Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterized by a series of pathological changes that range from simple fatty liver to nonalcoholic steatohepatitis (NASH). The objective of this study is to describe changes in global gene expression associated with the progression of human NAFLD. This study is focused on the expression levels of genes responsible for the absorption, distribution, metabolism, and elimination (ADME) of drugs. Differential gene expression between three clinically defined pathological groups—normal, steatosis, and NASH—was analyzed. Genome-wide mRNA levels in samples of human liver tissue were assayed with Affymetrix GeneChip Human 1.0ST arrays. A total of 11,633 genes exhibited altered expression out of 33,252 genes at a 5% false discovery rate. Most gene expression changes occurred in the progression from steatosis to NASH. Principal component analysis revealed that hepatic disease status was the major determinant of differential ADME gene expression rather than age or sex of sample donors. Among the 515 drug transporters and 258 drug-metabolizing enzymes (DMEs) examined, uptake transporters but not efflux transporters or DMEs were significantly over-represented in the number of genes down-regulated. These results suggest that uptake transporter genes are coordinately targeted for down-regulation at the global level during the pathological development of NASH and that these patients may have decreased drug uptake capacity. This coordinated regulation of uptake transporter genes is indicative of a hepatoprotective mechanism acting to prevent accumulation of toxic intermediates in disease-compromised hepatocytes.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a progressive disease of worldwide significance that currently afflicts 30 to 40% of the U.S. population (Ali and Cusi, 2009). Almost half of all individuals with NAFLD may actually have the more severe stage of nonalcoholic steatohepatitis (NASH) (Ali and Cusi, 2009). Globally, the prevalence of NAFLD is expected to approach that of the United States because of the spread of western lifestyle (Bellentani et al., 2010). The mechanisms responsible for the transition from simple fatty liver to NASH have yet to be elucidated. However, endeavors have characterized the progression of human NAFLD through a two-hit model of pathogenesis (Day and James, 1998). According to this model, NAFLD originates as steatosis through a “first hit” characterized by the abnormal accumulation of lipids within hepatocytes (Day, 2002; Rombouts and Marra, 2010). The “second hit” required for advancement to NASH is thought to be initiated by excess production of reactive oxygen species and proinflammatory cytokines (Pessayre et al., 2004). Gene expression studies of human NAFLD have been limited to themes of the first and second hit. There is a focus on obesity-related gene expression changes in NAFLD in addition to inflammatory and immune system components of the liver (Baranova et al., 2005; Younossi et al., 2005b; Bertola et al., 2010). However, there is a lack of studies on expression changes of genes related to the absorption, distribution, metabolism, and elimination (ADME) processes in progressive human NAFLD.
Gene expression changes associated with ADME processes may alter drug transport and distribution in NAFLD patients, resulting in an elevated risk of adverse drug reactions (ADRs) and altered drug bioavailability. The topic of ADME gene expression changes in the NAFLD patient population is of interest because this clinical group is often subjected to multiple prescription medications because of comorbidities related to the metabolic syndrome such as type 2 diabetes mellitus, hypertension, and dyslipidemia (Portincasa et al., 2006). Combination drug therapies put this patient population at significant risk of ADRs. ADRs are a frequent occurrence in today's medical field and represent a common cause of death in hospitalized patients (Lazarou et al., 1998).
The components of ADME genes include phase I and II drug-metabolizing enzymes (DMEs) as well as the so-called phase 0 uptake and phase III efflux transporters (Szakacs et al., 2008). Phase I and II DMEs modulate the pharmacokinetics of endogenous and exogenous compounds in the body. The expression of hepatic phase I DMEs, such as the cytochrome P450 (P450) family members, has been shown to be significantly altered in human NAFLD by previous studies (Fisher et al., 2009c). Transport proteins, considered the phase 0 and III components of ADME events, are recognized to have critical roles in the vectorial transport of drugs across cell membranes. Two main categories of transporters in the liver include uptake, generally known as the solute carrier transporters, and efflux, generally known as the ATP-binding cassette transporters. In the liver, efflux transporters residing on the sinusoidal and canalicular membranes of hepatocytes are responsible for substrate excretion into plasma and bile, respectively, whereas uptake transporters are primarily responsible for drug uptake into the hepatocyte from blood. Gene expression changes of solute carrier uptake transporter family members have previously been demonstrated in our laboratory using a rodent model of NAFLD (Fisher et al., 2009a). Other members of our laboratory have examined efflux transporter expression changes in a murine model of cholestasis. A hepatoprotective response to cholestasis was observed at the transcriptional level through the up-regulation of certain sinusoidal efflux transporters and down-regulation of specific uptake transporters (Lickteig et al., 2007b).
Patterns of gene expression changes provide evidence of transcriptional mechanisms of regulation. Coordinate regulation has been previously reported by investigators studying nuclear receptors (Maglich et al., 2002; Eloranta and Meier, 2005; Pascussi et al., 2008). Upon activation, nuclear receptors induce a small gene battery consisting of phase I and II metabolizing enzymes and phase 0 and III transporters (Kohle and Bock, 2007). Coordinate regulation at the global expression level for ADME genes has not been shown but could have profound effects upon hepatic function and toxicity (Kohle and Bock, 2009). Principal component analysis (PCA) is a method used to examine patterns of expression changes in genes. PCA simplifies complex gene array data by analyzing the variance. Patterns in ADME genes are analyzed in this study using PCA. The study presented here was designed to provide a comprehensive analysis of gene expression changes among DMEs and transporters across the progression of human NAFLD.
Materials and Methods
Human Liver Samples.
Human liver tissue was previously acquired from the National Institutes of Health-funded Liver Tissue Cell Distribution System at the University of Minnesota, Virginia Commonwealth University, and the University of Pittsburgh. Clinical and demographic information of these human liver samples has been described previously (Fisher et al., 2009c). The samples were diagnosed as normal (n = 19), steatotic (n = 10), NASH with fatty liver (n = 9), and NASH without fatty liver (n = 7). NAFLD activity scoring categorization was done by a Liver Tissue Cell Distribution System medical pathologist (Kleiner et al., 2003). Steatosis was diagnosed by >10% fat deposition within hepatocytes without inflammation or fibrosis. NASH with fatty liver was characterized by >5% fat deposition with accompanied inflammation and fibrosis. NASH without fatty liver was distinguished by <5% fat deposition and increased inflammation and fibrosis.
Total RNA Isolation.
Total RNA was isolated from the human liver samples using RNAzol B reagent (Tel-Test Inc., Friendswood, TX). Total RNA was purified using the RNeasy Mini Qiagen purification kit (QIAGEN, Valencia, CA) according to manufacturer's recommendations. RNA integrity was assessed using ethidium bromide staining after agarose gel electrophoresis. Concentration of the total RNA was determined using a Thermo Scientific Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The quality of the 18S and the 28S ribosomal bands were characterized on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) using the Eukaryotic Total RNA Nano Assay with a 1 μl volume of purified total RNA from each sample. The RNA integrity numbering (RIN) system was used to assess the quality of the total RNA samples, with a RIN number of 1 being the most degraded and unusable form of total RNA and a RIN number of 10 representing the most complete and non-degraded form of the RNA.
Microarray Gene Expression Analysis.
RNA was processed per manufacturer's protocol (Affymetrix GeneChip Whole Transcript Sense Target Labeling; Affymetrix, Santa Clara, CA) for hybridization onto microarrays (Affymetrix GeneChip Human 1.0 ST Arrays; Affymetrix). Array hybridization, washing, and scanning were performed according to manufacturer's recommendations. All microarray data, archiving, and analysis were generated by the Genomics Core Facility at the Arizona Cancer Center. The array data were deposited into the ArrayExpress public repository for microarray data and are accessible under the accession number E-MEXP-3291 (http://www.webcitation.org/5zyojNu7T). The differential expression of 33,252 global genes among three diagnosis groups (normal, steatosis, and NASH) was tested using the linear models for microarray data software package in Bioconductor (Smyth, 2011). Pairwise comparisons between diagnosis groups were performed using the linear models for microarray data software. The method of Benjamini and Hochberg (1995) was used to control the false discovery rate at the level of 0.05 to correct for multiple hypothesis testing. For the purpose of the statistical tests used in this study, NASH fatty and NASH not fatty samples were combined because of the lack of mechanistic differences between these two states despite histological differentiation. A pairwise comparison analysis of the gene expression at the 0.01 level of significance was performed to compare steatosis versus NASH fatty and NASH fatty versus NASH not fatty. This was done to demonstrate the lack of mechanistic differentiation between the two NASH categories and justify the combination of all NASH specimens into one group (Supplemental Fig. 3).
ADME Gene Analysis.
A list of 258 DMEs and 515 transporter genes was compiled from literature sources. Separate lists were compiled for the subsequent analysis of 437 uptake transporter genes, 60 efflux transporter genes, and 18 transporters classified as others. Differentially expressed genes are represented in Venn diagrams for each of the three pairwise comparisons for the global, DME, and transporter categories (Fig. 2). Distribution graphs were generated to show the proportion of genes tested among differentially expressed ADME gene categories. Categories were tested, and if they were found to have a proportion of genes that showed greater representation in up- or down-regulation compared with the proportion of a randomly tested set of genes the same size, then they were considered over-represented. The probability of acquiring a higher proportion of differentially expressed genes in a gene subset of DMEs or transporters was calculated from the simulated distribution (Fig. 3). Heat maps were generated to visualize hierarchical clustering between patient diagnosis groups and gene expression levels (Fig. 4). PCA was used to simplify the complex data sets of gene categories by analyzing the components with the greatest amount of variance. Graphical representations of the first and second components of the entire global gene set and each ADME gene category show the similarities and differences in gene expression for the different groups of diagnosis (Figs. 1 and 5).
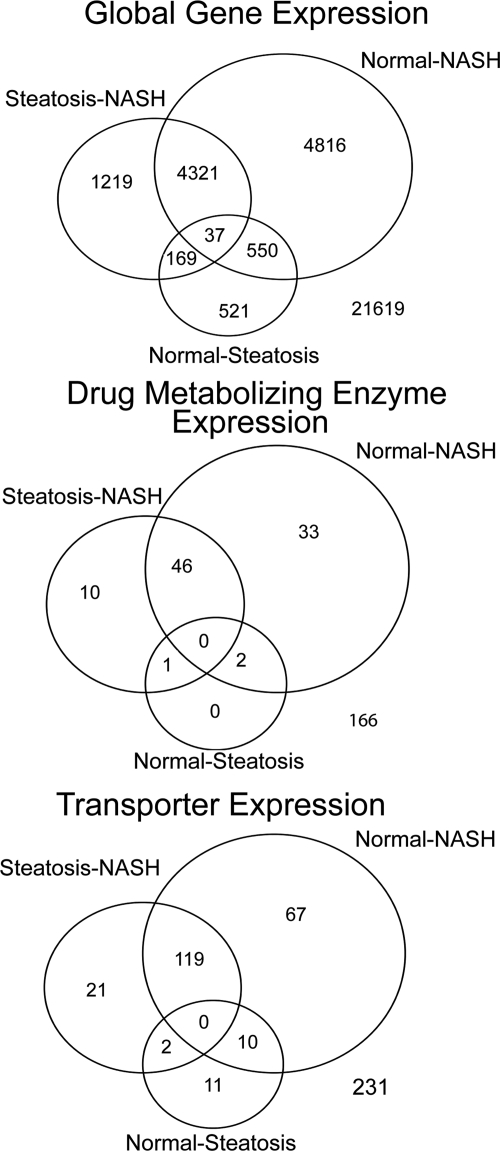
Fig. 2.
Differential gene expression in progressive human NAFLD. Venn diagrams summarize the magnitudes of genes differentially expressed for the global, DME, and transporter gene categories. Each circle represents one of the three pairwise comparisons between disease states (normal versus steatosis, steatosis versus NASH, and normal versus NASH).
Fig. 3.
Distribution histograms of gene expression. Up- and down-regulated gene expression is represented by red and green, respectively, in the barplots and distribution histograms for each of the three pairwise comparisons: normal versus steatosis, steatosis versus NASH, and normal versus NASH. Global gene expression changes are illustrated in the large barplot for all genes. Gene expression changes for DME, all transporters, efflux-only, and uptake-only transporter gene categories are represented by smaller barplots and distribution histograms. The vertical black bar in the distribution histograms represents the actual number of gene expression changes observed for a gene category as tested against an expected distribution of randomly chosen genes. N, normal; Ste, steatosis; NSH, NASH. Significance is determined by p < 0.05.
Fig. 4.
Hierarchical clustering. Heat maps were generated to represent hierarchical clustering data of differential gene expression for each gene category. Colored boxes represent up-regulation (red) and down-regulation (green). Tree structures matched to the two axes represent patterns between diagnosis groups and gene expression differences. Genes are represented on the x-axis and specimen diagnosis is represented on the y-axis (normal, yellow; steatosis, orange; NASH, red).
Fig. 1.
Principal component analysis of global gene expression. The first and second principal components used in this graphical representation account for 22 and 12%, respectively, of the total variance in the global gene expression. The PCA plot determines the factor that contributes to the variance. Sex and age do not appear to cause systematic changes in gene expression.
Fig. 5.
Principal component analysis of ADME gene expression. Principal component analysis of the categories for the DMEs, all transporters, uptake-only transporters, and efflux-only transporters are shown. The first two principal components representing most of the variance in the gene data are plotted to show similarities or differences in the expression changes for normal, steatosis, and NASH specimens.
Quantitative Reverse Transcription-Polymerase Chain Reaction (TaqMan) Validation Analysis.
Validation of CYP1A2, CYP2D6, CYP2E1, and CYP2A6 mRNA levels was conducted using cDNA from the same set of human liver samples plus additional samples of each disease state that were categorized in a previous study (Fisher et al., 2009c). Human liver tissue samples for these experiments were diagnosed as normal (n = 20), steatotic (n = 12), and NASH (n = 22). The cDNA was analyzed using gene-specific TaqMan primer/probe sets (Applied Biosystems, Foster City, CA). The ABI 7900HT real-time polymerase chain reaction system (Applied Biosciences) was used for these experiments using the manufacturer's protocol for the assays. Reactions with the specific primer/probes for the constitutively expressed glyceraldehyde-3-phosphate dehydrogenase gene were used as an endogenous control. The relation between the log2 microarray signal and raw −Ct values were plotted and examined for linearity for each selected gene (Supplemental Figs. 1 and 2).
Results
RINs for Human Liver Total RNA.
The Eukaryotic Total RNA Nano Assay using the Agilent Bioanalyzer 2100 revealed RIN values for normal liver samples to be in the range of 7.1 to 10. Values for steatosis samples ranged from 8.2 to 10. RIN values of 5.2 to 9.9 were observed for NASH fatty samples, and values of 5.5 to 8.1 were found for NASH not fatty samples. These RIN values were found to be adequate for application of these samples to the Affymetrix GeneChip Whole Transcript Sense Target Labeling protocol.
PCA of Global Gene Expression.
To reveal the major factors correlated with gene expression changes, PCA was used. The first two principal components demonstrated that 34% of all expression variance is correlated with disease diagnosis but not with sex or age (Fig. 1). In terms of the principal components, 22% of the total variance is represented by the first principal component and 12% by the second principal component (Fig. 1). For the purpose of this study, future analyses will only take into account the factor of diagnosis because this has been demonstrated by PCA to be the predominant contributor to the variance in the gene data set (Fig. 1).
Differential Gene Expression of Global and ADME Genes.
Gene expression analysis of the total 33,252 annotated and unannotated global genes revealed 11,633 differentially expressed genes at the 0.05 level of significance among three pairwise comparisons (normal versus steatosis, steatosis versus NASH, and normal versus NASH) (Table 1; Fig. 2). A total of 1277 gene expression alterations (3.8% of all global genes) were revealed in the normal versus steatosis comparison (Table 1; Fig. 2). A total of 5746 expression changes (17.3%) were discovered in the steatosis versus NASH comparison, whereas 9724 genes were differentially expressed (29.2%) in the normal versus NASH comparison (Table 1; Fig. 2). A total of 21,619 genes, or 65% of all global gene expression, was not changed (Table 1; Fig. 2). This analysis demonstrates a greater magnitude of global gene expression changes in the samples that have progressed to NASH (Table 1; Fig. 2).
TABLE 1.
The number and percentage of differentially expressed genes for each gene category is calculated from 33,252 global genes, 258 DME genes, and 515 transporter genes
| Gene Set | Normal vs. Steatosis | Steatosis vs. NASH | Normal vs. NASH |
|---|---|---|---|
| All genes | 1277 (3.8%) | 5746 (17.3%) | 9724 (29.2%) |
| DMEs | 3 (1.2%) | 57 (22.1%) | 81 (31.3%) |
| Transporters | 25 (4.8%) | 150 (29.1%) | 210 (40.8%) |
Analysis of 258 DME genes revealed only three differentially expressed genes (1.2% of DME genes) in the normal versus steatosis comparison (Table 1; Fig. 2; Supplemental Table 2). Although expression of 57 genes (22.1% of DME genes) was changed in the comparison between steatosis and NASH, 81 genes (31.3% of DME genes) were changed in expression between normal and NASH (Table 1; Fig. 2; Supplemental Table 2). The expression of a total of 166 DME genes (64.3%) remained unchanged (Table 1; Fig. 2). Tests for enrichment of the DME category for expression changes in up- or down-regulation revealed no over-representation in the distribution histogram (Fig. 3).
A sum of 25 transporter genes demonstrated expression alterations out of 515 (4.8% of transporter genes) in the normal versus steatosis comparison (Table 1; Fig. 2). Although 150 expression changes (29.1%) occurred in the steatosis versus NASH comparison, 210 expression changes in total (40.8%) were observed in the normal versus NASH comparison (Table 1; Fig. 2). The expression of 130 transporters (25.2%) remained unchanged (Table 1; Fig. 2). It is interesting to note that the transporter gene category as a whole was significantly over-represented in a test among genes down-regulated in expression for each of the NASH pairwise comparisons according to the transporter distribution histogram (Fig. 3).
In the analysis of the efflux transporter gene category, 17 efflux genes showed altered expression in the steatosis versus NASH comparison analysis, whereas 19 expression changes were observed in the normal versus NASH comparison. Only one efflux transporter was differentially expressed in the normal versus steatosis comparison (Supplemental Table 3). No over-representation was found for the 60 efflux transporter genes in up- or down-regulation (Fig. 3). For the uptake transporter gene category, 112 genes exhibited altered expression in the steatosis versus NASH comparison whereas only 22 were altered in expression in the normal versus steatosis comparison (Supplemental Table 2). A total of 162 uptake transporter genes were altered in the normal versus NASH comparison, of which many also revealed expression changes in the steatosis versus NASH comparison (Supplemental Table 2). The distribution analysis of 437 uptake transporter genes revealed a strong over-representation not seen in the analysis of the efflux transporter genes (Fig. 3). The over-representation among down-regulated uptake transporter genes was observed in the steatosis versus NASH and normal versus NASH comparisons (Fig. 3). The normal versus steatosis comparison did not demonstrate any over-representation of genes down-regulated in expression for the uptake or the efflux transporter categories (Fig. 3).
Hierarchical Clustering Analysis of ADME Gene Categories.
Hierarchical clustering revealed that the expression profile of transporter genes is capable of accurately assigning the pathological diagnosis of liver samples. Clustering of down-regulated genes is represented as green blocks in the hierarchical clustering diagrams whereas up-regulated genes are represented as red blocks (Fig. 4). The NASH samples in the hierarchical clustering model form two distinctive clusters within the heat map with only one outlying NASH sample. In contrast, the DME gene expression is not sufficient to separate out NASH samples from the normal and steatotic groups in the hierarchical clustering model. Neither gene set is sufficient to distinguish specimens diagnosed as normal from those with steatosis (Fig. 4).
PCA of ADME Gene Categories.
PCA of the transporter gene category shows a clear partition of NASH specimens from those diagnosed as normal and steatotic (Fig. 5). In contrast, more overlap is observed between the sample clusters in the DME PCA (Fig. 5). This overlap makes it difficult to distinguish some NASH specimens from normal and steatotic in the DME genes. PCA of both the uptake and efflux transporter gene categories demonstrates distinctive clustering and division of NASH samples from those diagnosed as normal and steatotic (Fig. 5).
Microarray Validation.
Microarray data were correlated against raw quantitative RT-PCR data generated from the same set of human samples for validation of the microarray data. CYP1A2, CYP2D6, CYP2E1, and CYP2A6 were chosen for validation on the basis of data from a previous study by our laboratory using the same set of human liver specimens (Fisher et al., 2009c). Normalized microarray expression (log2 signal) was compared to quantitative RT-PCR data. All of the P450-validated genes demonstrated a corresponding pattern of fold change for up- or down-regulation similar to that seen in the microarray. The microarray log2 signals and quantitative RT-PCR −Ct values from both sets of data were analyzed and found to be in good correlation for the four cytochromes differentially expressed in NASH (Supplemental Figs. 1 and 2).
Discussion
Human NAFLD has gained increasing attention as one of the more prominent chronic liver diseases of the decade, and it has raised clinical concerns because of its association with the pandemic of obesity and type 2 diabetes mellitus (de Alwis and Day, 2008; Ali and Cusi, 2009). The more severe stage of NAFLD has been associated with end-stage liver diseases such as cryptogenic cirrhosis and hepatocellular carcinoma. An estimated 3 to 15% of NASH patients will develop cirrhosis and liver failure (Ascha et al., 2010). Hepatic disease state is considered a primary factor in the altered disposition of many drugs (Lucena et al., 2003). NAFLD and, particularly, the pathophysiological stage of NASH is a relevant concern to health-care professionals because of the associated risk of ADRs that may accompany drug administration. The objective of the study presented here is to comprehensively analyze gene expression alterations of ADME genes in the progression of human NAFLD. The results demonstrate a coordinate regulation at the global level resulting in an enrichment of down-regulated uptake transporter genes in NASH. DME and efflux transport categories did not exhibit any enrichment for up- or down-regulation (Fig. 3).
Previous studies have examined gene expression changes in human NAFLD livers classified as steatotic (Greco et al., 2008) and in livers classified as NASH using various microarray platforms (Younossi et al., 2005a; Baranova et al., 2007; Rubio et al., 2007). These studies confirm that liver disease status alters gene expression changes. The microarray data presented here for the three separate pathological classifications (normal, steatosis, and NASH) reveal statistically significant global gene expression alterations, with most expression changes occurring in the pathological transition from steatosis to NASH and not from normal to steatosis (Fig. 2; Table 1). These data confirm previous studies that show an accumulation of gene expression changes in NASH (Rubio et al., 2007). The results we present in this study comprehensively analyze ADME gene expression alterations across each of the stages of human NAFLD and demonstrate that progression to NASH, with its accompanying features of the second hit, alters this critical category of genes. The limited number of human samples in this study demonstrated large variances in the ADME gene expression as observed in the PCA between samples diagnosed as steatotic and those diagnosed as NASH (Fig. 5). Despite the limited size of the sample groups, we show significant expression changes in ADME genes, specifically the transporters in the steatosis versus NASH and normal versus NASH comparisons. Other studies have reported P450 and transporter expression changes in patients with inflammatory disease states such as hepatitis and alcoholic liver disease (Morgan, 2001; Renton, 2005). More recently, our laboratory reported gene expression changes of cytochrome P450 enzymes in human NAFLD liver samples (Fisher et al., 2009c). Changes in DME expression may be important because as many as 75% of all drugs are biotransformed by the P450 enzymes alone in humans (Guengerich, 2008).
The pairwise comparison analysis for the DME gene category reflect upon findings of previous studies done in our laboratory that examined P450 enzyme activity and changes at the transcriptional and translational levels in this same set of human samples. Selected P450s demonstrated significant decreasing trends in mRNA levels with disease progression (Fisher et al., 2009c). Another study conducted by our laboratory demonstrated that the phase II conjugating glutathione transferase phase II enzymes exhibited an increasing trend in mRNA expression with progression of human NAFLD (Hardwick et al., 2010). The diversity in differential gene regulation within the DME category is clearly evident from these studies.
The pairwise comparison analysis of 515 transporter genes shows the distinctive roles of transporters in efflux or uptake (Figs. 2 and 3). The uptake transporter genes revealed a significant over-representation for genes down-regulated for both NASH pairwise comparisons (Fig. 3). These results extend the previous findings of our laboratory in a rodent NAFLD model to humans. Down-regulation of organic anion-transporting polypeptide (OATP) transporter gene expression in the rodent NAFLD model parallels the down-regulated uptake transporter expression seen in the human microarray data presented here (Fisher et al., 2009a). In that study, rat Oatp1a1, Oatp1a4, Oatp1b2, and Oatp2b1 mRNA expression levels were significantly decreased in the methionine- and choline-deficient diet rodent model of NASH, leading to a functional impairment in the uptake and subsequent elimination of bromosulfophthalein. On the basis of the similarity in decreased uptake transporter expression, the current data imply a similar functional impairment of the uptake transport process in human NASH (Fig. 3).
The over-representation of uptake transporter genes down-regulated (Fig. 3) suggests the presence of a coordinate transcriptional regulation in humans with NASH. Specifically, down-regulation of multiple uptake transporters could prevent the accumulation of xenobiotics and toxic intermediates in a diseased liver already compromised by oxidative stress. Investigations have revealed other expression alterations in hepatobiliary transporters in mouse models administered toxic doses of acetaminophen and carbon tetrachloride (Aleksunes et al., 2005). Coordinate gene expression regulation of ATP-binding cassette subfamily C (ABCC) efflux transporters in these mouse models contributed to a reduced chemical burden and hepatoprotection (Aleksunes et al., 2006).
Multiple mechanisms for a coordinated gene expression response have been identified in the altered regulation of key ADME genes (Kohle and Bock, 2009). Multiple phase I and II DMEs and transporters are coordinately regulated by nuclear receptors and transcription factors. This integrated biotransformation system includes such components as the aryl hydrocarbon receptor, constitutive androstane receptor, liver X receptor, and nuclear factor erythroid 2-related factor transcription factor. Studies of the phase I P450 DMEs, phase II conjugating enzymes, and efflux and uptake transporters in our laboratory support the theme of a coordinate regulation in progressive NAFLD (Lickteig et al., 2007a; Fisher et al., 2008, 2009a,c; Hardwick et al., 2010).
The remodeling of ADME gene expression in the progression of NAFLD is an important consideration in the diagnosis. The pathological stage of steatosis in our small sampling of human liver samples did not demonstrate significant expression changes from that of normal in this study. Therefore, the pathological staging of NAFLD is critical in identifying patients with alterations in the expression of ADME genes. The ADME gene expression changes we have presented in this study reveal an important down-regulatory function of uptake transport genes. Although this coordinated down-regulation of the ADME gene category is indicative of a hepatoprotective response, these findings may also have implications in drug dosing regimens. These implications should be taken into account by health-care practitioners and pharmaceutical investigators when making decisions regarding pharmacotherapy for the NAFLD patient population.
Supplementary Material
Acknowledgments
We express our sincere gratitude to Jose Munoz-Rodriguez and the Genomics Core Facility at the University of Arizona Cancer Center for the processing, archiving, and data acquisition of the arrays. We also thank the National Institutes of Health-sponsored Liver Tissue Cell Distribution System for assistance with the collection of liver samples from patients with all stages of NAFLD.
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK068039]; the National Institutes of Health National Institute of Allergy and Infectious Diseases Extramural Activities [Grant AI083927]; the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant HD062489] (to N.J.C.); the National Institutes of Health National Center for Complementary and Alternative Medicine [Grant AT002842] (to C.D.F.); and the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES006694]. The Liver Tissue Cell Distribution System was sponsored by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Contract no. N01-DK-7-0004/HHSN267200700004C].
Parts of this work were previously presented as follows: Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, and Cherrington NJ (2010). Analysis of global and ADME gene expression in the progressive stages of human non-alcoholic fatty liver disease. Society of Toxicology 49th Annual Meeting; 2010 Mar 7–11; Salt Lake City, Utah. Society of Toxicology, Reston, VA.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.111.040592.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- ADME
- absorption, distribution, metabolism, and excretion
- ADR
- adverse drug reaction
- DME
- drug-metabolizing enzyme
- P450
- cytochrome P450
- PCA
- principal component analysis
- RIN
- RNA integrity numbering
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- OATP
- organic anion-transporting polypeptide
- Ct
- cycle thresholds.
Authorship Contributions
Participated in research design: Lake, Billheimer, Klimecki, and Cherrington.
Conducted experiments: Lake and Hardwick.
Contributed new reagents or analytic tools: Fisher, Jackson, Billheimer, and Klimecki.
Performed data analysis: Novak, Lake, and Klimecki.
Wrote or contributed to the writing of the manuscript: Lake, Novak, Hardwick, Billheimer, Klimecki, and Cherrington.
References
- Aleksunes LM, Slitt AM, Cherrington NJ, Thibodeau MS, Klaassen CD, Manautou JE. (2005) Differential expression of mouse hepatic transporter genes in response to acetaminophen and carbon tetrachloride. Toxicol Sci 83:44–52 [DOI] [PubMed] [Google Scholar]
- Aleksunes LM, Scheffer GL, Jakowski AB, Pruimboom-Brees IM, Manautou JE. (2006) Coordinated expression of multidrug resistance-associated proteins (Mrps) in mouse liver during toxicant-induced injury. Toxicol Sci 89:370–379 [DOI] [PubMed] [Google Scholar]
- Ali R, Cusi K. (2009) New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD).Ann Med 41:265–278 [DOI] [PubMed] [Google Scholar]
- Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. (2010) The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology 51:1972–1978 [DOI] [PubMed] [Google Scholar]
- Baranova A, Schlauch K, Elariny H, Jarrar M, Bennett C, Nugent C, Gowder SJ, Younoszai Z, Collantes R, Chandhoke V, et al. (2007) Gene expression patterns in hepatic tissue and visceral adipose tissue of patients with non-alcoholic fatty liver disease. Obesity Surgery 17:1111–1118 [DOI] [PubMed] [Google Scholar]
- Baranova A, Schlauch K, Gowder S, Collantes R, Chandhoke V, Younossi ZM. (2005) Microarray technology in the study of obesity and non-alcoholic fatty liver disease. Liver Int 25:1091–1096 [DOI] [PubMed] [Google Scholar]
- Bellentani S, Scaglioni F, Marino M, Bedogni G. (2010) Epidemiology of non-alcoholic fatty liver disease. Dig Dis 28:155–161 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B Method 57:289–300 [Google Scholar]
- Bertola A, Bonnafous S, Anty R, Patouraux S, Saint-Paul MC, Iannelli A, Gugenheim J, Barr J, Mato JM, Le Marchand-Brustel Y, et al. (2010) Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP. (2002) Pathogenesis of steatohepatitis. Best Pract Res Clin Gastroenterol 16:663–678 [DOI] [PubMed] [Google Scholar]
- Day CP, James OF. (1998) Steatohepatitis: a tale of two “hits”? Gastroenterology 114:842–845 [DOI] [PubMed] [Google Scholar]
- Wilfred de Alwis NM, Day CP. (2008) Genes and nonalcoholic fatty liver disease. Curr Diab Rep 8:156–163 [DOI] [PubMed] [Google Scholar]
- Eloranta JJ, Meier PJ. (2005) Coordinate transcriptional regulation of transport and metabolism, in Methods in Enzymology Phase II Conjugation Enzymes and Transport Systems, pp 511–530, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- Fisher CD, Jackson JP, Lickteig AJ, Augustine LM, Cherrington NJ. (2008) Drug metabolizing enzyme induction pathways in experimental non-alcoholic steatohepatitis. Arch Toxicol 82:959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CD, Lickteig AJ, Augustine LM, Oude Elferink RP, Besselsen DG, Erickson RP, Cherrington NJ. (2009a) Experimental non-alcoholic fatty liver disease results in decreased hepatic uptake transporter expression and function in rats. European Journal of Pharmacology 613:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. (2009c) Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos 37:2087–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, et al. (2008) Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol 294:G1281–G1287 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2008) Cytochrome P450 and chemical toxicology. Chem Res Toxicol 21:70–83 [DOI] [PubMed] [Google Scholar]
- Hardwick RN, Fisher CD, Canet MJ, Lake AD, Cherrington NJ. (2010) Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos 38:2293–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta ML, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Yeh M, et al. (2003) Design and validation of a histologic scoring system for non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH). Hepatology 38:1313–1321 [DOI] [PubMed] [Google Scholar]
- Köhle C, Bock KW. (2007) Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol 73:1853–1862 [DOI] [PubMed] [Google Scholar]
- Köhle C, Bock KW. (2009) Coordinate regulation of human drug-metabolizing enzymes, and conjugate transporters by the Ah receptor, pregnane X receptor and constitutive androstane receptor. Biochem Pharmacol 77:689–699 [DOI] [PubMed] [Google Scholar]
- Lazarou J, Pomeranz BH, Corey PN. (1998) Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 279:1200–1205 [DOI] [PubMed] [Google Scholar]
- Lickteig AJ, Fisher CD, Augustine LM, Aleksunes LM, Besselsen DG, Slitt AL, Manautou JE, Cherrington NJ. (2007a) Efflux transporter expression and acetaminophen metabolite excretion are altered in rodent models of nonalcoholic fatty liver disease. Drug Metab Dispos 35:1970–1978 [DOI] [PubMed] [Google Scholar]
- Lickteig AJ, Slitt AL, Arkan MC, Karin M, Cherrington NJ. (2007b) Differential regulation of hepatic transporters in the absence of tumor necrosis factor-alpha, interleukin-1 beta, interleukin-6, and nuclear factor-kappaB in two models of cholestasis. Drug Metab Dispos 35:402–409 [DOI] [PubMed] [Google Scholar]
- Lucena MI, Andrade RJ, Tognoni G, Hidalgo R, Sanchez de la Cuesta F, and Spanish Collaborative Study Group on Therapeutic Management of Liver Diseases (2003) Drug use for non-hepatic associated conditions in patients with liver cirrhosis. Eur J Clin Pharmacol 59:71–76 [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. (2002) Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62:638–646 [DOI] [PubMed] [Google Scholar]
- Morgan ET. (2001) Regulation of cytochrome P450 by inflammatory mediators: Why and how? Drug Metab Dispos 29:207–212 [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Duret C, Daujat-Chavanieu M, Vilarem MJ, Maurel P. (2008) The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu Rev Pharmacol Toxicol 48:1–32 [DOI] [PubMed] [Google Scholar]
- Pessayre D, Fromenty B, Mansouri A. (2004) Mitochondrial injury in steatohepatitis. Eur J Gastroenterol Hepatol 16:1095–1105 [DOI] [PubMed] [Google Scholar]
- Portincasa P, Grattagliano I, Palmieri VO, Palasciano G. (2006) Current pharmacological treatment of nonalcoholic fatty liver. Curr Med Chem 13:2889–2900 [DOI] [PubMed] [Google Scholar]
- Renton KW. (2005) Regulation of drug metabolism and disposition during inflammation and infection. Expert Opin Drug Metab Toxicol 1:629–640 [DOI] [PubMed] [Google Scholar]
- Rombouts K, Marra F. (2010) Molecular mechanisms of hepatic fibrosis in non-alcoholic steatohepatitis. Dig Dis 28:229–235 [DOI] [PubMed] [Google Scholar]
- Rubio A, Guruceaga E, Vázquez-Chantada M, Sandoval J, Martínez-Cruz LA, Segura V, Sevilla JL, Podhorski A, Corrales FJ, Torres L, et al. (2007) Identification of a gene-pathway associated with non-alcoholic steatohepatitis. J Hepatol 46:708–718 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2011) Bioinformatics and computational biology solutions using R and bioconductor, in Limma: Linear Models for Microarray Data (Gentleman R, Carey V, Dudoit R, Irizarry W. eds) pp 397–420, Springer, New York [Google Scholar]
- Szakács G, Váradi A, Ozvegy-Laczka C, Sarkadi B. (2008) The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox). Drug Discov Today 13:379–393 [DOI] [PubMed] [Google Scholar]
- Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A, et al. (2005a) A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology 42:665–674 [DOI] [PubMed] [Google Scholar]
- Younossi ZM, Gorreta F, Ong JP, Schlauch K, Del Giacco L, Elariny H, Van Meter A, Younoszai A, Goodman Z, Baranova A, et al. (2005b) Hepatic gene expression in patients with obesity-related non-alcoholic steatohepatitis. Liver Int 25:760–771 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.