Abstract
The patterns of hybridization and asymmetrical gene flow among species are important for understanding the processes that maintain distinct species. We examined the potential for asymmetrical gene flow in sympatric populations of Eucalyptus aggregata and Eucalyptus rubida, both long-lived trees of southern Australia. A total of 421 adults from three hybrid zones were genotyped with six microsatellite markers. We used genealogical assignments, admixture analysis and analyses of spatial genetic structure and spatial distribution of individuals, to assess patterns of interspecific gene flow within populations. A high number of admixed individuals were detected (13.9–40% of individuals), with hybrid populations consisting of F1 and F2 hybrids and backcrosses in both parental directions. Across the three sites, admixture proportions were skewed towards the E. aggregata genetic cluster (x=0.56–0.65), indicating that backcrossing towards E. aggregata is more frequent. Estimates of long-term migration rates also indicate asymmetric gene flow, with higher migration rates from E. aggregata to hybrids compared with E. rubida. Taken together, these results indicate a greater genetic input from E. aggregata into the hybrid populations. This asymmetry probably reflects differences in style lengths (E. rubida: ∼7 mm, E. aggregata: ∼4 mm), which can prevent pollen tubes of smaller-flowered species from fertilizing larger-flowered species. However, analyses of fine-scale genetic structure suggest that localized seed dispersal (<40 m) and greater clustering between hybrid and E. aggregata individuals may also contribute to directional gene flow. Our study highlights that floral traits and the spatial distributions of individuals can be useful predictors of the directionality of interspecific gene flow in plant populations.
Keywords: hybridization, introgression, gene flow, mating patterns, fine scale spatial genetic structure, hybrid zones
Introduction
Natural interspecific hybridization occurs commonly in flowering plants (Mallet, 2005), and is thought to have an important role in their evolutionary history (Grant, 1981). In many cases, however, hybridization is asymmetrical, and one species is more often the maternal parent of hybrid offspring (Tiffin et al., 2001). In some cases, repeated backcrossing between hybrids and one of the parental species can lead to unidirectional introgression (Bacilieri et al., 1996). In particular, the directional bias in gene flow and infiltration of genes from one species into another can be important in determining the direction of evolutionary change and species succession (Bacilieri et al., 1996; Petit et al., 2004). Asymmetrical hybridization is also important in a conservation context, especially when hybrids exhibit high fitness and rare species exist in small populations. Under these conditions, frequent hybridization followed by unidirectional backcrossing towards the rare species, can further exacerbate their decline through a combination of pollen swamping and genetic assimilation (Rhymer and Simberloff, 1996). Determining which reproductive barriers influence asymmetrical gene flow is therefore important for understanding both the processes that facilitate speciation and maintain species identities.
Given the importance of asymmetrical hybridization and introgression, there has been a long standing interest in understanding the factors which drive these patterns in plants. Some of these underlying factors include: mating system variation (SI × SE rule; Lewis and Crowe, 1958), the relative proportions of parent species (Burgess et al., 2005) and differences in the fitness of reciprocal crosses (Tiffin et al., 2001). Flower size can also be a major structural barrier to hybridization that is often unilateral (Williams and Rouse, 1988; Gore et al., 1990). This trait is especially important because flower size often differs among hybridizing species. The genus Eucalyptus (Myrtaceae), which forms a dominant vegetative component of the Australian continent, provides a good model system for investigating hybridization dynamics in relation to flower size. On the basis of morphological patterns, interspecific hybridization is considered widespread in Eucalyptus (∼50% of species reportedly hybridize; Griffin et al., 1988) and several molecular studies have detected broad-scale introgression of maternally inherited markers among numerous sympatric species (McKinnon et al., 2001, 2010). In this genus, smaller flowered species are often unable to cross-fertilize with larger flowered species. This is because the pollen tubes of small flowered species are unable to grow the full length of the styles of larger flowered species (Gore et al., 1990). This potential difference in fertilization success provides a testable prediction regarding the expected direction of hybridization and introgression in natural hybrid zones consisting of small and large flowered Eucalyptus species.
The availability of highly polymorphic genetic markers (for example, microsatellites, amplified fragment length polymorphisms) and powerful statistical analyses (for example, Bayesian clustering) has greatly improved the detection of first generation and later generation hybrids. Apart from some notable exceptions (for example, Chung et al., 2005), few studies have utilized these statistical procedures to examine hybrid populations in the spatial context of their parental species. In particular, for plants with limited seed dispersal, this information can be important for inferring the direction of gene flow because early generation hybrids are expected to be located near their maternal parent. Moreover, greater spatial clustering of hybrids around one of the parental species may be important for reinforcing directional backcrossing and can contribute to subsequent asymmetrical introgression. When assessing patterns of hybridization and introgression, the spatial analysis of reproductive adult genotypes has several advantages over direct methods including reciprocal pollinations and assessments of introgression in open pollinated progeny arrays. First, for tall trees with a long generation time and time to first flowering such as Eucalyptus, crossing experiments are often impractical. Second, both direct techniques do not provide information regarding the establishment of backcrossed hybrids, a necessary pre-requisite for potential introgression between species.
Here, we investigate the potential for asymmetrical gene flow and introgression in putative hybrid zones of a common and a rare member of the genus Eucalyptus. In southeastern Australia, the uncommon forest tree Eucalyptus aggregata is restricted to the central and southern tablelands of New South Wales and a single locality in Victoria. This species occurs in small populations (n<1000; Field, 2008) in combination with more widespread species that belong to the same subgenus (Symphyomyrtus) and section (Maidenaria). Eucalyptus aggregata often co-occurs with E. rubida which is common across New South Wales, Victoria and Tasmania. These two species are well differentiated morphologically (Field et al., 2009) with E. aggregata assigned to series Faveolatae and E. rubida to series Viminales (Brooker, 2000). Morevoer, E. aggregata and E. rubida are well differentiated genetically on the basis of allozyme markers (FST=0.59; Field et al., 2009). There are also strong differences in habitat preference with E. aggregata found on poorly drained flats and hollows, whereas E. rubida is found on rocky and well drained skeletal soils of loams on clay subsoils (Cayzer, 1993). Where these species co-occur, putative adult hybrids are often observed, particularly along transition zones between the parental habitats. Both species are pollinated by generalists and substantial overlap in flowering times (October–January for E. rubida, December–February for E. aggregata) likely provides ample opportunities for interspecific pollen flow. Morphological and molecular evidence has identified substantial hybrid frequencies in the seed arrays of E. aggregata where the two species occur in sympatry (Mean 8.9% Field et al., 2009), and the presence of both F1 and later generation hybrids. As the more common E. rubida has larger flowers (style length ∼7 mm) than E. aggregata (style length ∼4 mm), and both species are pollinated by generalist insects (Field et al., 2008), floral morphology may be an important driver of asymmetrical gene flow. Despite the widespread occurrence of hybridization in Eucalyptus, little is known about the fine-scale patterns of hybridization and the factors driving asymmetrical gene flow in natural hybrid zones.
To assess the extent and direction of interspecific gene flow we use molecular markers and Bayesian analyses to examine the genomic composition of three hybrid zones consisting of E. rubida, E. aggregata and putative hybrids. A prevalence of F1 hybrids would indicate little opportunity for gene exchange, whereas a high frequency of backcross hybrids would suggest a high potential for introgression. Moreover, a skewed frequency of hybrid backcrosses in one parental direction would indicate the directionality of introgression. We also used coalescent-based methods to estimate long-term historical levels of gene flow between parental and hybrid populations. Given the differences in flower size, we predict that hybrid populations will exhibit directional gene flow towards E. aggregata. From these predictions we asked the following questions: (i) what is the frequency of introgression between the two species? and (ii) is asymmetrical gene flow in the direction of the smaller-flowered E. aggregata? In addition, we used spatial analyses and spatial autocorrelation methods to assess both the degree of spatial clustering between parental species and hybrids and the scale of seed dispersal. Given that seed dispersal is locally restricted in Eucalyptus (House, 1997) greater clustering between hybrids and one of the parental species could indicate which species is more often the maternal parent. Moreover, considering plant mating is more frequent among near neighbors, stronger spatial clustering between hybrids and one particular parental species may also contribute to asymmetrical gene flow. If restricted seed dispersal and spatial clustering is contributing to asymmetrical hybridization, we predict that spatial patterns should match the direction of asymmetrical gene flow. From this prediction, we ask the following questions: (iii) are hybrids spatially clustered around E. aggregata rather than E. rubida? and (iv) what is the scale of seed dispersal in E. aggregata, E. rubida and hybrid populations as indicated by fine-scale spatial genetic structure?
Methods
Study species and sampling
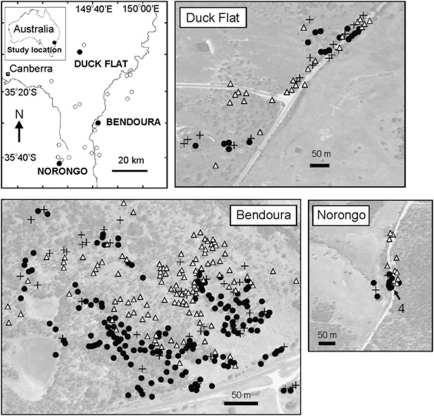
Three sites containing reproductive populations of E. aggregata, E. rubida and their putative hybrids were selected for this study on the southern tablelands of New South Wales, Australia (Figure 1). These included: Bendoura (35°30′ S, 142°42′ E), a relatively large and intact open woodland (N=337 adults); Duck Flat (33°28′ S, 150°02′ E), a medium sized remnant site (N=59); and Norongo (32°42′ S, 149°25′ E), a small and cluster of trees along a road side (N=25). At each of these sites, individual E. aggregata and E. rubida were easily distinguished on the basis of morphology. E. aggregata displays green elliptical-ovate-shaped leaves in alternating stem arrangements in the juvenile phase, and smaller lanceolate shaped leaves in the adult phase (12 cm long × 2.5 cm wide), brown and rough furrowed bark that is persistent into the upper branches, and has smaller flower buds (0.4 × 0.3 cm) in clusters of seven. In contrast, E. rubida displays round glaucous juvenile leaves in opposite arrangement on the stem in the juvenile phase, has larger lanceolate-shaped leaves in the adult phase (15 cm long × 2.4 cm wide), smooth white to reddish bark and larger flowers (0.7 × 0.4 cm) instead in groups of three (Brooker and Kleinig, 1999). Putative hybrids were identified on the basis of intermediate morphology between the parental species for each of the distinguishing characteristics including bark persistence, leaf size and flower size; however, hybrids always exhibited flowers in groups of seven. Adult populations were mapped using high-resolution aerial photos (1:1838; taken 2004, Lands and Property Information NSW) and by measuring the relative location of the individually-tagged trees on the ground.
Figure 1.
Map showing the location of three study sites, each consisting of populations of E. aggregata, E. rubida and their putative hybrids. Individual maps of each site indicate the location of adult E. aggregata (closed circles), E. rubida (open triangles), and hybrids (plus symbol). Arrow on Norongo map indicates the location of four hybrids.
To examine the genomic composition of the adult populations, mature leaves were collected for DNA and subsequent genotyping. A total of 421 trees were sampled, which included all observed reproductive adults of each parental species and putative hybrids displaying intermediate morphology from Duck Flat (N=59) and Norongo (N=25). At the Bendoura site, a 400 × 500 m area of the site containing the majority of reproductive trees was selected for sampling. Within this area, all reproductive E. aggregata and E. rubida, and all putative hybrids were sampled (N=337). The majority of the remaining trees at each site was the distantly related E. pauciflora, which is not reported to hybridize with either E. aggregata or E. rubida (Griffin et al., 1988).
Genetic analysis
Adult leaves were stored at −80 °C until freeze-dried for five days and then ground to powder using tungsten beads in a Mixer Mill MM 300 (Qiagen, Doncaster, Australia). DNA was extracted from up to 10 mg of the ground leaves using DNeasy 96 Plant extraction kits (Qiagen) as per the manufacturer's protocol. To reduce phenolic contaminates, 1% polyvinylpyrrolidone (molecular weight 40 000) and 5% sodium -Isoascorbate was added to the AP1 extraction buffer.
We initially screened a panel of 40 microsatellite primer pairs developed for Eucalyptus. Many microsatellites have been developed for Eucalyptus and cross-species amplification is often successful (for example, Byrne et al., 1996). We assayed adults for six microsatellite markers selected from the panel, including four markers developed for E. globulus: Eg65, Eg91, Eg96 (G Moran, unpublished) and Emcrc8 (Steane et al., 2001), and two markers developed for E. grandis and E. urophylla: Embra10 (Brondani et al., 1998), Fmrsa1 (van der Nest et al., 2000). All of these primers were directly labeled with fluorescent dyes (FAM, NED, VIC) and the SSR fragments were amplified by PCR in a mixture of 10 μl containing: 4.44 μl H20, 1 × Hotstar PCR buffer (1.5 m MgCl2, Tris-Cl, KCl, (NH4)2 SO4; Qiagen), 1.65 m MgCl2, 0.1 μ of the forward (fluorescently labelled) and reverse primer, 0.2 μ of each dNTP, 10 mg ml−1 bovine serum albumin, 0.05 U of Hotstar Taq (Qiagen), and 1 μl (2–15 ng) of DNA template. Amplification of SSR fragments were performed in 384-well plates (Applied Biosystems, Mulgrave, Australia) on a Hybaid express thermocycler with a step-down PCR program consisting of 94 °C for 4 min; 15 cycles at 94 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min 20 s; 30 cycles at 94 °C for 30 s, 50 °C for 30 s, 72 °C for 45 s. The final product was evaporated and 5 μl of combined loading dye (Hi-Di Formamide: Applied Biosystems) and an internal lane standard was added, followed by denaturing at 94 °C for 5 min. The final multi-mixed PCR products were separated by capillary electrophoresis on an ABI 3130 Genetic Analyser (Applied Biosystems) and their size determined in relation to an internal lane standard (GS-500 LIZ; Applied Biosystems) using Genemapper Version 4.0 (Applied Biosystems). Individuals with low or missing peaks were amplified and genotyped a second time. No evidence of null alleles was detected at Duck Flat or Norongo using the software MICROCHECKER (van Oosterhout et al., 2004). At Bendoura, there was a low probability (0.05) of a null allele at locus Fmrsa1. However, we included all loci as removing the Fmrsa1 locus had minimal effect on the following genetic analyses.
Microsatellite diversity
Genetic diversity parameters were assessed separately at each site for E. aggregata, E. rubida and hybrids classified using Bayesian analyses (see below). Observed number of alleles (A), unbiased genetic diversity (HE), and observed heterozygosity (HO) were calculated using the software GenAlEx version 6 (Peakall and Smouse, 2005). The fixation index (FIS) and departures from random mating were calculated using the software FSTAT version 2.9.3.2 (Goudet, 1995). To measure the average pair-wise level of genetic differentiation (FST; Wright (1965)) between populations of each species and the hybrids, according to the multi-locus methods implemented in FSTAT. Each pair-wise estimate and the 95% bootstrap confidence intervals (CIs) were estimated with 1000 replicates.
Patterns of hybridization
We used the Bayesian methods implemented in the program STRUCTURE version 2.2 (Pritchard et al., 2000; Falush et al., 2003) to distinguish hybrids from purebreds. This method uses a Markov chain Monte Carlo (MCMC) algorithm to assign individuals (as represented by their multi-locus genotypes) to K genetic clusters by minimizing within group linkage-disequilibrium and simultaneously assuming within group Hardy–Weinberg Equilibrium. Using this approach, individuals can be assigned largely to a single genetic cluster based on admixture values (for example, Q1=0.99, Q2=0.01) or jointly to two or more if their multi-locus genotype indicates admixture due to hybridization between clusters (for example, Q1=0.5, Q2=0.5). For all analyses we used STRUCTURE with the admixture model, no previous population information, correlated allele frequencies and a burn-in period of 20 000 and 200 000 MCMC. First we conducted four independent runs at each K from two to eight, to determine the most likely number of genetic clusters and test the stability of admixture values (r>0.98). For the final analysis we assumed K=2 (Q1=E. aggregata, Q2=E. rubida) as this agrees with the presence of two species and was the most likely number of genetic clusters identified (data not shown) using the post-hoc methods of Evanno et al. (2005).
Estimates of recent introgression
In order to assess the rate and direction of recent gene flow among populations at each site, we classified individuals further into genealogical classes using the methods implemented in the NEWHYBRIDS program (Anderson and Thompson, 2002). This analysis is a probability-based model, which computes through MCMC, the posterior probability, qi, of individuals belonging to distinct genealogical classes. We ran the software with the following parameters: a burn-in period of 20 000 generations and 200 000 MCMC, no previous population information, ‘Jeffery's like priors', and the default six genealogical classes that correspond to: (i) Pure E. aggregata, (ii) Pure E. rubida, (iii) F1 hybrid, (iv) F2 hybrid, (v) Bx aggregata (backcross towards E. aggregata), (vi) Bx rubida (backcross towards E. rubida). As suggested by Vähä and Primmer (2006), a posterior probability of >50% was used as a threshold for assigning an individual to a specific class. For broader classification of individuals into each of the three groups: E. rubida, E. aggregata and hybrids; the posterior probabilities of all hybrid classes were summed for an individual.
We used simulations to assess the efficiency (proportion of individuals in a group that were correctly identified) and accuracy (proportion of an identified group that truly belong to that category), of STRUCTURE for identifying hybrids and NEWHYBRIDS for delineating the six genealogical classes. Simulated E. aggregata, E. rubida, first-generation (F1) and second-generation (F2) hybrids and backcrosses with each parental were obtained using the microsatellite genotypes of real adults and the software HYBRIDLAB (Nielsen et al., 2006). As suggested by Vähä and Primmer (2006) we used reference parental genotypes of E. aggregata and E. rubida to obtain simulated genotypes. We used 40 E. rubida individual genotypes sampled from an allopatric population (Kings Hwy, Table 1 in Field et al., 2009) and 40 E. aggregata individuals from sites where E. aggregata was numerically dominant and hybridization was not detected (Field et al., 2009). Reference individuals were designated pure parentals only if initial runs with STRUCTURE indicated Q>0.95. Simulated E. aggregata (N=1000) and E. rubida (N=1000), and each hybrid generation described above (N=240) were analyzed using STRUCTURE and NEWHYBRIDS under the same conditions as the real dataset.
Table 1. Summary of genetic diversity measures and mean fixation (FIS) estimates in adult populations of Eucalyptus aggregata, E. rubida and their hybrids at each of three sites (Bendoura, Duck Flat, Norongo).
| Species/Site | N | A | HE (SE) | HO (SE) | FIS (95% CI) |
|---|---|---|---|---|---|
| E. aggregata | |||||
| Bendoura | 171 | 9.33 | 0.709 (0.019) | 0.702 (0.019) | 0.009 (−0.046, 0.064) |
| Duck Flat | 18 | 4.66 | 0.648 (0.024) | 0.711 (0.056) | −0.093 (−0.218, 0.032) |
| Norongo | 10 | 5.00 | 0.648 (0.049) | 0.733 (0.042) | −0.139 (−0.190, -0.087) |
| Hybrids | |||||
| Bendoura | 47 | 14.2 | 0.855 (0.010) | 0.823 (0.040) | 0.038 (−0.048, 0.125) |
| Duck Flat | 17 | 9.66 | 0.817 (0.018) | 0.862 (0.060) | −0.055 (−0.191, 0.082) |
| Norongo | 6 | 5.67 | 0.769 (0.023) | 0.889 (0.070) | −0.153 (−0.304, −0.002) |
| E. rubida | |||||
| Bendoura | 119 | 12.8 | 0.794 (0.027) | 0.738 (0.057) | 0.072 (−0.048, 0.188) |
| Duck Flat | 24 | 9.50 | 0.750 (0.048) | 0.736 (0.068) | 0.025 (−0.049, 0.100) |
| Norongo | 9 | 6.67 | 0.778 (0.018) | 0.907 (0.053) | −0.169 (−0.307, −0.030) |
Abbreviations: A, mean number of alleles per locus; CI, confidence interval; HE, Hardy–Weinberg mean expected heterozygosity; HO, observed heterozygosity; N, sample size; SE, standard error.
Estimates of long-term gene flow
To estimate migration rates and assess the potential for asymmetry in gene flow between the two species and the hybrid populations, we used a coalescent-based approach implemented in the program MIGRATE-N version 3.0 (Beerli and Felsenstein, 1999, 2001). This analysis uses maximum-likelihood or Bayesian inference to estimate the parameter M (M=m/u, where m is the migration rate, and u is the mutation rate), which describes the mutation-scaled long-term migration rate between populations. MIGRATE-N also jointly estimates Θ (Θ=4Neu), the mutation-scaled effective population size. Unlike F-statistics, this method allows for the possibility of asymmetrical gene flow between population pairs and differences in the effective population size.
For the purposes of assessing the rates of historical gene flow, individuals were assigned to one of three populations on the basis of admixture values implemented in STRUCTURE: E. aggregata (Q1 > 0.9), E. rubida (Q1 < 0.1) and hybrids (0.1 ⩽ Q1⩽0.9). For each site (Bendoura, Duck Flat and Norongo), we estimated Θ for each population and M between all pair-wise populations (3 × 3) using the approaches implemented in MIGRATE-N. We conducted four identical maximum-likelihood analyses under both the step-wise mutational model with the Brownian motion model and the infinite-allele model. Search parameters consisted of 10 short chains with an increment of 20 and sampling of 500; five long chains with an increment of 20 and sampling of 10000; and a burn-in of 5000. With each run we changed the random number seed and the starting values of Θ and M. The first run estimated Θ and M from FST values, and subsequent runs used the maximum-likelihood estimates of Θ and M from the previous run as starting parameters. We omitted locus Fmrsa1 from all analyses because of a high frequency of single base-pair mutations that do not adhere to the strict step-wise mutational model assumed by MIGRATE-N. Alleles occurring at moderate frequencies (>10%) at other loci also violated the strict step-wise mutational model and therefore we only report analysis using the infinite-allele model.
Spatial clustering
We used O-ring statistics to assess the spatial patterns among hybrids and parental species at Bendoura and Duck Flat. The Norongo site was excluded because of small sample size (N=25). We used Wiegand–Moloney's O-ring statistic (Wiegand and Moloney, 2004) to assess the spatial relationships of adults from three groups (E. aggregata, F1 hybrids and E. rubida) in two separate pair-wise analyses (E. aggregata vs hybrids, E. rubida vs hybrids). For each analysis, we calculated the bivariate O-ring statistic, Oij (r), from the number of individuals of group i, found in concentric rings of radius (r=3.5 m) about plants of group j. Values of Oij (r)=0, Oij (r) > 0, Oij (r) <0 correspond to spatial independence, spatial clustering and spatial repulsion, respectively. Tests for significance of spatial clustering against the null model (hybrids randomly dispersed around parent species) were performed by comparing the observed Oij (r) statistic with 95% confidence envelopes from multiple Monte Carlo simulations (n=1000) at each radius. All calculations and simulations were conducted using the software PROGRAMITA (Wiegand and Moloney, 2004).
Fine-scale spatial genetic structure (SGS)
SGS refers to the decrease of pair-wise relatedness among individuals with distance (that is, isolation-by-distance Wright, 1943). This can provide information regarding the scale of dispersal within populations and the potential for interspecific gene dispersal between sympatric species. To characterize SGS for E. aggregata, E. rubida and the hybrid populations at Bendoura and Duck Flat, we used the spatial autocorrelation procedures of Loiselle et al. (1995) and the software SPAGeDi version 1.2 (Hardy and Vekemans, 2002). Given the small sample size at Norongo (N=25), this site was excluded from the analysis. The average kinship coefficients (Fij) between pairs of individuals i and j were calculated for intervals of 15 m for distances between zero and 100 m, and intervals of 25 m for distances between 100 m and 450 m. Given the fragmented distribution of individuals at Duck Flat, here Fij was estimated to a maximum of 150 m. The SGS pattern was visualized separately for each group by plotting the mean Fij against distance. The significance of SGS at each distance interval was tested by permuting the spatial location of individuals 10 000 times to obtain 95% CIs about the null hypothesis of no SGS (Fij=0). We also calculated approximate standard errors (s.e.) for the mean Fij at each distance interval by jackknifing over loci.
Results
Microsatellite diversity
Across all adults (N=421) we detected a total of 105 alleles at six polymorphic loci. Among the adult populations, the widespread E. rubida tended to have the greater allelic diversity. A total of 77 alleles were recorded in E. rubida populations compared with 58 in the uncommon E. aggregata. The mean number of alleles per locus (A) was 12.8 at Bendoura, 9.5 at Duck Flat and 6.67 at Norongo for E. rubida, compared with 9.33 at Bendoura, 4.66 at Duck Flat and 5.0 at Norongo for E. aggregata (Table 1). Despite the lower allelic diversity in E. aggregata, each adult had a unique multi-locus genotype. Genetic diversity estimates (HE) were also consistently higher in E. rubida (Bendoura: HE=0.794, Duck Flat: HE=0.750, Norongo: HE=0.778) than E. aggregata (Bendoura: HE=0.709, Duck Flat: HE=0.648, Norongo: HE=0.648). The hybrids displayed the highest levels of genetic diversity at the two larger sites but not at Norongo given the small sample size (Bendoura: HE=0.855, Duck Flat: HE=0.817, Norongo: HE=0.769) (Table 1).
Wright's F-statistics
Mean fixation indices (FIS) were not significantly different from zero (random mating) for the E. aggregata, E. rubida or hybrid populations at the two largest sites Bendoura and Duck Flat. In contrast, significant departures from random mating were present for all three groups at the small remnant site Norongo, as FIS was significantly <0. Therefore, at Norongo heterozygosity was higher than expected among the E. aggregata (FIS=−0.139), E. rubida (FIS=−0.153) and hybrid populations (FIS= −0.169) (Table 1).
Genetic differentiation between E. aggregata and E. rubida populations was moderate and significant as shown by Wright's FST at Bendoura (FST=0.209), Duck Flat (FST=0.262) and Norongo (FST=0.171) (Table 2). This level of differentiation is consistent with the fact that allele frequencies at each locus were highly skewed towards either E. aggregata or E. rubida, rather than exhibiting fixed differences (data not shown). In contrast to interspecific genetic differentiation, the level of differentiation between E. rubida populations was very low (range FST; 0.035–0.059). For the uncommon E. aggregata, genetic differentiation between populations was higher than between populations of the widespread E. rubida (range FST; 0.067–0.119) (Table 2). Genetic differentiation between hybrids and E. aggregata (FST; 0.019–0.065) was lower at each site compared with differentiation between hybrids and E. rubida (FST; 0.082–0.119), but this was not significant as the 95% CIs overlapped (Table 2).
Table 2. Pair-wise tests for genetic differentiation (Wright's FST) among adult populations of E. aggregata, E. rubida and their hybrids at three sites (Bendoura, Duck Flat, Norongo).
| Pair-wise comparison | FST (95% CI) |
|---|---|
| Interspecific | |
| agg (BT) vs rub (BT) | 0.209 (0.170, 0.251) |
| agg (DF) vs rub (DF) | 0.262 (0.209, 0.312) |
| agg (NG) vs rub (NG) | 0.171 (0.117, 0.222) |
| Intraspecific | |
| agg (BT) vs agg (DF) | 0.067 (0.032, 0.103) |
| agg (BT) vs agg (NG) | 0.112 (0.080, 0.145) |
| agg (DF) vs agg (NG) | 0.119 (0.070, 0.169) |
| rub (BT) vs rub (DF) | 0.035 (0.022, 0.046) |
| rub (BT) vs rub (NG) | 0.038 (0.013, 0.069) |
| rub (DF) vs rub (NG) | 0.059 (0.034, 0.082) |
| Hybrids | |
| agg (BT) vs hyb (BT) | 0.051 (0.033, 0.070) |
| agg (DF) vs hyb (DF) | 0.065 (0.031, 0.110) |
| agg (NG) vs hyb (NG) | 0.019 (0.016, 0.051) |
| rub (BT) vs hyb (BT) | 0.082 (0.057, 0.107) |
| rub (DF) vs hyb (DF) | 0.105 (0.062, 0.153) |
| rub (NG) vs hyb (NG) | 0.119 (0.070, 0.169) |
Abbreviations: agg, Eucalyptus aggregata; rub, E. rubida; hyb, hybrids; BT, site Bendoura; DF, site Duck Flat; NG, site Norongo; CI, Confidence interval.
Simulations
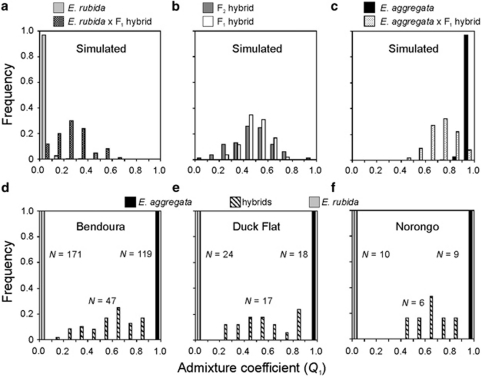
The admixture analysis performed with STRUCTURE had maximum accuracy when assigning individuals broadly to each of the three classes: E. aggregata, E. rubida and hybrids. With these classes, a threshold of Q=0.9 had the highest efficiency and accuracy for distinguishing hybrids from purebreds (Supplementary Figure S1). At this threshold, the number of apparent hybrids in the simulated population (Mean 7.1%) was the closest to the actual number of hybrids (7.4%). Most misidentified hybrids were backcrosses, with ∼10% of this group assigned incorrectly to a purebred class because of substantial overlap in admixture values (Figures 2a–c).
Figure 2.
Frequency distribution of admixture coefficients (Q1) of multilocus genotypes of simulated parental and hybrid individuals (a–c), and adult plants at each of three sites: (d) Bendoura, (e) Duck Flat and (f) Norongo. Adult plant assignment based on the following thresholds: Q1>0.9 as E. aggregata, Q1<0.1 as E. rubida and 0.1 ⩽Q1 ⩽ 0.9 as hybrids.
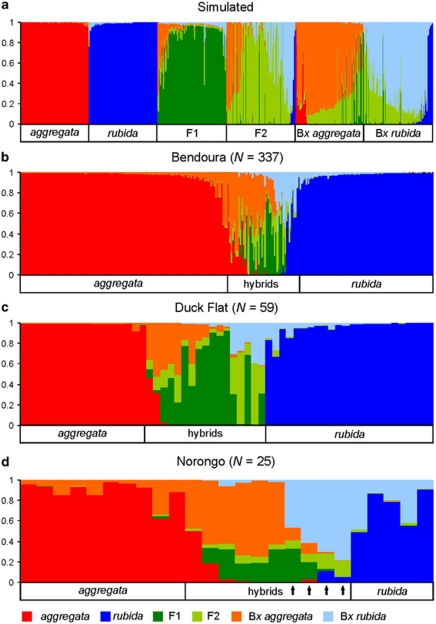
The assignment of simulated individuals using NEWHYBRIDS was mostly congruent with those obtained from STRUCTURE. All simulated purebred individuals were assigned with high posterior probability (qi >95%) to the same purebred category as STRUCTURE (Figure 3a). This analysis was most successful in identifying early generation hybrids. Using an assignment threshold of qi >50%, 93% of F1 hybrids were assigned to the correct class. NEWHYBRIDS performed poorly with the assignment of more advanced generation hybrids. A threshold of qi >50% had the highest efficiency and accuracy for assignment of simulated backcrosses with 75% of Bx aggregata and 77% of Bx rubida classes correctly assigned (Supplementary Figure S2). The majority of the misidentified backcrosses were mostly assigned as the parent species involved in the backcross.
Figure 3.
Bayesian assignment of multilocus genotypes of simulated purebred and hybrid individuals (a) and real adults (N=421) from three sympatric populations (b–d) using the software NEWHYBRIDS. Each line represents an individual's posterior probability of assignment (qi) to each of six genealogical classes including: purebred E. aggregata, purebred E. rubida and four hybrid classes (F1, F2, backcrosses towards each parent (Bx aggregata, Bx rubida)). Simulated individuals for each of the six classes were generated using reference genotypes and HYBRIDLAB. Arrows indicate individuals assigned as purebred E. rubida using the software STRUCTURE.
Frequency of hybridization
Analyses of admixture values calculated in STRUCTURE indicate that hybrids between E. aggregata and E. rubida are common, with 69 (16%) individuals displaying hybrid ancestry (that is, 0.1 ⩽Q1 ⩽ 0.9). The highest number of hybrids detected was at the fragmented site Duck Flat, where 17 (28.8%) of the trees were determined to be hybrids (Figure 2e). In comparison, 47 (13.9%) and 6 (24.0%) of the trees were identified as hybrids at Bendoura and Norongo, respectively (Figures 2d and f). When restricted to three classes (E. aggregata, E. rubida, hybrids), the NEWHYBRIDS analyses assigned individuals to the same classes at Bendoura and Duck Flat as the STRUCTURE analysis. In contrast, results from NEWHYBRIDS indicate that 10 (40%) of the trees at Norongo were hybrids. This was because four individuals assigned with STRUCTURE to E. rubida (Q1<0.1) exhibited above 80% probability of hybrid assignment when summed across all hybrid classes (Figure 3d). The discrepancy between the techniques for the four individuals at this site is probably because of the lower genetic differentiation between parental species at Norongo (Table 2) and the subsequent reduced power in distinguishing backcrosses from parentals at this site.
Nature of introgression
Assignment probabilities for genealogical classes (NEWHYBRIDS) indicate admixed individuals belong to a range of hybrid generations; however, the number of introgressed individuals is likely to be underestimated with these markers (see simulations above). Among the hybrid individuals, 20 of 47 (42.6%) at Bendoura (Figure 3b), 3 of 17 (17.6%) at Duck Flat (Figure 3c) and 6 of 10 (60%) at Noronogo (Figure 3d) displayed high probabilities (>50%) of belonging to a backcrossed generation. A skewed distribution in the admixture coefficients towards E. aggregata (STRUCTURE) suggests a greater genetic input from E. aggregata in the hybrid populations. We found 50 (72%) of the hybrids showed admixture coefficients closer to the E. aggregata cluster (0.5<Q1<0.9). This bias among the hybrid populations was evident at all three sites, with 35 (74%) at Bendoura (Figure 2d), 10 (58%) at Duck Flat (Figure 2e), and 5 (83%) at Norongo (Figure 2f) exhibiting admixture coefficients between 0.5 and 0.9. Similarly, the mean admixture values of hybrid populations (0.1<Q1<0.9) were skewed towards the E. aggregata cluster at Bendoura (Q1 x̃=0.59±0.03), Duck Flat (Q1 x̃=0.56±0.05) and Norongo (Q1 x̃=0.65±0.05).
Long-term estimates of gene flow
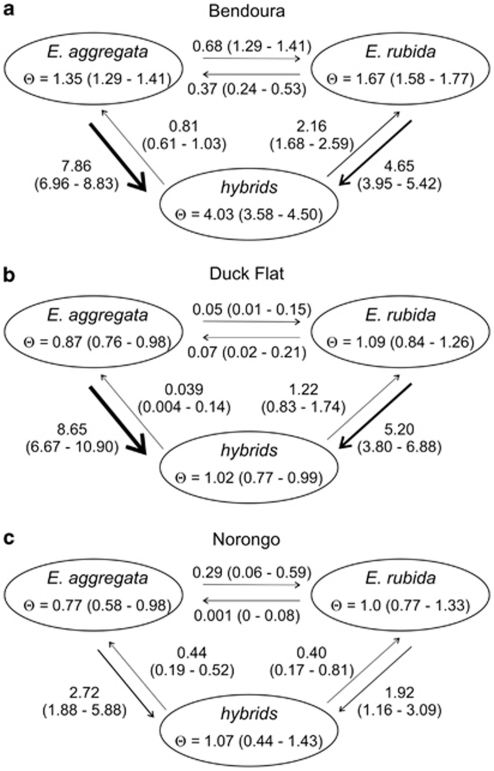
The maximum likelihood estimates of long-term gene flow using MIGRATE-N indicated low levels of migration between the two species. The migration rate M was <0.8 (Figure 4), suggesting low levels of historic gene exchange. There was also no evidence of asymmetrical gene flow between the two species (Figure 4) because of overlapping CIs for estimates of M in both directions at all three sites (for example, Bendoura, ME.aggregata → E.rubida=0.68, 95% CIs=(0.49, 0.93); ME.rubida → E.aggregata=0.37 (0.24, 0.53)). However, there was evidence that E. aggregata has a greater genetic input into the hybrid populations, with more gene flow detected from the E. aggregata to hybrid populations compared with E. rubida to the hybrid populations. Higher rates of gene flow from the E. aggregata into the hybrid population was most evident at Bendoura (ME.aggregata → hybrids=7.86, (6.96, 8.83); ME.rubida → hybrids =4.65 (3.95, 5.42)) (Figure 4a), and is consistent with the prediction of greater introgression towards the smaller-flowered species. Whereas higher maximum-likelihood esimates of gene flow from E. aggregata into the hybrid population was evident at the remaining sites, the 95% CIs marginally overlapped at both Duck Flat (ME. aggregata → hybrids=8.65, (6.67, 10.90); ME.rubida → hybrids=5.20 (3.80, 6.88)) (Figure 4b) and Norongo (ME.aggregata → hybrids=2.72, (1.88, 5.88); ME.rubida → hybrids=1.92 (1.16, 3.09)) (Figure 4c).
Figure 4.
Maximum-likelihood estimates of the long-term migration rates (M) and the mutation-scaled effective population size (Θ) in each of three Eucalyptus hybrid zones (a–c) using the methods of MIGRATE-N. Thickness of the arrows indicates the directional migration rate estimates (M=m/u, where m is the migration rate, and u is the mutation rate). Values in parenthesis indicate the 95% CIs.
Spatial clustering
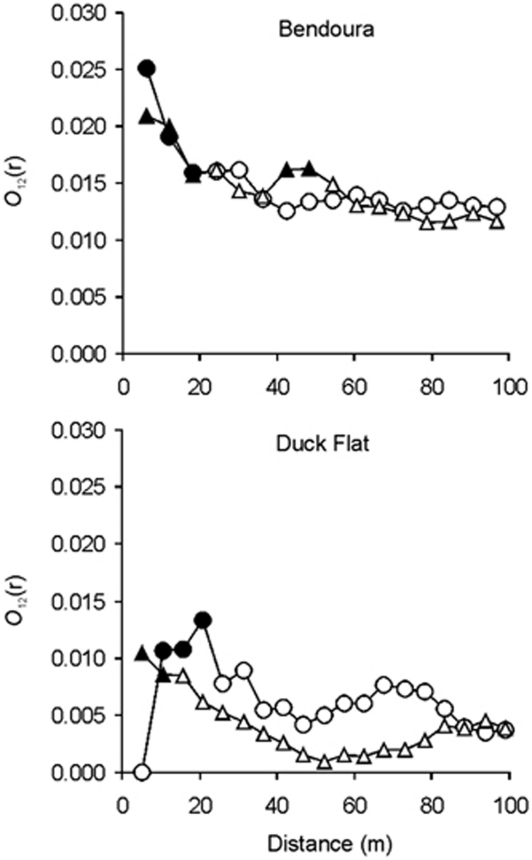
Bivariate O-ring analyses of the spatial distributions of individuals indicated significant spatial clustering between hybrids and both parental species at short distance intervals between 5 and 20 m (Figure 5). At greater distance intervals, there was little spatial clustering between these groups because the O12 (r) statistic was within the simulated 95% CIs (randomly dispersed). At short distance intervals, higher levels of the O12 (r) for E. aggregata vs hybrids comparison suggest hybrids are more spatially clustered with E. aggregata than E. rubida. At Bendoura, the higher spatial clustering between hybrids and E. aggregata was marginal and evident only at the smallest distance interval of 5 m (Figure 5). However, at Duck Flat, higher levels of clustering between hybrids and E. aggregata compared with hybrids and E. rubida persisted from 5 to 20 m (Figure 5).
Figure 5.
Wiegend and Moloney's O12 (r) statistics for bivariate analyses of adult populations of E. aggregata, E. rubida and hybrids at the continuous site Bendoura and remnant site Duck Flat. Circles and triangles represent bivariate analysis for E. aggregata vs hybrids and E. rubida vs hybrid individuals, respectively. Filled symbols indicate values significantly different from the null hypothesis as they lie outside the 95% CIs. The 95 % CIs were generated from 1000 Monte Carlo simulations of hybrid individuals randomly distributed around fixed E. aggregata and E. rubida distributions.
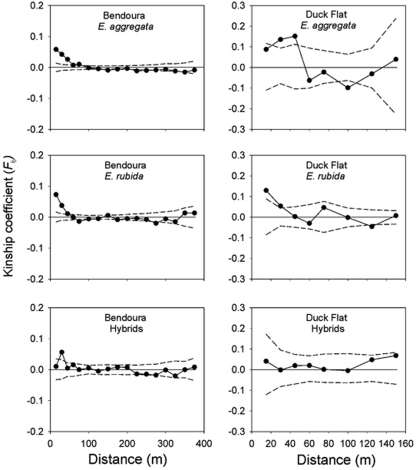
Fine-scale SGS
Fine-scale genetic structure was low to moderate in E. aggregata and E. rubida populations, with the mean kinship coefficient (Fij) significantly different from zero at several of the short distance classes (Figure 6). The Fij estimates were significant and positive at smaller distance intervals of <45 m for E. aggregata at Bendoura and Duck Flat and E. rubida at Bendoura. In comparison, the range of SGS was less for E. rubida at Duck Flat, because Fij was only significant and positive at distances <30 m. For the hybrid populations, significant SGS was only detected at Bendoura from 15 to 30 m (Fij=0.058, 95% CIs=(0.033, 0.085)) (Figure 6).
Figure 6.
Spatial autocorrelation analyses of pair-wise co-ancestry (Fij) estimated from multi-locus genotypes of Eucalyptus aggregata, E. rubida and hybrids at each site (Bendoura and Duck Flat). Dashed lines indicate upper and lower 95% CIs around the null hypothesis of Fij=0.
Discussion
The patterns of interspecific gene flow among species are important for understanding the processes that maintain distinct species. In this study, molecular evidence in each of the three hybrid zones of E. rubida and E. aggregata indicate that hybridization is frequent (14–40% of the adults), and introgression has occurred (5.1–24% of adults) with backcrosses in both directions. These results support previous molecular and morphological evidence of both F1 and backcross hybrids among progeny arrays collected from E. aggregata parents (Field et al., 2009). In addition, further genealogical assessments and measures of historical gene flow revealed greater genetic input from E. aggregata into the hybrid populations. These results fit our prediction that interspecific gene flow would be asymmetrical and directed towards the smaller flowered E. aggregata.
Extent of introgression
Detection of backcross hybrids and evidence of historic introgression suggest hybrid zones between E. aggregata and E. rubida are complex, and are not limited to the first generation. Hybridization beyond the F1 generation is commonly reported in plants, with many examples of advanced generation hybrids outnumbering early generations in hybrid populations (see review; Rieseberg and Carney, 1998). This is because bridging the initial gap between two species through the formation of F1 hybrids is generally considered the more difficult step (Manel et al., 2005). In Eucalyptus, asynchronous flowering is a strong reproductive barrier, with limited reports of hybridization among species with completely divergent flowering times (Potts et al., 2003). In our system, F1 hybrids produce viable seed and exhibit intermediate flowering times between the parental species (Field, 2008). Given the greater flowering synchrony between established F1 hybrids and parentals, early-generation hybrids may provide a bridge for the movement of genes between E. aggregata and E. rubida.
Frequent hybridization and introgression in adult populations (present study) and in seedling cohorts (Field et al., 2008) suggest that E. aggregata and E. rubida have maintained distinct morphological traits and habitat associations despite only partial reproductive isolation. This could reflect recent adaptive radiation of these species in the presence of ongoing gene flow, considering the low to moderate level of genetic differentiation between E. aggregata and E. rubida (FST: 0.17–0.26). There is a growing body of evidence of widespread introgression of cytoplasmic and nuclear DNA among distinct plant species (Mallet, 2005), suggesting that the speciation process can occur despite frequent and ongoing interspecific-gene flow (Lexer and Widmer, 2008). Fine-scale estimates of the frequency of introgressed individuals within mixed populations have varied widely among species pairs, from cases of low frequencies (0.01%) of advanced generation recombinants in Liparis (Chung et al., 2005) to moderate frequencies (14%) between the rare E. cordata and the widespread E. globulus (McKinnon et al., 2010). In the latter study, McKinnon et al., 2010 also found low levels of genetic differentiation between the rare and common species in mixed populations (FST=0.208) despite distinct morphological differences, habitat preferences and assignment to different series within section Maidenaria (series Obiculares and Globulares, respectively). Evidence of historical introgression among Eucalyptus species (McKinnon et al., 2001) and recent introgression in mixed populations of E. aggregata and E. rubida suggest frequent opportunities exist for gene exchange between distinct Eucalyptus species.
An important aspect in detecting introgression using nuclear DNA markers is being able to distinguish early-generation hybrids from backcrosses, with only the latter potentially resulting in the transfer of genetic material between species. In our study, simulations indicate that six microsatellite markers provide low accuracy (75–77%) in distinguishing backcross hybrids. This highlights an important limitation of using low numbers of codominant markers, as the power to accurately distinguish hybrid classes is dependent on both the number of loci and the level of genetic differentiation between the parentals. Vähä and Primmer (2006) found that a relatively high degree of accuracy (95%) in detecting backcross hybrids might require >48 codominant loci and high genetic divergence (FST=0.21). These requirements are often not practical in population studies of natural hybrid zones and are much higher than the number of loci commonly reported with codominant markers such as microsatellites (for example, Curtu et al., 2009; Valbuena-Carabana et al., 2007). Larger numbers of loci typically obtained using alternative techniques such as amplified fragment length polymorphism, may be the best strategy for detecting advanced hybrid generations with a high degree of confidence.
Direction of Introgression
Given the limitations in accurately detecting hybrid classes, what can we infer about the direction of introgression between E. aggregata and E. rubida? Several characteristics of the genomic composition of the hybrid populations and long-term estimates of gene flow suggest a higher contribution of genetic material from E. aggregata. First, lower genetic differentiation between hybrids and the uncommon E. aggregata, despite broad CIs, indicates gene flow between these populations is more frequent than between hybrids and the widespread E. rubida. This finding is consistent with long-term gene flow estimates from MIGRATE-N. Despite an overall low level of migration (M) between the purebred populations (M <1.0), we found evidence of asymmetrical gene exchange, with higher levels of migration between the hybrid populations and E. aggregata compared with E. rubida (Bendoura; Figure 4a). Although migration rates were not significantly asymmetrical at the remaining sites, broad CIs because of smaller sample sizes may have obscured the same underlying trend (Figures 4b and c). Contemporary approaches using NEWHYBRIDS analyses found a similar pattern, with more individuals assigned to the Bx aggregata class in each of the hybrid populations (Figure 3). This trend is also consistent with the results from STRUCTURE, with admixture values skewed towards E. aggregata and few hybrids exhibiting admixture values close to the E. rubida genetic cluster (Q1 0.05 to 0.375). Although this pattern could be a sampling artefact, the consistent trend across all three sites using contemporary approaches suggests backcrossing is probably more frequent in the direction of E. aggregata. This is in comparison with a dominance of F1 hybrids, which would suggest limited introgression. The complex genomic composition of the hybrid populations suggests a high potential for gene movement between E. aggregata and E. rubida through the hybrid populations.
Causes of asymmetric introgression
The greater degree of backcrossing towards E. aggregata observed within the three sympatric populations could be explained by several pre-zygotic and post-zygotic mechanisms. In natural plant populations, selection remains the primary mechanism implicated in determining patterns of hybridization and introgression (Lexer et al., 2005; Whitney et al., 2006). In this case, E. aggregata-like hybrids may possess a selective advantage over E. rubida-like hybrids in these mixed habitats. Alternatively, asymmetric introgression could reflect demographic processes related to species range expansion, which has been implicated as a major determinant of gene flow patterns in a wide array of plants and animals (Currat et al., 2008). Using simulations, Currat et al. (2008) demonstrated that range expansion can result in asymmetric introgression of neutral alleles from the local to the invading species. Under this model, expansion of the widespread E. rubida into E. aggregata habitat is predicted to result in more introgression of local E. aggregata alleles into the invading E. rubida and subsequent hybrid population, a pattern consistent with the higher frequency of introgression detected between E. aggregata and the hybrid populations at Bendoura. Although selection and demographic processes may have a role in determining the patterns observed in this system, the presence of unilateral pre-zygotic barriers may also contribute towards limiting the success of backcrossing in the direction of E. rubida. The most likely mechanism is a difference in flower size between the parental species. In Eucalyptus, this has been found to impede pollen tubes of small-flowered species from reaching the ovule of large-flowered species (Gore et al., 1990). This structural barrier may be reduced in F1 hybrids as they tend to exhibit intermediate flower size (Field, 2008). In this case, the presence of F1 hybrids may therefore reduce the strength of asymmetrical barriers to gene flow.
Interspecific gene flow between hybrids and E. rubida may also be impeded by the non-random distribution of individuals. At Bendoura and Duck Flat, O-ring analyses indicated that hybrids are more spatially clustered with E. aggregata compared with E. rubida. This spatial pattern could be because E. aggregata is more often the maternal parent of hybrid offspring combined with localized seed dispersal and recruitment of hybrids. Consistent with this prediction, our analyses of fine-scale SGS indicate seed dispersal and recruitment is restricted to <40 m around maternal plants. Eucalyptus seed are small (E. aggregata, 1–1.2 mm) and not adapted to wind dispersal, resulting in generally low dispersal distances (House, 1997). The seed dispersal radius inferred from the SGS analyses (<40 m), is consistent with direct measures of seed dispersal in similarly sized Eucalyptus (E. marginata; Cremer, 1977). Considering that plant mating is typically more frequent among nearest neighbors (for example, García et al., 2005; Stacy et al., 1996), this spatial pattern may further enhance positive assortative mating among hybrids and E. aggregata plants. Our data demonstrates that an initial bias in the maternal parent involved in F1-hybrid formation, coupled with localised pollen and seed dispersal, may be important contributing factors in determining the direction of backcrossing. This indicates that the local distribution of hybrids with respect to the parental species may be an important and under-appreciated process for reinforcing asymmetrical introgression in plant populations.
The complex genetic composition of hybrids and their persistence through a range of age cohorts (seed to adult populations) (Field et al., 2008), indicates that hybridization and introgression is a dynamic and ongoing process between E. aggregata and E. rubida. In this system, there are no differences in early hybrid fitness relative to purebreds (Field, 2008), but in general Eucalyptus hybrids tend to exhibit poorer fitness at later life-history stages (Lopez et al., 2000). Given that these species are associated with different habitats, selection may be strong against intermediate-hybrid genotypes that are less well adapted than the parental species in either local environment. It is therefore likely that selection, demography and floral architecture are contributing factors to the patterns of hybridization and introgression in this system.
Acknowledgments
We thank Linda Broadhurst, Judy Cassells and Bronwyn Matheson for assistance. We thank Melinda Pickup for comments that significantly improved the quality of the manuscript. This work was conducted while (DLF) was receiving a University of Wollongong Post-graduate Research Award, and ‘top-up' scholarship from CSIRO Plant Industry.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacilieri R, Ducousso A, Petit R, Kremer A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondani RPV, Brondani C, Tarchini R, Grattapaglia D. Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E urophylla. Theor Appl Genet. 1998;97:816–827. [Google Scholar]
- Brooker MIH. A new classification of the genus Eucalyptus L′Hér. (Myrtaceae) Aust Syst Bot. 2000;13:79–148. [Google Scholar]
- Brooker MIH, Kleinig DA.1999Field Guide to Eucalyptus South-Eastern Australia2nd edn.Bloomings Books: Hawthorn [Google Scholar]
- Burgess KS, Morgan M, Deverno L, Husband BC. Asymmetrical introgression between two Morus species (M. alba, M. rubra) that differ in abundance. Mol Ecol. 2005;14:3471–3483. doi: 10.1111/j.1365-294X.2005.02670.x. [DOI] [PubMed] [Google Scholar]
- Byrne M, Marquez-Garcia MI, Uren T, Smith DS, Moran GF. Conservation and genetic diversity of microsatellite loci in the genus Eucalyptus. Aust J Bot. 1996;44:331–341. [Google Scholar]
- Cayzer LW.1993. The investigation of phenotypic variation within and between three Eucalypt taxa occurring in the ACT. Honours Thesis, Australian National University.
- Chung MY, Nason JD, Chung MG. Patterns of hybridization and population genetic structure in the terrestrial orchids Liparis kumokiri and Liparis makinoana (Orchidaceae) in sympatric populations. Mol Ecol. 2005;14:4389–4402. doi: 10.1111/j.1365-294X.2005.02738.x. [DOI] [PubMed] [Google Scholar]
- Cremer KW. Distance of seed dispersal in eucalypts estimated from seed weights. Aust Forest Res. 1977;7:225–228. [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- Curtu AL, Gailing O, Finkeldey R. Patterns of contemporary hybridization inferred from paternity analysis in a four-oak-species forest. BMC Evol Biol. 2009;9:9. doi: 10.1186/1471-2148-9-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field DL.2008. The importance of ecological factors in determining the pattern of interspecific hybridisation in fragmented landscapes of Eucalyptus aggregata, PhD Thesis, University of Wollongong.
- Field DL, Ayre DJ, Whelan RJ, Young AG. Relative frequency of sympatric species influences rates of interspecific hybridization, seed production, and seedling performance in the uncommon Eucalyptus aggregata. J Ecol. 2008;96:1198–1210. [Google Scholar]
- Field DL, Ayre DJ, Whelan RJ, Young AG. Molecular and morphological evidence of natural interspecific hybridization between the uncommon Eucalyptus aggregata and the widespread Erubida and E viminalis. Conserv Genet. 2009;10:881–896. [Google Scholar]
- García C, Arroy JM, Godoy JA, Jordano P. Mating patterns, pollen dispersal, and the ecological maternal neighbourhood in a Prunus mahaleb L population. Mol Ecol. 2005;14:1821–1830. doi: 10.1111/j.1365-294X.2005.02542.x. [DOI] [PubMed] [Google Scholar]
- Gore P, Potts B, Volker P, Megalos J. Unilateral cross-incompatibility in Eucalyptus: the case of hybridisation between E globulus and E. nitens. Aust J Bot. 1990;38:383–394. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A computer program to calculate F- statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Grant V. Plant Speciation. Columbia University Press: New York; 1981. [Google Scholar]
- Griffin AR, Burgess IP, Wolf L. Patterns of natural and manipulated hybridization in the genus Eucalyptus. Aust J Bot. 1988;36:41–66. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- House SM.1997Reproductive biology of eucalyptsIn: Williams J, Woinarski J (eds)Eucalypt Ecology: Individuals to Ecosystems Cambridge University Press: Cambridge; 30–55. [Google Scholar]
- Lewis D, Crowe LK. Unilateral interspecific incompatibility in flowering plants. Heredity. 1958;12:233–256. [Google Scholar]
- Lexer C, Fay MF, Joseph JA, Nica MS, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): the role of ecology and life history in gene introgression. Mol Ecol. 2005;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. [DOI] [PubMed] [Google Scholar]
- Lexer C, Widmer A. The genic view of plant speciation: recent progress and emerging questions. Philos Trans R Soc B Biol Sci. 2008;363:3023–3036. doi: 10.1098/rstb.2008.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic-structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) Am J Bot. 1995;82:1420–1425. [Google Scholar]
- Lopez GA, Potts BM, Tilyard PA. F1 hybrid inviability in Eucalyptus: the case of E ovata x E. globulus. Heredity. 2000;85:242–250. doi: 10.1046/j.1365-2540.2000.00739.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Manel S, Oscar EG, Waples RS. Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol. 2005;20:136–142. doi: 10.1016/j.tree.2004.12.004. [DOI] [PubMed] [Google Scholar]
- McKinnon GE, Smith JJ, Potts BM. Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Mol Ecol. 2010;19:1367–1380. doi: 10.1111/j.1365-294X.2010.04579.x. [DOI] [PubMed] [Google Scholar]
- McKinnon GE, Vaillancourt RE, Jackson HD, Potts BM. Chloroplast sharing in the tasmanian Eucalyptus. Evolution. 2001;55:703–711. doi: 10.1554/0014-3820(2001)055[0703:csitte]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nielsen EE, Bach LA, Kotlicki P. HYBRIDLAB (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes. 2006;6:971–973. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Australian National University: Canberra, Australia; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytol. 2004;161:151–164. [Google Scholar]
- Potts BM, Barbour RC, Hingston AB, Vaillancourt RE. Genetic pollution of native eucalypt gene pools—identifying the risks. Aust J Bot. 2003;51:1–25. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu Rev Ecol Syst. 1996;27:83–109. [Google Scholar]
- Rieseberg LH, Carney SE. Plant hybridization. New Phytol. 1998;140:599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- Stacy EA, Hamrick JL, Nason JD, Hubbell SP, Foster RB, Condit R. Pollen dispersal in low-density populations of three neo-tropical tree species. Am Naturalist. 1996;148:275–298. [Google Scholar]
- Steane DA, Vaillancourt RE, Russel J, Powell W, Marshall D, Potts BM. Development and characterisation of microsatellite loci in Eucalyptus globulus (Myrtaceae) Silv Genet. 2001;50:89–91. [Google Scholar]
- Tiffin P, Olson MS, Moyle MC. Asymmetrical crossing barriers in angiosperms. Proc R Soc B. 2001;268:861–867. doi: 10.1098/rspb.2000.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Valbuena-Carabana M, Gonzalez-Martinez SC, Hardy OJ, Gil L. Fine-scale spatial genetic structure in mixed oak stands with different levels of hybridization. Mol Ecol. 2007;16:1207–1219. doi: 10.1111/j.1365-294X.2007.03231.x. [DOI] [PubMed] [Google Scholar]
- van der Nest MA, Steenkamp ET, Wingfield BD, Wingfield MJ. Development of simple sequence repeat (SSR) markers in Eucalyptus from amplified inter-simple sequence repeats (ISSR) Plant Breeding. 2000;119:433–436. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am Naturalist. 2006;167:794–807. doi: 10.1086/504606. [DOI] [PubMed] [Google Scholar]
- Wiegand T, Moloney KA. Rings, circles, and null-models for point pattern analysis in ecology. Oikos. 2004;104:209–229. [Google Scholar]
- Williams EG, Rouse JL. Disparate style lengths contribute to isolation of species in Rhododendron. Aust J Botany. 1988;36:183–191. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.