Abstract
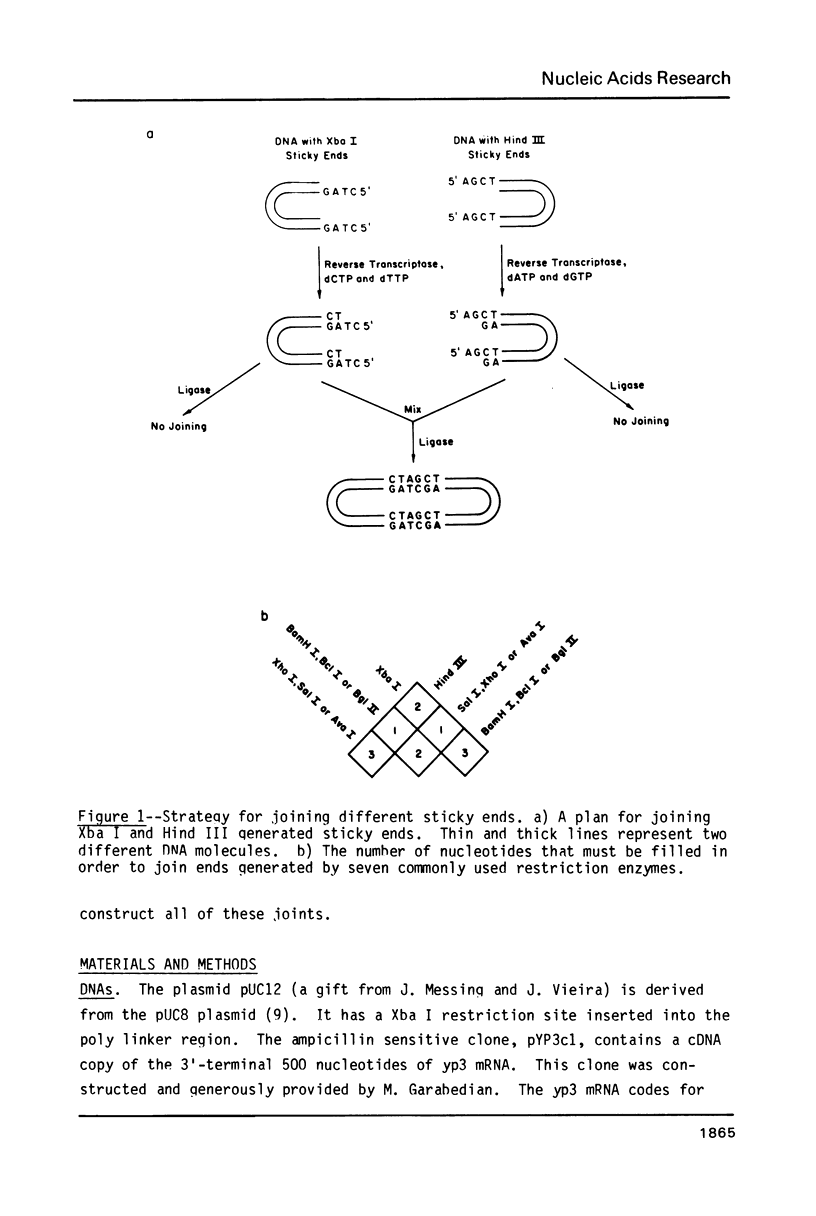
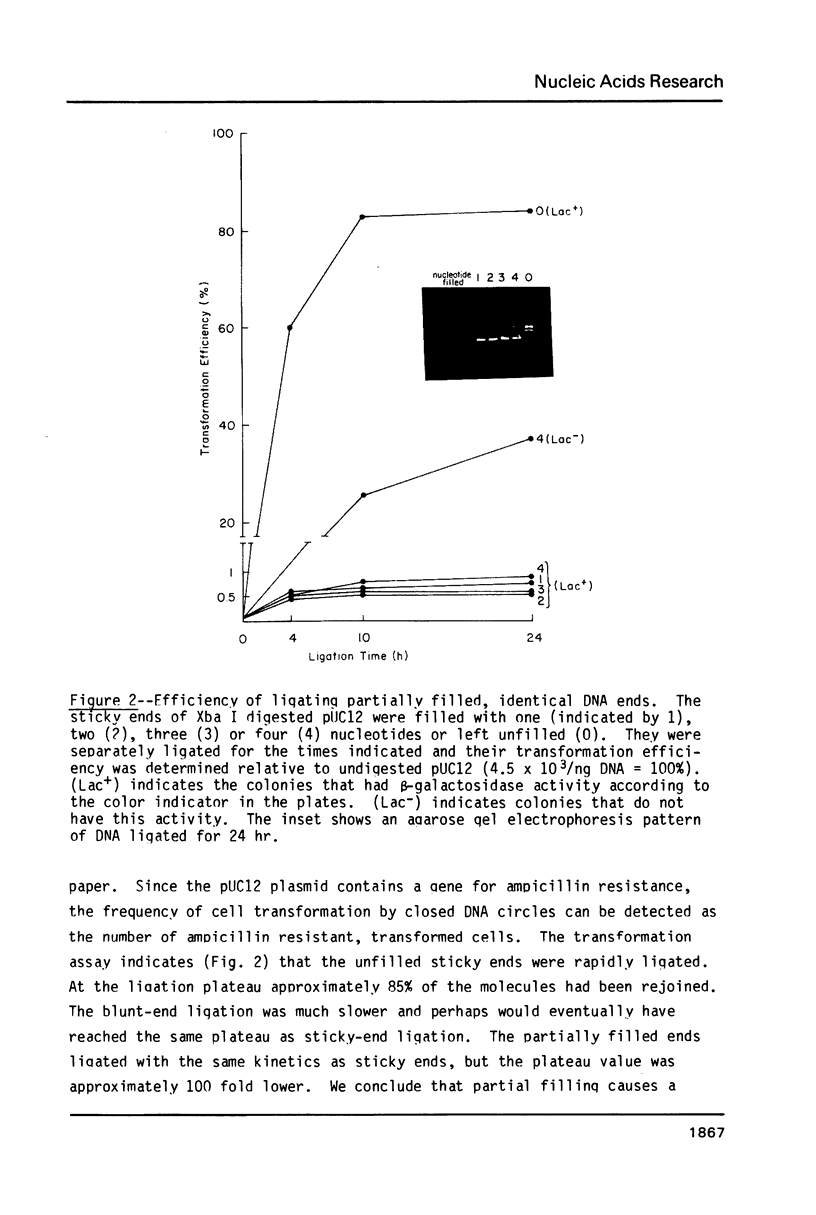
We describe a simple method for joining the 5'-protruding, single-stranded DNA ends generated by restriction enzymes. The method allows ends with different sequences to be joined and prevents identical ends from being joined. This is accomplished by partially filling the single strands in a controlled reverse transcriptase reaction. Partial filling can create new single-stranded ends that can be ligated to different, partially filled ends. In almost all useful cases, partial filling simultaneously eliminates the self-complementarity of identical ends and thus prevents them from being joined by DNA ligase. Although all possible combinations of partially filled ends were not tested, the tests performed indicate that the method is fairly general. We demonstrate that ends of the same length can be ligated with useful efficiency if they are: 1) one nucleotide long and complementary; 2) two nucleotides long and complementary or have a mismatch (dA:dC) at one position; or 3) three nucleotides long and, in our test, have a dT:dC mismatch at the middle position.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett T., Pachl C., Gergen J. P., Wensink P. C. The isolation and characterization of Drosophila yolk protein genes. Cell. 1980 Oct;21(3):729–738. doi: 10.1016/0092-8674(80)90436-5. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- HOGNESS D. S., SIMMONS J. R. BREAKAGE OF LAMBDA-DG DNA: CHEMICAL AND GENETIC CHARACTERIZATION OF EACH ISOLATED HALF-MOLECULE. J Mol Biol. 1964 Aug;9:411–438. doi: 10.1016/s0022-2836(64)80217-5. [DOI] [PubMed] [Google Scholar]
- Heyneker H. L., Shine J., Goodman H. M., Boyer H. W., Rosenberg J., Dickerson R. E., Narang S. A., Itakura K., Lin S., Riggs A. D. Synthetic lac operator DNA is functional in vivo. Nature. 1976 Oct 28;263(5580):748–752. doi: 10.1038/263748a0. [DOI] [PubMed] [Google Scholar]
- Lomant A. J., Fresco J. R. Structural and energetic consequences of noncomplementary base oppositions in nucleic acid helices. Prog Nucleic Acid Res Mol Biol. 1975;15(0):185–218. doi: 10.1016/s0079-6603(08)60120-8. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller R. H., Dickerson R. E., Boyer H. W., Riggs A. D., Itakura K. Chemical synthesis of restriction enzyme recognition sites useful for cloning. Science. 1977 Apr 8;196(4286):177–180. doi: 10.1126/science.847463. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Shine J., Martial J. A., Baxter J. D., Goodman H. M. Nucleotide sequence and amplification in bacteria of structural gene for rat growth hormone. Nature. 1977 Dec 8;270(5637):486–494. doi: 10.1038/270486a0. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Van de Sande J. H., Khorana H. G. Studies on polynucleotides, C. A novel joining reaction catalyzed by the T4-polynucleotide ligase. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1468–1475. doi: 10.1073/pnas.67.3.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]