Abstract
Genetic analyses of speciation have focused nearly exclusively on retrospective analyses of reproductive isolation between highly divergent species. Yet, a full understanding of the speciation process must encompass analysis of the consequences of genomic divergence in young lineages still capable of exchanging genes under natural conditions. The accumulation of conditionally neutral genetic variation may lead to the evolution of divergent gene networks. In a hybrid background, such mutations may no longer compensate one another, resulting in the appearance of selectively disadvantageous traits, including disruption of gene expression regulation. Here, we documented genome-wide patterns of gene expression divergence between young lineages of normal and dwarf lake whitefish and their backcross hybrids for which strong, yet incomplete post-zygotic isolation barriers exist. A significant proportion (33%) of backcross hybrids showed developmental abnormalities not seen in parental forms and eventually leading to death. Although the transcriptome of parental forms was nearly identical during embryonic development, suggesting a role for stabilizing selection, all hybrids displayed strongly divergent patterns of gene expression. By comparing healthy, surviving hybrids against moribund ones showing abnormal development, we observed that over 2000 genes were misregulated in these abnormal embryos. In particular, misregulation was significantly biased toward essential developmental genes, which were severely underexpressed. Furthermore, genes previously documented to be highly transgressive (exaggerated inter-individual variance) were almost invariably underexpressed in hybrids. Our results thus clearly showed a transcriptome-wide signature of hybrid breakdown in young, incipient species and demonstrated a persuasive link between misexpression of essential developmental genes and post-zygotic isolation.
Keywords: genetic incompatibility, hybridization, hybrid breakdown, microarray, reproductive isolation, speciation
Introduction
Despite ongoing efforts, the nature of the genomic changes underlying reproductive isolation and ultimately leading to speciation remain elusive (Coyne and Orr, 2004; Schluter, 2009; Presgraves, 2010). On the basis of our current comprehension of post-zygotic isolation, hybrid sterility and/or inviability arises as a consequence of incompatible allele combinations in hybrids (Dobzhansky–Muller incompatibilities) that have diverged between pure species or populations (Dobzhansky, 1937; Muller, 1942). These alleles, whether neutral or positively selected within their own genetic background interact negatively when brought together in inter-specific hybrid genomes. The break-up of co-adapted gene complexes will therefore generate mosaic chromosomes composed of the two diverging genomes and is expected to be more deleterious in later generation hybrids compared with F1 generations (that is, hybrid breakdown; Rieseberg et al, 1999; Coyne and Orr, 2004; Burton et al., 2006). It is believed that the most significant point in the speciation process is the initial development of reproductive isolation (Dettman et al., 2007; Via, 2009). By studying diverging lineages, which are only partially reproductively isolated, early barriers to gene flow and their underlying genetic basis can be identified before they become confounded with other differences that accumulate after speciation. Yet, studies of the effect of genomic incompatibilities on gene regulation have focused nearly exclusively on retrospective analyses of reproductive isolation between highly divergent species in which gene flow does not occur in natural conditions (Michalak and Noor, 2003; Ranz et al., 2004; Noor and Feder, 2006; Landry et al., 2007; Rottscheidt and Harr, 2007).
At the gene transcription level, genomic incompatibilities can lead to the disruption of the transcriptional machinery and the appearance of novel, unforeseen patterns of gene expression in hybrid lineages (Landry et al., 2007). This is often hypothesized to be a major factor underlying hybrid breakdown. Analyses of gene expression profiles using microarrays have shown that the combination of divergent regulatory elements within a common genetic background often results in the disruption of gene expression (reviewed by Landry et al., 2007; Ortiz-Barrientos et al., 2007). For example, the global expression profile of Drosophila melanogaster and Drosophila simulans are more closely related to each other than to their hybrid progeny (Ranz et al., 2004). Similarly, in sympatric but ecologically divergent populations of brook charr (Salvelinus fontinalis), dramatic breakdown of gene expression patterns in hybrids compared with their parental relatives was observed (Mavarez et al., 2009).
Lake whitefish species pairs represent excellent model species to study the early onset of reproductive isolation and its effect on genomic divergence (Lu and Bernatchez, 1998; Rogers and Bernatchez, 2007; Bernatchez et al., 2010). Following the last glacier retreat (<15 000 years ago), secondary contact of lake whitefish (Coregonus clupeaformis) evolutionary lineages isolated during the Pleistocene (100 000–200 000 years ago) has led to the parallel evolution of two morphologically and ecologically divergent sympatric species in several lakes of northeastern North America: benthic normal and limnetic dwarf whitefish (Bernatchez 2004). Furthermore, the ecological specificity of each lake seems to be the main driving isolation factor because, in certain lakes, secondary contact of these same lineages resulted in a hybrid swarm (Lu et al., 2001; Landry et al., 2007). As expected from a recent divergence event, the overall level of genetic differentiation between species pairs is relatively low and hybrids are still present at low frequency in natural populations (Falush et al., 2007). Moreover, early in ontogeny, normal and dwarf fish are phenotypically and ecologically indistinguishable (Chouinard and Bernatchez, 1998; Bernatchez, 2004). At the same time, hybrid whitefish experience striking fitness consequences of genomic incompatibilities and ongoing reproductive isolation (increased mortality, disruption of hatching time and reduced sperm performance, Rogers and Bernatchez, 2006; Whiteley et al., 2009; Bernatchez et al., 2010).
On the basis of this knowledge, we followed the ontogeny of pure dwarf and normal whitefish as well as their hybrids to identify the precise moment at which embryos started to show early signs of hybrid breakdown. By quantifying genome-wide levels of gene expression when developmental defects were the most extreme, we could identify transcriptome changes potentially associated with post-zygotic isolation. Most specifically, we tracked patterns of regulation for genes known to be essential for early fish development (Amsterdam et al., 2004). We predicted that these essential genes would be the most affected in hybrids because their expression is especially critical and has a severe cascading effect on embryonic development and fitness in general.
Materials and methods
Strains, crosses and fish maintenance
Eggs were obtained from outbred laboratory strains (normal whitefish originating from Aylmer Lake (45° 54′N, 71° 20′W), dwarf whitefish originating from Témiscouata Lake (47° 41′N, 68° 47′W), as detailed in Lu and Bernatchez, 1998) at the Laboratoire de Recherche en Sciences Aquatiques (LARSA, Université Laval, Quebec, Canada). We created half-sib families as follows. The dwarf group was created using one lab strain dwarf female crossed to five different dwarf males. The normal whitefish group was created by crossing one lab strain normal female to three separates normal males. Finally, an independent group of backcross individuals was obtained using an F1-hybrid female generated in the laboratory in a previous study (Rogers and Bernatchez, 2007) crossed to five normal males. As such, the backcross individuals are composed of a 75% normal and 25% dwarf genetic background. Fish were anesthetized with 0.001% eugenol solution whereupon eggs and semen were stripped and immediately fertilized in vitro. Fertilized eggs were disinfected (0.0001% iodine solution) and incubated on submerged grids in the same flow-through system (4.5–5.5 °C). To avoid contamination by fungi, dead eggs were removed on a daily basis and all samples were treated weekly with malachite green oxalate.
Sampling
We sampled embryos approximately 60 days post-fertilization (between 280 and 288 degree-days) in normal, dwarf and backcross families. Individually chosen eggs were preserved in RNA later (Ambion, Austin, TX, USA) and frozen at −20 °C for storage. Embryos sampled in the backcross family were separated into normally healthy looking ones, hereafter referred as backcross-healthy, and ones that showed developmental problems, hereafter referred as backcross-moribund (see results for the description of the developmental phenotype of each group). To avoid sampling dead eggs in which RNA degradation would affect gene expression measurements, 40 independent moribund embryos were observed daily to confirm that all were still alive at least several days following sampling (yolk sac of dead embryos changes from translucent to opaque). At the same time, these 40 moribund embryos as well as nearly all eggs classified as backcross-moribund died in the following weeks, before or just after hatching (at ∼100 days).
Experimental design
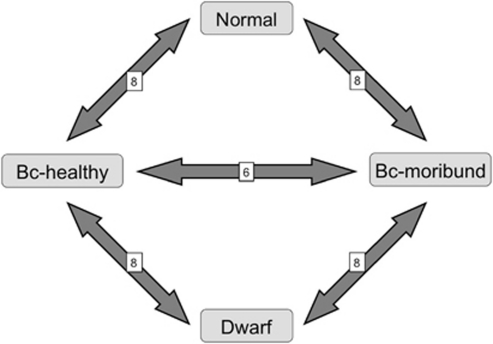
Gene expression analysis was performed using the 16 000 (v2.0) Salmon complementary DNA microarray provided by the cGRASP (consortium for Genomic Research on All Salmon Project, von Schalburg et al., 2005). More than 175 complementary DNA libraries constructed from a wide variety of tissues and different developmental stages (Atlantic salmon and rainbow trout) were used to produce the array. Furthermore, the validity of this array has been amply demonstrated in lake whitefish, whereas hybridization signal is the same as for Atlantic salmon (von Schalburg et al., 2005). Sequence divergence between normal and dwarf is also very low and therefore did not affect hybridization kinetics (Derome et al., 2006; Whiteley et al., 2008). Samples of dwarf, normal, backcross-healthy and backcross-moribund were hybridized in a loop design, involving eight biological replicates for the backcross-healthy and backcross-moribund comparison and six for the others. Dye swap was performed between each replicate. As a result, we obtained a final set of 32 microarray slides (Figure 1).
Figure 1.
Microarray design with the four experimental groups. Each double-headed arrow represents one microarray slide. Numbers written on arrows refer to the number of comparisons done between each group for a total of 32 comparison and 28 separate biological samples.
Total RNA was extracted using the Trizol Reagent protocol (Invitrogen, Carlsbad, CA, USA). A pool of five whole embryos preserved in RNA later was homogenized using a bead mill (Qiagen, Germantown, MD, USA) for each sample hybridized on the array. RNA pooling is a common practice when quantity of material is limiting and inference on gene expression patterns for most genes is not affected by this (Kendziorski et al., 2005). Crude total RNA was treated with DNase I for 15 min at room temperature (1 unit/ug, Invitrogen) and further cleaned by ultra filtration using microcon YM30 spin columns (Millipore, Billerica, MA, USA). Total RNA was eluted in pure water supplemented with Superase-In RNase Inhibitor (Ambion), quantified and quality checked using Experion RNA StdSens Analysis Kit (Bio-Rad, Hercules, CA, USA), then stored at −80 °C. Reverse transcription reactions were performed using Superscript II Kit (Invitrogen). Following this, Genisphere (Hatfield, PA, USA) 50Array Expression Array Detection Kit (Cy3/cy5) was used following the vendor's protocol (∼20 ug of complementary DNA material hybridized). Microarrays were scanned using a ScanArray Express scanner (Packard Bioscience, Wellesley, MA, USA) and spots were located and quantified using the histogram method in QuantArray 3.0 (Perkin Elmer, Waltham, USA). Gene expression data were deposited at Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, series accession GSE23095).
Data analysis
Unless stated otherwise, all analyses were performed in R (v.2.9.0. The R Foundation for Statistical Computing, 2009, 3-900051-07-0). First, local background was subtracted from spots. Only spots above the mean of empty (blank) spots plus 2 s.d., in at least one channel, were kept for further analysis. Missing data were imputed using the K-nearest neighbors (20 neighbors) and data were log2 transformed and normalized using Lowess algorithm. Data were fitted into a mixed model analysis of variance (R/maanova package v1.14, Kerr et al., 2000) to identify differential expression. The mixed model included the following terms as fixed sources of variance: group (dwarf, normal, backcross-healthy and backcross-moribund) and dye (two fluorescent dyes). Sample (28 biological samples) and array (32 individual microarrays) were included as random sources of variance. Finally, a surrogate variable analysis (Leek and Storey, 2007) enabled the identification of an eigengene explaining 68% of the residual variance and corresponding to a strong block effect (eight slides of the backcross-healthy/backcross-moribund comparisons done in August 2007, the rest of the slides done in June 2008). As a result, this block effect was also included in the mixed model as a random term. A permutation-based F-test was performed to test for statistically significant divergence in gene expression and restricted maximum likelihood was used to solve the mixed model equations (P-value <0.05, 1000 permutations, Fs-test). Following this, P-values were corrected for multiple hypotheses testing using q-value correction (q-value <0.05, Storey, 2002). To test for specific pairwise differences between groups (N, D, BC-healthy, BC-moribund), permutation-based t-tests between the four groups for all genes identified as differentially expressed in the analysis of variance (q-value <0.05, 1000 permutations, Fs-test) were performed. Finally, fold changes for the group effect (log2) were also extracted from the analysis for all significant transcripts. In order to remain conservative in interpreting the number of significant features, transcripts with <1% sequence divergence over 95% of the sequence printed on the array were also compressed into a single gene.
Best linear unbiased estimates of the dye, array and block effects (that is, technical variation) were subtracted from the normalized fitted values of gene expression data for each gene. Following this, each gene expression value was divided by the mean of its channel such that values should only represent the group and sample effects (that is, biological variation). Finally, as the same biological sample is sometimes represented on several arrays, we calculated mean gene expression for each of the 28 different samples used in the study. These values were then used to perform a hierarchical clustering analysis to construct a heatmap, with pairwise gene and sample distance matrices estimated from Pearson correlation coefficients.
Functional classification and assessment of significant differential representation of functional classes were performed using DAVID bioinformatics resources (v6.7, Huang et al., 2009). First, Unigen clusters were obtained from cGRASP for all annotated transcripts printed on the microarray. Then, overrepresented gene ontology (GO) classes (molecular function or biological process) among differentially expressed genes were identified (modified Fisher's test (Ease score) corrected for multiple hypothesis testing (Benjamini correction) <0.05). Finally, previously published data (Amsterdam et al., 2004) were used to identify patterns of gene expression for essential embryonic development genes in fish. Briefly, Amsterdam et al. (2004) identified a suite of 315 genes in early embryonic development in which knockdown mutations are lethal in zebrafish (Danio rerio). Furthermore, their study also confirmed that the propensity of being an essential gene is highly conserved throughout evolution. We then used BLASTn algorithm (Altschul et al., 1997) to match these essential genes to our own sequences printed on the array (e-value <1e-15).
Results
Development
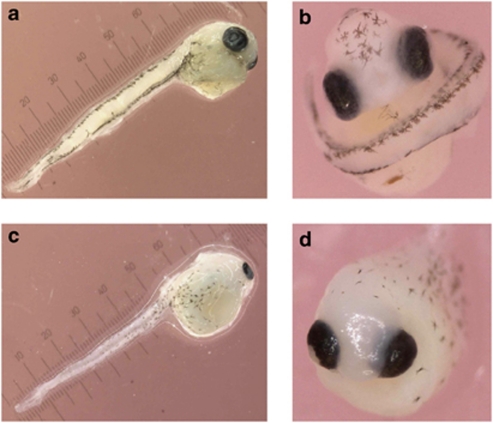
Approximately 60 days post-fertilization (between 280 and 288 degree-days) embryos were sampled in normal, dwarf and backcross families. At this stage, embryos have developed into the pharyngula period, which corresponds to the phylotypic stage (cf. Danio rerio developmental staging in Slack et al., 1993; Kimmel et al., 1995). Nearly all normal and dwarf embryos showed the same developmental phenotype, and only six embryos out of 104 (5.8%) assessed for developmental characteristics were abnormally developing in these pure crosses. It is noteworthy that these six embryos had major developmental defects that were substantially different (no axial body plan, ‘cyclops') from the backcross-moribund group described below. Normally developing pure and backcross embryos were phenotypically similar and characterized by: differentiated pigment cells, a beating heart pumping blood throughout the circulatory system, eyes pigmented and a fully mobile tail, detached from the yolk sac (Figures 2a and b). In the backcross family, a significant proportion of embryos (33%, 52 out of 156 embryos visually inspected (Fisher's exact test, P-value<0.0001)) showed either evident lag in development or atypical phenotype (small head, small eyes, deformed eye lens, heart not beating, reduced cell pigmentation, deformities of the tail) and were hereafter referred to as backcross-moribund embryos (Figures 2c and d). Thus, while not all backcross-moribund possessed exactly the same phenotype, they undoubtedly represented a discrete phenotype different from all other pure and healthy backcross embryos.
Figure 2.
(a, b) General developmental characteristics of normal, dwarf and backcross-healthy embryos (differentiated pigment cells, beating heart pumping blood throughout the circulatory system, eyes pigmented, eye lens completely formed and a fully mobile tail, detached from the yolk sac). (c, d) General phenotype of backcross-moribund embryos (small head, small eyes, deformed eye lens, reduced cell pigmentation, deformities of the tail). Note that egg chorion was removed (a, d) and embryos were manually unfolded with dissecting needles in a and c. Units on scale are 0.1 mm.
Gene expression regulation
Very little difference in gene expression was found between normal and dwarf whitefish, with only two transcripts differentially expressed (60s ribosomal protein L23a, hemoglobin subunit beta) after correction for multiple hypotheses testing and replicate spotting on the array, 0.1% of all transcripts expressed (false discovery rate q-value <0.05). This was in stark contrast with backcross-healthy hybrids for which 162 and 77 (3.0 and 1.5%) genes were differentially expressed compared with normal and dwarf, respectively. Even more strikingly, 2214 (39%) transcripts were differentially expressed between backcross-healthy and backcross-moribund, 1993 (35%) between normal and backcross-moribund and 1964 (35%) between dwarf and backcross-moribund (Figure 3, Supplementary Table 1).
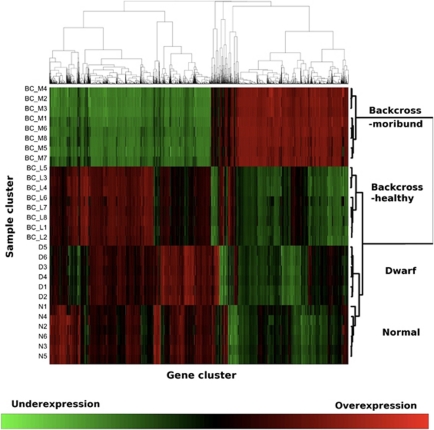
Figure 3.
Heatmap representing hierarchical clustering based on gene distance matrix (all genes differentially expressed between groups as identified by analysis of variance (ANOVA, q-value <0.05)) and sample distance matrix (28 biological samples).
Functional classification
Functional analyses using DAVID bioinformatic resources revealed that several biological processes and molecular functions were overrepresented among the lists of differentially expressed genes. First, translation and generation of precursor metabolites and energy GO terms were significantly overrepresented in all comparisons involving backcross-moribund individuals (Table 1). Comparatively, glycolysis, alcohol and carbohydrate catabolic processes were overrepresented in the list of genes differentially expressed between backcross-healthy and dwarf, while no GO terms were overrepresented in the backcross-healthy/normal or normal/dwarf categories. Transcripts differentially expressed in backcross-moribund were further divided into two categories: transcripts underexpressed and overexpressed compared with the average of all groups. In overexpressed genes, a total of 15 different GO terms, which were further grouped into 7 general categories, were overrepresented. These were mostly related to transport and energy metabolism functions. In underexpressed genes, 11 different GO terms, which were further grouped into six general categories, were overrepresented. These were related to completely distinct functional categories; mostly macromolecule metabolism and regulation of mRNA translation.
Table 1. Significant over-representation of GO categories (BP and MF) among genes, which showed different transcription levels between all genes expressed on the microarray and the different experimental groups.
| Comparison | Category | Descriptiona | Foldb | Count (%)c | P-valued | Number of GO termsa |
|---|---|---|---|---|---|---|
| Backcross-moribund/backcross-healthy | BP | Generation of precursor metabolites and energy | 1.74 | 67 (10.2%) | 0.000 | 1 |
| BP | Translational elongation | 1.91 | 28 (4.2%) | 0.066 | 1 | |
| MF | Structural molecule activity | 1.49 | 65 (9.8%) | 0.022 | 1 | |
| MF | Hydrogen ion transporter activity | 1.86 | 28 (4.2%) | 0.047 | 1 | |
| Backcross-moribund/dwarf | BP | Generation of precursor metabolites and energy | 1.72 | 60 (10.1%) | 0.000 | 1 |
| BP | Translation | 1.84 | 70 (11.7%) | 0.022 | 2 | |
| MF | Structural molecule activity | 1.58 | 62 (10.4%) | 0.006 | 1 | |
| Backcross-moribund/normal | BP | Generation of precursor metabolites and energy | 1.78 | 63 (10.3%) | 0.000 | 1 |
| BP | Translational elongation | 2.07 | 28 (4.6%) | 0.010 | 1 | |
| MF | Cytoskeletal protein binding | 1.75 | 38 (6.2%) | 0.021 | 1 | |
| Backcross-healthy/dwarf | BP | Glycolysis | 21.08 | 6 (22.2%) | 0.001 | 1 |
| BP | Alcohol catabolic process | 15.17 | 6 (22.2%) | 0.009 | 1 | |
| BP | Carbohydrate catabolic process | 14.59 | 6 (22.2%) | 0.006 | 4 | |
| Backcross-healthy/normal | — | |||||
| Dwarf/normal | — | |||||
| All genes differentially expressed and backcross-moribund overexpressed genes | BP | Generation of precursor metabolites and energy | 2.09 | 62 (18.5%) | 0.00 | 1 |
| BP | Oxidation reduction | 1.76 | 50 (14.9%) | 0.00 | 1 | |
| BP | Electron transport chain | 2.12 | 30 (8.9%) | 0.00 | 1 | |
| BP | Phosphorylation | 1.82 | 29 (8.6%) | 0.05 | 1 | |
| MF | Catalytic activity | 1.24 | 148 (44%) | 0.00 | ||
| MF | Magnesium ion binding | 2.05 | 21 (6.3%) | 0.01 | ||
| MF | Ion transporter activity | 1.80 | 28 (8.2%) | 0.02 | 9 | |
| All genes differentially expressed and | BP | Translation | 1.41 | 69 (15.4%) | 0.001 | 1 |
| backcross-moribund underexpressed genes | BP | Gene expression | 1.29 | 136 (30.3%) | 0.000 | 1 |
| BP | Macromolecule metabolic process | 1.26 | 182 (40.4%) | 0.00 | 4 | |
| BP | RNA metabolic process | 1.45 | 48 (10.7%) | 0.033 | 2 | |
| MF | Structural constituent of ribosome | 1.48 | 8.9 (40%) | 0.029 | 1 | |
| MF | Nucleic acid binding | 1.32 | 86 (19.2%) | 0.016 | 2 | |
| All genes differentially expressed and | BP | Translation | 3.91 | 22 (48.9%) | 0.000 | 1 |
| essential genes differentially expressed | BP | Gene expression | 2.64 | 32 (71.1%) | 0.000 | 1 |
| BP | Macromolecule biosynthetic process | 2.08 | 30 (66.7%) | 0.002 | 6 | |
| BP | Protein metabolic process | 1.97 | 32 (71.9%) | 0.001 | 3 | |
| MF | Structural constituent of ribosome | 5.20 | 16 (35.6%) | 0.000 | 1 | |
| MF | Structural molecule activity | 3.25 | 16 (35.6%) | 0.002 | 1 |
Abbreviations: BP, biological process; GO, gene ontology; MF, molecular function.
Similar GO terms were grouped together as one category using the most inclusive GO term provided (for example, macromolecule metabolic process grouped within macromolecule biosynthetic process). Number of original GO terms in each category was written in number of GO terms column.
Fold enrichment values given by DAVID as compared to the background group (either genes expressed or all genes differentially expressed).
Number of unigenes in GO category and percentage of total.
Note that when more than one GO term were included in the same category, the P-value (Ease score (Bonferroni correction implemented by Huang et al., 2009)) was the mean P-values of all the GO terms.
Of particular interest were 450 (3.2% of all transcripts printed on the array) transcripts, which matched (BLASTn e-value <1e-15) to genes essential for early fish development (Figure 4). Essential genes refer to those genes for which knockdown mutations are embryonic lethal in Danio rerio (Amsterdam et al., 2004). Among these essential transcripts, 336 were expressed and 204 differentially expressed. Both of these values were significantly higher compared with the proportion of all transcripts expressed and differentially expressed, respectively (χ2 test, P-value <0.01). Moreover, 81% (166 transcripts) of these essential transcripts were underexpressed in backcross-moribund compared with the average of the four groups and this was highly significant compared with the proportion of all transcripts underexpressed in these embryos (56% or 1445) (χ2 test, P-value <0.0001, Figure 4). We also identified several GO terms that were overrepresented among the lists of essential genes differentially expressed against all genes differentially expressed. These were related to different functional categories but mostly, macromolecule metabolism and regulation of mRNA translation (Table 1).
Figure 4.
Number of transcripts matching to essential early developmental genes. Note that transcripts were defined as essential if they matched (BLASTn e-value <1e-15) to the list of 315 genes identified as essential early developmental genes in zebrafish (Amsterdam et al., 2004) (χ2 test: *P-value <0.05, ***P-value <0.001).
Finally, in a previous study looking at gene expression differences in 30-day-old whitefish embryos, a suite of seven genes that showed highly transgressive patterns of expression in backcross hybrids and which also comprised three essential developmental genes was identified (Renaut et al., 2009). These genes did not differ in mean expression in 30 days old backcross embryos, but showed exaggerated inter-individual variance extending outside the range of both parents. In this study, 18 transcripts matched to these 7 transgressive genes and all except 1, were underexpressed in both backcross groups compared with the parents. Moreover, this effect was even more pronounced for backcross-moribund (Table 2).
Table 2. Relative gene expression for genes previously identified as highly transgressive in 30 days old backcross whitefish embryos (Renaut et al., 2009).
| Description | Fold change (N) | Fold change (D) | Fold change (Bc-h) | Fold change (Bc-m) | q-Value (Bc-m/Bc-h) | q-Value (Bc-m/D) | q-Value (Bc-m/N) | q-Value (Bc-h/D) | q-Value (Bc-h/N) | q-Value (N/D) |
|---|---|---|---|---|---|---|---|---|---|---|
| Asialoglycoprotein receptor 2 | 0.00 | 0.03 | 0.22 | 0.21 | NS | ** | ** | NS | * | NS |
| Protein kinase C | 0.00 | −0.23 | −0.27 | −0.23 | NS | NS | ** | NS | * | NS |
| Guanine nucleotide-binding protein | 0.00 | −0.29 | −0.42 | −0.39 | NS | NS | *** | NS | *** | NS |
| 40S ribosomal protein S11 (3 transcripts) | 0.00 | 0.06 | −0.03 | −0.22 | ** | *** | * | NS | NS | NS |
| Heat shock 70 kDa protein (10 transcripts) | 0.00 | −0.21 | −0.30 | −0.42 | NS | NS | *** | NS | NS | NS |
| Elongation factor 1 alpha | 0.00 | −0.36 | −0.39 | −0.56 | ** | ** | *** | NS | ** | NS |
| Fish-egg lectin | 0.00 | −0.07 | −0.27 | −0.95 | *** | *** | *** | NS | NS | NS |
Abbreviations: Bc-h, backcross-healthy; Bc-m, backcross-moribund; D, dwarf; N, normal; NS, nonsignificant.
q-values for the respective t-tests. *q-value <0.05, **q-value <0.01, ***q-value<0.001.
Genes products in bold are also essential developmental genes in fish embryos according to Amsterdam et al. (2004). Note that relative gene expression (log2 values) is expressed relative to the normal group. When several transcripts match to the same gene product, relative gene expression was calculated as the mean of all transcripts matching to that gene product.
Discussion
Our main objective was to quantify genome-wide levels of gene expression when developmental defects were the most extreme and thus potentially associate changes in expression to post-zygotic isolation. Here, we discuss the implications and inherent limitations of these results in light of our understanding of post-zygotic isolation in incipient species of lake whitefish and of the genetics of speciation in general.
A significant proportion of backcross embryos showed strong developmental defects. These developmental problems leading to a mortality rate of at least 33% clearly represent a severe fitness cost of producing hybrids between dwarf and normal whitefish in natural conditions and therefore undeniably act as a strong post-zygotic isolation mechanism. Admittedly, as these observations are drawn from five half-sib backcross families derived from a single hybrid female crossed to five different normal males, they should be interpreted cautiously. However, the fertilization success of those backcross half-sib families eggs was similar to the one observed for pure crosses, as also previously reported (Lu and Bernatchez, 1998; Rogers and Bernatchez, 2006). In addition, the distinct abnormal embryonic development phenotype observed in backcross embryos was not observed in any of the pure families. Moreover, our observations indicating that strong post-zygotic isolation barriers exist between normal and dwarf lake whitefish is corroborated by several lines of evidence previously documented. An elevated mortality rate around the same developmental time (Rogers and Bernatchez, 2006) as well as a reduced sperm performance (Whiteley et al., 2009) has been identified in independent backcross families. Elevated, albeit lower than for the backcross, mortality was also observed in F1 hybrids at the same developmental time (Lu and Bernatchez, 1998). Thus, increased mortality in hybrids has now been observed in three independent studies. Furthermore, strong segregation distortion for over 30% of mapped genetic markers reflected differential survival rates among backcross hybrids genotypes (Rogers and Bernatchez, 2007). For all these reasons, we propose that the developmental and expression differences observed here are direct consequences of hybridization between diverging lineages.
The specifics of hybrid breakdown on gene expression regulation
Using complementary DNA microarrays, we observed very little difference in gene expression between pure normal and dwarf whitefish. This lack of divergence between the transcriptome of the parental forms early in ontogeny is in line with the lack of differentiation in developmental time until emergence (Rogers and Bernatchez, 2006) and also corroborates previous findings whereas 30 days old embryos had 14 times less genes (n=5 genes) displaying significant regulatory divergence than 16 weeks old juvenile fish (Nolte et al., 2009; Renaut et al., 2009). In fact, dwarf and normal larval whitefish are nearly indistinguishable and their phenotypic and ecological divergence is expected to take place at the juvenile stage (Chouinard and Bernatchez, 1998). Embryos sampled here were in the phylotypic developmental stage corresponding to the pharyngula period in Danio rerio development (Kimmel et al., 1995). During this highly conserved stage of development, individuals are expected be more similar to each other than during any earlier or later developmental periods (Slack et al., 1993). This observation has frequently been made even for highly divergent taxa and should also apply for gene expression differences (Irie and Sehara-Fujisawa, 2007).
In stark contrast with parental forms, hybrids showed developmental problems that had far-reaching repercussions on gene expression regulation. Backcross hybrids all had strong divergent patterns of gene expression compared with parental crosses and this was especially true for moribund hybrids. The nature of breakdown in gene regulation observed in whitefish backcross hybrids is more likely a consequence of genomic incompatibilities rather than the mere result of a stochastic RNA degradation process. Genes were affected in a precise way depending on their functionality. This is also confirmed by functional analysis, whereby markedly different GO terms (whether biological process or molecular function) were overrepresented whether a gene was under or over expressed in the backcross-moribund group (Table 1). Therefore, developmental problems seem to be associated with this breakdown in gene regulation in hybrids. Nevertheless, the challenge will remain to establish whether this association is through causality and this remains an unavoidable limit of all gene expression studies of speciation (Noor and Feder, 2006).
As predicted, essential fish developmental genes were the most affected in hybrids. Their expression is especially critical and has a severe cascading effect on embryonic development and fitness in general. These genes, whose essentiality is highly conserved throughout evolution (Amsterdam et al., 2004), were not only more differentially expressed than expected but also strongly underexpressed in backcross compared with the average of the four groups (81% of those are underexpressed in backcross embryos, Figure 4). The functional analysis of GO terms also revealed that similar categories were overrepresented in essential genes differentially expressed as in backcross-moribund underexpressed genes and these were mostly genes involved in macromolecule metabolism and regulation of mRNA translation (Table 1).
Furthermore, the early signs of gene misexpression previously documented in 30 days old embryos (Nolte et al., 2009; Renaut et al., 2009) culminated in dramatic differences in development and gene expression regulation in the present study in 60 days old hybrid embryos, particularly so for genes known to be essential in early embryonic development (Table 2). For example, in zebrafish, knockdown mutation of Heat Shock Cognate 70 leads after 5 days to: pericardial edema, a necrotic head and a general degeneration of the body; and knockdown of elongation factor 1 alpha leads to a small head and eyes, rounder yolk and increased necrosis (Amsterdam et al., 2004). In lake whitefish backcross hybrids, both of these genes were among the most transgressive of all at 30 days (Renaut et al., 2009), were severely underexpressed in the present study (at 60 days, Table 2), and embryos showed similar general phenotype to zebrafish mutants (Figures 1c and d).
A remaining challenge is now to determine how many genomic regions actually contribute to reproductive barriers. One possibility is that this is attributed to the accumulation of conditionally neutral mutations throughout the genome. This is an attractive hypothesis as mutations accumulate throughout the genome at all times in all populations. As such, they provide different architectural starting points, which may subsequently be recruited by adaptive processes and potentially lead to different evolutionary trajectories, whether under stabilizing or divergent selection pressures (Lynch, 2007). Here, as very little difference was observed between normal and dwarf whitefish, post-zygotic reproductive isolation in these young species appears to involve mostly genes under stabilizing selection for gene expression, as it has been previously shown in drosophilids (Haerty and Singh, 2006) and brook charr (Mavarez et al., 2009).
The fact that in whitefish, over 30 % of mapped genetic markers show locus-specific deviations from expected Mendelian segregation (segregation distortion) tends to support the idea that many ‘ordinary loci' are associated with reproductive isolation (Rogers and Bernatchez, 2007). On the other hand, we cannot refute that one or a few genetic loci may underlie large-scale disruption of expression, especially so if those are associated with essential developmental genes, in which knockdown mutations are known to be lethal. Another explanation worth mentioning pertains to the fact that large-scale hybrid gene misexpression can also result from a variety of mechanisms related to the disrupting the chromatin integrity or the expression of non coding RNAs (Michalak, 2009). For example, miRNA networks, which have been shown to regulate gene expression in early development in many vertebrate species, can have a large cascading effect if disrupted and thus provide a likely ‘major gene' effect (Lee et al., 2007; Michalak, 2009). As such, all other differences in gene expression and phenotype would be a downstream effect of this major factor. Other minor incompatibilities could still exist, which would account for the differential expression of genes observed in the healthy backcross fish versus parental types. Along this line, we previously observed that a least one locus associated with embryonic mortality is also linked to a regulation (expression Quantitative Trait Locus, eQTL) hotspot and may harbor a major regulatory gene with strong pleiotropic effect on the expression of numerous genes (Bernatchez et al., 2010), thus representing a possible mechanism explaining the genome-wide disruption of regulation in hybrids.
Conclusion
By comparing healthy, surviving hybrids against moribund ones, our results identified a transcriptome-wide signature of hybrid breakdown in young, incipient species and revealed a persuasive link between misexpression of essential developmental genes and post-zygotic isolation. Our analysis of the genomic basis of ongoing speciation in young evolutionary fish lineages helps to bridge the gap between ecological studies of reproductive isolation with limited knowledge of their genetic basis, and genetic studies of speciation with an incomplete ecological perspective. Quite clearly, both are needed toward a truly general theory of the genetics of speciation.
Acknowledgments
We thank JC Therrien and S Higgins for their assistance in spawning and rearing whitefish, AW Nolte for suggestions on experimental design and analysis, J Jeukens, CR Landry, E Normandeau and J St-Cyr for comments on an earlier version of the paper. This research was funded by a Natural Science and Engineering Research Council of Canada (NSERC) and Canadian Research Chair in Genomics and Conservation of Aquatic Resources to LB and a NSERC postgraduate scholarship to SR. This paper is a contribution to the research program of Québec-Océan.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissean RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez L.2004Ecological theory of adaptive radiation: an empirical assessment from coregonine fishes (Salmoniformes)In: Hendry AP and Stearns SC (eds).Evolution Illuminated: Salmon and their Relatives Oxford Univ. Press: Oxford; 175–207. [Google Scholar]
- Bernatchez L, Renaut S, Whiteley AR, Derome N, Jeukens J, Landry L, et al. On the origin of species: insights from the ecological genomics of whitefish. Philos Trans R Soc Lond B Biol Sci. 2010;365:1783–1800. doi: 10.1098/rstb.2009.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am Nat. 2006;168:S14–S24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- Chouinard A, Bernatchez L. A study of trophic niche partitioning between larval populations of reproductively isolated whitefish (Coregonus sp.) ecotypes. J of Fish Biol. 1998;53:1231–1242. [Google Scholar]
- Coyne JA, Orr HA. Sinauer Associates. Sunderland, MA; 2004. [Google Scholar]
- Derome N, Duchesne P, Bernatchez L. Parallelism in gene transcription among sympatric lake whitefish (Coregonusclupeaformis Mitchill) ecotypes. Mol Ecol. 2006;15:1239–1249. doi: 10.1111/j.1365-294X.2005.02968.x. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Sirfusingh C, Kohn LM, Anderson JB. Incipient speciation by divergent adaptation and antagonistic epistastis in yeast. Nature. 2007;447:585–588. doi: 10.1038/nature05856. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press: New York; 1937. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Singh RS. Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol Biol Evol. 2006;23:1707–1714. doi: 10.1093/molbev/msl033. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocol. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irie N, Sehara-Fujisawa A. The vertebrate phylotypic stage and an early bilaterian-related stage in mouse embryogenesis defined by genomic information. BMC Biol. 2007;5:1. doi: 10.1186/1741-7007-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comp Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SE, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Devel Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Landry CR, Hartl DL, Ranz J. Genome clashes in hybrids: insights from gene expression. Heredity. 2007;99:483–493. doi: 10.1038/sj.hdy.6801045. [DOI] [PubMed] [Google Scholar]
- Lee C-T, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by ‘surrogate variable analysis. PLoS Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Bernatchez L. Experimental evidence for reduced hybrid viability between dwarf and normal ecotypes of Lake Whitefish (Coregonus clupeaformis Mitchill) Proc R Soc Lond Biol Sci. 1998;265:1025–1030. [Google Scholar]
- Lu G, Basley DJ, Bernatchez L. Contrasting patterns of mitochondrial DNA and microsatellite introgressive hybridization between lineages of lake whitefish (Coregonus clupeaformis); relevance for speciation. Mol Ecol. 2001;10:965–985. doi: 10.1046/j.1365-294x.2001.01252.x. [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Mavárez J, Audet C, Bernatchez L. Major disruption of gene expression in hybrids between young sympatric anadromous and resident populations of brook charr (Salvelinusfontinalis Mitchill) J Evol Biol. 2009;22:1708–1720. doi: 10.1111/j.1420-9101.2009.01785.x. [DOI] [PubMed] [Google Scholar]
- Michalak P. Epigenetic, transposon and small RNA determinants of hybrid dysfuntions. Heredity. 2009;102:45–50. doi: 10.1038/hdy.2008.48. [DOI] [PubMed] [Google Scholar]
- Michalak P, Noor MA. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol Biol Evol. 2003;20:1070–1076. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Nolte AW, Renaut S, Bernatchez L. Divergence in gene regulation at young life history stages of whitefish (Coregonus sp.) and the emergence of genomic isolation. BMC Evol Biol. 2009;9:925–936. doi: 10.1186/1471-2148-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Feder JL. Speciation genetics: evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Counterman BA, Noor MAF. Gene expression divergence and the origin of hybrid dysfunctions. Genetica. 2007;29:71–81. doi: 10.1007/s10709-006-0034-1. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Namgyal K, Gibson G, Hartl DL. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 2004;14:373–379. doi: 10.1101/gr.2019804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S, Nolte AW, Bernatchez L. Gene expression divergence and hybrid misexpression between Lake Whitefish species pairs (Coregonus spp. Salmonidae) Mol Biol Evol. 2009;26:925–936. doi: 10.1093/molbev/msp017. [DOI] [PubMed] [Google Scholar]
- Rogers SM, Bernatchez L. The genetic basis of intrinsic and extrinsic post-zygotic reproductive isolation jointly promoting speciation in the lake whitefish species complex (Coregonus clupeaformis) J Evol Biol. 2006;19:1979–1994. doi: 10.1111/j.1420-9101.2006.01150.x. [DOI] [PubMed] [Google Scholar]
- Rogers SM, Bernatchez L. The genetic architecture of ecological speciation and the association with signatures of selection in Natural Lake Whitefish (Coregonus sp. Salmonidae) species Pairs. Mol Biol Evol. 2007;24:1423–1438. doi: 10.1093/molbev/msm066. [DOI] [PubMed] [Google Scholar]
- Rottscheidt R, Harr B. Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics. 2007;177:1553–1567. doi: 10.1534/genetics.107.076190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Slack JMW, Holland PWH, Graham CF. The zootype and the phylotypic stage. Nature. 1993;361:490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
- Via S. Natural selection in action during speciation. Proc Natl Acad Sci USA. 2009;106:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schalburg KR, Rise ML, Cooper GA, Brown GD, Gibbs AR, Nelson CC, et al. Fish and chips: various methodologies demonstrate utility of a 16 006-gene salmonid microarray. BMC Genomics. 2005;15:126. doi: 10.1186/1471-2164-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AR, Derome N, Rogers SM, St-Cyr J, Nolte AW, Renaut S, et al. The phenomics and expression quantitative trait locus mapping of brain transcriptomes regulating adaptive divergence in Lake Whitefish species pairs (Coregonus sp.) Genetics. 2008;180:147–164. doi: 10.1534/genetics.108.089938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AR, Persaud KN, Derome N, Montgomerie R, Bernatchez L. Reduced sperm performance in backcross hybrids whitefish species-pairs (Coregonus sp.) Can J of Zoo. 2009;87:566–572. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.