Abstract
Characterization of residual plasma virus during antiretroviral therapy (ART) is a high priority to improve understanding of HIV-1 pathogenesis and therapy. To understand the evolution of HIV-1 pol and env genes in viremic patients under selective pressure of ART, we performed longitudinal analyses of plasma-derived pol and env sequences from single HIV-1 genomes. We tested the hypotheses that drug resistance in pol was unrelated to changes in coreceptor usage (tropism), and that recombination played a role in evolution of viral strains. Recombinants were identified by using Bayesian and other computational methods. High-level genotypic resistance was seen in ~70% of X4 and R5 strains during ART. There was no significant association between resistance and tropism. Each patient displayed at least one recombinant encompassing env and representing a change in predicted tropism. These data suggest that, in addition to mutation, recombination can play a significant role in shaping HIV-1 evolution.
Keywords: HIV-1 drug resistance, HIV-1 recombination, HIV-1 tropism
Introduction
HIV-1 replication may continue during antiretroviral therapy (ART) in some infected individuals resulting in the emergence of drug resistance mutations and viral evolution (Chun et al., 2005; Chun et al., 2008; Furtado et al., 1999; Zhang et al., 1999; Zhu et al., 2002; Martinez-Picado et al., 2000; Palmer et al., 2008; Shen and Siliciano, 2008; Marsden and Zack, 2008; Quan et al., 2008). Combination antiretroviral therapy (cART) has been successful in suppressing HIV-1 to undetectable levels and in reducing illness and death in many individuals (Hirsch et al., 2008). A significant fraction of patients, however, experience virologic failure while on cART (Ledergerber et al.,1999; Mezzaroma et al., 1999; Napravnik et al., 2005; Deeks et al.,2000). Characterizing the residual virus in plasma during cART is a high priority for improving the understanding of HIV-1 pathogenesis and therapy.
The selective pressures on HIV-1 are applied by ART and by the immune response (Napravnik et al., 2005; Goetz et al., 2006; Riddle et al., 2006; Shankarappa et al.,1999; Coffin, 1995; Kuiken et al.,2009; Hermankova et al., 2001; Harrington et al., 2007; Bagnarelli et al., 1999; Blay et al., 2006). Previous analyses of HIV-1 in different anatomic sites within an individual demonstrated compartmentalization, with viral sequences from each site distinct yet phylogenetically related (Kemal et al., 2007; Wong et al., 1997; Singh et al.,1999; Zhang et al., 2002; Kemal et al., 2003; Philpott et al., 2005; Nickle et al., 2003; De Pasquale et al., 2003).
Antiretroviral agents may differ in their availability or activity in different tissues and anatomic sites (Solas et al., 2003; Taylor et al., 1999; Nettles et al., 2006). Subtherapeutic drug levels in particular compartments may select for drug resistance mutations in those sites of viral replication (Hirsch et al., 2008; Johnson et al., 2007).
Coreceptor usage by HIV-1 (tropism) plays a critical role in pathogenesis, disease progression, and viral tropism for particular cell types (Ray and Doms, 2006; Lederman et al.,2006; Kreisberg et al., 2001; Björndal et al., 1997; Blaak et al., 2000; Cheng-Mayer et al., 2009; Weiser et al., 2008). In patients who have detectable CXCR4-tropic (X4) viruses in plasma, CCR5-tropic (R5) viruses usually coexist in the viral swarm (Scarlatti et al., 1997; Freel et al., 2003). We previously demonstrated that initiation of cART can lead to a preferential suppression of X4 strains, resulting in a residual virus population that predominantly uses coreceptor CCR5 (Philpott et al., 2001). In patients who have not received cART changes in tropism have been found to result from accumulated mutations in the HIV-1 envelope (Coetzer et al., 2008), and by recombination involving the V3 region of the envelope (Kemal et al., 2003).
Although much of HIV-1 variation stems from the accumulation of point mutations, recombination may contribute even more to viral evolution because it can lead to leaps in genetic evolution by bringing together two or more distinct beneficial mutations into a single genome (Malim and Emerman, 2001; Jung et al., 2002; Kemal et al., 2003; Philpott et al., 2005; Jetzt et al., 2000; Zhuang et al., 2002; Levy et al., 2004; Chen et al., 2005; Galetto et al., 2004; Shriner et al., 2004; Charpentier et al., 2006; Rousseau et al., 2007; Templeton et al., 2009). For recombination to occur, a host cell must be dually infected by different viruses, each producing progeny RNA’s. The distinct progeny RNA’s must then undergo packaging together in one virion. During the next round of replication, the reverse transcriptase (RT) switches from one template to another, resulting in a chimeric molecule composed of sequences from the two distinct parental genomes (Jetzt et al., 2000). Longitudinal studies of HIV-1 recombination in infected individuals have mostly focused on patients not receiving ART (Fang et al., 2004; Liu et al., 2002; Mild et al., 2007; van Rij et al., 2003).
The evolution of HIV-1 envelope (env) sequences in patients taking ART has been examined by using Bayesian methods (Kitchen et al., 2004) and heteroduplex tracking assays (Kitrinos et al., 2005). Most studies of the evolution of HIV-1 drug resistance mutations in patients taking ART however have examined only pol. Investigation of the evolution of drug resistance needs to examine env as well as pol, because HIV-1 tropism, encoded by the V3 region in gp120 (Hung et al., 1999; Resch et al., 2001; Jensen et al., 2003; Ho et al., 2005; Cardozo et al., 2007; Poveda et al., 2007), plays a role in directing HIV-1 to tissues and compartments that may have different ART activity and selective pressure than is present in blood. A recent paper examined drug resistance and tropism in plasma from viremic patients with subtype C HIV-1 who were taking non-suppressive ART. The methods involved assays of the viral population (Kassaye et al., 2009). Because HIV-1 exists as a swarm of related yet distinct viral species in infected people, in order to examine the relationship of pol and env sequences on the same viral genome it is necessary to study individual viral variants that span pol through gp120 (Kemal et al., 2007; Palmer et al., 2008; Salazar-Gonzales et al., 2008; Kieffer et al., 2004).
We aimed to address the following questions regarding the evolution of residual plasma virus in viremic patients taking cART:
Does all of the residual plasma virus obtained from patients taking cART harbor drug resistance-associated mutations?
Is the evolution of drug resistance-associated mutations in pol associated with HIV-1 tropism?
Does viral recombination play a significant role in the evolution of pol and env sequences in patients taking cART?
To address these questions we used a limiting dilution RT-PCR method to analyze pol and env sequences from individual HIV-1 subtype B variants obtained from serial plasma specimens from four viremic patients taking cART.
RESULTS
We aimed to examine the evolution of HIV-1 pol and env genomic sequences derived from serial patient plasma specimens. To achieve that end it is useful to study DNA fragments spanning pol through gp120 obtained from individual viral variants. We developed a limiting dilution approach to amplify individual HIV-1 variants from plasma, and validated its ability to isolate single viral variants for PCR amplification. We designed a 6.6kb RT PCR method that amplified the HIV-1 genome spanning pol through gp120 (see Materials and Methods).
We obtained HIV-1 strains from serial plasma specimens from four viremic subjects in the Women’s Interagency HIV Study (WIHS), a longitudinal investigation of HIV-1 infection of women (Anastos et al., 2000). At the start of the study, three of the four subjects (WC2, WC4, WC51) were untreated, and one (WC9) had received a two-drug antiretroviral regimen. At the outset none of the subjects had evidence of high level multidrug resistance, but over time they developed changes in HIV-1 drug resistance patterns following initiation or change of ART. The antiretroviral regimen varied over time for each patient, and the patients were prescribed different regimens.
Sequence analysis of each variant was first performed in the V3 region of gp120 in order to predict its tropism (Hung et al.,1999; Resch et al., 2001; Kemal et al., 2007; Cardozo et al., 2007). A total of 122 viral variants with predicted tropism were obtained from the four subjects. The sequences of pol (protease and reverse transcriptase) and gp120 were determined from each variant, and several variants were sequenced entirely from pol through gp120. Drug resistance-associated mutations were determined based on the protease and reverse transcriptase (RT) sequences (Johnson et al., 2007; http://hivdb.stanford.edu).
The virologic and clinical characteristics of subjects WC4 and WC2 at each timepoint are shown in Tables 1A (for WC4) and 1B (for WC2). Two other tables, from subjects WC9 and WC51, are shown in the Supplementary Material. The tables indicate at each timepoint the plasma HIV-1 RNA load, CD4+T cell count, ART prescribed, the number of viral variants predicted to use R5 or X4, and the drug resistance patterns. For each variant studied the sample identifier indicates the patient’s identifier, date, and variant number. The genotypic resistance-associated mutations and resistance interpretations are shown in Tables 1A and 1B (Johnson et al., 2007; http://hivdb.stanford.edu). A total of 25 timepoints were studied from the four subjects. The subjects were viremic at every timepoint, even though ART was prescribed at almost all timepoints. The plasma viral load ranged from 3.68 log–6.36 log HIV RNA copies/ml. The tables illustrate that at the start of the study all four subjects had a mixture of R5 and X4 strains of HIV-1, and there was no evidence of high level drug resistance-associated mutations in pol. Over time, however, multidrug resistance-associated mutations emerged in some variants from all subjects.
Table-1.
| Table-1A Virologic and clinical characteristics of subject WC4 | |||||
|---|---|---|---|---|---|
| Tropism of 6.6kb sequences, Nb |
Genotypic Resistance Associated Mutations | ||||
| Sample IDa / Variant | R5 | X4 | Protease | RT | Resistance interpretation |
| WC4P-0995: VL, log10 5.7; CD4, 28 cells/mm3; ART prescribed, None | |||||
| 9 | 3 | ||||
| WC4P-0995-3 | R5 | None | L74V | ddI-R | |
| WC4P-0995-8 | R5 | None | L74V | ddI-R | |
| WC4P-0995-9 | R5 | None | T69D, V179A | ddC-R | |
| WC4P-0995-5 | X4 | None | L74V | ddI-R | |
| WC4P-0995-6 | X4 | None | None | No resistance | |
| WC4P-0296: VL, log10 4.94; CD4, 42 cells/mm3; ART prescribed, 3TC | |||||
| 2 | 2 | ||||
| WC4P-0296-1 | R5 | None | None | No resistance | |
| WC4P-0296-3 | R5 | None | T69A, M184V | 3TC-R | |
| WC4P-0296-2 | X4 | None | L74V, M184V | ddI-R, 3TC-R | |
| WC4P-0296-4 | X4 | None | None | No resistance | |
| WC4P-0896: VL, log10 5.27; CD4, 25 cells/mm3; ART prescribed, d4T, 3TC | |||||
| 6 | 0 | ||||
| WC4P-0896-3 | R5 | None | L74V, M184V | ddI-R, 3TC-R | |
| WC4P-0896-4 | R5 | None | T69A | No resistance | |
| WC4P-0896-5 | R5 | None | T69D | ddC-R | |
| WC4P-0896-8 | R5 | None | L74V, M184V | ddI-R, 3TC-R | |
| WC4P-0397: VL, log10 5.61; CD4, 10 cells/mm3; ART prescribed, d4T, 3TC, IDV | |||||
| 6 | 3 | ||||
| WC4P-0397-1 | R5 | None | None | No resistance | |
| WC4P-0397-5 | R5 | None | T69A | No resistance | |
| WC4P-0397-6 | R5 | None | None | No resistance | |
| WC4P-0397-4 | X4 | None | None | No resistance | |
| WC4P-0397-8 | X4 | None | None | No resistance | |
| WC4P0897: VL, log10 5.81; CD4, 0 cells/mm3; ART prescribed, AZT, 3TC, NFV | |||||
| 6 | 3 | ||||
| WC4P-0897-1 | R5 | None | None | No resistance | |
| WC4P-0897-3 | R5 | None | None | No resistance | |
| WC4P-0897-5 | R5 | None | None | No resistance | |
| WC4P-0897-10 | X4 | None | None | No resistance | |
| WC4P-0897-14 | X4 | None | None | No resistance | |
| WC4P-0798: VL, log10 4.75; CD4, 97 cells/mm3; ART prescribed, AZT, 3TC, NFV | |||||
| 2 | 6 | ||||
| WC4P-0798-3 | R5 | D30N, N88D | K70R, M184V | NFV-R, 3TC-R | |
| WC4P-0798-8 | R5 | D30N, N88D | K70R, M184V | NFV-R, 3TC-R | |
| WC4P-0798-1 | X4 | D30N, N88D | K70R, M184V | NFV-R, 3TC-R | |
| WC4P-0798-5 | X4 | D30N, N88D | M184V | NFV-R, 3TC-R | |
| WC4P-0399: VL, log10 5.63; CD4, 108 cells/mm3 ART prescribed, AZT, 3TC NFV | |||||
| 2 | 8 | ||||
| WC4P-0399-3 | R5 | N88D | D67N, K70R, K103N, M184V, T215A, K219Q |
AZT-REVT, 3TC-R, NFV-R, NNRTI-R | |
| WC4P-0399-11 | R5 | N88D | D67N, K70R, M184V, K219Q | AZT-REVT, 3TC-R, NFV-R | |
| WC4P-0399-2 | X4 | D30N, N88D | K70R, M184V | 3TC-R, NFV-R | |
| WC4P-0399-5 | X4 | N88D | K103N, M184V | 3TC-R, NFV-R, NNRTI-R | |
| WC4P-0399-10 | X4 | N88D | K70R, V108I, M184V | 3TC-R, NFV-R, NNRTI-LLR | |
| WC4P-1299: VL, log10 5.5; CD4, 58 cells/mm3; ART prescribed, ABC, RTV, IDV, NVP | |||||
| 8 | 6 | ||||
| WC4P-1299-1 | R5 | None | None | No Resistance | |
| WC4P-1299-16 | R5 | None | None | No Resistance | |
| WC4P-1299-3 | X4 | None | None | No Resistance | |
| WC4P-1299-4 | X4 | None | None | No Resistance | |
| WC4P-1299-5 | X4 | None | None | No Resistance | |
| WC4P-1000: VL, log10 5.87; CD4, NAc; ART prescribed, ABC, d4T, RTV, IDV, NVP | |||||
| 8 | 2 | ||||
| WC4P-1000-1 | R5 | None | K103N | NNRTI-R | |
| WC4P-1000-2 | R5 | None | K103N | NNRTI-R | |
| WC4P-1000-5 | R5 | None | K103N, K219Q | NNRTI-R | |
| WC4P-1000-6 | R5 | None | K103N | NNRTI-R | |
| WC4P-1000-7 | R5 | None | K103N | NNRTI-R | |
| WC4P-1000-10 | R5 | None | K103N | NNRTI-R | |
| WC4P-1000-8 | X4 | None | K103N | NNRTI-R | |
| Table-1B Virologic and clinical characteristics of subject WC2 | |||||
|---|---|---|---|---|---|
| Tropism of 6.6kb sequences, Nb |
Genotypic Resistance Associated Mutations | ||||
| Sample IDa / Variant | R5 | X4 | Protease | RT | Resistance Interpretation |
| WC2P-0795: VL, log10 5.28; CD4, 6 cells/mm3; ART prescribed, None | |||||
| 8 | 1 | ||||
| WC2P-0795-4 | R5 | None | T69N, K70G | ddC-R | |
| WC2P-0795-8 | R5 | None | T69N, K70G | ddC-R | |
| WC2P-0795-9 | X4 | None | T69N, K70T | ddC-R | |
| WC2P-0296: VL, log10 4.88; CD4, 3 cells/mm3; ART prescribed, None | |||||
| 13 | 1 | ||||
| WC2P-0296-1 | R5 | None | T69D | ddC-R | |
| WC2P-0296-2 | R5 | None | T69N | ddC-R | |
| WC2P-0296-15 | X4 | None | T69N, V108I | ddC-R,NNRTI-LLR | |
| WC2P-0896: VL, log10 4.7; CD4, 74 cells/mm3; ART prescribed, AZT, 3TC | |||||
| 2 | 2 | ||||
| WC2P-0896-1 | R5 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0896-2 | R5 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0896-3 | X4 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0896-4 | X4 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0897: VL, log10 4.74; CD4, 90 cells/mm3; ART prescribed, d4T, 3TC | |||||
| 3 | 4 | ||||
| WC2P-0897-3 | R5 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0897-6 | R5 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0897-7 | R5 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0897-1 | X4 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0897-2 | X4 | None | T69N, K70R, M184V | ddC-R, 3TC-R | |
| WC2P-0897-4 | X4 | None | D67N, T69N, K70R, V108I, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R, NNRTI-LLR | |
| WC2P-0897-5 | X4 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-1297: VL, log10 5.43; CD4, NAc; ART prescribed, d4T, 3TC, SQV | |||||
| 8 | 2 | ||||
| WC2P-1297-1 | R5 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-1297-2 | R5 | None | D67S, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-1297-5 | X4 | None | D67A, T69N, K70R, V108I, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R, NNRTI-LLR | |
| WC2P-1297-14 | X4 | None | D67A, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0798: VL, log10 4.64; CD4, 188 cells/mm3; ART prescribed, ddI, NFV | |||||
| 7 | 3 | ||||
| WC2P-0798-1 | R5 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0798-2 | R5 | None | D67N, T69N, K70R, V118I, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0798-7 | R5 | None | D67N, T69N, K70R, V108I, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R, NNRTI-LLR | |
| WC2P-0798-5 | X4 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
| WC2P-0798-9 | X4 | None | D67N, T69N, K70R, M184V, T215F, K219E | AZT/d4T-R, ddC-R, 3TC-R | |
Sample ID: WC stands for Wadsworth Center, numbers indicate patient number, P stands for plasma, numbers following the dash are month and year sample collected; numbers following month and year of sample are variant numbers.
Numbers under R5 and X4 columns are the number of R5 and X4 variants as predicted by V3 sequences of the 6.6kb fragment. Sequences of pol and gp120 from the complete 6.6kb fragment were obtained only for those R5 and X4 variants listed.
NA, not available
Note. VL, viral load log10 RNA copies/ml; 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; d4T, stavudine; ddC, zalcitabine; ddI, didanosine; IDV, indinavir; NFV, nelfinavir; NVP, nevirapine; RTV, ritonavir; SQV, saquinavir; ART, antiretroviral therapy; NNRTI, non-nucleoside reverse-transcriptase inhibitors; RT, reverse transcriptase; R, high level resistance; LLR, low level resistance; REVT, revertant;
In total, 122 variants, 6.6kb in size, were characterized for predicted tropism and genotypic resistance from serial specimens from the four patients. Table 2 presents the results from each patient, indicating the number of variants of each tropism, R5 or X4, and the percentage of variants of each tropism that had high level genotypic drug resistance to any antiretroviral drug that targets pol. There were 55 strains from the four subjects that were predicted to be X4-tropic, 71 % of which had high level resistance mutations. There were 67 R5 variants from the four subjects, and 69% of them had high level resistance mutations. There was no significant association found between resistance mutations in pol and viral tropism by using Fisher’s Exact test and the binomial test for proportions.
Table 2.
Summary of tropism and resistance.
| Patient | X4 variants, n |
Resist. X4's, n |
Resist. X4's, % |
R5 variants, n |
Resist. R5's, n |
Resist. R5's, % |
|---|---|---|---|---|---|---|
| WC4P | 17a | 10 | 59 | 27 | 17 | 63 |
| WC2P | 12 | 12 | 100 | 14 | 14 | 100 |
| WC9P | 9 | 9 | 100 | 13 | 13 | 100 |
| WC51P | 17 | 8 | 47 | 13 | 2 | 15 |
| Total | 55 | 39 | 71 | 67 | 46 | 69 |
Numbers indicate individual HIV-1 variants obtained over time from each patient. The variants were sequenced in gp120 to predict tropism based on V3, and in pol to identify genotypic drug resistance mutations.
The lack of association between resistance mutations and tropism prompted us to examine the sequences of pol and gp120 by using computational methods. Figures 1 and 2 present phylogenetic trees of the protease and RT genes, and gp120 from patients WC4 and WC2 respectively. The entire 6.6kb pol-gp120 sequences obtained from patients WC4 and WC2 were also analyzed phylogenetically.
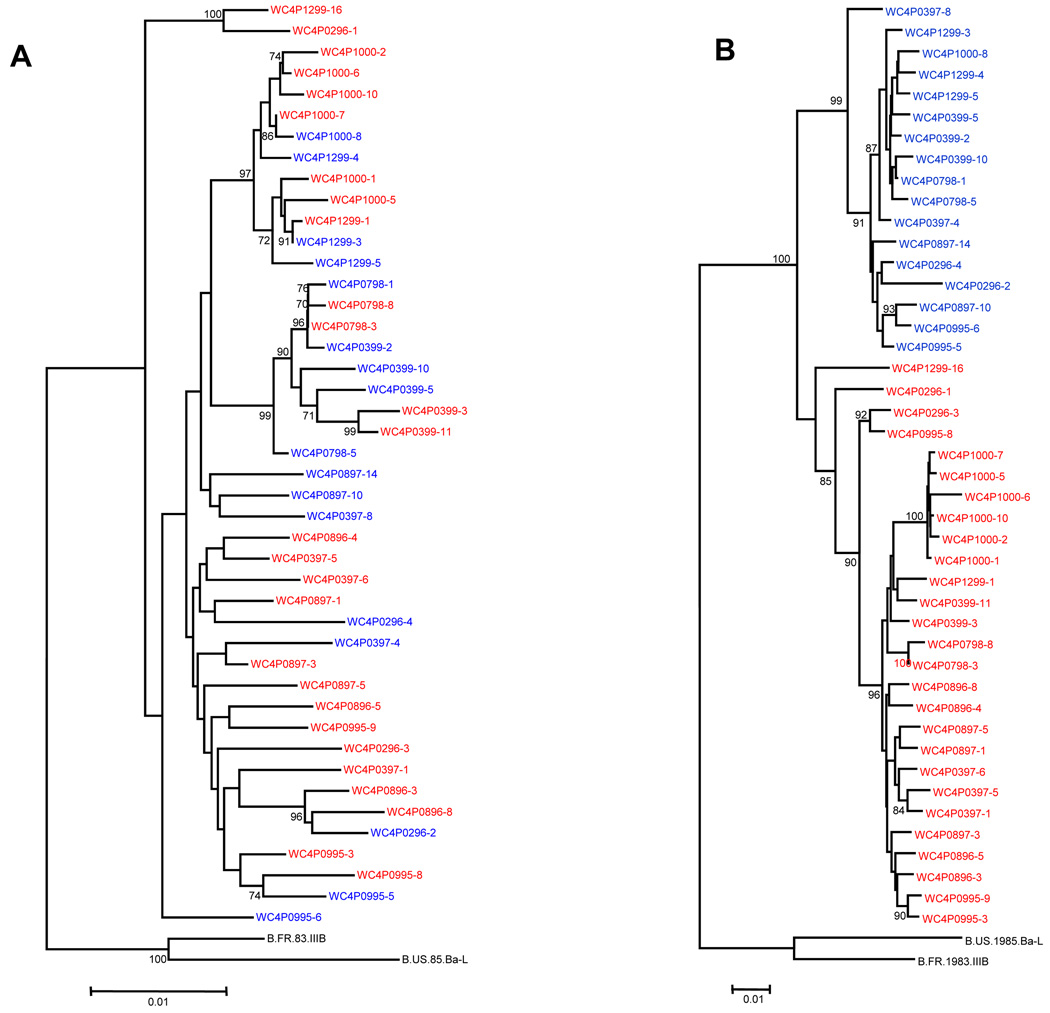
Figure 1. Phylogenetic trees of HIV-1 from patient WC4.
(A) Protease and RT genes from plasma. The labels in red indicate HIV-1 variants predicted to be R5-tropic based on the V3 sequence; branches in blue indicate X4-tropic variants. The dates of the serial sequences are indicated for each variant in its identifier. Each branch represents a variant that is listed in Table 1A. (B) Phylogenetic tree of HIV-1 gp120 gene from plasma.
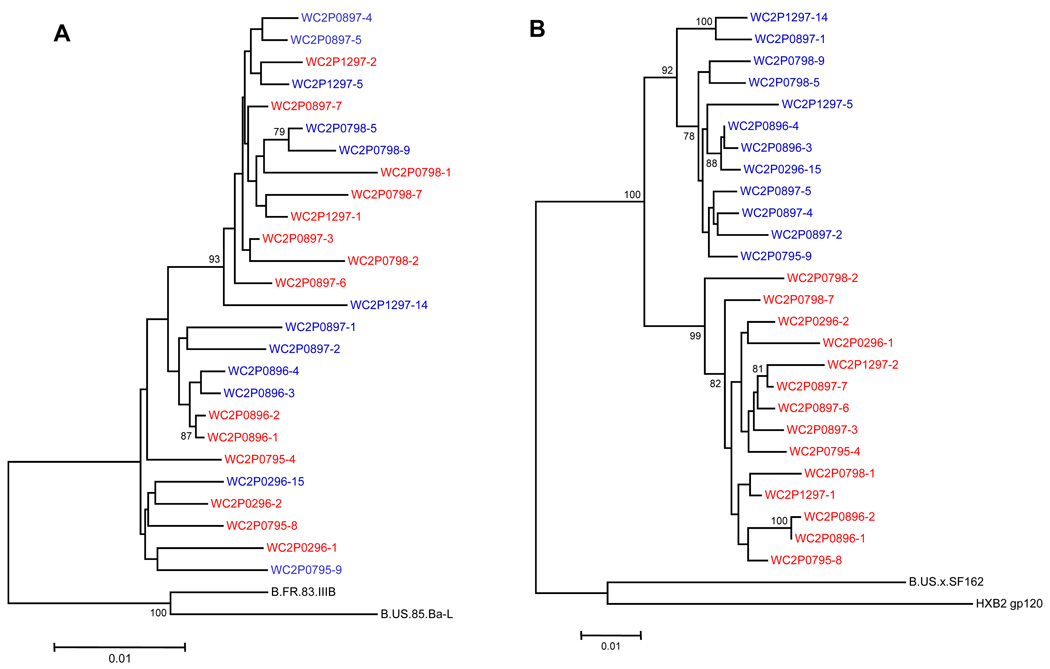
Figure 2. Phylogenetic trees of HIV-1 from patient WC2.
(A) Protease and RT genes from plasma. The labels in red indicate HIV-1 variants predicted to be R5-tropic based on the V3 sequence; branches in blue indicate X4-tropic variants. The dates of the serial sequences are indicated for each variant in its identifier. Each branch represents a variant that is listed in Table 1B. (B) Phylogenetic tree of HIV-1 gp120 gene from plasma.
Figures 1A and 1B illustrate phylogenetic trees of the protease-RT and gp120 sequences respectively, from patient WC4. The branches in red indicate HIV-1 variants predicted to be R5-tropic based on the V3 sequence; branches in blue indicate X4-tropic variants. The dates of the serial sequences are indicated for each variant in its identifier. Each branch represents a variant that is listed in Table 1A. The pol sequences seen in Fig. 1A were intermingled in regard to tropism, displaying a pattern of variation independent of tropism. Fig.1B illustrates the phylogenetic tree of gp120 from patient WC4; it includes the V3 sequence, and shows clustering by tropism. The envelope sequences still cluster by tropism even when the V3 sequences were removed to ensure against bias from convergent evolution (data with V3 sequences deleted not shown).
Figures 2A and 2B illustrate the trees from protease-RT and gp120 respectively, from patient WC2, with each branch representing a variant listed in Table 1B. As with WC4, the protease-RT sequences from WC2 showed a pattern of variation independent of tropism (Fig. 2A), while the gp120 sequences clustered according to tropism (Fig. 2B). The gp120 trees of all four patients generally demonstrated clustering of sequences by tropism, and all gp120 trees shown included V3 sequences. Tree topology of gp120 was similar for all subjects when the V3 loop was deleted (data not shown). The phylogenetic trees of pol and gp120 from patients WC9 and WC51 had similar patterns as the trees from the other patients and are shown in the Supplementary Material. We statistically tested for compartmentalization (clustering) by using the Slatkin-Maddison test and obtained a p<0.001 for compartmentalization.
The results of the phylogenetic analyses (Figs. 1 and 2) and the lack of association of high level drug resistance mutations with predicted tropism (Table 2) raised the question of whether recombination occurred between viral strains within individual patients. Identification of intrapatient HIV-1 recombinants has proved challenging due to the relatedness of the variants (Kemal et al., 2003; Fang et al., 2004; Philpott et al., 2005). Extensive sequence analysis of individual variants and advanced computational methods have now made it more feasible. To identify putative recombinants and the most likely parental sequences three computational methods were employed.
The Bayesian dual multiple change-point framework (Minin et al., 2005) allows for varying evolutionary rates, selective pressures, and phylogenetic trees along the sequence alignment, while providing estimates that include the site and number of recombination events.
The SimPlot (Lole et al., 1999) compares related sequences to a sequence under investigation.
The GARD method uses a genetic algorithm to search a multiple-sequence alignment to detect putative recombinant breakpoints (Pond et al., 2006).
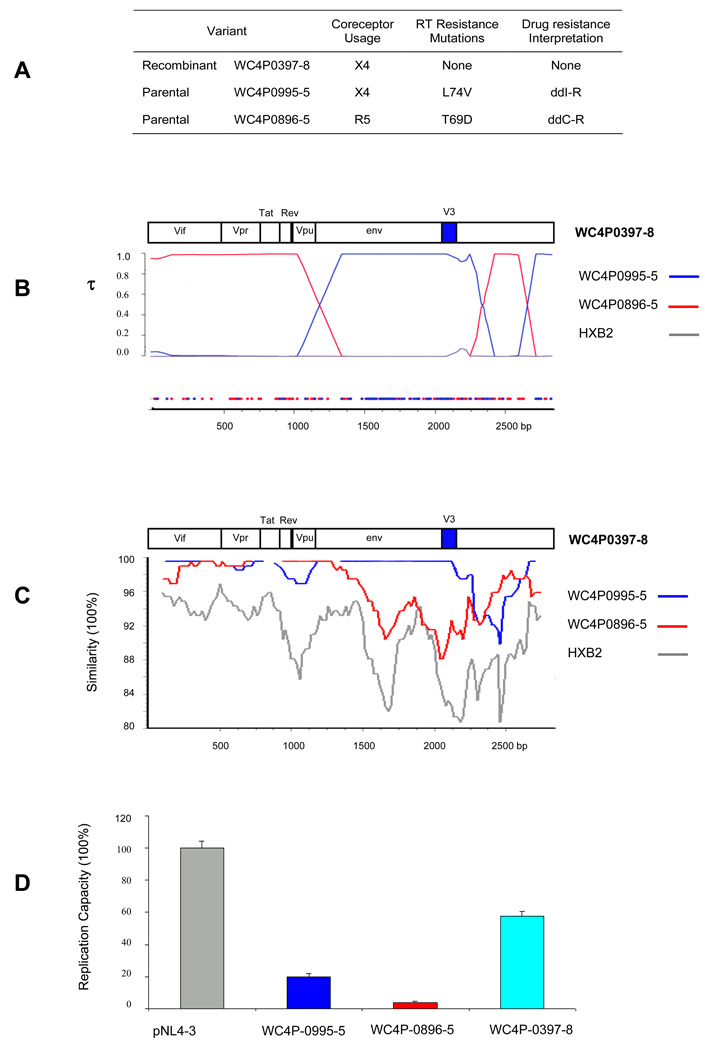
Figure 3 presents an analysis of a putative recombinant, the sequence of an HIV-1 strain obtained from the plasma of patient WC4 in March, 1997 (WC4P0397-8: X4-tropic). A table in the figure (Figure 3A) shows the most likely parental sequences that gave rise to the recombinant; they were identified as coming from that patient in September, 1995 (WC4P0995-5: X4-tropic), and August, 1996(WC4P0896-5: R5-tropic). Viral tropism and drug resistance-associated mutations are also shown in the table (Figure 3A). The recombinant and the parental sequences had very few or no drug resistance-associated mutations in the protease and RT genes. Figure 3B shows the Bayesian dual change-point analysis with recombination breakpoints in gp120. In addition, the sequence was identified as a recombinant by using GARD (data not shown). The recombination breakpoints that were identified by Bayesian analysis were also supported by informative site analysis (Fig. 3B). Phylogenetically informative sites are plotted by color indicating which parental strains have the highest posterior support (Fig. 3B). Figure 3C presents a SimPlot analysis of the same putative recombinant variant, WC4P0397-8. The SimPlot supports the view that recombination occurred in gp120 including the V3 region from parental sequences that differed in tropism. The recombination resulted in a change in predicted tropism compared to the tropism of one of the two most likely parental sequences.
Figure 3. HIV-1 recombination in patient WC4.
(A) Recombinant WC4P0397-8 and its most likely parental sequences are shown in the table at the top of the figure with predicted tropism and drug resistance mutations. (B) A Bayesian analysis of the envelope region of HIV-1 DNA sequences is shown below the table. Sequences that are predicted to be R5-tropic are shown in red; sequences predicted to be X4-tropic are in blue. An informative site analysis with similar color coding is shown at the bottom of the figure. (C) SimPlot of sequences of the envelope region of recombinant WC4P0397-8 and its most likely parentals. (D) Replication capacity assay of the recombinant WC4P0397-8 and its parentals in comparison to the control HIV-1 strain pNL4-3.
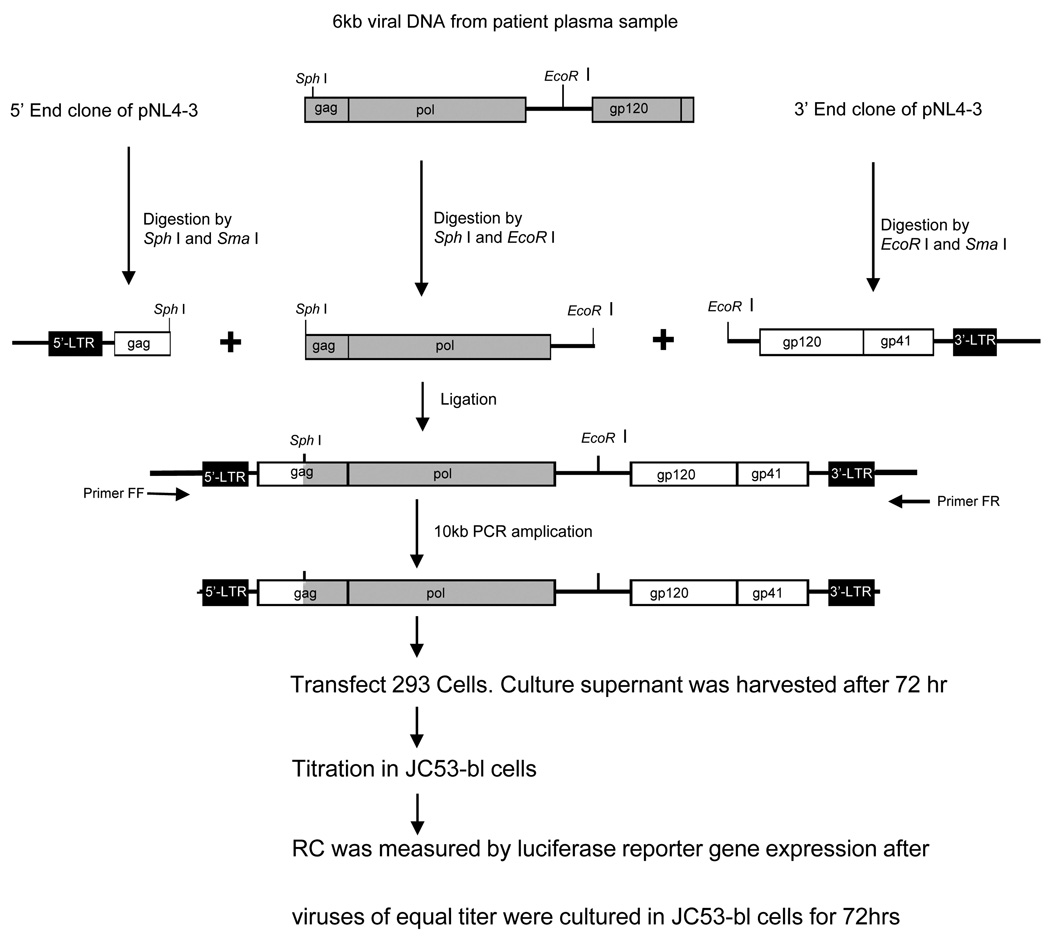
Figure 3D shows the results of replication capacity (RC) measurements performed by using the most likely parental and putative recombinant sequences shown in Fig. 3A and B, as well as a positive control (HIV-1 pNL4-3). We developed an HIV-1 RC assay to be able to measure the RC of different viral variants based on the pol gene amplified from the patient. The RC assay involved construction of 10kb chimeric HIV-1 molecules followed by transfection into a cell line. The chimeric molecules were formed by using ligation-mediated recombination PCR. We used HIV-1 pNL4-3 as a backbone with insertion of the entire pol gene amplified from plasma-derived virus by RT-PCR (Fig. 4, Materials and Methods). The putative recombinant, WC4P0397-8, had no resistance-associated mutations in pol, while the two most likely parental strains had resistance-associated mutations in RT (Fig. 3A). The recombinant had a much greater RC than the parentals. The significantly greater RC of the recombinant compared to the parental sequences supports the interpretation that WC4P0397-8 was a genuine in vivo recombinant that had a survival advantage over the parentals.
Figure 4. Replication capacity (RC) assay.
The RC assay was based on transfection of a 10kb chimeric molecular clone, which was constructed by using ligation-mediated recombination PCR. HIV-1 pNL4-3 was used as a backbone with insertion of the entire pol gene amplified from plasma-derived virus by RT-PCR.
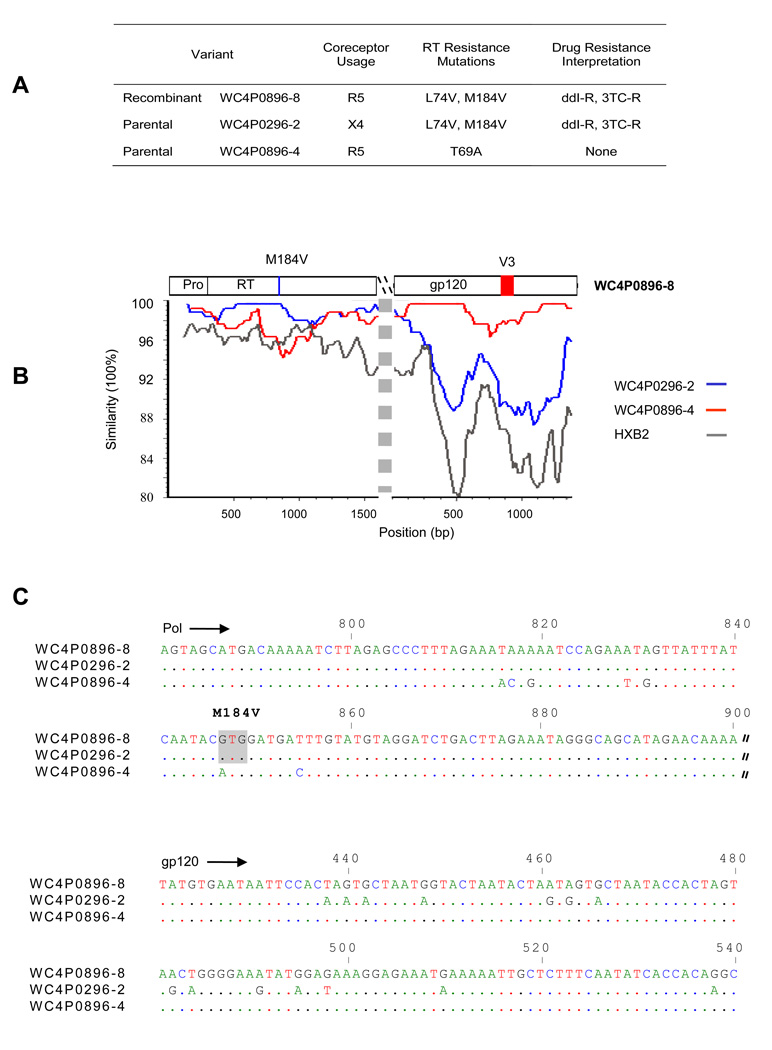
Figure 5 presents a SimPlot and sequence alignment to identify another recombinant variant, WC4P0896-8, and its most likely parental sequences from the same patient, WC4. Fig. 5A is a table listing the putative recombinant and the likely parental sequences, their predicted tropism, and drug resistance mutations. Recombination was confirmed by using Bayesian analysis (data not shown). The SimPlot (Fig. 5B) and sequence alignment (Fig. 5C) show shared mutations, indicating that the virus with the M184V drug resistance mutation in pol and R5 tropism in the V3 region of env (WC4P0896-8: R5-tropic) was most likely derived from two viruses, one with the M184V mutation (WC4P0296-2:X4-tropic) and one with the R5 tropism (WC4P0896-4: R5-tropic). More shared mutations are seen in the sequence alignment further illustrating recombination. The recombinant WC4P0896-8 shared 8 other base changes in pol with the parental sequence WC4P0296-2, in addition to the M184V codon (6 of the base changes are shown in the figure). In the V3 region of gp120, WC4P0896-8 shared many other base changes with the parental WC4P0896-4 besides the ones relevant for coreceptor use. These data illustrate recombination breakpoints between pol and env.
Figure 5. Additional HIV-1 recombination in patient WC4.
(A) Recombinant WC4P0896-8 and its most likely parental sequences are shown in a table with predicted tropism and drug resistance mutations. (B) SimPlot of pol and env sequences of HIV-1 DNA fragments is shown below the table. (C) A sequence alignment of recombinant WC4P0896-8 and its most likely parental sequences in portions of pol and gp120 is shown below the SimPlot.
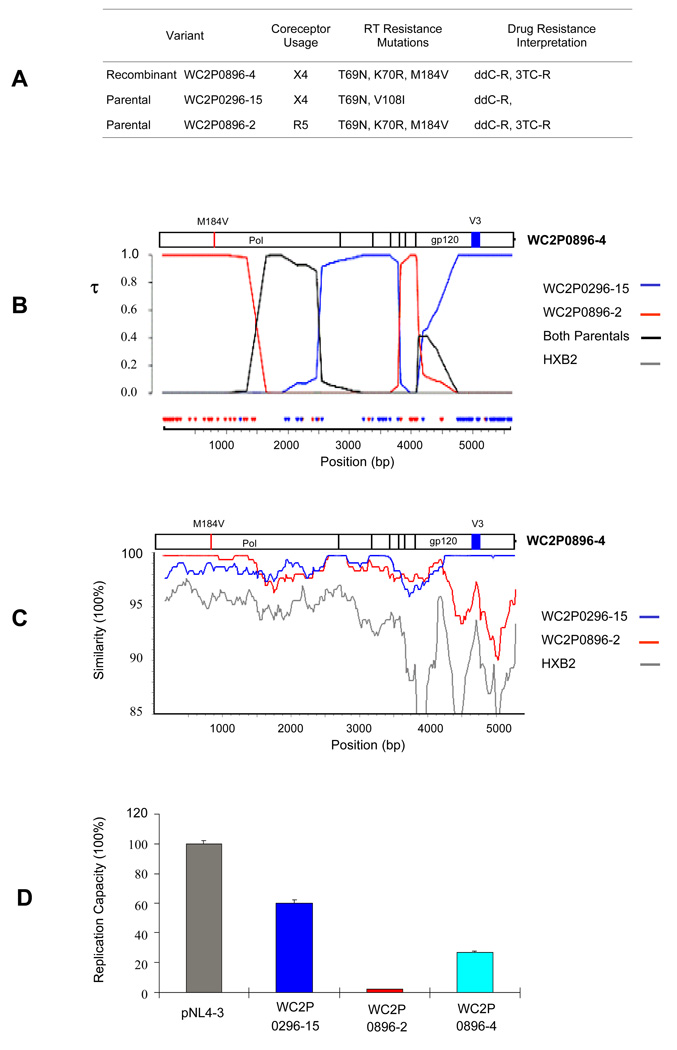
Figure 6 presents analyses from another patient, WC2, of the sequence of a putative recombinant (WC2P0896-4: X4-tropic) and its most likely parental sequences (WC2P0296-15: X4-tropic; and WC2P0896-2: R5-tropic). Figure 6A shows a table with the tropism and drug resistance mutations of the parental and recombinant variants. In Fig. 6B a Bayesian analysis illustrates that WC2P0896-4 is a recombinant formed most likely from the identified parental sequences with breakpoints in pol and env Recombination was supported by a color-coded informative site analysis at the bottom of Fig. 6B. Recombination was also identified by using GARD (data not shown). The SimPlot shown in Fig. 6C shows breakpoints in pol and gp120. The recombinant WC2P0896-4 appears to have acquired drug resistance-associated mutations in pol from one parental (WC2P0896-2 ) and the tropism from the other parental sequence (WC2P0296-15). Figure 6D shows the RC of the most likely parental and recombinant sequences, with the recombinant having an RC approximately 7 times that of one parental (WC2P0896-2) and 40% that of the other parental sequence (WC2P0296-15). Both parental sequences had drug resistance-associated mutations, but the parental with the higher RC (WC2P0296-15) had two drug resistance mutations in the RT conferring resistance to only one antiretroviral drug, ddC. The other parental sequence (WC2P0896-2) and the recombinant, WC2P0896-4, had the same three resistance-associated mutations, and they were interpreted as being resistant to two drugs, ddC and 3TC. The parental strain (WC2P0896-2) had a very low RC, possibly due, at least in part, to the drug resistance mutations. The recombinant, which had the same resistance mutations, had a higher RC, possibly due to other sequences in pol that were not in the drug resistant parental but were acquired from the other parental by recombination. The recombinant is likely to have a survival advantage over the parental with the low RC; this finding is consistent with variant WC2P0896-4 being a real recombinant strain formed in vivo.
Figure 6. HIV-1 recombination in patient WC2.
(A) Recombinant WC2P0896-4 and its most likely parental sequences are shown in the table with predicted tropism and drug resistance mutations. (B) A Bayesian analysis of pol and env sequences of HIV-1 DNA is shown below the table. Sequences that are predicted to be R5-tropic are shown in red; sequences predicted to be X4-tropic are in blue. An informative site analysis with similar color coding is shown at the bottom of the figure. Tau (τ) represents the marginal posterior probabilities of the most probable tree topology. That is, the probability of the putative recombinant clustering with tree i at each position. For this case, we used a 5 taxa tree so there is a topology (which is called “both parentals” which refers to the recombinant clustering with parentals 1 and 2 which had higher posterior support than (1,5) or (2,5). This is most likely due to the fact that both of the parental strains are very similar in this region and we could not distinguish between parentals 1 and 2. (C) SimPlot of pol and env sequences of the recombinant WC2P0896-4 and its parentals. The window size is 300bp, and the step size is 20bp. (D) Replication capacity assay of the recombinant WC2P0896-4 and its parentals in comparison to the control HIV-1 strain pNL4-3.
The data suggests that all four patients in this study had recombinant virus. Data regarding viral recombination in subjects WC9 and WC51 is presented in the Supplementary Material. These data include tables of virologic and clinical characteristics, phylogentic trees, SimPlots, and sequence alignments, and show that these subjects, in addition to subjects WC2 and WC4, also had recombinant HIV-1. All of the HIV-1 variants described as putative recombinants were confirmed as recombinants by using the Bayesian dual multiple change-point framework. At least one recombinant was identified from each patient that encompassed env and conferred a change in predicted tropism compared to at least one of its most likely parental strains.
DISCUSSION
In order to improve the therapy of HIV-1 infection it is necessary to develop a better understanding of the residual virus that persists in patients who experience virologic failure while taking cART. We asked whether the residual plasma virus harbored drug resistance mutations in pol and whether the emergence of drug resistance mutations was associated with tropism. We further evaluated whether viral recombination played a significant role in the evolution of pol and env in individuals taking cART.
This study examined individual viral variant genomic sequences derived from serial plasma specimens from four patients who failed to fully suppress viremia while cART was prescribed. The viral loads remained high, perhaps due to inadequate adherence with the therapy by the patients. At the outset all patients had a mixture of R5 and X4 strains of HIV-1 detected without high level multidrug resistance mutations detected in pol. Over time, multidrug resistance-associated mutations did emerge in all patients.
To perform these analyses on single viral variants we developed a limiting dilution long RT-PCR method that was characterized and shown to isolate individual viral variant genomes for direct sequencing. This technique helped avoid selection bias and PCR artifacts. Several measures were taken to avoid potential artifacts including study of single variants from serial samples, analysis of only one variant at a time (Kemal et al., 2007), and procedures to minimize PCR-mediated recombination (Fang et al., 1998). By using these methods we obtained 122 unique 6.6kb amplicons from the four subjects over time and sequenced the pol and gp120 genes. Approximately 70 percent of the variants had high level drug resistance mutations, whether they were predicted to use X4 or R5. There was no significant association between high level drug resistance in pol and tropism. These results indicate that it is not possible to predict whether the residual plasma virus is drug resistant or has a particular tropism without performing an analysis of the specimen. It also suggests that the evolution of pol and env genes in a viral genome can occur relatively independently. It likely reflects the different selective pressures exerted by antiretroviral agents which target pol, and the host environment. A recent paper used multiplex PCR to examine single-genome sequences from a different compartment, peripheral blood mononuclear cells (Wagner and Frenkel, 2008). It reported that 69% of X4-tropic strains had drug resistance mutations, while R5-tropic strains had 48%. In our study of the residual plasma virus, approximately 70% of X4-tropic strains had resistance mutations.
Phylogenetic trees analyzing pol sequences found them to be intermingled in regard to tropism, while trees analyzing gp120 sequences found them to cluster according to predicted tropism. The clustering of gp120 trees according to tropism suggests that the cellular environment which supports replication of viral strains of one tropism may exert particular evolutionary pressures on gp120.
In this study viral recombinants were identified by computational analysis of the sequences. Recombinants obtained from the same patient at different times were found to be derived from a common ancestor in that patient. The recombinants identified from patient WC4 in October,2000 (WC4P1000-8), March, 1997 (WC4P0397-8), and August,1996 (WC4P0896-8), were derived from a common ancestor sequence (Figs. 1A and B). This finding supports the view that the sequences identified as recombinants arose in the patient.
The RC assay gave results supporting the identification of recombinants. In Figure 3D the significantly greater RC of the recombinant, compared to that of its most likely parental sequences, supports the interpretation that the sequence WC4P0397-8 was a genuine recombinant formed in the patient and was likely to have a survival advantage over the parental sequences. The enhanced RC of that recombinant may have been due, at least in part, to having no resistance-associated mutations in pol, while two of the most likely parental strains had a mutation.
The sequences that were obtained were analyzed by using phylogenetic trees and three computational methods to detect recombination: Bayesian dual multiple change-point framework, SimPlot, and GARD (Kitchen et al., 2004; Lole et al., 1999; Pond et al., 2006). The Bayesian phylogenetic-based approach proved to be the best in delineating recombination breakpoints, showing high accuracy. This approach has a great advantage over the other approaches in detecting ancient recombination events, as the prediction accuracy was the least dependent on the extent of subsequent substitution. As suggested by Chan et al., 2006, we used several methods to screen for recombinants. SimPlot was chosen as it is a widely available and easy to use screening tool and has nice visually appealing output. GARD was chosen as it is one of the newer models available and there is a very nice web-based interface. If these two programs were in agreement regarding a recombination event, then the sequences were analyzed using the more computationally intensive but accurate Dual Brothers. The main crux of the analysis was based on Dual Brothers. This was done out of completeness and thoroughness and not out of heterogeneity in the results. These methods do not identify with certainty the parentals of a putative recombinant. Although several groups, including ours, are working to solve this problem, the methods are not currently available. For the analysis, sequence sets found to contain recombination identified in SimPlot were used for further analysis. The permutations of these sequences that were time-consistent, that is the possible parentals must have been older than the putative recombinant, were chosen for analysis using the Bayesian dual change-point model. It is possible that there were unidentified variants that were not sequenced that were the true parentals. However these true parentals would be evolutionarily related to the parentals identified in the analysis.
Previous studies of HIV-1 recombination documented frequent recombination, and found intrasubtype recombinants in individuals living in parts of the world where multiple subtypes were found (Malim and Emerman, 2001; Jung et al., 2002; Kemal et al., 2003; Fang et al., 2004; Philpott et al., 2005; Jetzt et al., 2000; Zhuang et al., 2002; Levy et al., 2004; Chen et al., 2005; Galetto et al., 2004; Shriner et al., 2004; Charpentier et al., 2006; Rousseau et al., 2007; Templeton et al., 2009; Kuiken et al., 2009). The presence of different subtypes facilitates the identification of recombinants. However, relatively few studies have investigated viral recombination longitudinally in individuals infected with a single subtype of HIV-1 (Fang et al., 2004; Liu et al., 2002; Mild et al., 2007; van Rij et al., 2003; Templeton et al., 2009).
One possible factor contributing to the frequency of recombination may be certain resistance mutations such as the M184I mutation in the RT gene. This mutation has been found to be associated with an increase in the rate of RT template switching, resulting in increased HIV-1 recombination (Nikolenko et al., 2004). We identified the M184I mutation in a recombinant ((WC9P0896-9) and one of its parental sequences (WC9P089605) from patient WC9 (data not shown, available on request).
These analyses of HIV-1 recombination provided some insight into the frequency of recombination in individual patients. We studied 4 patients infected with subtype B over a period of approximately 1–5 years and identified recombinants from all four patients. The recombinants often had drug resistance associated mutations in pol. In each patient we found at least one recombinant that encompassed env and conferred a change in predicted tropism compared to a parental strain. These data demonstrate that switches in tropism may occur commonly and may result from recombination as well as from accumulated mutations (Coetzer et al., 2008).
The finding of frequent recombination between R5 and X4 viral strains indicates that the strains infect some of the same cells, at least on occasion. This may reflect the fact that primary lymphocytes and macrophages, the main targets of infection in vivo, can express coreceptors CCR5 and CXCR4 (Goodenow and Collman, 2006).
CONCLUSIONS
We demonstrated that changes in viral tropism and the development of drug resistance mutations occurred independently of each other. Additionally, we found that viral recombination occurred in all patients studied, suggesting that recombination played a role in the evolution of HIV-1 strains. Approximately 70% of the serial plasma viral strains from viremic patients prescribed cART had high level drug resistance mutations, whether the strains were R5 or X4 tropic.
Future studies should examine serial specimens from patients to better define the role that recombination plays in pathogenesis, the development of drug resistance, tropism switches, and viral evolution.
MATERIALS AND METHODS
Study population
We obtained serial plasma specimens from participants in the Bronx-Manhattan site of the Women’s Interagency HIV Study (WIHS), a NIH multicenter study of the natural history of HIV-1 infection in women (Anastos et al., 2000). Specimens from some of these subjects had previously been examined in a previous study of coreceptor usage (Philpott et al., 2001). The patient identifiers in the previous study were coded differently than in the present study. In the present study we examined specimens from patients WC2, WC4, WC9, and WC51; the initials WC stands for Wadsworth Center. In the previous study the same patients were studied and their code numbers were 2 (WC2 in this study), 7(WC4), 5(WC9), and 4(WC51). For the present study, subjects were chosen who had serial specimens with viral loads of at least 4,000 RNA copies/ml, and viral populations that were known to harbor both X4 and R5 strains of HIV-1. The earliest specimens from these subjects were screened for the presence of high level HIV-1 multidrug resistance (MDR), and only those subjects without evidence of MDR at the initial time point were selected for longitudinal investigation.
To initially screen these specimens for antiretroviral drug resistance mutations, total RNA was extracted from 140 µl of plasma by using the QIAamp® Viral RNA kit (Qiagen, Inc. Valencia, CA) according to the manufacturer’s instructions. Genotyping of the pol region was performed by using the TruGene™ HIV-1 Genotyping Kit (Siemens Healthcare Diagnostics, Deerfield, IL) according to the manufacturer's recommendations.
Amplification
Reverse transcription (RT) and long RT-PCR were performed by using methods established by our lab (Fang et al., 1998) to minimize the potential for PCR-mediated recombination. We also used end-point limiting dilution methods to avoid artifactual PCR recombination and resampling (Kemal et al., 2007; Liu et al., 1996). TRIzol LS Reagent (Invitrogen, Carlsbad, CA) was used in RNA extraction. Following the manufacturer’s instructions, 250 µl patient plasma was used in each experiment and finally RNA was dissolved in 20 µl of RNase-free H2O.
The SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) was used. The reverse transcription included 5 µl RNA, 2U SuperScript™III, and 1 µl 50 µM oligo (dT)20 as RT primer in a total 20 µl reaction. The RT reaction was carried out at 50°C for 60min. The long RT time was chosen to yield complete cDNA. 21 Following the manufacturer’s instructions, cDNA was treated with 1µl RNase H before the next PCR step, or it was stored at −20°C.
A nested PCR was used to amplify a 6,625bp HIV-1 DNA fragment which starts before the pol gene and ends after gp120 (1319 – 7944 nt positions, according to HXB2). The first PCR primer pair was F1309 (forward) 5’-GCATTATCAGAAGGAGCCAC-3’ and R8065 (reverse) 5’-CAACTAGCATTCCAAGGCAC-3’; the second PCR primer pair was F1319 (forward) 5’-AAGGAGCCACCCCACAAG -3’ and R7944 (reverse) 5’-TGATGCCCCAGACTGTGAG-3’. The GeneAmp XL PCR kit and a hot start PCR with AmpliWax PCR Gem100 (Applied Biosystems, Foster City, CA) were used following the manufacturer’s instructions. The final primer concentration was 0.2µM, and the optimized Mg(OAc)2 concentration was 0.875mM. Thermal cycling parameters were as follows: 94°C, 1 minute for preheat; then 20 cycles of 94°C for 15 seconds, 60°C for 6 minutes; then another 15 cycles of 94°C for 15 seconds, 60°C for 6 minutes with 10 second extension increase per cycle; with final extension at 72°C for 10 minutes, then hold at 4°C. Extra extension time was added after 20 cycles to ensure the synthesis of the full length amplicon. The first PCR used 3 µl of cDNA at end-point dilution as template. For the second PCR, 3µl of the first PCR product was used as template.
Limiting dilution PCR
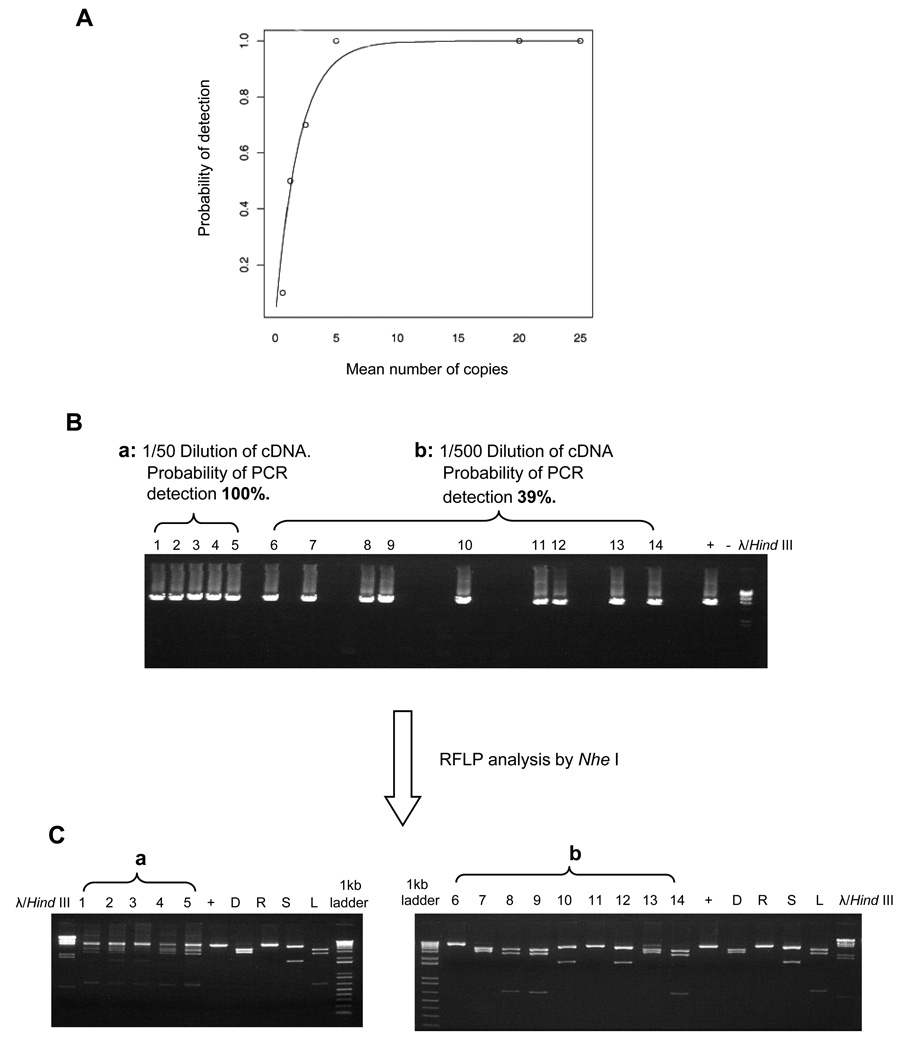
We developed a limiting dilution approach to amplify individual HIV-1 variants. To estimate the sensitivity of this 6.6kb RT-PCR, we applied the parametric model (Hughes and Totten, 2003). Serial endpoint dilution PCR was performed by using known copy numbers of HIV-1 pNL4-3 as template, and a sensitivity curve was drawn (Fig.7A). The parametric model was plotted along with the empirical sensitivity at each concentration. The parametric model showed a very good fit with our experimental data. Based on the model we found that when the 6.6kb PCR sensitivity approaches a single copy of template, the probability of a positive reaction is 0.405.
Figure 7. Analysis of limiting dilution PCR using a parametric model.
(A) The sensitivity curve from the parametric model is shown in the solid line, and the empirical sensitivity of PCR at each dilution is shown in circles. (B) End-point limiting dilution PCR. Four HIV-1 strains that differ in restriction fragment length polymorphisms (RFLP) when their PCR product was digested by the restriction enzyme Nhe I (HIV-1 strains LAV, SF162, RF-V82F/I84V, and patient donor E) were mixed in known quantities. The DNA was subjected to 1% agarose gel electrophoresis. The left end of the gel (a: lanes1–5) shows the 4 HIV-1 strains after a minimal 1/50 dilution. The right end of the gel (b: lanes 6–14) illustrates an end-point dilution (1/500 dilution of the cDNA mixture), yielding a positive PCR in less than 40% of equivalent dilutions. (C) RFLP. The diluted cDNA was digested with Nhe I and subjected to 1% agarose gel electrophoresis. Lanes 1–5 (a) illustrate the gel pattern of lanes containing the mixed strains at the 1/50 dilution after digestion by Nhe I . Lanes 6–14(b) indicate, based on the gel pattern after Nhe I digestion, that the final 1/500 dilutions of the original mixture of 4 strains contained only one viral strain per lane.
To evaluate the ability of the limiting dilution PCR method to isolate single viral genomes for amplification, differences in restriction fragment length polymorphism (RFLP) patterns among known strains of HIV-1 were examined. Four HIV-1 strains, LAV, SF162, RFV82F/I84V, and Donor E, were chosen because their 6625bp RT PCR product had different RFLP patterns after digestion by restriction endonuclease NheI. The strains were obtained from the NIH AIDS Reference and Reagent Program (www.aidsreagent.org). A mixture of equal quantities of these four virus strains was prepared. After RNA extraction and RT, limiting dilution PCR was performed by using the RT product as template. The positive PCR products were gel purified, and then 200ng of purified PCR product was digested with 5U of restriction enzyme NheI (New England Biolabs, Ipswich, MA) at 37°C for 1 hour. RFLPs were analyzed by using 1% agarose gel electrophoresis.
Figure 7B shows the examples of amplification of a 6.6kb PCR when using template from two different dilutions of the mixed HIV-1 strains. The DNA was subjected to 1% agarose gel electrophoresis. Lanes 1–5, amplified from a relatively high concentration of the virus mixture, had a 100% probability of positive reactions, and lanes 6–14, amplified from a lower concentration of the virus mixture, had a 39% probability of detection (end point dilution). The corresponding positive PCR products in each lane were then digested with Nhe I (Fig. 7C) and subjected to 1% agarose gel electrophoresis. Lanes 1–5 illustrate the RFLP pattern of samples containing the mixed strains. Lanes 6–14 show 23 the RFLP pattern of individual strains, which indicates that the template in this PCR experiment was from a single HIV genome.
Sequence analysis
We studied unique 6.6kb amplicons extending from pol through gp120 obtained from single HIV-1 genomes by using limiting dilution RT-PCR. Sequences were determined by direct sequencing from PCR products. Direct sequencing was used instead of molecular cloning to avoid the potential inefficiency that could result from attempting to clone such large fragments. Efforts were also made to avoid PCR-mediated recombination and other artifacts (Fang et al.,1998; Fang et al., 2004; Kemal et al., 2003; Kemal et al., 2007; Philpott et al., 2005). All sequence data were carefully checked for the presence of multiple peaks. In rare cases, when multiple peaks were found, those sequences were excluded from the final analysis. Computational analyses, including a BLAST search, and phylogenetic analysis of the sequences showed no evidence of contamination, and open reading frames were intact, with no significant deletions, insertions, or nonsense mutations. All sequences were identified as HIV-1 subtype B.
Coreceptor usage prediction algorithm
To predict HIV-1 coreceptor usage (tropism), we applied three different algorithms:
The method validated in our lab on the basis of the overall charge of the V3 loop and the presence of basic or acidic residues at positions 275 and 287 of the env gene as described (Bhattacharyya et al., 1996; Hung et al., 1999; Philpott et al., 2001).
A modified prediction method based on the 3D molecular modeling of the HIV-1 V3 loops (Cardozo et al., 2007). The study by Cardozo et al. showed a 94% concordance between genotypic prediction and cell-based phenotypic analysis.
A web based bioinformatic method based on scoring of position-specific scoring matrices (PSSM) (Jensen et al., 2003).
Genotypic drug resistance
Drug resistance was predicted by using the Stanford HIV drug-resistance database (http://hivdb.stanford.edu) for interpretation of genotype data. Results reported here are based on February, 2008 updates. In addition to the Stanford database, we have also applied the International AIDS Society-USA Drug Resistance Mutations Group guidelines for interpretation of resistance mutations (Hirsch et al., 2008; Johnson et al. 2007).
HIV-1 replication capacity assay
We developed an HIV-1 replication capacity (RC) assay to be able to measure the RC of different viral variants. The RC assay was based on construction of 10kb chimeric HIV-1 molecules followed by transfection into a cell line. The chimeric molecules were formed by using ligation-mediated recombination PCR. We used HIV-1 pNL4-3 as a backbone with insertion of the entire pol gene amplified from plasma-derived virus by RT-PCR (Fig. 4). To perform a 3 way ligation-mediated recombination PCR, two fragments were made from HIV-1 pNL4-3 serving as a backbone. The 5’ end fragment was constructed by digestion of the plasmid pNL-5END (a pNL4-3 5’end clone, includes the 5’LTR to 2048nt, according to HIV-1 HXB2) by restriction enzymes Sph I and Sma I. The 3’ END fragment was prepared by digestion of pNL-3END (a pNL4-3 3’end clone, includes 5245 nt to the 3’LTR, according to HXB2) by EcoR I and Sma I. The third fragment containing the full length HIV-1 pol gene was obtained from patient plasma viral RNA by RT-PCR. The 6kb RT-PCR products were double digested by both Sph I and EcoR I and then gel purified. The ligation product was used as template for a 10kb PCR to generate chimeric HIV-1 complete genomic DNA. The insertion was confirmed by sequencing the cross-over region of the 10 kb PCR product. Cultured 293T cells (Invitrogen) were transfected with purified 10 kb DNA by using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis,IN) following the manufacturer’s instructions. After 72 h in culture, the supernatant was aliquoted, and stored at −80°C until use.
Virus production from the transfected 293T cells was quantitated in JC53-bl cells. JC53-bl is a HeLa-based cell line that has been genetically engineered to stably express high levels of CD4, CXCR4, and CCR5 and is susceptible to infection by diverse isolates of HIV-1 (www.aidsreagent.org). The β-Gal and luciferase genes were introduced into JC53-bl cells separately, each under transcriptional control of the HIV-1 long terminal repeat. The β-Gal reporter was included to allow direct enumeration of infectious viral units by counting β-Gal expression-positive infected cell colonies under a microscope. The luciferase reporter enables automated quantitation of HIV infection. JC53-bl cells were previously analyzed for their susceptibility to infection with primary HIV-1 isolates. Compared with phytohemagglutinin (PHA)-stimulated cultures of peripheral blood mononuclear cells (PBMC), the JC53-bl cells were at least 2–5 fold more sensitive to infection with primary HIV-1 isolates than PHA-stimulated PBMCs (Wei et al., 2002).
In our experiments, the infectious titer of the virus in the supernatant obtained from the transfected 293T cells was determined by infecting JC53-bl cells with a range of viral dilutions in a 24-well plate. The cells were stained for β-galactosidase expression 48 h postinfection by using the In Situ β-Galactosidase Staining Kit (Stratagene, La Jolla,CA). Viral infectious units were determined by counting the number of blue cells under a microscope, using dilutions that gave between 200 and 600 stained cells (Kimpton and Emerman,1992).
Recombinant virus variants of the same infectious titer were used to infect 1 × 104 JC53-bl cells in 24-well tissue culture plates. At 72 hours postinfection, the supernatant was removed and the cells were lysed by using a luciferase assay system kit (Promega, Madison, WI.). The light intensity of each cell lysate was measured on a TD-20/20 luminometer (Turner Biosystems, Sunnyvale, CA). Mock-infected wells were used to determine background luminescence, which was subtracted from the sample wells. This 10 kb chimeric HIV-1 DNA produced recombinant HIV-1 virus after transfection of 293T cells. We measured the replication capacity of the recombinant virus in the indicator cell line JC53-bl in the absence of antiretroviral agents by measuring expression of the luciferase reporter gene and comparing it to luciferase expression by the control HIV-1 strain pNL4-3 (Barbour et al., 2002).
Computational methods
Multiple alignments of sequences from patients plus suitable reference sequences were assembled with MAFFT and/or CLUSTAL-W, checked for quality and hand-edited, if necessary, by using BioEdit software: (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Prior to phylogenetic tree construction, columns of the alignment containing gaps were removed. Maximum likelihood and neighbor-joining phylogenetic trees were produced using PAUP with the general time reversible model of evolution and a gamma distribution of site-specific rates of evolution. Bootstrap replications (B=100) were used to assess tree robustness. To ensure against bias from convergent evolution, envelope trees were also constructed using sequences that had the V3 loop deleted. The gp120 trees that had the V3 loop deleted were not shown in the paper or in the Supplementary Material.
Intrapatient recombination was detected by using SimPlot (Lole et al., 1999), the Los Alamos National Laboratory (LANL) HIV Database Highlighter tool, and GARD. The GARD method uses a genetic algorithm to search a multiple-sequence alignment to detect putative recombinant breakpoints (Pond et al., 2006). A more rigorous recombination analysis was done by using a Bayesian dual change-point model using Dual Brothers software (Minin et al., 2005).
There are many tools to analyze recombination, each with their own specific strengths and drawbacks and limitations. In this case we were specifically interested in identifying the breakpoints in the putative recombinants. The main tool of our analysis was Dual Brothers, the Bayesian phylogenetic-based approach. In Chan et al., 2006, the authors examined the predictive accuracy of several recombination detection programs and showed that Dual Brothers, in particular, were less sensitive to parameter setting than other methods and had higher accuracy in locating recombination breakpoints compared to other approaches.
Sequences were aligned by using Clustal-W and hand edited as necessary. For this analysis the V3 loops were included. Phylogenetically informative sites were plotted beneath the posterior probability of tree support.
Statistics
Fisher’s exact test and the binomial test for proportions were used to assess significance. All p-values are two-sided and a p-value < 0.05 was considered significant.
Nucleotide sequences
The sequences have been contributed to GenBank and the following accession numbers were assigned: FJ861709-FJ861830 for pol gene sequences from subjects WC2, WC4, WC9, WC51; and FJ861831-FJ861952 for gp120 sequences from subjects WC2, WC4, WC9, WC51. For 6kb sequences from patient WC4 the accession numbers were FJ861953-FJ861964, and for 6 kb sequences from patient WC2 the numbers were GQ358530-GQ358532.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the study subjects for their participation and the Wadsworth Center Applied Genomics Technologies Core for DNA sequencing. We thank the NIH AIDS Reference and Reagent Program for HIV-1 strains and cell lines. This study was supported by NIH grants RO1-AI52015 and UO1-AI35004.
Role of the funding source
The funding source (NIH) had no involvement in the conduct of the research or in the preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anastos K, Gange SJ, Lau B, Weiser B, Detels R, Giorgi JV, Margolick JB, Cohen M, Phair J, Melnick S, Rinaldo CR, Kovacs A, Levine A, Landesman S, Young M, Muñoz A, Greenblatt RM. Association of race and gender with HIV-1 RNA levels and immunologic progression. J Acquir Immune Defic Syndr. 2000;24:218–226. doi: 10.1097/00126334-200007010-00004. [DOI] [PubMed] [Google Scholar]
- Bagnarelli P, Mazzola F, Menzo S, Montroni M, Butini L, Clementi M. Host-specific modulation of the selective constraints driving human immunodeficiency virus type 1 env gene evolution. J Virol. 1999;73:3764–3777. doi: 10.1128/jvi.73.5.3764-3777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour JD, Wrin T, Grant RM, Martin JN, Segal MR, Petropoulos CJ, Deeks S. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J Virol. 2002;76:11104–11112. doi: 10.1128/JVI.76.21.11104-11112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Brooks BR, Callahan L. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res Hum Retroviruses. 1996;12:83–90. doi: 10.1089/aid.1996.12.83. [DOI] [PubMed] [Google Scholar]
- Björndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyö EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H, van't Wout AB, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in a simian/human immunodeficiency virus-infected macaques. J Virol. 2006;80:999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23:415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA. Detecting recombination in evolving nucleotide sequences. BMC Bioinformatics. 2006;7:412. doi: 10.1186/1471-2105-7-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006;80:2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rhodes T, Hu W. Comparison of the genetic recombination of human immunodeficiency virus type 1 in macrophages and T cells. J Virol. 2005;79:9337–9340. doi: 10.1128/JVI.79.14.9337-9340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng-Mayer C, Tasca S, Ho SH. Coreceptor switch in infection of nonhuman primates. Curr HIV Res. 2009;7:30–38. doi: 10.2174/157016209787048500. [DOI] [PubMed] [Google Scholar]
- Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O’Shea MA, Hallahan CW, Daucher M, Ward DJ, Moir S, Mullins JI, Kovacs C, Fauci AS. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;181:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- Coetzer M, Nedellec R, Salkowitz J, McLaughlin S, Liu Y, Heath L, Mullins JI, Mosier DE. Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008;83:11758–11766. doi: 10.1128/JVI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Barbour JD, Martin JN, Swanson MS, Grant RM. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J Clin Infect Dis. 2000;181:946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- De Pasquale MP, Leigh Brown AJ, Uvin SC, Allega-Ingersoll J, Caliendo AM, Sutton L, Donahue S, D'Aquila RT. Differences in HIV-1 pol sequences from female genital tract and blood during antiretroviral therapy. J Acquir Immune Defic Syndr. 2003;34:37–44. doi: 10.1097/00126334-200309010-00005. [DOI] [PubMed] [Google Scholar]
- Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J. Virol. Methods. 1998;76:139–148. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- Fang G, Weiser B, Kuiken C, Philpott SM, Rowland-Jones S, Plummer F, Kimani J, Shi B, Kaul R, Bwayo J, Anzala O, Burger H. Recombination following superinfection by HIV-1. AIDS. 2004;18:153–159. doi: 10.1097/00002030-200401230-00003. [DOI] [PubMed] [Google Scholar]
- Freel SA, Fiscus SA, Pilcher CD, Menezes P, Giner J, Patrick E, Lennox JL, Hicks CB, Eron JJ, Jr, Shugars DC. Envelope diversity, coreceptor usage and syncytium-inducing phenotype of HIV-1 variants in saliva and blood during primary infection. AIDS. 2003;17:2025–2033. doi: 10.1097/00002030-200309260-00003. [DOI] [PubMed] [Google Scholar]
- Furtado MR, Callaway DS, Phair JP, Kunstman KJ, Stanton JL, Maken CA, Perelson AS, Wolinsky SM. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- Galetto R, Moumen A, Giacomoni V, Véron M, Charneau P, Negroni M. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol Chem. 2004;279:36625–36632. doi: 10.1074/jbc.M405476200. [DOI] [PubMed] [Google Scholar]
- Goetz MB, Holodniy M, Poulton JS, Rodriguez FH, Rigsby MO. Utilization and access to antiretroviral genotypic resistance testing and results within the US Department of Veterans Affairs. J Acquir Immune Defic Syndr. 2006;41:59–62. doi: 10.1097/01.qai.0000187445.76579.08. [DOI] [PubMed] [Google Scholar]
- Goodenow MN, Collman RG. HIV-1 coreceptor preference is distinct from target cell tropism: a dual parameter nomenclature to define viral phenotypes. J Leukoc Biol. 2006;80:965–972. doi: 10.1189/jlb.0306148. [DOI] [PubMed] [Google Scholar]
- Harrington PR, Nelson JA, Kitrinos KM, Swanstrom R. Independent evolution of human immunodeficiency virus type 1 env V1/V2 and V4/V5 hypervariable regions during chronic infection. J Virol. 2007;81:5413–5417. doi: 10.1128/JVI.02554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermankova M, Ray SC, Ruff C, Powell-Davis M, Ingersoll R, D'Aquila RT, Quinn TC, Siliciano JD, Siliciano RF, Persaud D. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA. 2001;286:196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, Hammer SF, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, Yeni PG, Jacobsen DM, Richman DD. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- Ho SH, Shek L, Gettie A, Blanchard J, Cheng-Mayer C. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J Virol. 2005;79:12296–123303. doi: 10.1128/JVI.79.19.12296-12303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JP, Totten P. Estimating the accuracy of polymerase chain reaction-based tests using endpoint dilution. Biometrics. 2003;59:505–511. doi: 10.1111/1541-0420.00060. [DOI] [PubMed] [Google Scholar]
- Hung CS, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Li FS, van 't Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetzt AE, Yu H, Klarman J, Ron Y, Preston BD, Dougherty JP. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VA, Brun-Vézinet F, Clotet B, Günthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: 2007. Topics in HIV Medicine. 2007;15:119–124. [PubMed] [Google Scholar]
- Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A. Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- Kassaye S, Johnston E, McClogan B, Kantor R, Zijenah L, Katzenstein D. Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-Infected individuals receiving nonsuppressive antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;50:9–18. doi: 10.1097/QAI.0b013e31818ffdff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, Kitchen C, Philpott SM, Gao W, Robison E, Holman S, Dehner C, Beck S, Meyer WA, 3rd, Landay A, Kovacs A, Bremer J, Weiser B. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci USA. 2003;100:12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Burger H, Mayers D, Anastos K, Foley B, Kitchen C, Huggins P, Schroeder T, Picchio G, Back S, Gao W, Meyer WA, III, Weiser B. HIV-1 drug resistance in variants from the female genital tract and plasma. J Infect Dis. 2007;195:535–545. doi: 10.1086/510855. [DOI] [PubMed] [Google Scholar]
- Kieffer TL, Finucane MM, Nettles RE, Quinn TC, Broman KW, Ray SC, Persaud D, Siliciano RF. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis. 2004;189:1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen CM, Philpott S, Burger H, Weiser B, Anastos K, Suchard MA. Evolution of human immunodeficiency virus type 1 coreceptor usage during antiretroviral Therapy: a Bayesian approach. J Virol. 2004;78:11296–11302. doi: 10.1128/JVI.78.20.11296-11302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitrinos KM, Nelson JA, Resch W, Swanstrom R. Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J Virol. 2005;79:10627–10637. doi: 10.1128/JVI.79.16.10627-10637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3108. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Kreisberg JF, Kwa D, Schramm B, Trautner V, Connor R, Schuitemaker H, Mullins JI, van't Wout AB, Goldsmith MA. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J Virol. 2001;75:8842–8847. doi: 10.1128/JVI.75.18.8842-8847.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Wolinsky S, Korber B. Theoretical Biology and Biophysics Group. Los Alamos, NM: Los Alamos National Laboratory; 2009. HIV Sequence Compendium. [Google Scholar]
- Ledergerber B, Egger M, Opravil M, Telenti A, Hirschel B, Battegay M, Vernazza P, Sudre P, Flepp M, Furrer H, Francioli P, Weber R Swiss HIV Cohort Study. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Lancet. 1999;353:863–868. doi: 10.1016/s0140-6736(99)01122-8. [DOI] [PubMed] [Google Scholar]
- Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci. 2004;101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Mittler JE, Nickle DC, Mulvania TM, Shriner D, Rodrigo AG, Kosloff B, He X, Corey L, Mullins JI. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J Virol. 2002;76:10674–10684. doi: 10.1128/JVI.76.21.10674-10684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- Marsden MD, Zack JA. Eradication of HIV: current challenges and new directions. J Antimicrob Chemother. 2008 Nov. 4 doi: 10.1093/jac/dkn455. (Epub). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, Rosenberg E, Gunthard HF, Sutton L, Savara A, Petropoulos CJ, Hellmann N, Walker BD, Richman DD, Siliciano R, D'Aquila RT. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc Natl Acad Sci USA. 2000;97:10948–10953. doi: 10.1073/pnas.97.20.10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzaroma I, Carlesimo M, Pinter E, Muratori DS, Di Sora F, Chiarotti F, Cunsolo MG, Sacco G, Aiuti F. Clinical and immunologic response without decrease in virus load in patients with AIDS after 24 months of highly active antiretroviral therapy. Clin Infect Dis. 1999;29:1423–1428. doi: 10.1086/313520. [DOI] [PubMed] [Google Scholar]
- Mild M, Esbjörnsson J, Fenyö EM, Medstrand P. Frequent intrapatient recombination between human immunodeficiency virus type 1 R5 and X4 envelopes: implications for coreceptor switch. J Virol. 2007;81:3369–3376. doi: 10.1128/JVI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin VN, Dorman KS, Fang F, Suchard MA. Dual multiple change-point model leads to more accurate recombination data. Bioinformatics. 2005;13:3034–3042. doi: 10.1093/bioinformatics/bti459. [DOI] [PubMed] [Google Scholar]
- Napravnik S, Edwards D, Stewart P, Stalzer B, Matteson E, Eron JJ., Jr HIV-1 drug resistance evolution among patients on potent combination antiretroviral therapy with detectable viremia. J Acquir Immune Defic Syndr. 2005;40:34–40. doi: 10.1097/01.qai.0000174929.87015.d6. [DOI] [PubMed] [Google Scholar]
- Nettles RE, Kieffer TL, Parsons T, Johnson J, Cofrancesco J, Jr, Gallant JE, Carson KA, Siliciano RF, Flexner C. Marked intraindividual variability concentrations may limit the utility of therapeutic drug monitoring. Clin Infect Dis. 2006;42:1189–1196. doi: 10.1086/501458. [DOI] [PubMed] [Google Scholar]
- Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, Mittler JE, Mullins JI. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J Virol. 2004;78:8761–8770. doi: 10.1128/JVI.78.16.8761-8770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott S, Weiser B, Anastos K, Kitchen CM, Robison E, Meyer WA, 3rd, Sacks HS, Mathur-Wagh U, Brunner C, Burger H. Preferential suppression of CXCR4-specific strains of HIV-1 by antiviral therapy. J Clin Invest. 2001;107:431–437. doi: 10.1172/JCI11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott S, Burger H, Tsoukas C, Foley B, Anastos K, Kitchen C, Weiser B. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J Virol. 2005;79:353–363. doi: 10.1128/JVI.79.1.353-363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Poveda E, Briz V, de Mendoza C, Benito JM, Corral A, Zahonero N, Lozano S, Gonzalez-Lahoz J, Soriano V. Prevalence of X4 tropic HIV-1 variants in patients with differences in disease stage and exposure to antiretroviral therapy. J. Med Virol. 2007;79:1040–1046. doi: 10.1002/jmv.20900. [DOI] [PubMed] [Google Scholar]
- Quan Y, Brenner BG, Dascal A, Wainberg MA. Highly diversified multiply drug resistant HIV-1 quasispecies in PBMC’s: a case report. Retrovirology. 2008;5:43. doi: 10.1186/1742-4690-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N, Doms RW. HIV-1 coreceptors and their inhibitors. Curr Topics Microbiol Immunol. 2006;303:97–120. doi: 10.1007/978-3-540-33397-5_5. [DOI] [PubMed] [Google Scholar]
- Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- Riddle TM, Shire NJ, Sherman MS, Franco KF, Sheppard HW, Nelson JA. Sequential turnover of human immunodeficiency virus type 1 env throughout the course of infection. J Virol. 2006;80:10591–10599. doi: 10.1128/JVI.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau CM, Learn GH, Bhattacharya T, Nickle DC, Heckerman D, Chetty S, Brander C, Goulder PJ, Walker BD, Kiepiela P, Korber BT, Mullins JI. Extensive intrasubtype recombination in South African human immunodeficiency virus type 1 subtype C infections. J Virol. 2007;81:4492–4500. doi: 10.1128/JVI.02050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzales JF, Bailes E, Pham KT, Salazar MG, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, Fenyö EM, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Siliciano RF. Viral Reservoirs, residual viremia and the potential of highly active antiretroviral therapy to eradicate HIV infection. J Allergy Clin Immunol. 2008;122:22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Rodrigo AG, Nickle DC, Mullins JI. Pervasive genomic recombination of HIV-1 in vivo. Genetics. 2004;167:1573–1583. doi: 10.1534/genetics.103.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Besson G, Mobasher A, Collman RG. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J Virol. 1999;73:6680–6690. doi: 10.1128/jvi.73.8.6680-6690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solas C, Lafeuillade A, Halfon P, Chadapaud S, Hittinger G, Lacarelle B. Discrepancies between protease inhibitor concentrations and viral load in reservoirs and sanctuary sites in human immunodeficiency virus-infected patients. Antimicrob Ag Chemother. 2003;47:238–243. doi: 10.1128/AAC.47.1.238-243.2003. Stanford University HIV Drug Reference Database. Available at http://hivdb.stanford.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Back DJ, Workman J, Drake SM, White DJ, Choudhury B, Cane PA, Beards GM, Halifax K, Pillay D. Poor penetration of the male genital tract by HIV-1 protease inhibitors D. AIDS. 1999;13:859. doi: 10.1097/00002030-199905070-00017. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Kramer MG, Jarvis J, Kowalski J, Gange S, Schneider MF, Shao Q, Khang GW, Yeh M, Tsai H, Zhang H, Markham RB. Multiple-infection and recombination in HIV-1 within a longitudinal cohort of women. Retrovirology. 2009;6:54. doi: 10.1186/1742-4690-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Worobey M, Visser JA, Schuitemaker H. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology. 2003;314:451–459. doi: 10.1016/s0042-6822(03)00454-9. [DOI] [PubMed] [Google Scholar]
- Wagner TA, Frenkel LM. Potential limitation of CCR5 antagonists: drug resistance more often linked to CXCR4-utilizing than to CCR5-utilizing HIV-1. AIDS. 2008;22:2393–2395. doi: 10.1097/QAD.Ob013e328312c72c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser B, Philpott S, Klimkait T, Burger H, Kitchen C, Burgisser P, Gorgievski M, Perrin L, Piffaretti JC, Ledergerber B the Swiss HIV Cohort Study. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression combination antiretroviral therapy. AIDS. 2008;22:469–479. doi: 10.1097/QAD.0b013e3282f4196c. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson AS, Korber BT, Markowitz M, Guo Y, Duran M, Hurley A, Tsay J, Huang YC, Wang CC, Ho DD. Human Immunodeficiency Virus Type 1 in the Semen of Men Receiving Highly Active Antiretroviral Therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- Zhang L, Rowe L, He T, Chung C, Yu J, Yu W, Talal A, Markowitz M, Ho DD. Compartmentalization of surface envelope glycoprotein of human immunodeficiency virus type 1 during acute and chronic infection. J Virol. 2002;76:9465–9473. doi: 10.1128/JVI.76.18.9465-9473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Muthui D, Holte S, Nickle D, Feng F, Brodie S, Hwangbo Y, Mullins JI, Corey L. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus patients on highly active antiretroviral therapy. J Virol. 2002;76:707–716. doi: 10.1128/JVI.76.2.707-716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Jetzt AE, Sun G, Yu H, Klarmann G, Ron Y, Preston BD, Dougherty JP. Human Immunodeficiency Virus Type 1 Recombination: Rate, Fidelity, and Putative Hot Spots. J Virol. 2002;76:11273–11282. doi: 10.1128/JVI.76.22.11273-11282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.