Abstract
Many species with restricted gene flow repeatedly respond similarly to local selection pressures. To fully understand the genetic mechanisms behind this process, the phylogeographic history of the species (inferred from neutral markers) as well as the loci under selection need to be known. Here we sequenced an intron in the arginine kinase gene (Ark), which shows strong clinal variation between two locally adapted ecotypes of the flat periwinkle, Littorina fabalis. The ‘small-sheltered' ecotype was almost fixed for one haplotype, H1, in populations on both sides of the North Sea, unlike the ‘large-moderately exposed ecotype', which segregated for ten different haplotypes. This contrasts with neutral markers, where the two ecotypes are equally variable. H1 could have been driven to high frequency in an ancestral population and then repeatedly spread to sheltered habitats due to local selection pressures with the colonization of both sides of the North Sea, after the last glacial maximum (∼18 000 years ago). An alternative explanation is that a positively selected mutation, in or linked to Ark, arose after the range expansion and secondarily spread through sheltered populations throughout the distribution range, causing this ecotype to evolve in a concerted fashion. Also, we were able to sequence up to four haplotypes consistently from some individuals, suggesting a gene duplication in Ark.
Keywords: Littorina spp., selective sweep, concerted ecotype evolution, parallel evolution, local adaptation, intron
Introduction
Understanding the genetic basis of adaptation and speciation is an important topic in evolutionary biology. Repeated evolution of phenotypic traits is common whenever demographically independent populations are exposed to similar ecological conditions, suggesting a major role for natural selection in adaptation to local environments (Rundle et al., 2000; Schluter, 2001, 2009; McKinnon et al., 2004). The genetic mechanisms behind this process are in many cases unclear but possible scenarios (reviewed in Johannesson et al., 2010) include: (1) parallel evolution, that is, the independent evolution of homologous loci that fulfil the same function in two or more lineages (Wood et al., 2005), (2) secondary contact after an initial allopatric divergence (Wilding et al., 2001), (3) evolution from standing genetic variation (Campbell and Bernatchez, 2004; Schluter et al., 2004; Barrett and Schluter, 2008) and (4) evolution in concert where positively selected alleles, directly after their appearance, spread among demographically independent populations by migration (Rieseberg and Burke, 2001; Morjan and Rieseberg, 2004; Johannesson et al., 2010). Distinguishing between these scenarios is possible but requires the detailed and joint analyses of both neutral and selected loci (Johannesson et al., 2010). An increasingly popular approach to identify loci under selection is to look for outliers in genome scans (see Nosil et al., 2009 for a review). Here, simultaneous analysis of a large number of loci is used to find loci with higher than expected differentiation among populations, measured by FST. These loci mark genomic regions that presumably contribute to local adaptation, but further characterization of them is necessary in order to fully understand the forces shaping the variation around them. This is often difficult if sequence information around the loci cannot readily be obtained (Wood et al., 2008), such as in non-model organisms where the whole genome has not been sequenced.
An often neglected track to identify candidate genes is through allozymes, revealed by horizontal starch gel electrophoresis. As the allozymes are usually well characterized, obtaining sequence information around them may potentially be easier compared with novel outliers detected by genome scans. Here, we used a degenerate primer approach to sequence and analyse genetic differentiation in an intron of arginine kinase (Ark), a locus that, according to previous allozyme studies (Tatarenkov and Johannesson 1994, 1999), shows consistent allele frequency differences between locally adapted ecotypes of the marine intertidal gastropod Littorina fabalis (one of two species of the flat periwinkles) and thus is likely to be influenced by differential selection.
L. fabalis is widely distributed along the NE Atlantic coasts, from Portugal to the White Sea and Iceland. It lives in the intertidal zone and grazes microepiphytes on fucoid macroalgae. Gene flow among populations is restricted as crawl-away juveniles hatch directly from benthic egg masses and the net movement of adult individuals is no more than a few metres per generation (Williams, 1990; Tatarenkov and Johannesson, 1998). Adult snails living on moderately exposed shores (hereafter simply exposed) are about 25% larger than snails living on sheltered shores, even at sites <10 m from each other. A series of manipulative selection experiments has demonstrated that an increased risk of being dislodged from the algae (which constitute refuges from crab predation) selects for large size in exposed habitats because large shell size protects snails from crab predation when they inhabit the seafloor beneath the algae (Kemppainen et al., 2005). Transplant experiments have shown that adult size is largely genetically determined (Tatarenkov and Johannesson, 1998). Microsatellite genotypes group individuals by geographic location rather than ecotype (Kemppainen et al., 2009), but among 30 allozyme loci, Tatarenkov and Johannesson (1994, 1999) found one, arginine kinase (Ark), that varied predictably in Wales (GB), Brittany (France) and Sweden, such that strong clines (sometimes <10 m wide) in allele frequencies are produced going from sheltered to exposed habitats (the middle of the cline being of intermediate exposure). Ark plays a central role in both temporal and spatial adenosine triphosphate buffering in cells that display high and variable rates of energy turnover (Wyss et al., 1992). For marine intertidal gastropods, attachment to the substrate in order not to be dislodged by waves is crucial and, for species that live in heterogeneous environments with respect to wave exposure, Ark may potentially be under differential selection with different alleles favoured in different microhabitats (Tatarenkov and Johannesson, 1994). In addition to the substructuring of the Ark variation in exposed and sheltered habitats, in three locations on the west coast of Sweden, one random amplified polymorphic DNA (RAPD) locus (out of 19) was closely associated with size (ecotype) and with the Ark allozyme variation in snails from sites of intermediate exposure where both Ark genotypes and all size classes were present (Johannesson and Mikhailova, 2004). This strong linkage disequilibrium (LD) could be due to some mechanism that restricts recombination, such as a chromosomal inversion (Tatarenkov and Johannesson, 1999; Johannesson and Mikhailova, 2004), a selective sweep where the RAPD allele hitchhiked to high frequency along with the potentially selected Ark locus (or another selected gene closely linked to both the RAPD and the Ark loci), or simply a result of a balance between divergent selection influencing both loci and gene flow between the different habitats (Barton and Gale, 1993).
Thus, the two ecotypes of L. fabalis represent a typical example of repeated and adaptive evolution of morphological and genetic traits, but where the genetic basis of this adaptation is unclear. In order to understand this in more detail, we sequenced an intron of Ark for periwinkles of the small-sheltered (SS) ecotype and the large-moderately exposed (LM) ecotype from their whole distribution range along with new molecular analyses of Ark using allozyme electrophoresis from a location where the association between size and Ark was less clear as in other locations. We report that although both ecotypes are equally variable in neutral markers (Tatarenkov and Johannesson, 1994; Kemppainen et al., 2009), the SS ecotype is essentially fixed for one haplotype of Ark whereas the LM ecotype segregates for ten different haplotypes, indicating an ecotype-specific selective sweep. We also found that up to four haplotypes could be obtained from each of a number of individuals, suggesting the presence of a duplicate gene.
Materials and methods
Sampling
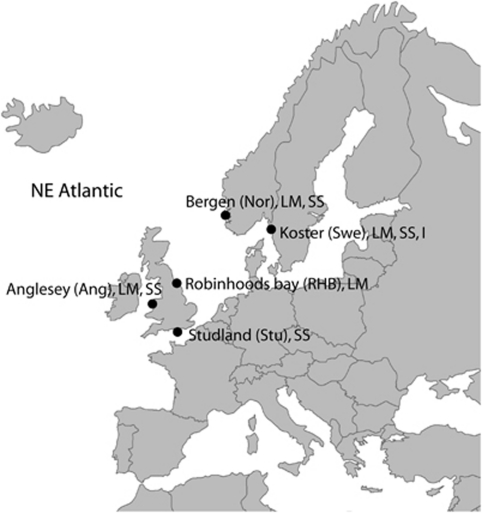
L. fabalis was sampled between 2004 and 2007 from five sites throughout its distribution range from distinct patches of either exposed or sheltered habitat. From three locations, Bergen (Norway), Kosterfjord (Sweden) and Anglesey (GB), both ecotypes were collected from sites 20–100 m from each other, while in Robin Hood's Bay (GB) only the LM ecotype was present and in Studland (GB) only the sheltered ecotype could be found (Figure 1). The ecotype distinctions were made based on habitat characteristics and adult size; only large individuals (width of the aperture measuring 10–13 mm) from exposed habitats were defined as LM ecotypes, and small individuals (width of the aperture measuring 4–7 mm) from sheltered habitats were defined as SS ecotypes. A broader size range of snails was however collected from Bergen (Norway), where the association between size and Ark was less clear. The populations in Bergen (Norway) and Anglesey (GB) were collected from distinct patches of sheltered and exposed shore, respectively, whereas the Koster (Sweden) population was collected from opposite ends of a cline, going from sheltered to exposed habitat (from the same location that was used in a previous Ark allozyme study by Tatarenkov and Johannesson, 1998).
Figure 1.
Sampling locations. Abbreviations of sites used throughout this study are given in parentheses. In all locations a distinction was made between large ecotypes in moderately exposed habitats (the LM ecotype) and small, sheltered ecotypes (the SS ecotype) for Littorina fabalis (but not for L. obtusata). In Kosterfjord, individuals from an intermediate site (I) were also used.
The closely related L. obtusata was included as an outgroup. No Ark clines have been described in the NE Atlantic for L. obtusata (but see Schmidt et al., 2007 for possible selection on Ark in L. obtusata in the NW Atlantic) and therefore no distinction was made between exposed and sheltered individuals of this species.
Allozyme electrophoresis (Bergen population)
Structuring of Ark allozyme genotypes across habitats is known to be present in Kosterfjord (Sweden), Wales and Brittany (Tatarenkov and Johannesson, 1994, 1999). In these locations, Ark100 and Ark80 dominate among large individuals in exposed habitats (the LM ecotype) whereas Ark120 is confined to small snails in sheltered habitats (the SS ecotype; Tatarenkov and Johannesson, 1994). In Bergen, we also found a clear size difference between snails from exposed and sheltered locations (Kemppainen et al., 2009), but initial analyses showed that the association between Ark and size was less clear here than in Sweden and Great Britain. To investigate this, we scored snails from Bergen for Ark allozyme variation using protein electrophoresis prior to sequencing the same individuals. Horizontal starch-gel electrophoresis was conducted for snail homogenates from one exposed shore (n=32) and one sheltered shore (n=32), and we stained for Ark using the protocol from Manchenko (1994; method 2, p 160) following the procedure in Tatarenkov and Johannesson (1994). Positive controls from Kosterfjord with known allele sizes were used throughout the different runs. The width of the aperture was used as a proxy for size.
Sequencing
The degenerate primers CK6-5′ and ARK7-3′ (Palumbi, 1996) were initially used to amplify across an intron in the Ark locus. As these primers were not specific, the different bands obtained were cloned and sequenced (C Gio Gatta, unpublished). From resulting sequences that matched Ark in GenBank (http://www.ncbi.nlm.nih.gov), new primers were designed (Ark 1F: 5′-CAGAAGGTCAGGTAGCCCAG-3′ and Ark 1R: 5′-ATGCAGCAGGGCGGTG-3′) that amplified a 287–308 bp fragment of DNA. It was not possible to sequence longer fragments due to large indels and complicated compound microsatellites in the remaining intron sequence, which is about 700 bp in total.
We also sequenced individuals from an intermediate location in Kosterfjord (a location between the exposed and sheltered location described above), for which the Ark allozyme genotypes were already known (M Fokin, unpublished; Table 1; for individuals in Bergen the allozyme genotypes were scored specifically for this study, see above).
Table 1. Sequences for individuals for which allozyme genotypes for the Ark are known (from Kosterfjord (M Fokin, unpublished) and from Bergen (this study)).
| Samplea | Allele 1 | Allele 2 | Genotype | No. of sequences | Haplotype no. as in Figure 2 |
|---|---|---|---|---|---|
| Kosterfjord intermediate location (Sweden) | |||||
| FSwe1 (3) | 100 | 120 | ES | 2 | 1,3 |
| FSwe2 | 70 | 120 | ES | 2 | 1 |
| FSwe3 | 100 | 120 | ES | 1 | 1 |
| FSwe4 (1) | 100 | 120 | ES | 8 | 2,3 |
| FSwe5 (2) | 100 | 120 | ES | 4 | 1,2,3b |
| FSwe6 | 100 | 100 | EE | 2 | 3 |
| FSwe7 | 120 | 120 | SS | 3 | 1 |
| FSwe8 | 120 | 120 | SS | 1 | 1 |
| FSwe9 | 120 | 120 | SS | 8 | 1 |
| FSwe10 | 120 | 120 | SS | 1 | 1 |
| FSwe11 (27) | 70 | 120 | ES | 13 | 1,4 |
| FSwe12 | 80 | 120 | ES | 1 | 1 |
| Bergen sheltered location (Norway) | |||||
| FNorS1 (11) | 100 | 120 | ES | 3 | 1,2 |
| FNorS2 (12) | 100 | 100 | EE | 3 | 2,3,4b |
| FNorS3 (13) | 100 | 100 | EE | 4 | 2,4 |
| FNorS4 | 120 | 120 | SS | 2 | 1 |
| FNorS5 (14) | 120 | 120 | SS | 2 | 1,4 |
| FNorS6 | 120 | 120 | SS | 4 | 1 |
| FNorS8 (15) | 100 | 100 | EE | 4 | 2,3 |
| FNorS9 | 120 | 120 | SS | 4 | 1 |
| FNorS12 | 120 | 120 | SS | 4 | 1 |
| FNorS13 (30) | 100 | 100 | EE | 3 | 2,11 |
| FNorS14 | 120 | 120 | SS | 62 | 1 |
| FNorS15 (16) | 100 | 100 | EE | 4 | 2,3 |
DNA was extracted as in Kemppainen et al. (2009). Due to multiple indels, direct sequencing was not possible. Instead, we used the mark-recapture cloning method (MR cloning) as described by Bierne et al. (2007); PCR products from individual samples were marked with 49 unique combinations of tagged primers (additional nucleotides were added to the 5′ end of the primer sequence; 7 forward and 7 reversed primers in total) then pooled and finally cloned in one reaction. The PCR products were ‘recaptured' by sequencing positive clones with universal primers annealing to the plasmid sequence; the unique combination of tags revealed the identity of the sample. PCR reactions were performed with 1 μl DNA template (10–20 ng μl–1), 1.3 μl buffer (10 × ), 1 μl of dNTP mix (10 m, 0.78 μl of MgCl2 (1.5 m, 1 μl of forward and reversed primers (5 m) and 0.052 μl of taq (Takara rTaq; Takara Bio Inc., Otsu, Japan; 5 U μl–1) in 13 μl reactions. Cycling parameters were 95 °C for 5 min, 34 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C 2 min and a final elongation at 72 °C for 15 min. PCR products of similar quantities were pooled and cloned (with TOPO TA cloning kit for sequencing; Invitrogen, Life Technologies, Carlsbad, CA, USA). Typically, twice as many positive clones as PCR products were sequenced, according to the guidelines in Bierne et al. (2007) using M13 (-20) universal primers. PCR products were used as templates, after treatment with ExoSap-IT (USB; Cleveland, OH, USA), for sequencing with the Beckman Coulter seq 8000 series Genetic Analysis System (Beckman Coulter, Brea, CA, USA). Only the forward primer was used as the sequence was short and only haplotypes occurring in at least three individuals were used in the subsequent analyses (one haplotype (no. 12) in this study is only represented by one individual, but this haplotype also occurred in individuals from populations that were not included in this study). To obtain an estimate of the point mutation rate caused by the Taq polymerase, DNA from one homozygous SS individual was amplified in nine independent PCR reactions. The PCR products were cloned and between four and eight clones were sequenced for each cloning reaction.
When sequencing with the MR cloning protocol, we cloned one population at a time as only a minute contamination of primer with the wrong 5′ tag may yield incorrect identification of an individual. Nevertheless, in some instances, we repeatedly recovered more than two haplotypes for some individuals. Although we have never encountered more than two alleles per individual for Ark using allozyme electrophoresis, sequencing data across a wide group of invertebrates (including the molluscs Nautilus pompilius and Crassostrea gigas) suggest that duplications of Ark have occurred independently at least four times in this group (reviewed in Uda et al., 2006). To exclude the possibility of contamination of primers with incorrect 5′ tags or contamination of template DNA, some of the individuals, from which more than two haplotypes were found, were sequenced a second time. Furthermore, in order to exclude the possibility that the template itself contained DNA from more than one individual, DNA was re-extracted from the eight individuals from which more than two haplotypes were obtained, and these were cloned individually instead of using the MR cloning protocol (to exclude the possibility of contamination of tagged primers).
Data analysis
For each sequence, the MR tags were used to identify the individual in (; Madison, WI, USA) and all plasmid sequences were discarded. Sequences were aligned in () and a 95% parsimony haplotype network was created with version 1.21 (Clement et al., 2000). The presence of recombination was detected using the four-gamete test in sp (Hudson and Kaplan, 1985; Rozas et al., 2003), the NSS (Jakobsen and Easteal, 1996), Max χ2 (Smith, 1992) and Φw methods (Bruen et al., 2006) implemented in the program PhiPack (TC Bruen, University of California, CA, USA), and by visual inspection of homoplasy and loops in the haplotype networks created by . As our results below indicate that Ark has been duplicated, nucleotide and haplotype diversities cannot be accurately estimated (this would require data that separate the duplicate copies). Therefore, only the number of haplotypes is presented as a measure of genetic diversity, although one needs to be aware that the numbers of haplotypes increase with sample sizes.
Results
Ark electrophoresis (Bergen population)
Both the Ark100 allele, typical for the LM ecotype, and Ark120, typical for the SS ecotype in Sweden, France and United Kingdom (Tatarenkov and Johannesson, 1994, 1999), were found to be common in the Bergen (Norway) population. However, differences between the exposed and sheltered sites were not as large as in, for example, Sweden (although statistically significant: P<0.05, Fisher's exact test); the exposed site was fixed for the Ark100 allele, but the same allele was also common in the sheltered site (60%). This is despite the fact that this site is ecologically very similar to Swedish sheltered sites with dense populations of both Ascophyllum nodosum and L. obtusata, which, in both Sweden and Norway, usually indicates protection from wave action. The sheltered Bergen location had a deficiency of Ark100/Ark120 heterozygotes (P<0.05, χ2 test, d.f.=1), which is also common in the Kosterfjord (Sweden; Tatarenkov and Johannesson, 1994).
In addition, the sheltered Bergen population did not show the expected size difference (t-test; P=0.20) between individuals that were homozygous for the Ark100 allele (n=16, mean size=6.9 mm, s.d.=1.2) and the Ark120 homozygotes (n=10, mean size=6.3, s.d.=0.89). Nevertheless, the Ark100 homozygotes in the sheltered site (n=11, mean size=6.9 mm, s.d.=1.2) were clearly smaller than the Ark100 homozygotes in the exposed site (n=32; mean size=8.2 mm, s.d.=0.83, t-test; P<0.001).
Ark intron sequencing
In total, 355 sequences of 287–308 bp were obtained from 88 individuals in the MR cloning. From the sequencing of nine cloning reactions from one homozygous individual (4–8 clones were sequenced for each reaction), we found that the number of unique haplotypes caused by polymerase error was high: 19 in a total of 61 sequences. These haplotypes were caused by 25 point mutations, giving an error rate of 1.4 nucleotides per 1000 bp, which is normal for standard Taq (Palumbi and Baker, 1994). These artefact mutations were easy to detect in our data set and they were pruned according to the method outlined in Supplementary Appendix S1. In addition, unpublished sequences using Phusion High-Fidelity Taq (Finnzymes, Espoo, Finland) gave similar results to the pruned data set, showing that the pruning is not creating any substantial bias. Two individuals with unique indels were completely excluded from the data set.
Analysis of recombination was performed on the pruned data set and 13 pairs of sites with 4 gametic types were detected, giving a minimum number of recombination events of 3, according to the four-gamete test. In addition, Φw but not NSS or Max χ2 detected significant recombination. However, all these estimates exclude sites with gaps, and when gaps were treated as a fifth state in the haplotype network, one indel (involving sites 283–288) appeared in five different positions in the network, which strongly indicates that recombination has occurred. This made it impossible to reconstruct a haplotype network without loops and we therefore excluded all nucleotides after position 283. This reduced the number of sites with four gametic types to four and only one recombination event was suggested by the four-gamete test, the Φw test was no longer significant (P>0.05) and all loops in the haplotype network were now resolved. By this procedure, we lost 20 bp (including a 5-bp gap) but only 2 out of 14 parsimony informative sites. All subsequent analyses were made on this modified data set.
DNA polymorphism
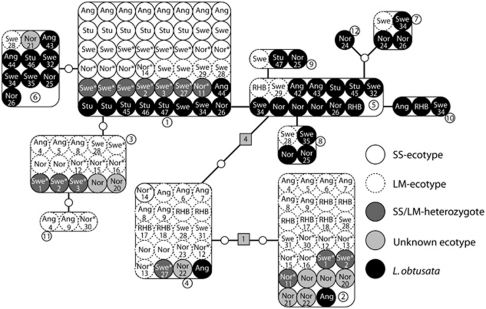
After pruning all likely artefact mutations (Supplementary Appendix S1) and discarding 20 bp from the end of the Ark intron sequence, all redundant sequences from the MR cloning (that is, similar sequences from one individual) were excluded. The data on which all subsequent analyses were conducted contained 152 sequences from 100 individuals of both L. obtusata and the LM and SS ecotypes of L. fabalis (Table 2), and among these, 12 haplotypes were found (GenBank accession numbers: HM446636–HM446647). Interestingly, one haplotype completely dominated the SS ecotypes (haplotype H1 in Figure 2), and only one individual (from Bergen) out of a total of 30 SS ecotypes had a different haplotype (H4; Figure 2).
Table 2. Haplotype counts for Littorina obtusata, the small-sheltered (SS) and the large-moderately exposed ecotypes (LM) of L. fabalis as well as heterozygotes between alleles typical for the LM and SS ecotypes (LM/SS, see also Table 1).
| No. of sequences | No. of individuals | No. of haplotypes | |
|---|---|---|---|
| Anglesey, Wales, UK (Ang) | |||
| L. fabalis LM | 16 | 8 | 4 |
| L. fabalis SS | 8 | 8 | 1 |
| L. obtusata | 8 | 5 | 6 |
| Studland, UK (Stu) | |||
| L. fabalis SS | 7 | 7 | 1 |
| L. obtusata | 9 | 6 | 4 |
| Robin Hood's Bay, UK (RHB) | |||
| L. fabalis LM | 10 | 8 | 3 |
| L. obtusata | 2 | 2 | 2 |
| Kosterfjord, Sweden (Swe) | |||
| L. fabalis LM | 14 | 7 | 9 |
| L. fabalis LMa | 1 | 1 | 1 |
| Sum | 15 | 8 | 9 |
| L. fabalis SS | 4 | 4 | 1 |
| L. fabalis SSa | 4 | 4 | 1 |
| Sum | 7 | 7 | 1 |
| L. fabalis LM/SSa | 11 | 7 | 3 |
| L. obtusata | 10 | 4 | 6 |
| Bergen, Norway (Nor) | |||
| L. fabalis LM | 6 | 5 | 3 |
| L. fabalis LMa | 11 | 5 | 4 |
| Sum | 17 | 10 | 4 |
| L. fabalis SS (LM)b | 9 | 6 | 4 |
| L. fabalis SSa | 7 | 6 | 2 |
| L. fabalis LM/SSa | 2 | 1 | 2 |
| L. obtusata | 13 | 7 | 7 |
| All exposed L. fabalis | 60 | 34 | 10 |
| All sheltered L. fabalis | 30 | 28 | 2 |
| All L. obtusata | 43 | 24 | 10 |
| All individuals | 152 | 101 | 12 |
Indicates that allozyme results exist showing that individuals indeed are homozygotes for either the exposed Ark alleles (Ark100, Ark80 or Ark70) or sheltered ark alleles (Ark120) or heterozygotes between these groups of alleles.
This subsample from Bergen represents the initial sample of small individuals that did not amplify the ‘sheltered' haplotype 1 (H1; Figure 2), which warranted us to genotype all individuals in Bergen for Ark allozymes (see text for details).
Abbreviations used for the locations throughout this article are given in parentheses.
Figure 2.
Haplotype network of an intron of arginine kinase (Ark). Large boxes are haplotypes, small circles represent individual sequences (site abbreviations are given in Figure 1), lines are mutations, small empty circles represent missing hypothetical haplotypes and boxes are indels with the number of mutation involved indicated by a number. The legend explains the identity of the different individuals. Individuals, for which allozyme genotypes are known, are indicated with an asterisk (see Table 1). Samples for which multiple haplotypes were obtained are numbered 1–47, such that it is possible to see which haplotypes were sequenced from each individual.
Of the 30 SS ecotypes that were sequenced for the Ark intron in this study, 12 have also been geneotyped for the Ark allozyme (Kosterfjord; M Fokin, unpublished, Bergen; this study), and of these, all were homozygous for the sheltered allele, Ark120 (Table 1). In addition, although there was no significant association between size and Ark allozyme genotype in the sheltered location in Bergen, from five out of six individuals that were homozygous for the sheltered Ark120 allele, only the H1 haplotype was obtained. In contrast, none of the individuals from this location, which were homozygous for the exposed Ark100 allele, contained this haplotype.
Completely different haplotypes were obtained from two independent cloning reactions for individual 28 in Figure 2 (Table 3), indicating that indeed mixing of samples/template DNA had occurred. Nevertheless, more than two haplotypes were also found in five of eight samples that were cloned individually (that is, not using the MR cloning procedure). In addition, from four of these eight individuals, exactly the same haplotypes were obtained from at least two independent cloning reactions, even after re-extracting the DNA, which was done to exclude contamination of the DNA itself (Table 3). This shows that the recovery of more than two haplotypes from one individual is not entirely due to mistakes during the MR cloning procedure, PCR artefacts or contamination of the DNA, but instead suggests the presence of gene duplication.
Table 3. Summary of sequencing of individuals with more than two haplotypes.
| Individual | Species |
First MR cloning |
Second MR cloning |
After re-extraction and individual cloning |
|||
|---|---|---|---|---|---|---|---|
| No. of sequences | Haplotypes | No. of sequences | Haplotype(s) | No. of sequences | Haplotypes | ||
| 4 | Fab LM | 5 | 2,3,11 | 14 | 2,3,11 | 6 | 2,3 |
| 8 | Fab LM | 9 | 2,3,4 | 3 | 2,4 | 9 | 2,3,4 |
| 9 | Fab LM | 4 | 2,4,11 | 0 | NA | 4 | 2,11 |
| 25 | Obt | 4 | 6,8,9 | 0 | NA | 9 | 6,8,9 |
| 26 | Obt | 4 | 5,6,7 | 0 | NA | 5 | 1,5,6 |
| 28 | Fab LM | 6 | 1,6,8a | 0 | NA | 5 | 2,3,4a |
| 34 | Obt | 7 | 1,5,6,10 | 1 | 7a | 11 | 1,5,6,10 |
Abbreviations: Fab, L. fabalis; LM, large-moderate ecotype; MR, mark-recapture; NA, not available; Obt, L. obtusata.
Likely cases of well-to-well contamination prior to MR cloning.
Individual and haplotype numbers refer to the numbers given in Figure 2.
Individuals for which at least three haplotypes could be recovered from a minimum of two independent clonings are indicated in bold.
Discussion
Ecotype-specific selective sweep
Four alternative (not mutually exclusive) genetic mechanisms have been suggested for the repeated evolution of phenotypic traits (‘parallel evolution') in pairs of contrasting ecotypes (see Johannesson et al., 2010). (1) The mutation responsible for the local adaptation could be due to parallel evolution in the strict sense; that is, independent mutations controlling the locally adapted traits could have arisen and been driven to fixation in different geographic locations (Rolan-Alvarez et al., 2004; Panova et al., 2006; Quesada et al., 2007; Galindo et al., 2009). (2) The ecotypes could initially have evolved allopatrically and the current sympatric or parapatric distribution could be the result of secondary contact and introgression (Wilding et al., 2001). (3) The locally adapted alleles could have been present in an ancestral population as standing genetic variation (see, for example, Barrett and Schluter, 2008 and references therein) and only later been driven to high frequency repeatedly by local selection pressures. (4) Locally adapted characters could have evolved in a concerted fashion; that is, the alleles responsible for trait differences have each arisen once and thereafter they have spread by spatial selective sweeps to similar microhabitats in other locations. This idea assumes that gene flow among ecotypes is high enough to allow the spread of advantageous alleles at a rate that at least overrides the rate by which these alleles would have arisen by repeated new mutations in each local site (for a similar mechanism explaining the collective evolution of species, see Rieseberg and Burke, 2001; Morjan and Rieseberg, 2004; Johannesson et al., 2010).
To discriminate fully between these scenarios, the phylogeographic history of the species in general (inferred from neutral markers) as well as the histories of specific loci under selection need to be known. In this study, we sequenced an intron to Ark in the SS and LM ecotypes of L. fabalis from five different locations throughout their distribution range in order to infer Ark's phylogeographic history. The Ark intron sequence revealed that the small-sized ecotype found in sheltered microhabitats (SS ecotype) in L. fabalis was almost fixed for haplotype 1 (H1; Figure 2) in four locations, on both sides of the North Sea, and only one, out of 30 SS ecotype individuals, contained a different haplotype (H4). In contrast, the large-sized ecotype, present in exposed locations (LM ecotype), displayed similar levels of polymorphism and population structure to its sister species L. obtusata. Re-analyses of microsatellites from Kemppainen et al. (2009; Supplementary Appendix S2), mitochondrial DNA cyt-b sequence data (Kemppainen et al., 2009; no significant frequency difference could be found between the ecotypes in three different locations) and allozymes (Tatarenkov and Johannesson, 1994; apart from Ark no loci showed any consistent differences between the ecotypes) all show that the SS ecotype is not generally less genetically variable than the LM ecotype and that genetically, populations always group by geographic location rather than by ecotype.
The most obvious signature of positive selection is the reduction of physically linked neutral variation around the selected mutation as it ‘hitchhikes' to high frequency along with the new selected allele in the affected population (Maynard-Smith and Haigh, 1974; Kaplan et al., 1989; Stephan et al., 1992). Although the H1 haplotype is nearly fixed in the SS ecotype, it also exists among LM ecotypes (but only in Scandinavia, see below). A likely explanation for this is that a mutation advantageous only in the SS ecotype arose recently, at a locus very closely linked to both the H1 haplotype and the mutation responsible for the Ark120 allozyme variant. This mutation then drove the H1 haplotype and the Ark120 allozyme variant to high frequency in the sheltered habitats by means of an ecotype-specific selective sweep. As the association between size and Ark exists on both sides of the North Sea, we can consider two scenarios for the possible spread of the selected allele: either it was already present in an ancestral population, possibly at the end of the last glaciation, around 10 000 years ago (that is, before both sides of the North Sea were colonized; alternative 3 above—standing variation), or the mutation arose after the post-glacial separation of the North Sea populations, on one or the other side of the North Sea, and then spread to all other populations (alternative 4—evolution in concert).
Both microsatellites (Supplementary Appendix S2; Kemppainen et al., 2009) and mitochondrial cytochrome b sequence data (Kemppainen et al., 2009) show clear differentiation between populations on different sides of the North Sea and thus gene flow between these geographic areas is at present likely to be restricted, but unlikely to be completely absent. Using data from Tatarenkov and Johannesson (1999), we estimated the selection coefficient(s) that is necessary to maintain two different Ark clines, at Jutholmen and Lökholmen (on the west coast of Sweden), to be 0.019 and 0.0018, respectively, assuming a sharp habitat boundary, no dominance in fitness (Barton and Gale, 1993) and an average migration rate of 2.12 m per generation (Tatarenkov and Johannesson, 1998; Supplementary Appendix S3). From the model of Slatkin (1976), even very low levels of migration (Nm=0.1) are sufficient for the spread of strongly positively selected alleles (s=0.05) through a structured population in <10 000 generations. As L. fabalis dwell and lay their egg masses on fucoid macroalgae, which occasionally get ripped off by storms and transported to other locations by ocean currents, some long-distance dispersal is expected. It is therefore possible that the strongly selected Ark allele could have spread from one location to all others after colonization of both sides of the North Sea. Because, in the LM ecotype, the H1 haplotype has so far only been found in Scandinavia, it is likely that the mutation driving the selective sweep of the H1 haplotype in the SS ecotype also arose somewhere in this region. However, 10 000 years is not long for variation to recover around a locus that has undergone a selective sweep (see below). Therefore, we cannot exclude the possibility that the adaptive allele was already present in an ancestral SS ecotype population invading the North Sea area. In this case, the H1 haplotype in the LM ecotype (not linked to any selected mutation) must have, by chance, only spread to exposed habitats in Scandinavia. Nevertheless, assuming that similar selection pressures with respect to the present-day SS and LM ecotypes also existed during past glacial periods, it is likely that an ecotype-specific selective sweep has occurred at some point of time (either before or after re-colonization of the North Sea).
Is there any support for a chromosomal rearrangement?
A previous study demonstrated a strong association between one RAPD locus and the Ark locus (and consequently also size) in intermediate habitats from three different clines from two islands on the west coast of Sweden (Johannesson and Mikhailova, 2004). As size has a strong heritable component (Tatarenkov and Johannesson, 1998), this implies LD between Ark, the RAPD locus and at least some quantitative trait loci for size. This LD could be a result of a very close physical linkage on the same chromosome, or some mechanism that restricts recombination over larger chromosomal blocks, such as a pericentric inversion (Tatarenkov and Johannesson, 1999; Johannesson and Mikhailova, 2004), and this hypothesis is readily testable. An alternative hypothesis is that selection independently maintains differences in size and Ark (and the RAPD locus is physically close to either Ark or a strong quantitative trait locus for size) between exposed and sheltered sites and that, in intermediate locations within clines (Tatarenkov and Johannesson, 1999), LD is maintained by gene flow from both sides of the cline (Barton and Gale, 1993). Interestingly a strong association between Ark and size has been found in all locations where these ecotypes have been studied in detail (Sweden, Wales and the Britannic peninsula; Tatarenkov and Johannesson, 1998) except in Bergen, Norway (this study). In Bergen, SS and LM ecotypes were not collected from a cline but from two distinct patches of either sheltered or moderately exposed shores located approximately 100 m from each other (in contrast to all other locations where Ark and size has been studied). In the ‘sheltered' Bergen population, both the exposed Ark100 allele (60%) and the sheltered Ark120 allele (40%) were common, and both very small (equivalent to the SS ecotype) and very large snails (equivalent to the LM ecotype) could be found (see Kemppainen et al., 2009 for details of the size distribution), suggesting that this specific location is effectively intermediate with respect to selection on both size and Ark. Thus, the explanation for a lack of an association between size and Ark in this location could be the lack of gene flow from exposed and sheltered locations, respectively, which weakens the support for chromosomal rearrangement as a general explanation for the LD between a strong quantitative trait locus for size and Ark. More data are needed to further test this hypothesis.
Evidence for gene duplication, heterozygote deficiency and shared haplotypes between L. fabalis and L. obtusata
In this study, we were consistently able to sequence up to four different haplotypes from some individuals (even after re-extraction of the same individuals; Table 3), although interestingly none of these individuals were SS ecotypes. Although this was only fully demonstrated in four individuals (Table 3), it should be noted that these were the four individuals with the highest number of sequenced clones. For most other individuals in our study only a few clones were sequenced, reducing the chance of detecting more than two haplotypes. PCR-mediated recombination (Meyerhans et al., 1990) could be excluded as a possible source of excess haplotypes as no signs of recombination were detected in the final data set. Thus, one possibility is that Ark and/or its intron has undergone gene duplication. Although more than two bands have never been encountered in one individual following allozyme electrophoresis (personal observation and personal communication with A Tatarenkov and M Fokin), this could be related to the fact that (1) no additional non-synonymous polymorphism has occurred between the parental and the duplicated gene copy or (2) only one copy is expressed. Because of the lack of deep divergence in the total data set, there is reason to believe that this putative gene duplication is relatively young, perhaps even unique to this taxon, unless gene conversion has restricted divergence between the gene copies (Beisswanger and Stephan, 2008). It is surprising that, despite this putative gene duplication, the SS ecotype has remained monomorphic for the Ark intron sequence, but the explanation for this will have to await further studies.
Heterozygote deficiency (essentially fewer Ark120/100 genotypes than expected) is substantial in both Kosterfjord (Tatarenkov and Johannesson, 1994) and Bergen populations (this study). Tatarenkov and Johannesson (1994) suggested either selection against heterozygotes or a Wahlund effect (mixing of Ark120/120 and Ark100/100 genotypes by gene flow from cline edges). The isolated location of the ‘sheltered' Bergen population precludes Wahlund mixing, at least in this location. It is possible that the heterozygote deficiency can more readily be explained once we know more details about the putative gene duplication.
The extensive haplotype sharing between L. fabalis and L. obtusata did not come as a surprise as these species share all common haplotypes in the mitochondrial cyt-b gene as well (Kemppainen et al., 2009). The sharing of cyt-b haplotypes was inferred to be due to a short divergence time relative to the effective population size, that is, incomplete lineage sorting, and some mitochondrial introgression. However, no sign of introgression was detected in nuclear microsatellite markers (Kemppainen et al., 2009) and thus, it is likely that the haplotype sharing of Ark between L. fabalis and L. obtusata is solely due to incomplete lineage sorting. Although the presence of the H1 haplotype in L. obtusata suggests that this haplotype has existed in both species for a long time (L. fabalis and L. obtusata are thought to have diverged about 1 Myr ago; Kemppainen et al., 2009), the closely linked, presumably exonic mutation, which ultimately drove the selective sweep in the SS ecotype, could nevertheless be much younger.
Conclusions
The genetic mechanisms behind repeated (‘parallel') ecotype evolution, and, in particular, the idea of parallel trends in selection pressures acting on allelic variation already present in the population, have recently received much attention (see, for example, Schluter et al., 2004; Campbell and Bernatchez, 2004; Barrett and Schluter, 2008). One potential origin of such variation, as suggested in this study and in Johannesson et al. (2010), is ecotype evolution in concert, that is, new positively selected alleles directly sweep to high frequency in a specific habitat (ecotype) over large parts of a species' distribution. This explanation is similar to the ‘transporter mechanism', as suggested by Schluter and Conte (2009), in which selection in one habitat repeatedly acts on standing genetic variation that is maintained in another habitat by export of alleles adaptive to the first habitat from elsewhere in the range. However, the ‘transporter mechanism' does not define the origin and initial spread of the selected allele. As recombination and mutation will, over time, erode the region of reduced polymorphism around a positively selected allele, a selective sweep can only be traced over a relatively short period of time (about 0.1 Ne generations (Kim and Stephan (2000))—in the case of L. fabalis: roughly 100 000 years; data from Kemppainen et al., 2009). This will make it difficult to evaluate the general importance of ecotype evolution in concert. Nevertheless, with support from the present case study we suggest that this mechanism is potentially important for the evolution of local adaptation in species where migration introduces new genetic variation into geographically distant populations more frequently than mutations.
Acknowledgments
We thank Andrei Tatarenkov for valuable comments on the manuscript and Thomas Dahlgren, Erik Boström, Ricardo Pereyra, Anders Erikson, Carl André, Pieternella Luttikhuizen and Carles Vilá for the discussions that ultimately have led to this publication. We also thank three anonymous reviewers for comments that have improved this manuscript dramatically. This work was performed within the Linnaeus Centre for Marine Evolutionary Biology (http://www.cemeb.science.gu.se/) supported by the Swedish Research Councils VR and Formas. Financial support was also provided by a grant from Kungliga Fysiografiska sällskapet i Lund (Nilsson Ehle donationen).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Barton NH, Gale KS.1993Genetic analysis of hybrid zonesIn: Harris R (ed).Hybrid Zones and the Evolutionary Process Oxford University Press: New York [Google Scholar]
- Beisswanger S, Stephan W. Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila. Proc Natl Acad Sci USA. 2008;115:5447–5452. doi: 10.1073/pnas.0710892105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Tanguy A, Faure M, Faure B, David E, Boutet I, et al. Mark-recapture cloning: a straightforward and cost-effective cloning method for population genetics of single-copy nuclear DNA sequences in diploids. Mol Ecol Notes. 2007;7:562–566. [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Bernatchez L. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol Biol Evol. 2004;21:945–956. doi: 10.1093/molbev/msh101. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Galindo J, Moran P, Rolan-alvarez E. Comparing geographical genetic differentiation between candidate and noncandidate loci for adaptation strengthens support for parallel ecological divergence in the marine snail Littorina saxatilis. Mol Ecol. 2009;18:919–930. doi: 10.1111/j.1365-294X.2008.04076.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- Johannesson K, Mikhailova N. Habitat-related genetic substructuring in a marine snail (Littorina fabalis) involving a tight link between an allozyme and a DNA locus. Biol J Linnean Soc. 2004;81:301–306. [Google Scholar]
- Johannesson K, Panova M, Kemppainen P, André C, Rolán-Alvarez E, Butlin RK. Repeated evolution of reproductive isolation in a marine snail: unveiling mechanisms of speciation. Phil Trans R Soc B. 2010;365:1735–1747. doi: 10.1098/rstb.2009.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The hitchiking effect revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen P, Panova M, Hollander J, Johannesson K. Complete lack of mitochondrial divergence between two species of NE Atlantic marine intertidal gastropods. J Evol Biol. 2009;22:2000–2011. doi: 10.1111/j.1420-9101.2009.01810.x. [DOI] [PubMed] [Google Scholar]
- Kemppainen P, van Nes S, Ceder C, Johannesson K. Refuge function of marine algae complicates selection in an intertidal snail. Oecologia. 2005;143:402–411. doi: 10.1007/s00442-004-1819-5. [DOI] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics. 2000;155:1415–1427. doi: 10.1093/genetics/155.3.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchenko G. A Handbook of Detection of Enzymes on Electrophoretic Gels. SRS Press: Florida; 1994. [Google Scholar]
- Maynard-Smith J, Haigh J. The hitchhiking effect of a favorable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McKinnon JS, Mori S, Blackman BK, David L, Kingsley DM, Jamieson L, et al. Evidence for ecology's role in speciation. Nature. 2004;429:294–298. doi: 10.1038/nature02556. [DOI] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Palumbi SR.1996Nucleic acids ll: the polymerase chain reactionIn: Hills DH, Moritz C, Mable B (eds).Molecular Systematics Sinauer Associations, Inc.: Sunderland, MA [Google Scholar]
- Palumbi SR, Baker CS. Contrasting population-structure from nuclear intron sequences and mtDNA of humpback whales. Mol Biol Evol. 1994;11:426–435. doi: 10.1093/oxfordjournals.molbev.a040115. [DOI] [PubMed] [Google Scholar]
- Panova M, Hollander J, Johannesson K. Site-specific genetic divergence in parallel hybrid zones suggests non-allopatric evolution of reproductive barriers. Mol Ecol. 2006;15:4021–4031. doi: 10.1111/j.1365-294X.2006.03067.x. [DOI] [PubMed] [Google Scholar]
- Quesada H, Posada D, Caballero A, Moran P, Rolan-Alvarez E. Phylogenetic evidence for multiple sympatric ecological diversification in a marine snail. Evolution. 2007;61:1600–1612. doi: 10.1111/j.1558-5646.2007.00135.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Burke JM. The biological reality of species: gene flow, selection, and collective evolution. Taxon. 2001;50:47–67. [Google Scholar]
- Rolan-Alvarez E, Carballo M, Galindo J, Moran P, Fernandez B, Caballero A, et al. Nonallopatric and parallel origin of local reproductive barriers between two snail ecotypes. Mol Ecol. 2004;13:3415–3424. doi: 10.1111/j.1365-294X.2004.02330.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nagel L, Boughman JW. Natural selection and parallel speciation in sympatric sticklebacks. Science. 2000;287:306–308. doi: 10.1126/science.287.5451.306. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proc Natl Acad Sci USA. 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS. Parallel evolution and inheritance of quantitative traits. Am Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Phifer-Rixey M, Taylor GM, Christner J. Genetic heterogeneity among intertidal habitats in the flat periwinkle, Littorina obtusata. Mol Ecol. 2007;16:2393–2404. doi: 10.1111/j.1365-294X.2007.03323.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. The Rate of Spread of an Advantageous Allele in a Subdivided Population. Academic Press, Inc.: New York; 1976. [Google Scholar]
- Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Stephan W, Wiehe THE, Lenz MW. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor Popul Biol. 1992;41:237–254. [Google Scholar]
- Tatarenkov A, Johannesson K. Habitat related allozyme variation in a microgeographical scale in the marine snail Littorina mariae (Prosobranchia: Littorinacea) Biol J Linn Soc. 1994;53:105–125. [Google Scholar]
- Tatarenkov A, Johannesson K. Evidence of a reproductive barrier between two forms of marine periwinkle Littorina fabalis (Gastropoda) Biol J Linn Soc. 1998;63:349–365. [Google Scholar]
- Tatarenkov A, Johannesson K. Micro- and macrogeographical allozyme variation in Littorina fabalis; do sheltered and exposed forms hybridize. Biol J Linn Soc. 1999;67:199–212. [Google Scholar]
- Uda K, Fujimoto N, Akiyama Y, Mizuta K, Tanaka K, Ellington WR, et al. Evolution of the arginine kinase gene family. Comp Biochem Physiol Part D-Genomics Proteomics. 2006;1:209–218. doi: 10.1016/j.cbd.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Wood HM, Grahame JW, Humprey JR, Butlin RK. Sequence differentiation in regions identified by a genome scan for local adaptation. Mol Ecol. 2008;17:123–3135. doi: 10.1111/j.1365-294X.2008.03755.x. [DOI] [PubMed] [Google Scholar]
- Wood TE, Johne MB, Rieseberg LH. Parallel genotypic adaptation: when evolution repeats itself. Genetica. 2005;123:157–170. doi: 10.1007/s10709-003-2738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding CS, Butlin RK, Grahame J. Differential gene exchange between parapatric morphs of Littorina saxatilis detected using AFLP markers. J Evol Biol. 2001;14:611–619. [Google Scholar]
- Williams GA. Littorina mariae—a factor structuring low shore communities. Hydrobiol. 1990;193:139–146. [Google Scholar]
- Wyss M, Smeitink J, Wevers RA, Wallimann T. Mitochondrial creatine-kinase: a key enzyme of aerobic energy-metabolism. Biochim Biophys Acta. 1992;1102:119–166. doi: 10.1016/0005-2728(92)90096-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.