Abstract
Background
Mitochondrial ferritin is a nuclear encoded iron-storage protein localized in mitochondria. It has anti-oxidant properties related to its ferroxidase activity, and it is able to sequester iron avidly into the organelle. The protein has a tissue-specific pattern of expression and is also highly expressed in sideroblasts of patients affected by hereditary sideroblastic anemia and by refractory anemia with ringed sideroblasts. The present study examined whether mitochondrial ferritin has a role in the pathogenesis of these diseases.
Design and Methods
We analyzed the effect of mitochondrial ferritin over-expression on the JAK2/STAT5 pathway, on iron metabolism and on heme synthesis in erythroleukemic cell lines. Furthermore its effect on apoptosis was evaluated on human erythroid progenitors.
Results
Data revealed that a high level of mitochondrial ferritin reduced reactive oxygen species and Stat5 phosphorylation while promoting mitochondrial iron loading and cytosolic iron starvation. The decline of Stat5 phosphorylation induced a decrease of the level of anti-apoptotic Bcl-xL transcript compared to that in control cells; however, transferrin receptor 1 transcript increased due to the activation of the iron responsive element/iron regulatory protein machinery. Also, high expression of mitochondrial ferritin increased apoptosis, limited heme synthesis and promoted the formation of Perls-positive granules, identified by electron microscopy as iron granules in mitochondria.
Conclusions
Our results provide evidence suggesting that Stat5-dependent transcriptional regulation is displaced by strong cytosolic iron starvation status induced by mitochondrial ferritin. The protein interferes with JAK2/STAT5 pathways and with the mechanism of mitochondrial iron accumulation.
Keywords: mitochondrial ferritin, iron acculumation, hereditary sideroblastic anemia, refractory anemia with ringed sideblasts, ROS, iron starvation, JAK2/STA5
Introduction
Mitochondrial ferritin (FtMt) is an iron-storage protein localized to the mitochondrial matrix.1,2 The human protein is encoded by a nuclear intronless gene that is transcribed into a mRNA similar to H-and L-ferritin mRNA, but it lacks the typical consensus iron-responsive element (IRE) sequence for iron-dependent regulation.3 FtMt has a three-dimensional structure4 and biochemical properties analogous to those of human H-ferritin. FtMt rapidly incorporates and oxidizes iron in vitro5 and its expression strongly affects cellular iron homeostasis as, revealed by several studies of cellular and animal models.6–10 The protein drives iron into mitochondria, induces cytosolic iron starvation and, depending on cellular growth conditions, improves or limits mitochondrial Fe/S enzymatic activities.8 FtMt, through its iron-sequestering capacity, inhibits the development of radical oxygen species (ROS) after treatment with oxidative insults or in enhanced respiratory conditions, suggesting that its primary function is linked to the control of ROS formation.7,8 Accordingly, the expression of FtMt in a frataxin-deficient yeast model,7 in fibroblasts from patients with Freidreich’s ataxia8 and in neuroblastoma cell lines,11 as well as silencing of FtMt in HeLa cells12 showed an important role of the protein in protecting cells from oxidative damage. Studies of mice demonstrated a tight tissue-specific pattern of FtMt expression which confirmed the anti-oxidant role of the protein.13 In fact, FtMt was found to be preferentially expressed in cells characterized by high-energy consumption and is not present in iron storage tissues.13 Moreover, in mutant SOD1 transgenic mouse, a model of amyotrophic lateral sclerosis, an increase of FtMt was observed in the spinal cord, both in motor neurons and astrocytes.14 In humans, the protein is highly expressed in spermatozoa,15 in neurons16 and, at low levels, in other tissues, such as the kidney, and thymus.15 It was also found to be associated with pathological conditions. FtMt is highly expressed in sideroblasts of patients affected by inherited X-linked sideroblastic anemia (XLSA) (OMIM #300751) and by refractory anemia with ringed sideroblasts (RARS), in whom it was demonstrated to be a specific marker of this subtype of myelodysplastic syndrome.17,18 In RARS, the protein is expressed at a very early stage of erythroblast differentiation, its level continuously increases during differentiation and it is apparently linked to cytochrome c release,19 suggesting that it may be involved in the development of ineffective erythropoiesis. Recently FTMT−/− mice were obtained and subjected to vitamin B6 (pyridoxine) deprivation to induce sideroblast/siderocyte formation.20 The authors did not observe significant defects in FTMT−/− mice; however, neither wild-type nor mutant mice showed bone marrow ringed sideroblasts, confirming the limitations of using the mouse as a model for sideroblastic anemia.20 Further investigations are, therefore, needed to establish whether FtMt has an effect on the pathogenesis of these diseases.
FtMt has the property of limiting ROS production and this may have an effect on the signal transduction pathways in which ROS have been described as second messengers such as the JAK/STAT pathways.21–23 In particular, erythropoiesis depends on signal transduction through the erythropoietin receptor (EpoR)-Janus kinase 2 (Jak2)-signal transducer and activator of transcription 5 (Stat5) axis.24,25 Analysis of Stat5−/− mice established a link between EpoR/JAK/STAT signaling and iron metabolism. Stat5−/−mice suffered from microcytic hypochromic anemia and showed reduced transcription of TfR1 mRNA and iron regulatory protein 2 (IRP2), the major translational regulator of transferrin receptor 1 (TfR1) mRNA stability in erythroid cells, indicating that both genes were transcriptional targets of Stat5.26 Stat5 also controlled iron regulatory protein 1 (IRP1) transcription in nitric oxide-treated cells;27 alteration of Stat5 activity may, therefore, have critical consequences on cellular iron homeostasis.
The aim of the present study was to clarify whether the expression of FtMt has an effect on the regulation of erythropoiesis.
Design and Methods
Cell cultures
The human erythroleukemic (K562) and murine erythroleukemia (MEL) cell lines and CD34+ bone marrow cells were separated, grown and treated following standard procedures. Details are provided in the Online Supplementary Design and Methods. Informed consent was obtained from the subjects and the study followed the guidelines of the research ethical committee of the IRCCS Policlinico San Matteo in Pavia.
Plasmid and viral constructs
The pcDNA3-FtMt plasmid, encoding the entire FtMt protein precursor, has been described previously.6 The FtMtΔfeox mutant (E62K, H65G, H-chain numbering, previously described as FtMt222) with inactivated ferroxidase activity was described by Corsi et al.6 The FtMt and FtMtΔfeox coding regions were subcloned under the control of the ubiquitous PGK promoter into the pCCL.sin.cPPT.eGFP.WPRE vector28 using BglIII and XbaI restriction sites to generate the lentiviral expression constructs pPGKMT and pPGKMTΔfeox, respectively.
Transient transfections, lentiviral vector stock preparation and K562 cell transduction
MEL cells were transiently transfected using Lipofectamine 2000 (Invitrogen, San Giuliano Milanese, Italy) following the procedure recommended by the manufacturer. As previously described, lentiviral vectors pPGKMT (LV-FtMt), pPGKMTΔfeox (LV-FtMtΔfeox) and the control pPGKGFP (LV-GFP) were produced by transient co-transfection with four plasmids in 293T cells.28,29 Different vector dilutions were used to transduce target cells K562 and CD34+ cells. After 48 h, green fluorescent protein (GFP)-fluorescent cells were visualized using a fluorescence microscope to test transduction efficiency. Cells transduced with the virus dilution that showed the best result for GFP were tested for ferritins expression.
Preparation of total RNA, reverse transcription, and quantitative real-time polymerase chain reaction
Total RNA was extracted from cells with the RNeasy kit (Qiagen, Milano, Italy). Next, 1 μg of total RNA was incubated with 1 U DNaseI (Invitrogen), which was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Monza, Italy). Samples without reverse transcriptase were included as negative controls. Primers were designed to specifically amplify human Bcl-xL (F 5′-ATTGTGGCCTTTTTCTCCTT-3′, R 5′-GTTCCA-CAAAAGTACCCAG-3′) and transferrin receptor 1 (TfR1) (F 5′-GCAACAGTTACTGGTAAACTG-3′, R 5′-AGCATTTGCAAC-CTTTTCTG3′) cDNA. Real-time polymerase chain reaction (PCR) was performed using the SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7900HT fast-real time PCR system. The mRNA/cDNA abundance of each gene was calculated relative to the expression of a housekeeping gene, glyceralde-hyde-3-phosphate-dehydrogenase (GAPDH). For IRP1 (Hs00158095_m1) and IRP2 (Hs00386293_m1), TaqMan gene expression assays (Applied Biosystems) were utilized, and ACTB (Hs99999903_m1) was considered as the housekeeping gene.
Western blot analysis
For each sample, 25 μg of soluble protein were loaded onto 10% or 12% gels for sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS–PAGE), separated and then transferred to nitro-cellulose membranes. After transfer, the filters were blocked and then incubated with primary antibodies. Anti β-actin antibody was purchased from Sigma, anti-TfR1 antibody from Zymed (San Francisco, CA, USA), anti-PARP from Promega (Milan, Italy), anti-Stat5 from BD Biosciences (Buccinasco, Italy), anti-phospho-Stat5 from Upstate (Temecula, CA, USA), and anti-IRP2 from Santa Cruz Biotechnology (Heidelberg, Germany). Monoclonal anti-ferritin, polyclonal anti-mitochondrial ferritin and polyclonal anti-IRP1 antibodies were produced in our laboratory.8,30,31 Primary antibodies were revealed with horseradish peroxidase-labeled secondary antibodies (Sigma) and a chemiluminescence kit (ECL) (GE Healthcare, Milan, Italy).
Iron proteins and iron content analysis
Methods for quantification of ferritin, the electromobility shift assay, cellular 55Fe incorporation, heme and total protein content are detailed in the Online Supplementary Design and Methods.
Detection of reactive oxygen species
Cells (1×106) were incubated with 30 μM dihydrorhodamine 123 (DHR 123) (Molecular Probes, Invitrogen) in Hanks’ balanced saline solution supplemented with 10 mM glucose for 15 min at 37°C. After washing cells were seeded onto 96-well plates, then fluorescence was read on a Victor3 Multilabel Counter (Wallac, Perkin Elmer) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm in the basal condition or after incubation of cells with 0.3 mM H2O2 for 30 min.
Fluorescence analysis and immunocytochemistry
Cells (5×105) were fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton x-100. To visualize nuclei, the fixed cells were stained with 1.5 μg/mL of 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) for 2 min. To reveal the presence of mitochondrial ferritin, they were incubated with polyclonal anti-FtMt (diluted 1:500) in phosphate-buffered saline, 1% bovine serum albumin followed by anti-IgG TRITC-labeled antibodies (DAKO, Milano, Italy) or by anti-IgG alkaline phosphatase-labeled antibodies (Sigma). May Grünwald Giemsa staining was performed on cytospins prepared at various time-points. Apoptosis was measured by means of the nuclear DNA fragmentation assay, using a terminal-deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) technique. An alkaline phosphatase anti-fluorescein antibody was then used. The “In situ Cell Death Detection Kit” (Roche Diagnostics GmbH, Mannheim, Germany) was used according to the manufacturer’s instructions. The apoptotic index was expressed as the percentage of positive nuclei for at least 500 counted cells at a magnification of 1000x. For simultaneous detection, within the same cell, of FtMt accumulation and apoptosis, the “EnVision G/2 Doublestain System, Rabbit/Mouse Kit” (Dakopatts, Glostrup, Denmark) was used according to the manufacturer’s instructions.
Perls’ staining, electron microscopy and electron spectroscopic imaging/electron energy loss spectroscopy
Cells were grown for 6 days in the presence of 20 mM ferric ammonium citrate (FAC) or 20 mM iron transferrin (FeTf) and then stained for iron content with Perls’ Prussian blue reaction by incubation for 30 min in 1% potassium ferrocyanide, 1% hydrochloric acid in distilled water. To perform electron microscopy K562 cells and clones were fixed for 15 min at 4 °C with 4% paraformaldehyde and 2.5% glutaraldehyde in 125 mM cacodylate buffer and centrifuged at high speed. The pellet was post-fixed (1 h) with 2% OsO4 in 125 mM cacodylate buffer, washed, dehydrated and embedded in Epon. Conventional thin sections were collected on uncoated grids, stained with uranyl and lead citrate and examined in a Leo912 electron microscope (Zeiss, Arese, Italy). Electron spectroscopic imaging/electron energy loss spectroscopy (ESI/EELS) analyses were performed on the same electron microscope. Briefly, the patterns of net iron distribution were obtained by computer-assisted processing of two images collected below (651 and 683 eV) and one beyond the Fe-L3 absorption edge at 719 eV. The final iron map (coded in pseudocolors) was then superimposed on the ultrastructural organization of the same field obtained at 250 eV (i.e., at an energy loss at which most of the elements contribute to the image).
Statistical analyses
Data are reported as mean values ± standard deviation or as representative of at least three independent experiments. The data for ROS, heme content, ferritin quantification by enzyme-linked immunosorbent assay (ELISA), Bcl-xL-RT, TfR1-RT, IRP1-RT and IRP2-RT assays were analyzed using Student’s t test, which was considered statistically significant when the P value was less than 0.05.
Results
Production and characterization of K562 stable clones expressing mitochrondrial ferritin
To verify the hypothesis that the ROS-limiting properties of FtMt may affect the JAK2/STAT5 pathway, we took advantage of the constitutive phosphorylation of Stat5 present in the K562 cell line. We developed K562 stable clones, expressing FtMt and its mutant lacking ferroxidase activity (FtMtΔfeox, previously described as FtMt222). It was previously shown that the expression of FtMtΔfeox in cells does not alter cellular iron homeostasis.6 We, therefore, used it to distinguish between the effects due to the iron-chelation capacity of FtMt from those due to the presence of the protein itself. As a negative control, we used GFP, an unrelated protein. We produced lentiviral vectors carrying cDNA of human mitochondrial ferritins and GFP downstream of the human phosphoglycerate kinase (PGK) promoter. Varying the amount of viral particles, we obtained a transduction efficiency of more than 90%, with cells expressing different amounts of FtMt and FtMtΔfeox. The homogeneity of exogenous protein expression in the cell population was obtained by cloning the transduced cells by sequential dilution. By specific ELISA we selected three clones with different amounts of FtMt expression: clone Mt1=6.2 (±4.1), clone Mt4=58.2 (±28.3), and clone Mt8=334 (±114) ng of FtMt per mg of total proteins. Furthermore, we selected two FtMtΔfeox-expressing clones: clone MtΔfeox5=165 (± 47.1) and clone MtΔfeox7=201 (±60.0) ng of FtMtΔfeox per mg of total proteins. The stabilized clones were grown and analyzed by immunocytochemistry with a polyclonal anti-FtMt antibody (Figure 1A). The results showed homogeneity of stain intensity in all cells with typical mitochondrial dotted staining and a proportional increase of FtMt expression in the three different clones (Figure 1A) and similarly for the two FtMtΔfeox clones (data not shown). The FtMt, FtMtΔfeox clones and two controls, K562 and K562-GFP, were grown for 18 h in the presence of 1 μM 55Fe(III)-citrate and 10 μM ascorbic acid. Cells were then collected and lysed to obtain soluble proteins. The mitochondrial and cytosolic ferritins were separated on native PAGE, and their iron content was revealed by autoradiography (Figure 1B). As expected, 55Fe was associated only with the cytosolic ferritin band in FtMtΔfeox clones, GFP and K562 cells, while radiolabeled iron distribution between cytosolic and mitochondrial ferritins varied in the three FtMt clones. The low amount of FtMt in the Mt1 clone induced only a slight allocation of iron inside mitochondria. In Mt4, the amount of iron was equally shared between the two proteins; however, in Mt8, the high amount of FtMt abolished the cytosolic ferritin iron band, indicating cytosolic iron starvation.
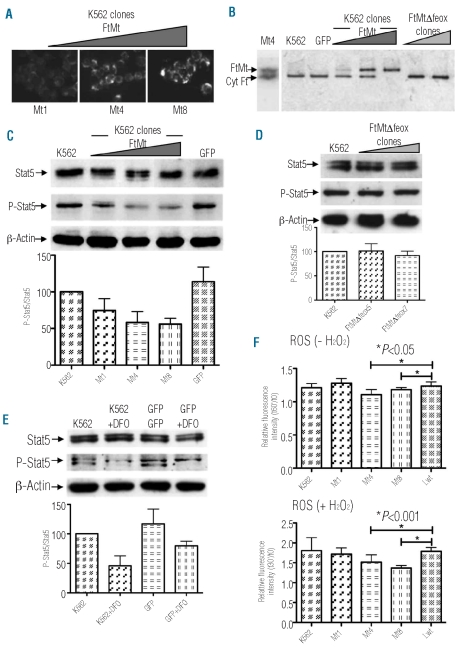
Figure 1.
Effect of FtMt expression on K562 clones. (A) Immunostaining of the three selected clones (Mt1, Mt4 and Mt8) with the antibody specific for human FtMt. (B) Control and transduced K562 cells were grown in the presence of 1 μM 55Fe(III)-citrate and 10 μM ascorbic acid for 18 h. Soluble lysates were separated on 7% non-denaturing PAGE, dried and exposed to autoradiography. Positions of immunodection of Mt4 clone, probed with the specific anti-mitochondrial and anti-cytosolic ferritin antibodies by western blotting, and iron associated with mitochondrial (FtMt) and cytosolic (Cyt Ft) ferritins are indicated by the arrows. One representative experiment of three independent experiments is shown. (C) Stable clones and control K562 soluble cell lysates (25 μg per lane) were separated by 7.5% SDS-PAGE, blotted onto a nitrocellulose filter and probed with antibodies specific for total Stat5, phosphorylated Stat5 (P-Stat5) and β-actin. Arrows indicate positions of protein bands. Plots show band intensities of P-Stat5 relative to total Stat5 representative of three independent experiments. Triangles indicate increasing amount of expressed protein. (D) Stable FtMtΔfeox clones and control K562 soluble cell lysates were analyzed as in panel (C). (E) Control, GFP transduced and DFO-treated K562 cells were analyzed as in panel (C). (F) Control and transduced K562 cells were loaded with a ROS-sensitive fluorescent probe DHR123 and treated (+ H2O2) or not (− H2O2) with H2O2 for 30 min. Plots represent mean (±SD) of ROS production after 30 min of incubation with or without H2O2 treatment (t30′) relative to basal ROS level (t0). Three independent experiments, were performed each in octuplicate. Significant changes are marked.
Effect of mitochondrial ferritin on Stat5 phosphorylation and production or reactive oxygen species in K562 clones
K562 clones were analyzed for the effect of FtMt and its mutant on Stat5 phosphorylation. Minor, not significant changes were sometimes observed in the amount of total Stat5. Conversely, the expressed FtMt limited the phosphorylation of Stat5 (both the a and b isoforms) down to about 50% compared to controls, in a dose-dependent manner (Figure 1C). Stable FtMtΔfeox clones were shown to have no effect on the phosphorylation of Stat5 (Figure 1D). The null effect of the FtMtΔfeox suggested that the reduction of P-Stat5 was linked to the ability of FtMt to oxidize and incorporate iron.6 Thus, we treated the K562 and GFP-expressing cells with the iron-chelator desferrioxamine (DFO; 0.1 mM for 18 h) to verify the iron-dependency of the decrease in P-Stat5. The treatment resulted in a reduced P-Stat5, down to 50–60% compared to the controls (Figure 1E), similarly to the FtMt-expressing clones. As a result, FtMt iron sequestration capacity appears responsible for this effect, probably limiting ROS formation. We, therefore, tested the level of ROS in the clones; cells were loaded with the redox sensitive fluorescent probe dihydrorhodamine 123 (DHR123) and ROS production was evaluated by fluorescence. To avoid interference with fluorescence analyses, K562-LV-GFP was replaced by K562-LV-FtL, which over-expresses wild-type L-ferritin (Lwt)28 and was shown to have no effect on cellular iron metabolism. In basal conditions, the cells expressing a high amount of FtMt (Mt4 and Mt8) showed a significant decrease in ROS production, although only 5% lower than that in control cells (Figure 1F, upper panel). We, therefore, treated the cells with 0.3 mM H2O2 for 30 min and the results showed that FtMt further reduced the level of ROS compared to the controls (Figure 1F, bottom panel). In the latter condition, the ROS reduction appeared proportional to the amount of FtMt, with significant decreases of 15% and 25% for the Mt4 and Mt8 clones, respectively.
Effect of mitochondrial ferritin expression on transcription of Stat5 target genes
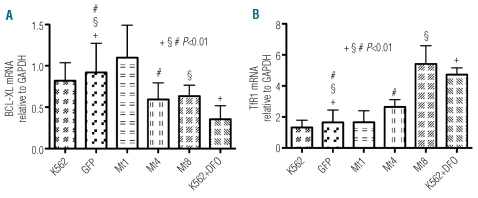
To verify the effect of reduced Stat5 phosphorylation at a transcriptional level, we performed real-time quantitative PCR on TfR1, IRP1 and IRP2, and on the Stat5-regulated anti-apoptotic gene Bcl-xL. Clones and controls were grown for 3 days, and K562 cells treated with 1 mM DFO for 48 h were used as a positive control because DFO treatment has an effect on apoptosis32 and on iron-dependent TfR1 translation.33 In the Mt1 clone, the Bcl-xL transcript was not significantly different; however, it was significantly reduced in Mt4 and Mt8 clones (approximately 35%) and in DFO-treated cells (approximately 60%) with respect to the Bcl-xL transcript of the control cells (Figure 2A). In contrast, as expected by the iron starvation cellular status, TfR1 mRNA increased by 1.6-, 3.3- and 2,8-fold in Mt4, Mt8 clones and DFO-treated cells, respectively. Also, no difference relative to the controls appeared in the Mt1 clone (Figure 2B). There were not statistically significant differences in IRP1 and IRP2 transcripts between control cells and clones (data not shown).
Figure 2.
Effect of FtMt expression on transcription of Stat5 target genes in K562 clones. Control, transfected and DFO-treated K562 cells were harvested for isolation of total RNA. Levels of mRNA expression were determined by quantitative real-time PCR for Bcl-xL (A) and TfR1 (B) and then normalized to mRNA expression of GAPDH. Results are presented as mRNA amount relative to GAPDH (±SD). Four independent experiments were performed each in triplicate. Significant changes compared to GFP control cells are marked.
Effect of mitochondrial ferritin on iron metabolism proteins in K562 clones
The effect of FtMt over-expression on iron metabolism has been studied in different cell lines;6–11 however no data are available on the erythroid compartment. We, therefore, determined whether FtMt maintains its avidity with regards to iron incorporation in erythroleukemia cells. We evaluated the amount of cytosolic ferritins, TfR1, and IRP1 and measured IRP1/2 binding activities. Quantification of cytosolic ferritins in cell soluble homogenates by ELISA showed that the presence of FtMt in the clones resulted in a down-regulation of both chains of cytosolic ferritin, FtH and FtL, compared to controls (Online Supplementary Figure S1A). The homogenates were also analyzed by western blotting after SDS-PAGE separation of the soluble proteins, and anti-TfR1 specific antibodies revealed that TfR1 was particularly increased in the Mt8 clone (Online Supplementary Figure S1B). Electromobility shift assay showed that the binding activities of IRP1/2 increased only in Mt4 and Mt8, while Mt1 maintained a binding activity similar to that of controls even if the amount of IRP was lower than that of the other clones (Online Supplementary Figure S1C). Similar results were also obtained for IRP2 activity, which can be measured as an IRE-IRP2 complex in electromobility shift assay, which was super-shifted with the anti-IRP2 antibody (Online Supplementary Figure S1D). The antibody specific for IRP1 confirmed a decrease of this protein in the Mt1 clone; however, no variation occurred in the IRP1 level of Mt4 and Mt8 compared to controls (Online Supplementary Figure S1E). These results suggested that Mt4 and Mt8 clones showed a phenotype compatible with cellular iron deficiency.
Effect of mitochondrial ferritin on apoptosis in K562 clones and in CD34+ cells
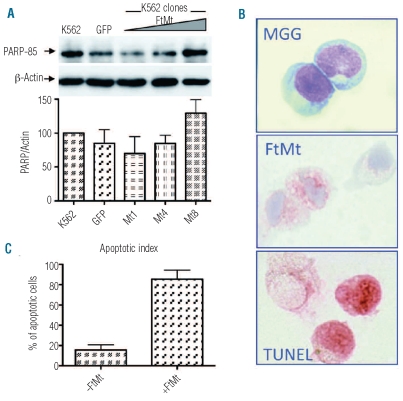
Based on the previous results, we expected that the high expression of FtMt would produce a phenotype with enhanced apoptosis and mitochondrial iron overload. DAPI staining revealed that the FtMt clones did not show an increase in apoptotic cells with respect to the control cells in the basal condition (data not shown). Thus, the cells were treated with 1 mM DFO for 48 h to induce apoptosis. DAPI staining and analysis of the presence of the p85 fragment of poly ADP ribose polymerase (PARP) by specific antibodies in western blotting were performed. DAPI staining resulted in a minor increase of apoptotic cells without differences between the FtMt clones and control cells (data not shown). However, western blotting analysis showed that the amount of p85 fragment increased in parallel to FtMt expression and appeared enriched in the Mt8 clone, which had a higher level of FtMt (Figure 3A). To study apoptosis further, we tested the effect of FtMt over-expression in human erythroid progenitors. CD34+ cells were transduced using LV-FtMt and after 14 and 21 days of culture collected and analyzed for morphological analysis, FtMt expression and apoptosis (Figure 3B). Many intermediate and late erythroblasts were seen in both transduced and non-transduced samples. Immunostaining with polyclonal anti-FtMt revealed that the efficiency of transduction was only about 30% at day 14. FtMt expression remained throughout the culture period. Due to the limit of transduction efficiency in primary cells, we determined the percentage of apoptotic cells in FtMt-expressing and non-expressing cells by double immunostaining. More than 80% of the FtMt-expressing cells underwent apoptosis, while only 20% of the FtMt non-expressing cells showed positivity in the TUNEL staining (Figure 3C). Similar findings were observed at day 21. Control CD34+ cells and CD34+ cells transduced using LV-GFP showed an apoptotic index similar to that of the FtMt non-expressing cells (data not shown). This preliminary result strongly supports the involvement of FtMt in inducing apoptosis in human CD34+ cells.
Figure 3.
Effect of FtMt expression on apoptosis in K562 and CD34+ cells. (A) Stable clones and control K562 were treated with DFO, soluble cell lysates (25 μg per lane) were separated by 10% SDS PAGE, blotted onto a nitrocellulose filter and probed with antibodies specific for the p85 PARP fragment (PARP-85) and β-actin. Plots show band intensities of the PARP fragment relative to β-actin. Results are representative of three independent experiments. Arrows indicate positions of protein bands. Triangles indicate increasing amount of expressed protein. (B) CD34+ bone marrow cells were isolated from normal donors, transduced with LV-FtMt and cultured for 21 days. After 14 and 21 days, cells were analyzed for morphology by May Grünwald Giemsa staining (MGG), for FtMt expression by immunocytochemistry (FtMt) and for apoptosis by a TUNEL assay (TUNEL). Representative samples at day 14. (C) Plots represent the apoptotic index of FtMt expressing (+FtMt) and non-expressing (−FtMt) cells, evaluated by a double immunocytochemistry technique, at day 14.
Effect of mitochondrial ferritin on mitochondrial iron overload
To check mitochondrial iron accumulation in K562 cells, Mt8 and MtΔfeox7 clones were grown for 6 days in medium supplemented with 20 μM of FAC or 20 μM Fe-Tf and then stained for iron content and FtMt expression (Figure 4A). The Perls’ staining of FAC-treated cells revealed the presence of iron granules in 12% of Mt8 cells (data not shown), while no positive iron staining was revealed in the case of MtΔfeox7 expression (data not shown). The effect was more evident in Mt8 cells treated with 20 μM Fe-Tf where almost all the cells showed the presence of Perls’-positive granules, with a distribution similar to that of FtMt staining (Figure 4A, lower panels), while the MtΔfeox7 clone remained negative (Figure 4A, upper panels). The Fe-Tf treated samples were also subjected to electron microscopy, which confirmed the presence of dense granules in mitochondria in the Mt8 clone (Figure 4B, both middle panels) and not in MtΔfeox7 and K562 cells (Figure 4B, upper panels). To confirm the chemical composition of these granules we used an ESI/EELS approach. Consistent ESI signals were observed in correspondence to the granules in the majority of mitochondria (Figure 4B, bottom panels) and the EELS spectrum analyses confirmed the presence of iron (data not shown).
Figure 4.
Effect of FtMt expression on mitochondrial iron load and heme synthesis. (A) K562 Mt8 and MtΔfeox7 clones were grown for 6 days in the presence of 20 μM Fe-Tf and then stained for iron content with Perls’ reaction (left panels) or immunostained with an antibody specific for FtMt (right panels). The arrows point to granules positive for iron staining. (B) K562 cells and clones treated as in (A) were analyzed by electron microscopy. Upper panels show the controls, MtΔfeox7 and K562 cells, stained with uranyl and lead citrate. The second row shows a portion of Mt8 cells with a group of mitochondria stained with uranyl and lead citrate (left panel). The area selected in the white square is enlarged in the right panel, the arrows point to electrondense spots.
The panels in the third row show the presence of granules in unstained Mt8 cells; the square selected in white is enlarged in the right panel. The bottom panels show the conventional ultrastructural organization (left panel) and the iron map (red color) superimposed on the ultra-structural organization of the same field obtained at 250 eV (right panel) obtained by ESI (see Design and Methods section). Bars indicate 500 nm. (C) Control and transfected MEL cells were grown for 3 days in the presence of 1.5% DMSO, and heme production was evaluated by a chemical method. Plots represent mean (±SD) of heme content relative to DMSO-treated control cells. Three independent experiments, were performed each in duplicate. Significant changes are marked.
Heme production in murine erythroleukemia cells over-expressing mitochondrial ferritin
K562 cells and clones after dimethylsulfoxide (DMSO) or hemin treatment did not synthesize large amounts of heme, demonstrating incomplete differentiation. We, therefore, chose the more suitable MEL cells to evaluate the effect of FtMt expression on the production of heme. MEL cells were transfected either with pcDNA3-FtMt or with the empty plasmid, obtaining an efficiency of approximately 50% as determined by immunofluorescence with anti-FtMt (data not shown). Next, the cells were treated for 3 days with 1.5% DMSO to promote differentiation. Cells were lysed in formic acid, and the heme content was measured by absorbance at 400 nm. The cells differentiated, as evidenced by the 2.9-fold increased heme production. The results indicated that the heme content was significantly diminished (approximately 30%) in cells expressing FtMt compared to in the control cells (Figure 4C), even when the cell population was not homogenous in FtMt expression.
Discussion
Erythropoiesis depends on the JAK2/STAT5 pathway and requires a large amount of iron to maintain heme synthesis. FtMt avidly sequesters iron inside the mitochondria (this work and 6,8) and is up-regulated in XLSA and RARS,17,18 disorders characterized by ineffective erythropoiesis. The presence of FtMt distinguishes sideroblasts, whereas it is absent in normal erythroblasts, suggesting that it may be, at least in part, involved in the pathophysiology of this subtype of low-risk myelodysplastic syndrome.17 Previous data on erythroblast cultures from bone marrow cells of patients with RARS indicated that FtMt expression occurs at an early stage of differentiation, before the iron burden develops, and parallels the apoptotic signals and the block of normal erythropoiesis.19 The authors hypothesized that the abnormal mitochondrial iron accumulation might have triggered apoptosis via ROS production, with consequent mitochondrial damage and arrest of erythropoietic maturation.19 However, the presence of FtMt in these cells suggests that mitochondrial iron was not available for ROS formation, being sequestered in the protein. In fact, one of the better characterized properties of FtMt is its ability to control the development of ROS, as this work and several other cellular studies have pointed out.7,8,11 Consequently, we hypothesized that the protein may be the inducer of the cascade of events leading to defects in erythroid maturation.
To evaluate this hypothesis, we chose erythroleukemia cells that spontaneously develop characteristics similar to early-stage erythroblasts and show constitutive enhanced Stat5 phosphorylation. FtMt expression experiments and DFO treatment of K562 cells indicated that: (i) FtMt specifically reduces P-Stat5 in a dose-dependent manner; (ii) FtMt ferroxidase activity is necessary to exploit this property; (iii) iron chelator molecules can mediate the P-Stat5 decrease. From these data we argue that variations in the amount of iron, induced by iron-sequestering agents, might be associated with the decrease of Stat5 phosporylation and this might be mediated by a decrease in ROS. In fact, in the FtMt stable clones the reduction of P-Stat5 was paralleled by a tendency of ROS levels to decrease, which became statistically significant in the clones expressing more than 60 ng/mg of total protein both in the basal condition and after an oxidative insult. It appears, therefore, that FtMt creates a relationship between the control of ROS development and the level of P-Stat5, indicating that FtMt may be considered an inhibitor of Stat5 phosphorylation as previously reported for ROS scavengers.21–23 Interestingly, a recent report on gene expression profiling of erythroblasts from RARS patients detected a significant down-regulation of STAT5B gene,34 giving an in vivo confirmation of the importance of Stat5 in this disease and supporting our in vitro results.
A large amount of FtMt has a pro-apoptotic effect, as shown by the decrease of Bcl-xL transcript. Although K562 is an apoptosis-resistant cellular model due to malignant outgrowth characteristics, the presence of caspase 3 proteolytic peptide revealed that the apoptosis pathway was triggered in the Mt8 clone. Interestingly, FtMt strongly promoted apoptosis when over-expressed in the primary CD34+ cells, demonstrating that the presence of a high level of FtMt induced cellular death in erythroid progenitors. These data are in accordance with those of a previous study19 in which a correlation between cytochrome c and FtMt expression was observed in CD34+ cells from RARS patients. Here we further studied the putative mechanism leading to apoptosis and found that it was apparently due to deregulation of Stat5 transcriptional activity, which was caused primarily by cytosolic iron deficiency.
The FtMt clones were representative of different cellular iron distribution in the cytosolic and mitochondrial compartments. These data confirmed the cytosolic iron depriving action of FtMt in erythroleukemia cells, as previously shown in epithelial HeLa cells.6,8 The different protein content in clones reveals a complex interplay between the Stat5 transcriptional effect and the cellular response to iron deprivation. Despite the fact that FtMt was able to reduce P-Stat5, the effect on the decrease of Stat5-dependent transcription was detectable only for the Bcl-xL gene, a gene unrelated to iron metabolism. At difference from the findings in Stat5−/− mice,26 no measurable decrease was observed in the genes involved in maintaining iron homeostasis, such as TfR1 and IRP1/2. Probably, the limited reduction of P-Stat5 obtained in our model, compared to the null expression in Stat5−/− mice, was not sufficient to determine a transcriptional effect on iron-related genes. In contrast, we detected a significant increase of TfR1 transcript levels in Mt4 and Mt8. This finding suggests that the effect of Stat5 on transcriptional regulation of TfR1 is overcome by the cytosolic iron starvation induced by FtMt. Cytosolic iron deprivation stimulates the IRE/IRP machinery with consequent inhibition of ferritin translation and greater stability of TfR1 mRNA. Hence, the Stat5 regulatory effect on TfR1 transcription appears to be prevented by cellular iron requirements.
Iron-burdened mitochondria are a characteristic of sider-oblasts, the hallmark of inherited and acquired sideroblastic anemias.35 In our Mt8 clone we succeeded in obtaining characteristics similar to those present in these diseases, as witnessed by dotted Perls’-positive staining. Electron microscopy and ESI/EELS localized these electron dense spots within the mitochondria and identified them as iron deposits, which occurred only if the FtMt was functional in iron incorporation. This finding strongly supports the conclusion that the presence of FtMt is sufficient to reproduce this hallmark of the disease. Interestingly, most cells were positive after incubation with Fe-Tf; however, only 12% were positive after addition of FAC. This observation supports data indicating that, at least in erythroleukemic cells, iron is preferentially delivered to mitochondria via TfR1 endocytosis.36
The high amount of FtMt in the Mt8 clone is most likely a non-physiological state; however, it can be found in pathological conditions such as XLSA and RARS, in which the protein is up-regulated.17 In patients with inherited XLSA, the presence of iron-overloaded mitochondria, which are usually observed in late erythroblasts, can be explained by the fact that iron cannot be used for heme synthesis due to metabolic defects. While iron accumulates in mitochondria in inherited XLSA, the cause of the anomalous mitochondrial iron deposition in RARS is still unknown. Considering that FtMt is expressed in CD34+ cells of RARS patients,19 we suggest that its abnormal presence may trigger the pathogenic mechanism that leads to sideroblast formation (Online Supplementary Figure S2). The increased level of FtMt causes cytosolic iron starvation, which induces activation of the iron sensor IRP, which results in increased cellular iron uptake and leads to iron accumulation in mitochondria. Simultaneously, FtMt may stimulate apoptosis by reducing P-Stat5 and affect heme synthesis, sequestering iron at the mitochondrial level. This cascade of events may result in a decline of normal maturation of erythroblasts and promote the formation of sideroblasts (Online Supplementary Figure S2). According to this hypothesis, the initial event of sideroblast formation in RARS would result in the up-regulation of FtMt. Unfortunately, the regulation of the expression of this protein is far from clear. A previous report identified the FTMT gene as one of the genes that contain non-X-linked, bona fide promoter CpG islands, which are densely methylated in normal somatic tissues.37 This finding suggests that the regulation of FtMt expression may depend on epigenetic events. Interestingly, in the last years, it has been shown how abnormal epigenetic modulation may play a crucial role in the pathogenesis and biological evolution of myelodysplastic syndrome.38 The lack of gene silencing of FtMt, due to epigenetic alterations, may, therefore, be the key point in the development of myelodysplastic syndromes characterized by the presence of sideroblasts.
In conclusion, our data suggest that FtMt acts in different ways depending on its amount. In physiological conditions, the protein is expressed at a very low level13 and most likely does not alter iron homeostasis and P-Stat5, its role being limited to the regulation of ROS formation at a mitochondrial level. Indeed, the expression of FtMt is tightly regulated in normal tissues, probably because its antioxidant role is fundamental only in cells with high oxygen consumption. In contrast, when the expression of FtMt is high, the protein can acquire pathological effects. In particular, in erythroid progenitors the up-regulation of FtMt may interfere with either the JAK2/STAT5 regulatory pathway or heme synthesis, contributing to ineffective erythropoiesis.
Acknowledgments
The authors would like to thank Clara Camaschella for her critical reading of the manuscript, the Alembic facility of San Raffaele Scientific Institute for the electron microscopy, Fabio Grohovaz and Maria Carla Panzeri for helping with the ESI/EELS analysis and Giulia Della Chiara and Fanis Missirlis for useful discussion.
Footnotes
Funding: this work was partially supported by a Fondazione CARIPLO-2007 grant to SL and by a grant from IRCCS San Matteo Foundation, Pavia, Italy to RI; AC was supported by the University Vita-Salute San Raffaele and the Italian Institute of Technology PhD program.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, Sanford D, et al. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem. 2001;276(27):24437–40. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 2.Arosio P, Levi S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta. 2010;1800(8):783–92. doi: 10.1016/j.bbagen.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Drysdale J, Arosio P, Invernizzi R, Cazzola M, Volz A, Corsi B, et al. Mitochondrial ferritin: a new player in iron metabolism. Blood Cells Mol Dis. 2002;29(3):376–83. doi: 10.1006/bcmd.2002.0577. [DOI] [PubMed] [Google Scholar]
- 4.Langlois d'Estaintot B, Santambrogio P, Granier T, Gallois B, Chevalier JM, Precigoux G, et al. Crystal structure and biochemical properties of the human mitochondrial ferritin and its mutant Ser144Ala. J Mol Biol. 2004;340(2):277–93. doi: 10.1016/j.jmb.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Bou-Abdallah F, Santambrogio P, Levi S, Arosio P, Chasteen ND. Unique iron binding and oxidation properties of human mitochondrial ferritin: a comparative analysis with human H-chain ferritin. J Mol Biol. 2005;347(3):543–54. doi: 10.1016/j.jmb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Corsi B, Cozzi A, Arosio P, Drysdale J, Santambrogio P, Campanella A, et al. Human mitochondrial ferritin expressed in HeLa cells incorporates iron and affects cellular iron metabolism. J Biol Chem. 2002;277(25):22430–7. doi: 10.1074/jbc.M105372200. [DOI] [PubMed] [Google Scholar]
- 7.Campanella A, Isaya G, O'Neill HA, Santambrogio P, Cozzi A, Arosio P, et al. The expression of human mitochondrial ferritin rescues respiratory function in frataxin-deficient yeast. Hum Mol Genet. 2004;13(19):2279–88. doi: 10.1093/hmg/ddh232. [DOI] [PubMed] [Google Scholar]
- 8.Campanella A, Rovelli E, Santambrogio P, Cozzi A, Taroni F, Levi S. Mitochondrial ferritin limits oxidative damage regulating mitochondrial iron availability: hypothesis for a protective role in Friedreich ataxia. Hum Mol Genet. 2009;18(1):1–11. doi: 10.1093/hmg/ddn308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie G, Sheftel AD, Kim SF, Ponka P. Overexpression of mitochondrial ferritin causes cytosolic iron depletion and changes cellular iron homeostasis. Blood. 2005;105(5):2161–7. doi: 10.1182/blood-2004-07-2722. [DOI] [PubMed] [Google Scholar]
- 10.Nie G, Chen G, Sheftel AD, Pantopoulos K, Ponka P. In vivo tumor growth is inhibited by cytosolic iron deprivation caused by the expression of mitochondrial ferritin. Blood. 2006;108(7):2428–34. doi: 10.1182/blood-2006-04-018341. [DOI] [PubMed] [Google Scholar]
- 11.Shi Z, Nie G, Duan XL, Rouault T, Wu WS, Ning B, et al. Neuroprotective mechanism of mitochondrial ferritin on 6-hydroxy-dopamine induced dopaminergic cell damage: implication for neuroprotection in Parkinson's disease. Antioxid Redox Signal. 2010;13(6):783–96. doi: 10.1089/ars.2009.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanella I, Derosas M, Corrado M, Cocco E, Cavadini P, Biasiotto G, et al. The effects of frataxin silencing in HeLa cells are rescued by the expression of human mitochondrial ferritin. Biochim Biophys Acta. 2008;1782(2):90–8. doi: 10.1016/j.bbadis.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Santambrogio P, Biasiotto G, Sanvito F, Olivieri S, Arosio P, Levi S. Mitochondrial ferritin expression in adult mouse tissues. J Histochem Cytochem. 2007;55(11):1129–37. doi: 10.1369/jhc.7A7273.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong SY, Rathore KI, Schulz K, Ponka P, Arosio P, David S. Dysregulation of iron homeostasis in the CNS contributes to disease progression in a mouse model of amyotrophic lateral sclerosis. J Neurosci. 2009;29(3):610–9. doi: 10.1523/JNEUROSCI.5443-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi S, Arosio P. Mitochondrial ferritin. Int J Biochem Cell Biol. 2004;36(10):1887–9. doi: 10.1016/j.biocel.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Snyder AM, Wang X, Patton SM, Arosio P, Levi S, Earley CJ, et al. Mitochondrial ferritin in the substantia nigra in restless legs syndrome. J Neuropathol Exp Neurol. 2009;68(11):1193–9. doi: 10.1097/NEN.0b013e3181bdc44f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cazzola M, Invernizzi R, Bergamaschi G, Levi S, Corsi B, Travaglino E, et al. Mitochondrial ferritin expression in erythroid cells from patients with sideroblastic anemia. Blood. 2003;101(5):1996–2000. doi: 10.1182/blood-2002-07-2006. [DOI] [PubMed] [Google Scholar]
- 18.Della Porta MG, Malcovati L, Invernizzi R, Travaglino E, Pascutto C, Maffioli M, et al. Flow cytometry evaluation of erythroid dysplasia in patients with myelodysplastic syndrome. Leukemia. 2006;20(4):549–55. doi: 10.1038/sj.leu.2404142. [DOI] [PubMed] [Google Scholar]
- 19.Tehranchi R, Invernizzi R, Grandien A, Zhivotovsky B, Fadeel B, Forsblom AM, et al. Aberrant mitochondrial iron distribution and maturation arrest characterize early erythroid precursors in low-risk myelodysplastic syndromes. Blood. 2005;106(1):247–53. doi: 10.1182/blood-2004-12-4649. [DOI] [PubMed] [Google Scholar]
- 20.Bartnikas TB, Campagna DR, Antiochos B, Mulhern H, Pondarré C, Fleming MD. Characterization of mitochondrial ferritin-deficient mice. Am J Hematol. 2010;85(12):958–60. doi: 10.1002/ajh.21872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattler M, Winkler T, Verma S, Byrne CH, Shrikhande G, Salgia R, et al. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 1999;93(9):2928–35. [PubMed] [Google Scholar]
- 22.McCormick J, Barry SP, Sivarajah A, Stefanutti G, Townsend PA, Lawrence KM, et al. Free radical scavenging inhibits STAT phosphorylation following in vivo ischemia/reperfusion injury. Faseb J. 2006;20(12):2115–7. doi: 10.1096/fj.06-6188fje. [DOI] [PubMed] [Google Scholar]
- 23.Iiyama M, Kakihana K, Kurosu T, Miura O. Reactive oxygen species generated by hematopoietic cytokines play roles in activation of receptor-mediated signaling and in cell cycle progression. Cell Signal. 2006;18(2):174–82. doi: 10.1016/j.cellsig.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93(5):841–50. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 25.Grebien F, Kerenyi MA, Kovacic B, Kolbe T, Becker V, Dolznig H, et al. Stat5 activation enables erythropoiesis in the absence of EpoR and Jak2. Blood. 2008;111(9):4511–22. doi: 10.1182/blood-2007-07-102848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerenyi MA, Grebien F, Gehart H, Schifrer M, Artaker M, Kovacic B, et al. Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1. Blood. 2008;112(9):3878–88. doi: 10.1182/blood-2008-02-138339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starzynski RR, Goncalves AS, Muzeau F, Tyrolczyk Z, Smuda E, Drapier JC, et al. STAT5 proteins are involved in down-regulation of iron regulatory protein 1 gene expression by nitric oxide. Biochem J. 2006;400(2):367–75. doi: 10.1042/BJ20060623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–22. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 29.Cozzi A, Rovelli E, Frizzale G, Campanella A, Amendola M, Arosio P, et al. Oxidative stress and cell death in cells expressing L-ferritin variants causing neuroferritinopathy. Neurobiol Dis. 2010;37(1):77–85. doi: 10.1016/j.nbd.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Cozzi A, Levi S, Bazzigaluppi E, Ruggeri G, Arosio P. Development of an immunoassay for all human isoferritins, and its application to serum ferritin evaluation. Clin Chim Acta. 1989;184(3):197–206. doi: 10.1016/0009-8981(89)90052-1. [DOI] [PubMed] [Google Scholar]
- 31.Campanella A, Levi S, Cairo G, Biasiotto G, Arosio P. Blotting analysis of native IRP1: a novel approach to distinguish the different forms of IRP1 in cells and tissues. Biochemistry. 2004;43(1):195–204. doi: 10.1021/bi035386f. [DOI] [PubMed] [Google Scholar]
- 32.Jin H, Terai S, Sakaida I. The iron chelator deferoxamine causes activated hepatic stellate cells to become quiescent and to undergo apoptosis. J Gastroenterol. 2007;42(6):475–84. doi: 10.1007/s00535-007-2020-5. [DOI] [PubMed] [Google Scholar]
- 33.Recalcati S, Minotti G, Cairo G. Iron regulatory proteins: from molecular mechanisms to drug development. Antioxid Redox Signal. 2010;13(10):1593–616. doi: 10.1089/ars.2009.2983. [DOI] [PubMed] [Google Scholar]
- 34.Camaschella C. Hereditary sideroblastic anemias: pathophysiology, diagnosis, and treatment. Semin Hematol. 2009;46(4):371–7. doi: 10.1053/j.seminhematol.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Nikpour M, Pellagatti A, Liu A, Karimi M, Malcovati L, Gogvadze V, et al. Gene expression profiling of erythroblasts from refractory anaemia with ring sideroblasts (RARS) and effects of G-CSF. Br J Haematol. 2010;149(6):844–54. doi: 10.1111/j.1365-2141.2010.08174.x. [DOI] [PubMed] [Google Scholar]
- 36.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 2007;110(1):125–32. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 37.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007:2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musolino C, Sant'antonio E, Penna G, Alonci A, Russo S, Granata A, et al. Epigenetic therapy in myelodysplastic syndromes. Eur J Haematol. 2010;84(6):463–73. doi: 10.1111/j.1600-0609.2010.01433.x. [DOI] [PubMed] [Google Scholar]