determine deuterium uptake level for both the low (folded) and high (unfolded and labeled) mass populations in the EX1 unfolding event, the spectra were fitted with two Gaussian distributions (Fig. 3C). Deuterium uptake for both isotopic distributions was plotted individually (Fig. 3C, bottom). There was one region (residues 72–82) which only showed an EX1 signature in lipid-free MSP1D1 (Fig. 3D). Peptides containing residues 72–82 had an unfolding half-life of between ∼1 and 5 min. The final region (residues 60–72) displayed an EX1 signature in both lipid-free and lipid-associated MSP1D1 (data for residues 58–71 are shown in Fig. 3E). The half-life of unfolding was different under the two conditions: without lipid it was ∼1 min whereas with lipid it was slowed to greater than 5 min. All of the EX1 unfolding information is summarized in Fig. 4.
Fig. 4.
Location and magnitude of EX1-related cooperative unfolding in MSP1D1. A, Diagram showing the spatial relationship between unstructured regions (purple) and EX1 unfolding regions (colored according to the half-life of unfolding, as shown) in lipid-free apoA-1 (top) and apoA-1 in discoidal HDL particle model (bottom). The 10 amphipathic helices (denoted by numbers above and below the colored bars) and predicted salt bridges (colored lines in the +lipid panel, bottom) for apoA-1 are shown. Two of the salt bridges (black) are most likely not present because of the unstructured nature of MSP1D1, which differs from apoA-1 in the N terminus (see text). B, Locations of EX1 unfolding plotted on a space-filling model of lipid-free apoA-1 (28), PDB 2A01. Regions that exhibited EX1 unfolding are colored with respect to the magnitude of the half-life of unfolding. Regions colored gray did not undergo EX1 unfolding. C, Regions of EX1 unfolding plotted on a space-filling model of apoA-1 in the discoidal double belt model (32). Regions that exhibited EX1 kinetics are colored as in panel B. The top copy of MSP is shown in gray and the bottom copy in blue (regions from the bottom copy are shown with asterisks). The lipid bilayer is shown in green.
Without lipid, EX1-related partial cooperative unfolding was limited to the N-terminal half of MSP1D1 (Fig. 4A and 4B, see also supplementary Movie S1). The unfolding was in the third and fourth helices of the apoA-1 crystal structure. Helix 3 coincides with the end of the highly disordered region (the N terminus of MSP1D1 and the second helix in the apoA-1 structure, as previously mentioned). We speculate that the lack of structure in the N terminus of MSP1D1 destabilized these regions of helix 3–4, causing them to undergo cooperative unfolding. In lipid-associated MSP1D1, as in lipid-free MSP1D1, the N-terminal region with EX1 kinetics coincided with the end of the highly disordered N terminus (Fig. 4A and 4C, see also supplementary Movie S2). In the case of MSP1D1 in the nanodisc, the disordered N terminus did not extend as far in the protein sequence so the unfolding started earlier in the MSP1D1 sequence (residue 60 without lipid and 40 with lipid). The last region in lipid-associated MSP1D1 which unfolded (171–178, found in helix 8) was in the only part of the double-belt model which would not be paired via salt bridges to the second copy of MSP1D1 in the nanodisc (Fig. 4A and 4C, see also supplementary Movie S2). One predicted salt bridge (Fig. 4A, black) between helix 1 of the first copy of MSP and helix 8 of the second copy of MSP most likely does not form as a result of the high degree of disorder in the N-terminal region of MSP1D1. The dynamic nature of the N terminus of lipid-associated MSP1D1 may be responsible for the EX1 unfolding of helix 8 through the absence of a stabilizing salt bridge between the two copies of MSP in this region. An alternative explanation for the observation of the EX1 signatures is that the two copies of the MSP are asymmetric—meaning one is in one conformational state and the other is in a different state. Although this possibility might explain the EX1 signatures found in the assembled nanodisc, it does not seem possible that it explains the EX1 signatures in the lipid-free MSP1D1 where all molecules should be in the same environment. Given that the region of EX1 unfolding in the lipid-free and lipid associated states are similar, and right near the N terminus in both cases, we do not favor an asymmetry explanation.
CONCLUSIONS
This work provides a catalog of the expected peptic peptides of MSP1D1 in a nanodisc-HX MS experiment and their deuterium uptake signatures. During nanodisc assembly, these data can be used to ensure that the discs have formed correctly because properly formed nanodiscs should reproducibly display protection from exchange (e.g. <30% relative deuteration signatures, Fig. 2, bottom) compared with lipid-free MSP1D1. Also as shown here, as nanodiscs are increasingly used to characterize membrane-associated proteins embedded in them, it is important to keep in mind that they are not static objects. They have a very dynamic nature and this may well have an effect on the lipid environment that an embedded protein experiences.
Several other conclusions about MSP1D1 and nanodiscs can be drawn from this work. In solution, MSP1D1 most likely does not assume the N-terminal 4-helix bundle shown in the crystal structure of apoA-1 (28) (PDB 2A01). The disorder in the first 60 residues of lipid-free MSP1D1, as detected by rapid deuteration in this area, is not indicative of a region stabilized by the many hydrogen bonds required in such a long helix as well as the hydrophobic interactions between the four helices of the putative four-helix bundle. In our results, very little of the lipid-free form maintained significant protection after 5 min of deuteration, only one segment (residues 127–137) maintained <45% relative deuteration throughout the time course. In contrast, MSP1D1 in nanodiscs was highly protected (<15% relative deuteration) at the short time points. However, deuterium uptake reached greater than 50% relative deuteration (not corrected for back-exchange) in all peptides from MSP1D1 in nanodiscs by 4 h of labeling. In order for this to occur, many localized unfolding events (either EX2 or EX1 kinetics) resulting in exchange competent (i.e. broken) hydrogen bonds in the helices surrounding the lipid bilayer must have occurred. Such results indicate that these helices are dynamic in the nanodisc structure. This could be because of flexing of the nanodisc or helices actually unraveling and exposing backbone amide hydrogens to solution.
The EX1 kinetics seen in both lipid-free MSP1D1 and MSP1D1 in the nanodisc were likely a function of the particular MSP1D1 construct, and may not be present in native lipid-free apoA-1 or discoidal HDL particles. In both lipid-free MSP1D1 and MSP1D1 in nanodiscs, the unfolding in the N terminus coincided with the end of a highly disordered region (residues 1–60 and 1–40 in lipid-free and lipid-associated MSP1D1, respectively). A portion of the disordered region in the lipid-free state (residues 41–60) was stabilized by lipid-association; in the lipid-free form this region was maximally (>75%) deuterated after 5 min of labeling whereas in the presence of lipid the deuterium level stayed below 75% even after 4 h of labeling. These disordered segments destabilized 32 residues further in the sequence both with and without lipid. The unfolding near the C terminus of MSP1D1 in the nanodisc was likely because of the absence of one intermolecular salt-bridge between helix 1 of one copy of MSP and helix 8 of the copy of MSP. The histidine tag and linker in MSP1D1 replaced much of helix 1 in apoA-1 which, if present, would stabilize helix 8 through this interaction. Although many times EX1 unfolding can signify an interesting multiconformational event with biological relevance, it is unclear if this is true for MSP1D1, a protein that does not occur in living systems.
Lastly, this work provides a template for investigation of HDL particles and, in particular, apoA-1 mutants that cause reduced HDL cholesterol levels and lead to coronary heart disease [genetic variation which leads to disease is reviewed in (78)]. HDL does not act alone in cholesterol efflux; many other proteins associate with HDL particles and the interactions of these proteins on HDL particles could also be probed with this method. The nanodisc-HX MS method (22) combined with this work show that HX MS is a viable tool for determining the location and magnitude of both structural and dynamic changes in lipoprotein particles.
Acknowledgments
We thank Stephen G. Sligar and James H. Morrissey at the University of Illinois at Urbana-Champaign and David L. Straight at the University of North Carolina at Chapel Hill for many useful discussions.
Footnotes
* This work was supported by the National Institutes of Health (grants GM086507 to J. R. E., HL48318 to D. W. S.), the Danish Council for Independent Research in Natural Sciences (FNU 09-063876 to K. D. R.), and the Waters Corporation (J. W. J. and J. R. E.). C. M. H. was supported in part by a fellowship from Merck Research Laboratories. This is contribution 990 from the Barnett Institute.
 This article contains supplemental Figs. S1 to S3 and Movies S1 and S2.
This article contains supplemental Figs. S1 to S3 and Movies S1 and S2.
1 The abbreviations used are:
- apoA-1
- apolipoprotein A-1
- MSP
- membrane scaffold protein
- HX
- hydrogen exchange
- HDL
- high-density lipoprotein
- MSP1D1
- membrane scaffold protein 1D1
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine.
REFERENCES
- 1. Seddon A. M., Curnow P., Booth P. J. (2004) Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 1666, 105–117 [DOI] [PubMed] [Google Scholar]
- 2. Gohon Y., Popot J. L. (2003) Membrane protein-surfactant complexes. Curr. Opin. Colloid In 8, 15–22 [Google Scholar]
- 3. le Maire M., Champeil P., Moller J. V. (2000) Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508, 86–111 [DOI] [PubMed] [Google Scholar]
- 4. Garavito R. M., Ferguson-Miller S. (2001) Detergents as tools in membrane biochemistry. J. Biol. Chem. 276, 32403–32406 [DOI] [PubMed] [Google Scholar]
- 5. Rigaud J. L., Pitard B., Levy D. (1995) Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223–246 [DOI] [PubMed] [Google Scholar]
- 6. Morera F. J., Vargas G., González C., Rosenmann E., Latorre R. (2007) Ion-channel reconstitution. Methods Mol Biol 400, 571–585 [DOI] [PubMed] [Google Scholar]
- 7. Bayburt T. H., Grinkova Y. V., Sligar S. G. (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters 2, 853–856 [Google Scholar]
- 8. Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 9. Shih A. Y., Arkhipov A., Freddolino P. L., Sligar S. G., Schulten K. (2007) Assembly of lipids and proteins into lipoprotein particles. J. Phys. Chem. B 111, 11095–11104 [DOI] [PubMed] [Google Scholar]
- 10. Bayburt T. H., Sligar S. G. (2003) Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12, 2476–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bayburt T. H., Sligar S. G. (2010) Membrane protein assembly into Nanodiscs. FEBS Lett. 584, 1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denisov I. G., Sligar S. G. (2011) Cytochromes P450 in nanodiscs. Biochim. Biophys. Acta 1814, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsukamoto H., Sinha A., DeWitt M., Farrens D. L. (2010) Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 399, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katayama H., Wang J., Tama F., Chollet L., Gogol E. P., Collier R. J., Fisher M. T. (2010) Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc. Natl. Acad. Sci. U.S.A. 107, 3453–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raschle T., Hiller S., Yu T. Y., Rice A. J., Walz T., Wagner G. (2009) Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J. Am. Chem. Soc. 131, 17777–17779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borch J., Hamann T. (2009) The nanodisc: a novel tool for membrane protein studies. Biol. Chem. 390, 805–814 [DOI] [PubMed] [Google Scholar]
- 17. Nath A., Atkins W. M., Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 18. Glück J. M., Koenig B. W., Willbold D. (2011) Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Anal. Biochem. 408, 46–52 [DOI] [PubMed] [Google Scholar]
- 19. Kijac A., Shih A. Y., Nieuwkoop A. J., Schulten K., Sligar S. G., Rienstra C. M. (2010) Lipid-protein correlations in nanoscale phospholipid bilayers determined by solid-state nuclear magnetic resonance. Biochemistry 49, 9190–9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nath A., Trexler A. J., Koo P., Miranker A. D., Atkins W. M., Rhoades E. (2010) Single-molecule fluorescence spectroscopy using phospholipid bilayer nanodiscs. Methods Enzymol. 472, 89–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alvarez F. J., Orelle C., Davidson A. L. (2010) Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 132, 9513–9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hebling C. M., Morgan C. R., Stafford D. W., Jorgenson J. W., Rand K. D., Engen J. R. (2010) Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal. Chem. 82, 5415–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rothblat G. H., Phillips M. C. (2010) High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 21, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. (2008) Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lund-Katz S., Phillips M. C. (2010) High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51, 183–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito H., Dhanasekaran P., Nguyen D., Deridder E., Holvoet P., Lund-Katz S., Phillips M. C. (2004) Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 279, 20974–20981 [DOI] [PubMed] [Google Scholar]
- 27. Rogers D. P., Brouillette C. G., Engler J. A., Tendian S. W., Roberts L., Mishra V. K., Anantharamaiah G. M., Lund-Katz S., Phillips M. C., Ray M. J. (1997) Truncation of the amino terminus of human apolipoprotein A-I substantially alters only the lipid-free conformation. Biochemistry 36, 288–300 [DOI] [PubMed] [Google Scholar]
- 28. Ajees A. A., Anantharamaiah G. M., Mishra V. K., Hussain M. M., Murthy H. M. (2006) Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. U.S.A. 103, 2126–2131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Phillips J. C., Wriggers W., Li Z., Jonas A., Schulten K. (1997) Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys. J. 73, 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tricerri M. A., Behling Agree A. K., Sanchez S. A., Bronski J., Jonas A. (2001) Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry 40, 5065–5074 [DOI] [PubMed] [Google Scholar]
- 32. Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. (1999) A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274, 31755–31758 [DOI] [PubMed] [Google Scholar]
- 33. Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. (2010) Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas M. J., Bhat S., Sorci-Thomas M. G. (2008) Three-dimensional models of HDL apoA-I: implications for its assembly and function. J. Lipid Res. 49, 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 36. Davidson W. S., Hilliard G. M. (2003) The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass spectrometry study. J. Biol. Chem. 278, 27199–27207 [DOI] [PubMed] [Google Scholar]
- 37. Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. (2005) A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry 44, 8600–8607 [DOI] [PubMed] [Google Scholar]
- 38. Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. (2011) Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol., doi:10.1038/nsmb.2028 [DOI] [PMC free article] [PubMed]
- 39. Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. (2007) The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 40. Rand K. D., Zehl M., Jensen O. N., Jørgensen T. J. (2009) Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 81, 5577–5584 [DOI] [PubMed] [Google Scholar]
- 41. Zehl M., Rand K. D., Jensen O. N., Jørgensen T. J. (2008) Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J. Am. Chem. Soc. 130, 17453–17459 [DOI] [PubMed] [Google Scholar]
- 42. Shih A. Y., Sligar S. G., Schulten K. (2008) Molecular models need to be tested: the case of a solar flares discoidal HDL model. Biophys. J. 94, L87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gogonea V., Wu Z., Lee X., Pipich V., Li X. M., Ioffe A. I., Didonato J. A., Hazen S. L. (2010) Congruency between biophysical data from multiple platforms and molecular dynamics simulation of the double-super helix model of nascent high-density lipoprotein. Biochemistry 49, 7323–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. (2009) Double superhelix model of high density lipoprotein. J. Biol. Chem. 284, 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones M. K., Zhang L., Catte A., Li L., Oda M. N., Ren G., Segrest J. P. (2010) Assessment of the validity of the double superhelix model for reconstituted high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285, 41161–41171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Skar-Gislinge N., Simonsen J. B., Mortensen K., Feidenhans'l R., Sligar S. G., Lindberg Møller B., Bjørnholm T., Arleth L. (2010) Elliptical structure of phospholipid bilayer nanodiscs encapsulated by scaffold proteins: casting the roles of the lipids and the protein. J. Am. Chem. Soc. 132, 13713–13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Kijac A. Z., Sligar S. G., Rienstra C. M. (2006) Structural analysis of nanoscale self-assembled discoidal lipid bilayers by solid-state NMR spectroscopy. Biophys. J. 91, 3819–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shih A. Y., Denisov I. G., Phillips J. C., Sligar S. G., Schulten K. (2005) Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys. J. 88, 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L., Pan H., Smith D. L. (2002) Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell Proteomics 1, 132–138 [DOI] [PubMed] [Google Scholar]
- 50. Wales T. E., Fadgen K. E., Gerhardt G. C., Engen J. R. (2008) High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 80, 6815–6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Plumb R. S., Johnson K. A., Rainville P., Smith B. W., Wilson I. D., Castro-Perez J. M., Nicholson J. K. (2006) UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 52. Houde D., Berkowitz S. A., Engen J. R. (2011) The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J. Pharm. Sci. 100, 2071–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weis D. D., Engen J. R., Kass I. J. (2006) Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass. Spectrom. 17, 1700–1703 [DOI] [PubMed] [Google Scholar]
- 54. Schrodinger LLC. (2010) The PyMOL Molecular Graphics System, Version 0.98 [Google Scholar]
- 55. Weis D. D., Wales T. E., Engen J. R., Hotchko M., Ten Eyck L. F. (2006) Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 17, 1498–1509 [DOI] [PubMed] [Google Scholar]
- 56. Chen S., Brier S., Smithgall T. E., Engen J. R. (2007) The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 16, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoofnagle A. N., Resing K. A., Ahn N. G. (2003) Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 32, 1–25 [DOI] [PubMed] [Google Scholar]
- 58. Tsutsui Y., Wintrode P. L. (2007) Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr. Med. Chem. 14, 2344–2358 [DOI] [PubMed] [Google Scholar]
- 59. Wales T. E., Engen J. R. (2006) Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 60. Brier S., Engen J. R. (2008) Hydrogen Exchange Mass Spectrometry: Principles and Capabilities. In: Chance M. R. ed. Mass Spectrometry Analysis for Protein-Protein Interactions and Dynamics, pp. 11–43, Blackwell Publishing [Google Scholar]
- 61. Konermann L., Pan J., Liu Y. H. (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 [DOI] [PubMed] [Google Scholar]
- 62. Englander S. W., Kallenbach N. R. (1983) Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16, 521–655 [DOI] [PubMed] [Google Scholar]
- 63. Hvidt A., Nielsen S. O. (1966) Hydrogen exchange in proteins. Adv. Protein Chem. 21, 287–386 [DOI] [PubMed] [Google Scholar]
- 64. Woodward C., Simon I., Tüchsen E. (1982) Hydrogen exchange and the dynamic structure of proteins. Mol. Cell Biochem. 48, 135–160 [DOI] [PubMed] [Google Scholar]
- 65. Engen J. R., Smith D. L. (2000) Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol. Biol. 146, 95–112 [DOI] [PubMed] [Google Scholar]
- 66. Zhang Z., Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miranker A., Robinson C. V., Radford S. E., Aplin R. T., Dobson C. M. (1993) Detection of transient protein folding populations by mass spectrometry. Science 262, 896–900 [DOI] [PubMed] [Google Scholar]
- 68. Arrington C. B., Teesch L. M., Robertson A. D. (1999) Defining protein ensembles with native-state NH exchange: kinetics of interconversion and cooperative units from combined NMR and MS analysis. J. Mol. Biol. 285, 1265–1275 [DOI] [PubMed] [Google Scholar]
- 69. Chetty P. S., Mayne L., Lund-Katz S., Stranz D., Englander S. W., Phillips M. C. (2009) Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. (2007) Electron paramagnetic resonance spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apoA-I in solution. J. Biol. Chem. 282, 9143–9149 [DOI] [PubMed] [Google Scholar]
- 71. Oda M. N., Forte T. M., Ryan R. O., Voss J. C. (2003) The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat. Struct. Biol. 10, 455–460 [DOI] [PubMed] [Google Scholar]
- 72. Kroon D. J., Kupferberg J. P., Kaiser E., Kezdy F. J. (1978) Mechanism of lipid-protein interaction in lipoproteins: a synthetic peptide:lecithin vesicle model. J. Am. Chem. Soc. 100, 5975–5978 [Google Scholar]
- 73. Srinivas R. V., Venkatachalapathi Y. V., Rui Z., Owens R. J., Gupta K. B., Srinivas S. K., Anantharamaiah G. M., Segrest J. P., Compans R. W. (1991) Inhibition of virus-induced cell fusion by apolipoprotein A-I and its amphipathic peptide analogs. J. Cell. Biochem. 45, 224–237 [DOI] [PubMed] [Google Scholar]
- 74. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 75. Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., Anantharamaiah G. M. (1998) Studies of synthetic peptides of human apolipoprotein A-I containing tandem amphipathic alpha-helixes. Biochemistry 37, 10313–10324 [DOI] [PubMed] [Google Scholar]
- 76. Fang J., Rand K. D., Beuning P. J., Engen J. R. (2011) False EX1 signatures caused by sample carryover during HX MS analyses. Int. J. Mass Spectrom. 302, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. (2003) Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278, 23227–23232 [DOI] [PubMed] [Google Scholar]
- 78. Klos K. L., Kullo I. J. (2007) Genetic determinants of HDL: monogenic disorders and contributions to variation. Curr. Opin. Cardiol. 22, 344–351 [DOI] [PubMed] [Google Scholar]