Background: Expression of the yeast HO gene is tightly regulated, with a large complex promoter.
Results: The Rpd3(L) histone deacetylase is recruited twice by distinct factors to different promoter regions.
Conclusion: Chromatin represses the HO promoter, making multiple coactivators required for gene expression.
Significance: Multiple factors contribute to the establishment of a tightly repressed, cell cycle-regulated promoter.
Keywords: Cell Cycle, Chromatin, Gene Regulation, Transcription Coactivators, Yeast Transcription
Abstract
The yeast HO gene is tightly regulated, with multiple activators and coactivators needed to overcome repressive chromatin structures that form over this promoter. Coactivator binding is strongly interdependent, as loss of one factor sharply reduces recruitment of other factors. The Rpd3(L) histone deacetylase is recruited to HO at two distinct times during the cell cycle, first by Ash1 to the URS1 region of the promoter and then by SBF/Whi5/Stb1 to URS2. SBF itself is localized to only a subset of its potential binding sites in URS2, and this localization takes longer and is less robust than at other SBF target genes, suggesting that binding to the HO promoter is limited by chromatin structures that dynamically change as the cell cycle progresses. Ash1 only binds at the URS1 region of the promoter, but an ash1 mutation results in markedly increased binding of SBF and Rpd3(L) at URS2, some 450 bp distant from the site of Ash1 binding, suggesting these two regions of the promoter interact. An ash1 mutation also results in increased coactivator recruitment, Swi/Snf and Mediator localization in the absence of the normally required Gcn5 histone acetyltransferase, and HO expression even in the presence of a taf1 mutation affecting TFIID activity that otherwise blocks HO transcription. Ash1 therefore appears to play a central role in generating the strongly repressive environment at the HO promoter, which limits the binding of several coactivators at URS2 and TATA region.
Introduction
Chromatin is generally repressive, blocking access of factors to DNA and preventing gene transcription. Many chromatin factors can promote transcriptional activation by changing the post-translational modification status of nucleosomes, altering the positioning of nucleosomes, or by evicting nucleosomes from the DNA (1, 2).
The promoter of the yeast HO gene is under extremely tight transcriptional repression (3). HO encodes an endonuclease that initiates mating type switching by causing a double strand break at the MAT locus, and inappropriate expression of an endonuclease could be toxic. Three distinct coactivator complexes, Swi/Snf, SAGA, and Mediator, are each required for activation of HO transcription. Genetic studies have identified an array of other important factors that regulate HO, including the Sin3/Rpd3 histone acetylase (4, 5).
In contrast to most yeast genes that have small compact promoters, regulatory elements at HO extend to −1900 bp from the transcription start site, and the next upstream open reading frame is at −3084 bp. The promoter has been divided into two regions, URS1 from −1900 to −1000 and URS2 from −900 to −200 (Fig. 1) (6). HO is cell cycle-regulated, and HO activation is complex, involving the sequential localization of DNA-binding factors and transcriptional coactivators (3, 7, 8). The Swi5 DNA-binding protein acts first, entering the nucleus as cells pass anaphase in mitosis then binding to two sites at −1816 and −1305 within URS1. Swi5 recruits three transcriptional coactivators, Swi/Snf, SAGA, and Mediator, and Swi5 is then rapidly degraded, leading to rapid loss of Swi5 and coactivator localization at the HO promoter. Subsequently, the SBF activator, composed of the Swi4 and Swi6 subunits, binds to the URS2 region and recruits the Swi/Snf, SAGA, and Mediator coactivators to this promoter region. The later events are dependent on the earlier ones, but it is not known whether the coactivators recruited to URS2 are the same complexes recruited to URS1.
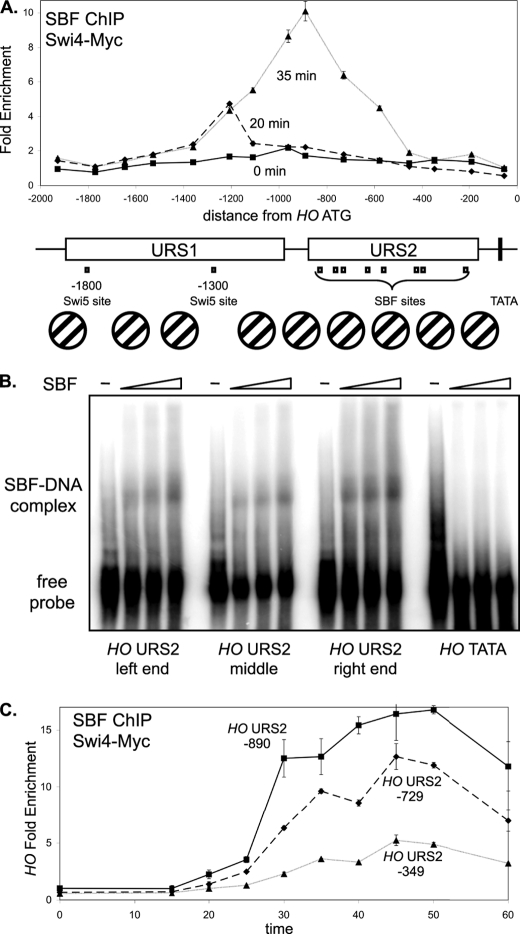
FIGURE 1.
SBF binding is strongest to sites at the left end of URS2. A, DY12794 cells (GALp::CDC20 SWI4-Myc) with a GALp::CDC20 allele were synchronized in mitosis by removing galactose, followed by release by addition of galactose (t = 0). The CDC20 arrest is at the G2/M transition, and HO expression at 40 min following release corresponds to late G1 phase. SBF binding was measured by Swi4-Myc ChIP using samples taken at t = 0, 20, and 35 min following the release. ChIP samples were analyzed with 15 sets of PCR primers across the HO promoter, with an average PCR product size of 208 bp. URS1-, URS2-, Swi5- and SBF-binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20. Nucleosome positions along the HO promoter, determined by micrococcal nuclease mapping and by H3 ChIP (3), are shown. B, electrophoretic mobility shift assays examine in vitro binding of recombinant SBF to sites at the left (−857), middle (−461) and right (−246) parts of URS2, and the TATA region as a negative control. C, DY12794 cells (GALp::CDC20 SWI4-Myc) were synchronized and SBF binding by Swi4-Myc ChIP during the time course to three regions of HO URS2 at −890, −729, and −349, where the numbers refer to the center of the amplified region.
Sequential changes in chromatin structure occur during the cell cycle at the HO promoter, well before transcription occurs (3). Initially, nucleosomes are positioned at the promoter, with the two Swi5-binding sites at −1816 and −1305 in nucleosome-free regions. Similar to some other promoters, nucleosome eviction from the HO promoter begins before transcriptional activation (9–12). The situation at HO is unusually complex as it involves ordered waves of eviction. Nucleosomes are lost first from URS1, and this event requires Swi5 and Swi/Snf. Later in the cell cycle, nucleosomes are evicted from the left end of URS2, and this requires the earlier events at URS1 and also the FACT complex, which is recruited by SBF. Next, nucleosome loss occurs at the right end of URS2, requiring the Asf1 H3/H4 histone chaperone as well as the earlier events. Finally nucleosomes are lost at the TATA region coincident with binding of RNA polymerase II and transcriptional activation. This dependent series of events reveals the presence of several distinct chromatin structures at this promoter and their importance in maintaining strict repression of HO expression.
The ASH1 gene was identified as a negative regulator of HO expression (13, 14). Budding yeast divides asymmetrically, producing larger mother cells and smaller daughter cells at mitotic division. HO has the unusual property of being expressed only in mother cells. The Ash1 protein is responsible for this asymmetry, with Ash1 protein accumulating primarily in daughter cells (15). Ash1 has been identified as a component of the Rpd3(L) complex (16), which is one of two complexes containing the Rpd3 histone deacetylase, Rpd3(L) and Rpd3(S). Rpd3(L) functions primarily at promoters, whereas Rpd3(S) is recruited to methylated histone H3-K36 marked by the elongating RNA polymerase II complex and functions at open reading frames to repress cryptic transcription (17–19). Ash1 shows similarity to the GATA family of DNA-binding proteins and has been suggested to target Rpd3(L) to some promoters (16). In addition to repressing HO expression, Ash1 is required for efficient activation of the FLO11 gene required for pseudohyphal growth (20, 21) and may regulate CLN3 expression in daughter cells (22). Ash1 is therefore another regulator of HO transcription that acts by altering the structure of chromatin, in this case by assisting the recruitment of the Rpd3(L) histone deacetylase complex. Although the majority of Ash1 accumulates in daughter cells, the protein is also present in mother cells (13), and we have shown that the regulatory properties of the HO gene in mother cells is markedly affected by the Ash1 present in mothers.
In this study, we describe new complexities at the HO promoter. The recruitment of each coactivator to HO is dependent on the action of other coactivators, and thus coactivator binding is strongly interdependent. We show that the Rpd3(L) histone deacetylase is sequentially recruited to two different promoter regions during the cell cycle. Ash1 recruits Rpd3(L) only transiently to a site in URS1, but an ash1 mutation allows long lasting consequences on factor binding at distant promoter sites, suggesting that Ash1 generates repressive chromatin that persists long after Ash1 is degraded. The repressive nature of this promoter, combined with a requirement for interdependent coactivator recruitment at sequentially acting promoter elements, provides a mechanism for preventing unintentional expression of this endonuclease.
EXPERIMENTAL PROCEDURES
All yeast strains used are listed in supplemental Table S1 and are isogenic in the W303 background (23). Standard genetic methods were used for strain construction (24, 25). The ho(ΔURS2[−929 to −172]) mutant strain was constructed by transforming DY5401, which has URA3 inserted into the HO promoter at −1496 to −1222, with NsiI-PstI digested plasmid C539 (6), kindly provided by Kim Nasmyth, and selecting on 5-fluoroorotic acid. The HO(−1166-m) mutation was generated by PCR-directed mutagenesis, converting the sequence “AATCGCGTAAAA” to “AATATT,” and using 5-fluoroorotic acid selection to insert this sequence into the promoter of strain DY7874, which has URA3 inserted into the HO promoter at −1496 to −1222, as well as a KanMX marker inserted downstream of the 3′UTR. Promoter mutations were verified by direct sequencing. Plasmids used are listed in supplemental Table S2. Details of plasmid construction are available upon request.
Cell cycle synchronization was performed by galactose withdrawal and re-addition with a GALp::CDC20 strain grown at 25 °C in YEP medium containing 2% galactose and 2% raffinose (26). A high degree of synchrony was demonstrated by flow cytometry analysis, budding indices, and analysis of cell cycle regulated mRNAs (data not shown). In other experiments, cells were grown in YEPD medium at 30 °C (25), unless otherwise noted. Cells containing the YCp-KanMX(HA-CDC28) plasmid were grown in media supplemented with 0.2 mg/ml G418.
Chromatin immunoprecipitations (ChIPs) were performed as described previously (26, 27) using 9E11 (Abcam) or 4A6 (Upstate) monoclonal antibody to the Myc epitope, monoclonal antibody to the HA epitope (12CA5, University of Utah Bioprocessing Resource), and antibody-coated magnetic beads (Pan Mouse IgG beads, Dynal Biotech). ChIP assays were analyzed by real time PCR as described previously (28). PCR primers are listed in supplemental Table S3. Each ChIP sample was first normalized to an input DNA sample and then to the ChIP signal for a control region on chromosome I. Error bars in ChIP assays reflect the standard deviation of three replicate PCRs. For cell synchrony experiments, the ChIP values were normalized so that the zero time point was 1.0. For each epitope-tagged strain used for ChIP analysis during logarithmic growth, a genetically identical untagged strain was used as control.
RT-qPCR3 was used to measure mRNA levels as described previously (27) using primers listed in supplemental Table S3, using either ACT1 or RDN25 RNA as the internal control. Error bars in RT-qPCR assays reflect the standard deviation of three replicate PCRs. Electrophoretic mobility shift assays were performed using in vitro translated Swi4 and recombinant Swi6 as described previously (29). Swi4 and Swi6 were expressed from plasmids pBD972 and pBD400, respectively, kindly provided by Linda Breeden. Immunoprecipitations were performed as described previously (30) using anti-HA antibody, and blots were probed with anti-Myc and anti-HA antibodies and scanned using a Odyssey Infrared Imaging System (Li-Cor Biosciences).
RESULTS
SBF Binds to HO URS1 before URS2
The SBF factor (Swi4–Swi6) is required for expression of the HO gene. We therefore used chromatin immunoprecipitation (ChIP) assays to measure Swi4-Myc binding to the HO promoter. Cells were synchronized by GALp::CDC20 arrest and release, which typically results in HO expression at 40 min after the release (3). PCR primers tiled across the HO promoter were used to measure SBF binding at 0, 20, and 35 min following release (Fig. 1A). At 20 min, SBF binds to the URS1 region at approximately −1200 from the ATG, although at 35 min binding is seen at URS2. There are several surprises in these observations. First, SBF binds to URS1, although there are no recognizable SBF-binding sites in this region. Second, although SBF sites are distributed throughout URS2 (see map in Fig. 1A), strong binding at 35 min is only seen near the SBF sites in the left half of URS2. Although each SBF-binding site in URS2 has the same CACGAAAA sequence, it is possible that surrounding DNA sequences play an important role in determining binding affinity. To test this idea, we performed electrophoretic mobility shift assays (EMSA) to examine in vitro binding of SBF to sites at the left (−857), middle (−461), and right (−246) parts of URS2. The EMSA signals in this assay are dependent on the presence of the recombinant Swi4 and Swi6 proteins (29). This experiment shows SBF has similar affinity for the three sites in vitro (Fig. 1B), despite the ChIP results showing strong SBF binding in vivo to the −857 region and very little binding to the −246 region (Fig. 1A). This differential binding of SBF to the sites along URS2 may be due to the nucleosome eviction at the left end of URS2 that occurs before the chromatin changes occur at the right end (3). Interestingly, time course ChIP experiments examining three regions of URS2 show that SBF binding at −729 is somewhat reduced compared with −890, and binding to the right end of URS2 (−349) is both weak and slow (Fig. 1C). This gradient of SBF binding across URS2 is consistent with the changes in chromatin structure that sweep across HO URS2 during the cell cycle (3), revealing the importance of chromatin structure in regulating SBF binding.
To explain SBF binding at URS1, we looked for potential SBF-binding sites within this region. Although there is no sequence that matches the original CACGAAAA consensus (31), the sequence CGCGTAAAA present at −1166 in URS1 (supplemental Fig. S1A) is a reasonable match to the CgCGAAA consensus derived by both biochemical and informatics approaches (32). Consistent with our results, Taba et al. (33) detected SBF binding to a 482-bp RsaI fragment from −1352 to −870. We have mutated this potential site at −1166, and SBF does not bind to URS1 in vivo in a strain with this mutation (supplemental Fig. S1B), whereas SBF binding to URS2 (supplemental Fig. S1C) and CLN2 (supplemental Fig. S1D) are largely unaffected in this same strain. HO expression levels are normal in this HO(−1166-mut) mutant, so reduced binding of SBF to URS1 does not lead to diminished transcription from this promoter (supplemental Fig. S1E). We next investigated the effect of deleting the URS2 region of HO. We constructed strains lacking the URS2 region (−929 to −172 from the ATG) in the chromosomal HO gene. ChIP experiments show equivalent Swi4 binding to URS1 at native HO and the modified HO(ΔURS2) gene (supplemental Fig. S1F), demonstrating that URS2 is not needed for SBF binding to the site at −1166. We next measured expression of this HO(ΔURS2) gene in wild type and swi6 mutant cells. The HO(ΔURS2) gene is expressed at normal levels in both cases, and thus Swi6 is not required for expression of HO(ΔURS2) (supplemental Fig. S1G), as reported previously (31). These experiments show that although SBF binding does occur at URS1, it is not required for HO activation, and SBF binding at URS2 is needed to overcome the inhibitory effect of URS2.
Coactivator Binding to the HO Promoter Is Interdependent
The Swi5 DNA-binding protein binds to HO URS1 in late M/early G1 and recruits three coactivator complexes, Swi/Snf, SAGA, and Mediator, to this far upstream region of the promoter (3). Govind et al. (34) showed that the Gcn4 activator simultaneously recruits Swi/Snf, SAGA, and Mediator to the ARG1 promoter and that mutation of one coactivator sharply reduces recruitment of the other coactivators. We therefore examined the situation for Swi5 and found that the coactivators are largely dependent on one another for stable binding to HO URS1 (Fig. 2A). A swi5 gene disruption eliminates binding of all three coactivators, consistent with recruitment by the Swi5 DNA-binding protein (3). A swi2(E834K) mutation affecting Swi/Snf reduces binding of both SAGA and Mediator, disruption of the GCN5 subunit of SAGA reduces binding of both Swi/Snf and Mediator, and disruption of the GAL11 subunit of Mediator reduces binding of both Swi/Snf and SAGA. (We previously showed that a gcn5 mutation reduces Swi/Snf binding (35).) Thus, in addition to Swi5 being required to recruit the coactivators, any single coactivator mutation reduces binding of the other coactivators. We conclude that binding of these coactivators is interdependent at the HO promoter.
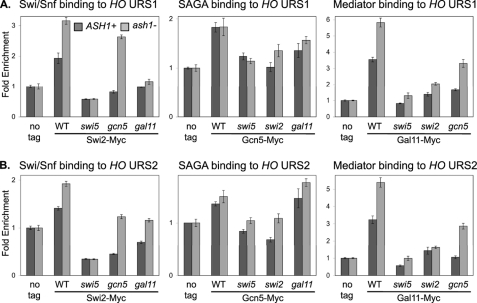
FIGURE 2.
Coactivator mutations reduce coactivator binding to HO. ChIP experiments were performed with logarithmically growing untagged control strains (DY150, wild type; DY4394, ash1; DY161, swi5; DY4846, swi5 ash1; DY12761, swi2(E834K); DY9711, swi2(E834K) ash1; DY5925, gcn5; DY7387, gcn5 ash1; DY5628, gal11; and DY14144, gal11 ash1), SWI2-Myc strains (DY6151, wild type; DY7403, ash1; DY9395, swi5; DY6378, swi5 ash1; DY8738, gcn5; DY8736, gcn5 ash1; DY14147, gal11; and DY14148, gal11 ash1), GCN5-Myc strains (DY7196, wild type; DY7194, ash1; DY6067, swi5; DY14137, swi5 ash1; DY12861, swi2(E834K); DY11278, swi2(E834K) ash1; DY7240, gal11; and DY14140, gal11 ash1), and GAL11-Myc strains (DY6148, wild type; DY6739, ash1; DY6197, swi5; DY6741, swi5 ash1; DY12729, swi2(E834K); DY14186, swi2(E834K) ash1; DY12711, gcn5; DY14142, gcn5 ash1). A, binding to URS1 was measured with primers that amplify from −1429 to −1139. B, binding to URS2 was measured with primers that amplify from −825 to −489.
We considered the possibility that coactivators are needed to facilitate Swi5 binding to a chromatin template. By this model, Swi5 interacts with coactivators as it attempts to bind to HO, and the coactivators assist Swi5 in binding. To test this idea, we examined the ability of Swi5 to bind in strains with coactivator mutations, but we found that Swi5 displayed no defect in binding either to HO URS1 or to another Swi5-dependent gene, PIR1, in coactivator mutant strains (supplemental Fig. S2). Thus, Swi5 is able to bind without assistance from coactivators.
ash1 Mutation Affects Coactivator Recruitment to HO
An ash1 mutation can suppress transcriptional defects at HO, allowing expression despite loss of Swi2 or Gcn5 coactivator functions (35). We therefore investigated the effects of an ash1 mutation on coactivator binding. By comparing the wild type and ash1 mutants, we saw a marked increase in Swi/Snf and Mediator binding to HO URS1 in the ash1 mutants (Fig. 2A, compare dark and light gray bars). This increased binding could be due, at least in part, to loss of Ash1 repression of HO expression in daughter cells (15). We next examined whether ash1 can suppress the defects in coactivator binding caused by other coactivator mutants. Although a gcn5 mutation reduces binding of both Swi/Snf and Mediator to HO URS1, Swi/Snf and Mediator do bind to HO in the gcn5 ash1 double mutant, and thus ash1 suppresses the gcn5 defect (Fig. 2A, compare dark and light gray bars). In contrast, ash1 does not suppress the defects in coactivator binding caused by swi2 or gal11 mutations. Finally, an ash1 mutation cannot suppress a swi5 disruption eliminating the DNA-binding protein that recruits the coactivators. Gcn5 is a histone acetyltransferase and Ash1 recruits the Rpd3(L) histone deacetylase to HO URS1 (described below), and these results are consistent with histone acetylation playing a role in facilitating binding of Swi/Snf and Mediator to HO URS1. We conclude that Ash1 inhibits Swi/Snf and Mediator binding to HO, binding that is stimulated by Gcn5-dependent histone acetylation.
We also examined coactivator binding to the URS2 region of the HO promoter in various mutants (Fig. 2B). Disruption of the SWI5 gene encoding the DNA-binding protein that recognizes sites in URS1 eliminates coactivator binding to URS2, as do mutations in other coactivators. This is expected, as these mutations eliminate coactivator recruitment to URS1, and events at URS1 are a prerequisite for factor recruitment to URS2 (3). An ash1 mutation results in increased Swi/Snf and Mediator binding to URS2 and also suppresses the defects in Swi/Snf and Mediator binding at URS2 caused by a gcn5 mutation. This suggests that increased coactivator recruitment at URS1 also results in increased coactivator binding at URS2.
Ash1 Recruits Rpd3(L) to the URS1 Region of the HO Promoter
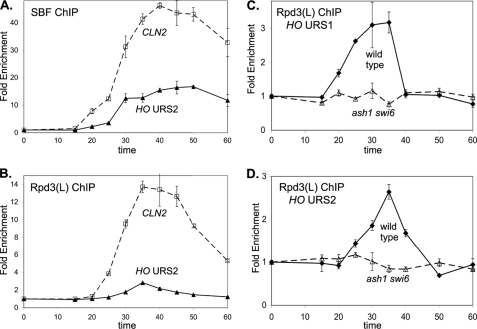
Mutations in subunits of the Rpd3(L) complex allow HO transcription in the absence of normally required activators (5, 36). Additionally, mass spectroscopic analysis of purified Rpd3(L) complex identified peptides corresponding to Ash1 (16). Based on these results, we performed ChIP assays with synchronized Ash1-Myc and Sds3-Myc strains to examine binding of Ash1 and Rpd3(L) to the HO promoter. Sds3 is a subunit of Rpd3(L) but is absent from the Rpd3(S) complex (17). Ash1 and Rpd3(L) bind to HO URS1 with similar kinetics during the cell cycle (Fig. 3A). Rpd3(L) is recruited to the CLN1 and CLN2 genes (37), but Ash1 does not bind to these promoters (supplemental Fig. S3A) or to HO URS2 (Fig. 3A). To identify where Ash1 and Rpd3(L) bind at HO, we performed ChIP assays using log phase cells and analyzed binding using PCR primers throughout the HO promoter (Fig. 3B). Binding for each protein is centered at approximately −1200 in URS1. Binding of Rpd3(L) at the −1000 to −900 region is stronger than Ash1 binding, and we therefore examined Rpd3(L) ChIP in synchronized cells. Rpd3(L) binding to URS1 can be seen at 25 min following release, whereas binding at −900 in URS2 is only modest until 30 min (Fig. 3C). Additionally, this Rpd3(L) binding at −900 corresponds to the left half of URS2 where SBF binds (Fig. 1A), consistent with SBF recruiting Rpd3(L) to promoters (see below). Importantly, Rpd3(L) binds to HO URS1 slightly earlier in the cell cycle than to URS2 (compare Figs. 3E and 4A, wild type).
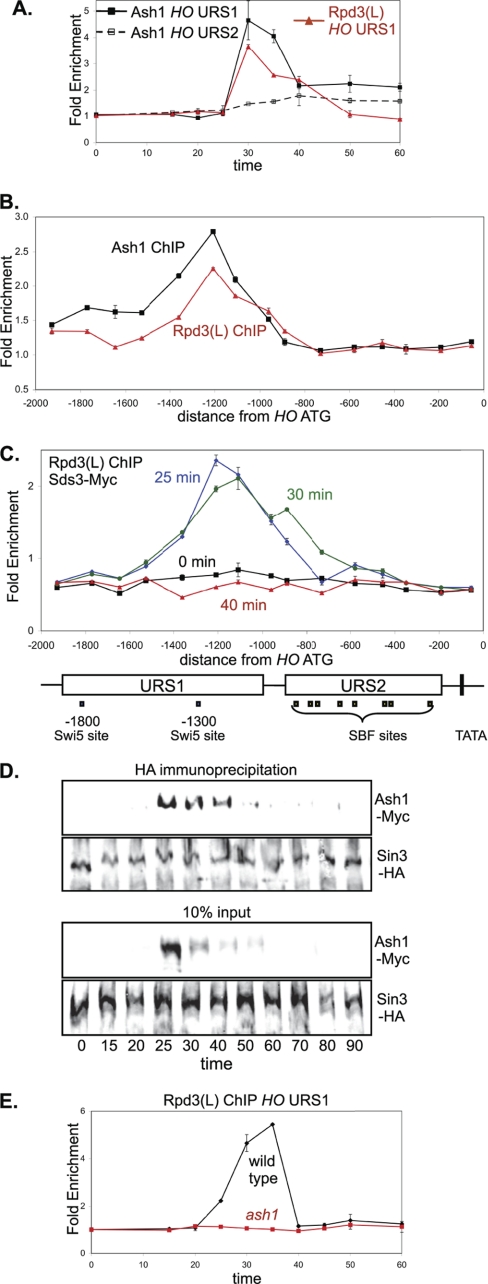
FIGURE 3.
Ash1 recruits Rpd3(L) to HO URS2. A, strains DY8312 (GALp::CDC20 ASH1-Myc) and DY12247 (GALp::CDC20 SDS3-Myc) were synchronized, and ChIP experiments were performed to measure Ash1 and Rpd3(L) binding to the HO promoter. Binding to URS1 and URS2 was measured with primers that amplify from −1429 to −1139 and from −825 to −489, respectively. Sds3 is a subunit specific to Rpd3(L). B, ChIP samples were prepared from logarithmically growing strains DY8312 (ASH1-Myc) and DY12247 (SDS3-Myc), and binding was analyzed with 15 sets of PCR primers across the HO promoter. C, strain DY12247 (GALp::CDC20 SDS3-Myc) was synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Binding was analyzed with 15 sets of PCR primers across the HO promoter. URS1-, URS2-, Swi5-, and SBF-binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20. D, strain DY13197 (GALp::CDC20 Ash1-Myc SIN3-HA) was synchronized, and protein samples were taken at various times following release from CDC20 arrest. Sin3-HA was immunoprecipitated with anti-HA antibody and analyzed on Western blots, along with controls corresponding to 10% of the input before immunoprecipitation, and the blots were probed with anti-HA and anti-Myc antibodies. E, strains DY12247 (GALp::CDC20 SDS3-Myc) and DY12251 (GALp::CDC20 SDS3-Myc ash1) were synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Binding to HO URS1 was measured with primers that amplify from −1429 to −1139.
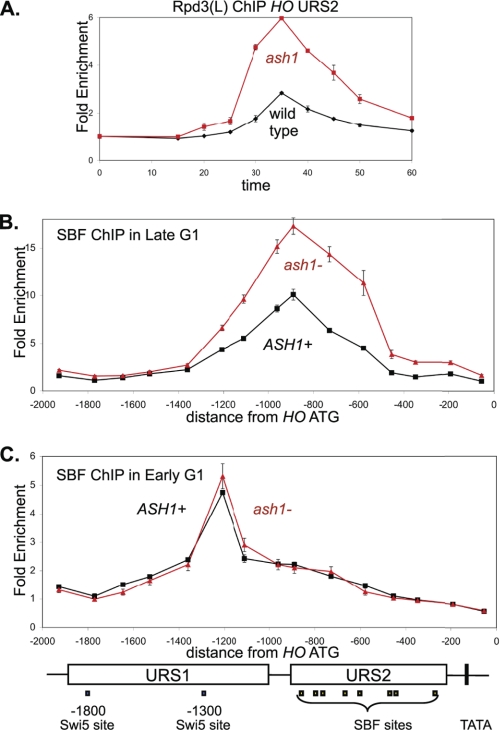
FIGURE 4.
ash1 mutation results in increased SBF and Rpd3L binding. A, strains DY12247 (GALp::CDC20 SDS3-Myc) and DY12251 (GALp::CDC20 SDS3-Myc ash1) were synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Rpd3(L) binding to HO URS2 was measured with primers that amplify from −825 to −489. Sds3 is a subunit specific to Rpd3(L). B, strains DY12794 (GALp::CDC20 SWI4-Myc) and DY12795 (GALp::CDC20 SWI4-Myc ash1) were synchronized, and ChIP samples were taken at 35 min following release from CDC20 arrest, when cells are in late G1. SBF binding was analyzed with 15 sets of PCR primers across the HO promoter. C, strains DY12794 (GALp::CDC20 SWI4-Myc) and DY12795 (GALp::CDC20 SWI4-Myc ash1) were synchronized, and ChIP samples were taken at 20 min following release from CDC20 arrest, when cells are in early G1. SBF binding was analyzed with 15 sets of PCR primers across the HO promoter. URS1-, URS2-, Swi5-, and SBF-binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20.
The ASH1 gene is cell cycle regulated (14), and the protein is rapidly degraded (38, 39). Thus, it seemed unlikely that Ash1 associates with Rpd3(L) throughout the cell cycle, and we investigated this question with coimmunoprecipitation experiments. We used synchronized cells to examine the interaction of Ash1-Myc with the Sin3-HA subunit of Rpd3(L) during the cell cycle (Fig. 3D). The immunoprecipitation shows the strongest Ash1-Rpd3(L) interaction at 25–30 min following release from a CDC20 arrest, shortly before both proteins were detected at URS1. The results suggest that Ash1 recruits Rpd3(L) to URS1, and Rpd3(L) binding to URS1 ends when Ash1 is degraded. In support of this idea, we find that an ash1 mutation eliminates Rpd3(L) binding to URS1 (Fig. 3E). Rpd3(L) is recruited to other genes, including CLN1 and CLN2 (37), but an ash1 mutation does not alter Rpd3(L) binding to these promoters (supplemental Fig. S3B). We conclude that Ash1 is required for Rpd3(L) binding to HO URS1 and likely recruits Rpd3(L) to this promoter region.
ash1 Mutation Affects SBF and Rpd3(L) Binding to URS2
An ash1 mutation results in a strong increase in Rpd3(L) binding to HO URS2 (Fig. 4A). Additionally, SBF binding to HO URS2 assayed by ChIP is markedly increased in the ash1 mutant (Fig. 4B), but ash1 does not affect SBF binding to the −1166 site in HO URS1 (Fig. 4C), CLN1, or CLN2 (supplemental Fig. S4A). There are two surprising results here. First, Ash1 does not bind to URS2 but seems to be responsible for decreasing the interaction of SBF and Rpd3(L) with URS2. Second, Ash1 recruits Rpd3(L) to URS1, but it prevents recruitment of Rpd3(L) to URS2.
An ash1 mutation results in increased binding of Swi/Snf and Mediator to HO URS1 and URS2 (Fig. 2), and we hypothesized that increased Swi/Snf at HO could result in more SBF binding. SBF binding to URS2 is markedly reduced in cells with a mutation in the Swi2 subunit of the Swi/Snf chromatin remodeler (supplemental Fig. S4B), suggesting that Swi/Snf is required for SBF binding to this promoter. In contrast, the swi2 mutation has no effect on SBF binding at CLN2 (supplemental Fig. S4C), although SBF does recruit Swi/Snf to the CLN2 promoter (supplemental Fig. S4D). Thus Swi/Snf is required for SBF to bind at the URS2 region of the HO promoter, but SBF binding to CLN2 is independent of Swi/Snf. In all likelihood, the chromatin at HO is inhibitory to SBF binding. Finally, an ash1 mutation allows SBF to bind to HO despite a mutant Swi/Snf chromatin remodeling factor (supplemental Fig. S4B), but the ash1 mutation has no effect on SBF binding at CLN2 (supplemental Fig. S4C) or on CLN2 expression (supplemental Fig. S4E). These results suggest that the chromatin at HO somehow inhibits SBF from binding and that SBF binding is regulated in opposing fashion by Swi/Snf and Ash1-Rpd3(L).
SBF and Rpd3(L) Binding to HO URS2 Is Weak and Delayed Compared with CLN2
We have shown that there is increased binding of SBF and Rpd3(L) to URS2 in the ash1 mutant. Additionally, SBF and Rpd3(L) appear to bind sooner to URS2 in the ash1 mutant compared with wild type. To investigate this further, we performed ChIP experiments with synchronized cells to compare the kinetics of SBF and Rpd3(L) binding to two SBF-dependent promoters, HO and CLN2. Importantly, HO is repressed by Ash1, but CLN2 is not regulated by Ash1. Ash1 does not bind to CLN2 (supplemental Fig. S3A), and an ash1 mutation does not affect CLN2 expression (supplemental Fig. S4E), binding of SBF to CLN2 (supplemental Fig. S3C), or binding of Rpd3(L) to CLN2 (supplemental Fig. S4A). The ChIP experiments clearly show that SBF (Fig. 5A) and Rpd3(L) (Fig. 5B) binding to HO URS2 is substantially delayed and reduced compared with their binding to CLN2. Taken with the results in Fig. 4, this suggests that Ash1 inhibits SBF binding to HO URS2, resulting in binding that is both delayed and reduced. This is in stark contrast with HO URS1, where Ash1 is responsible for recruiting Rpd3(L), revealing complex roles for Ash1 at different sites within the same promoter.
FIGURE 5.
SBF and Rpd3(L) binding to HO URS2 is weak compared with binding at CLN2. A, strain DY12794 (GALp::CDC20 SWI4-Myc) was synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. SBF binding to HO URS2 was measured with primers that amplify from −825 to −489 and binding to CLN2 with primers that amplify from −661 to −379. B, strain DY12247 (GALp::CDC20 SDS3-Myc) was synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. SBF binding to HO URS2 was measured with primers that amplify from −825 to −489 and binding to CLN2 with primers that amplify from −661 to −379. Sds3 is a subunit specific to Rpd3(L). C, strains DY12247 (GALp::CDC20 SDS3-Myc) and DY12830 (GALp::CDC20 SDS3-Myc ash1 swi6) were synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Rpd3(L) binding to HO URS1 was measured with primers that amplify from −1429 to −1139. D, strains DY12247 (GALp::CDC20 SDS3-Myc) and DY12830 (GALp::CDC20 SDS3-Myc ash1 swi6) were synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Rpd3(L) binding to HO URS2 was measured with primers that amplify from −825 to −489.
SBF and Whi5 Recruit Rpd3(L) to the URS2 Region of the HO Promoter
Rpd3(L) is recruited to URS1 by Ash1 (Fig. 3E), but Rpd3(L) recruitment to URS2 is independent of Ash1 (Fig. 4A). SBF is a good candidate for a DNA-binding factor that recruits Rpd3(L) to HO URS2, as a SWI6 gene disruption affecting SBF eliminates Rpd3(L) binding to SBF-dependent genes CLN1 and CLN2 (37). According to this view, Ash1 recruits Rpd3(L) to URS1 and subsequently SBF recruits Rpd3(L) to URS2. This predicts that Rpd3(L) should not bind at all to any part of the HO promoter in an ash1 swi6 double mutant. Indeed, ChIP experiments show no Rpd3(L) binding to either URS1 (Fig. 5C) or URS2 (Fig. 5D) of the HO promoter in ash1 swi6 cells.
Stb1 and Whi5 interact with SBF and are described as inhibitors of activation by SBF (40–42). We have shown that Rpd3(L) binding to the CLN1 and CLN2 SBF-dependent genes is lost in a whi5 stb1 double mutant (37). Using synchronized cells, we examined the effect of stb1 and whi5 mutations on Rpd3(L) binding to the HO promoter (supplemental Fig. S5). Although the stb1 and whi5 single mutations have only a minor effect on Rpd3(L) binding at HO URS1, they each reduce Rpd3(L) binding to HO URS2, especially whi5. Importantly, Rpd3(L) binding to HO URS2 is eliminated in the stb1 whi5 double mutant strain, whereas binding to HO URS1 is largely unaffected. We conclude that SBF and Whi5 are required to recruit Rpd3(L) to the URS2 region of the HO promoter.
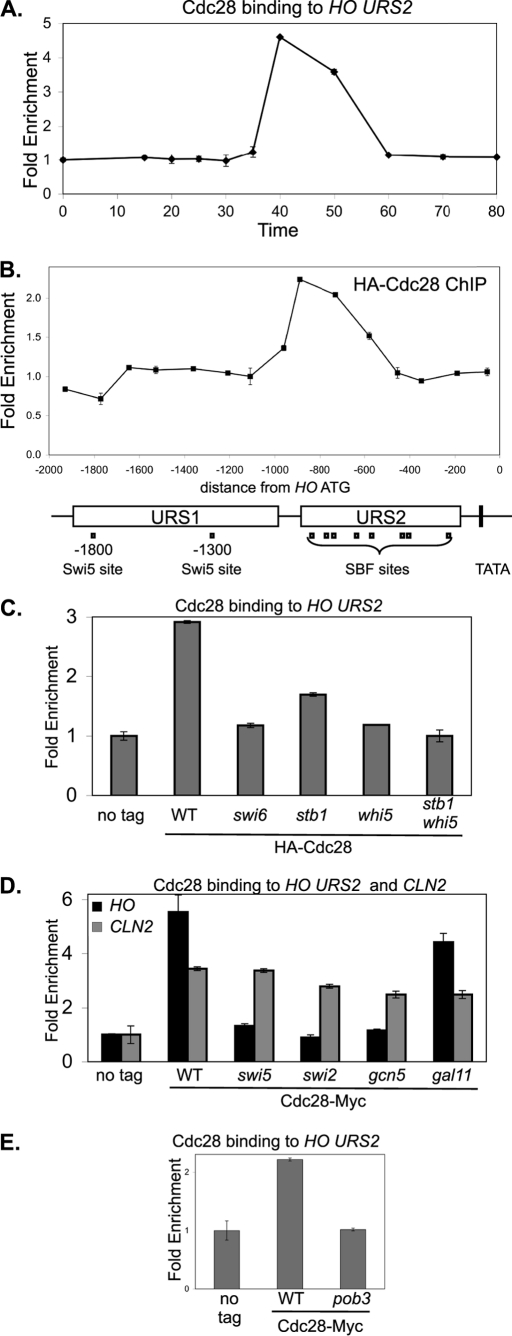
Cdc28 Kinase Is Recruited to HO URS2
The Cdc28 CDK has long been implicated in transcriptional activation by SBF and MBF (43), and Cdc28 is recruited to SBF- and MBF-dependent promoters by Swi6 and Whi5 (37). Using ChIP assays to examine Cdc28 binding to the HO promoter in synchronized cells, we find Cdc28 binds to HO URS2 at 40 min following release from a CDC20 arrest (Fig. 6A). ChIP assays from asynchronous cells show that Cdc28 binds to the left half of URS2 (Fig. 6B); this corresponds well with the time and place of SBF binding (Fig. 1).
FIGURE 6.
Cdc28 kinase is recruited to HO URS2. A, strain DY6669 (GALp::CDC20 SWI4-Myc) with HA-CDC28 on a YCp-KanMX plasmid was synchronized, and ChIP samples were taken at various times following release from CDC20 arrest. Cdc28 binding to HO URS2 was measured with primers that amplify from −825 to −489. B, ChIP samples prepared from logarithmically growing strain DY150 with either a YCp-KanMX(HA-CDC28) plasmid or an empty YCp-KanMX vector were synchronized, and Cdc28 binding was analyzed with 15 sets of PCR primers across the HO promoter. URS1-, URS2-, Swi5-, and SBF-binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20. C, ChIP experiments were performed with logarithmically growing untagged control strains (DY150, wild type; DY6999, swi6; DY13454, stb1; DY9559, whi5; and DY13640, whi5 stb1) and CDC28-Myc strains (DY13020, wild type; DY13723, swi6; DY13729, stb1; DY13731, whi5; and DY13733, stb1 whi5). Cdc28 binding to HO URS2 was measured with primers that amplify from −825 to −489. D, ChIP experiments were performed with logarithmically growing strains (DY150, wild type; DY161, swi5; DY5270, swi2Δ; DY5925, gcn5; and DY5628, gal11) containing either a YCp-KanMX(HA-CDC28) plasmid or an empty YCp-KanMX vector. Cdc28 binding to HO URS2 was measured with primers that amplify from −825 to −489 and binding to CLN2 with primers that amplify from −661 to −379. E, ChIP experiments were performed with logarithmically growing untagged control strains (DY150, wild type; DY7379, pob3(L78R)) and CDC28-Myc strains (DY13020, wild type; DY13527, pob3(L78R)), grown at 25 °C. Cdc28 binding to HO URS2 was measured with primers that amplify from −825 to −489.
We also examined which factors are required for Cdc28 recruitment to HO URS2. We previously showed that a swi6 mutation (affecting SBF) or a whi5 mutation eliminates recruitment of Cdc28 to SBF-dependent promoters such as CLN2, whereas a stb1 mutation has only a modest effect (37). A similar effect is also seen at HO, although the stb1 mutation causes a greater effect at HO than at CLN2 (Fig. 6C). Mutations affecting the Swi5 DNA-binding protein and the Swi/Snf remodeler eliminate SBF binding to HO (3), and the swi5 and swi2 mutations also eliminate Cdc28 recruitment to HO (Fig. 6D). Interestingly, a gcn5 mutation affecting the histone acetyltransferase in SAGA eliminates Cdc28 recruitment (Fig. 6D) but only slightly reduces SBF binding to HO (3). Finally, a gal11 mutation affecting Mediator has only modest effects on either SBF binding (3) or Cdc28 recruitment (Fig. 6D). Importantly, these mutations do not affect Cdc28 recruitment to CLN2 (Fig. 6D).
Finally, we examined a mutation affecting the FACT chromatin reorganizing complex (44). FACT binds to HO URS2 and is required for both nucleosome eviction and gene expression (3). POB3 encodes one of the subunits of FACT, and we find that a pob3 mutation eliminates Cdc28 recruitment (Fig. 6E). A pob3 mutation also reduces SBF binding to HO URS2 (3). We conclude that factors required for SBF binding to HO are required for recruitment of the Cdc28 cyclin-dependent kinase. Whi5 and Stb1, which bind to SBF, are also required for Cdc28 recruitment.
ash1 Mutation Can Affect Events at the Distant TATA Element
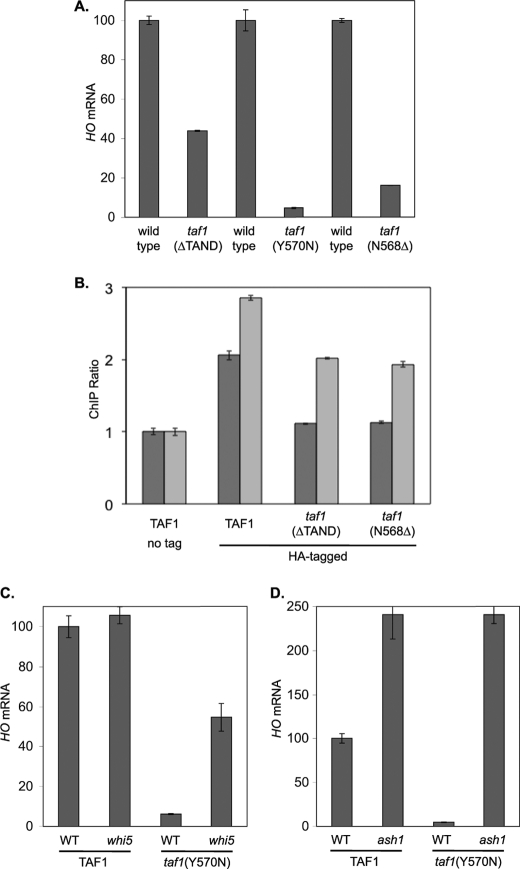
The TFIID complex contains the TATA-binding protein and also TATA-binding protein-associated factors (TAFs). Some promoters are defined as TAF-dependent because they require TFIID for activation (45). In another report, the physical interaction between Swi6 and TAFs was reported (46). To determine whether HO is a TAF-dependent promoter, we examined HO expression in strains with taf1 mutations (Fig. 7A). The taf1(ΔTAND), taf1(Y570N), and taf1(N568Δ) mutations all show defects in HO expression, indicating that HO is TAF-dependent. ChIP experiments with HA-tagged TAF1 show that TFIID binds to the HO TATA element and that this binding is lost in strains with taf1 mutations (Fig. 7B).
FIGURE 7.
Defect in HO expression caused by a taf1 mutation can be suppressed by whi5 or ash1. A, HO mRNA levels were measured by RT-qPCR from cells growing logarithmically at 33 °C: DY150, wild type; DY10598, taf1(ΔTAND); DY11348, taf1Δ YCp(TRP1)-TAF1; DY11349, taf1Δ YCp(TRP1)-TAF1(Y570N); DY12303, taf1Δ YCp(HIS3)-TAF1-HA; and DY12305, taf1Δ YCp(HIS3)-TAF1(N568Δ)-HA. B, ChIP experiments were performed from cells growing logarithmically at 33 °C: DY10204 (taf1Δ YCp(TRP1)-TAF1) and DY10366 (taf1Δ YCp(TRP1)-TAF1 ash1) as untagged controls. YCp(HIS3) plasmids expressing TAF1-HA, TAF1(T657K)-HA, or TAF1(N568Δ)-HA were shuffled into strains DY10204 (ASH1) and DY10366 (ash1), and ChIP experiments were performed with the HA-tagged TAF1 as the only copy in the cell. Taf1 binding to the HO TATA was measured with primers that amplify from −43 to +175. C, HO mRNA levels were measured by RT-qPCR from cells growing logarithmically at 33 °C: DY11348, taf1Δ YCp(TRP1)-TAF1; DY11574, taf1Δ YCp(TRP1)-TAF1 whi5; DY11349, taf1Δ YCp(TRP1)-TAF1(Y570N); and DY11575, taf1Δ YCp(TRP1)-TAF1(Y570N) whi5. D, HO mRNA levels were measured by RT-qPCR from cells growing logarithmically at 33 °C: DY11348, taf1Δ YCp(TRP1)-TAF1; DY11570, taf1Δ YCp(TRP1)-TAF1 ash1; DY11349, taf1Δ YCp(TRP1)-TAF1(Y570N); and DY11571, taf1Δ YCp(TRP1)-TAF1(Y570N) ash1.
We next constructed strains to see if mutations in negative regulators could suppress the taf1(Y570N) defect in HO expression. A whi5 mutation allows HO expression despite the taf1(Y570N) mutation (Fig. 7C). In wild type cultures, an ash1 mutation increases HO expression (Fig. 7D) and also increases TFIID binding to the TATA (Fig. 7B), possibly because HO is expressed in both mothers and daughters (13, 14). Importantly, the defects in both HO expression (Fig. 7B) and TFIID binding (Fig. 7D) in the taf1(Y570N) mutant are suppressed by an ash1 mutation. This result is noteworthy as Ash1 binds more than 1 kb upstream of the TATA element, at a time before when TFIID is recruited (Fig. 3). We conclude that the transient binding of Ash1 to the upstream region of the HO promoter affects a distant region of the promoter, making the core promoter TAF-dependent.
DISCUSSION
Studies of the budding yeast HO gene have been very productive in elucidating novel mechanisms in gene regulation (3, 7, 26, 47). The gene encodes an endonuclease that must be tightly regulated as inappropriate expression could be deleterious to the cell. The promoter has evolved to be tightly repressed, with multiple activators and coactivators required to overcome this repression. HO has two distinct promoter regions that function sequentially during the cell cycle (3). At URS1, the Swi5 DNA-binding factor recruits the Swi/Snf, SAGA, and Mediator coactivators that affect changes in chromatin structure. These effects at URS1 are somehow then transmitted to the downstream URS2 region, where chromatin changes permit the SBF DNA-binding factor to bind, recruit coactivators, and finally activate transcription. SBF is held in an inactive form by the Whi5 and Stb1 corepressors (40–42); this inhibition is overcome by the Cdc28 cyclin-dependent kinase, which we show is recruited to the HO promoter by its substrates Whi5 and Stb1 (Fig. 6).
HO expression is under three distinct types of regulation, as the gene is cell cycle regulated, it is expressed in haploid cells but not in diploids, and it is expressed in mother cells but not in daughter cells. This asymmetric expression of HO only in mother cells is determined by the Ash1 protein, which accumulates primarily in daughter cells and blocks HO expression there (13, 14). However, Ash1 does not accumulate exclusively in daughter cells, and regulation of HO in mother cells is significantly different in ASH1 and ash1 cells (35).
The Rpd3(L) histone deacetylase has long been known to negatively regulate HO expression (5), and the identification of Ash1 as a component of the purified Rpd3(L) complex (16) links these two repressive factors. However, Ash1 is cell cycle regulated (14, 38), making it unlikely that Ash1 associates with Rpd3(L) throughout the cell cycle. We used coimmunoprecipitation experiments with synchronized cells to demonstrate that Ash1 associates with Rpd3(L) for only a brief period during G1, and ChIP experiments show Ash1 recruits Rpd3(L) to HO URS1 at this time (Fig. 3). Additionally, we show that Rpd3(L) is recruited twice to the HO promoter during the cell cycle, first to URS1 by Ash1, and subsequently to URS2 by Whi5 and Stb1, which interact with the SBF DNA-binding protein. Rpd3(L) recruitment to the two promoter regions is independent, as an ash1 mutation only eliminates binding to URS1, whereas Rpd3(L) binding to URS2 is specifically eliminated in a whi5 stb1 mutant.
SBF is thought to be the proximal transcription factor for HO activation. Here, we show several unusual features of SBF binding to the URS2 region of the HO promoter. First, although there are eight binding sites for SBF distributed throughout the 700-bp URS2 region, ChIP experiments show that in vivo SBF binds primarily to sites only at the left end of URS2 (Fig. 1). Electrophoretic mobility shift assays show that SBF binds equally well in vitro to sites at the left and right ends of URS2, suggesting that chromatin structure restricts access to the right half of URS2. This idea is consistent with the sequential chromatin changes that occur along the promoter during the cell cycle, with eviction of nucleosomes from the left end of URS2 occurring before eviction at the right end (3). Second, SBF binding to HO is much slower and weaker than at CLN2, another SBF-dependent promoter (Fig. 5). These results are consistent with chromatin inhibiting SBF binding to HO, especially because the SBF-binding sites at the CLN2 promoter are in a nucleosome-depleted region (48). Finally, URS2 binding by SBF, as well as by the Rpd3(L) corepressor recruited by SBF, is markedly increased in an ash1 mutant (Fig. 4). Ash1 binds at approximately −1200 bp in URS1, yet it markedly affects SBF binding at URS2, at a region from −850 to −650 bp. Thus Ash1, bound at URS1, affects SBF binding at URS2, some 450 bp away, suggesting interactions between these two regions of the promoter.
The first event in HO activation is binding of the Swi5 DNA-binding protein to two sites in URS1. Swi5 recruits three coactivator complexes, Swi/Snf, SAGA, and Mediator, causing changes in chromatin structure. We show that mutations affecting any one of the coactivators reduces binding of the other two, and thus binding of the coactivators is interdependent (Fig. 2). Such interdependence of coactivator binding has been seen at other promoters (12, 49). We suggest that chromatin structure at HO is strongly inhibitory, and mutual action by the coactivators is needed to overcome the repressive environment. Interestingly, Ace2, which has a DNA binding specificity nearly identical to that of Swi5 and binds to the HO promoter in vitro, does not bind to HO in vivo and thus does not activate HO expression (27, 51). This result is consistent with the idea of an unusually repressive chromatin structure at HO, as it suggests that the binding and activation capabilities of Swi5 require more than accessibility of its binding site. Binding of Cat8 to some promoters is reduced by coactivator mutations (12), and thus we considered the idea that coactivators assist Swi5 in binding at HO, whereas Ace2 cannot bind because it does not get coactivator help. However, Swi5 binding to HO is unaffected by mutations in these coactivators (supplemental Fig. S2). Finally, it is believed that Mediator serves as a bridge between upstream activator proteins and the basic polymerase II machinery (52, 53), and thus a role for Mediator at URS1 at a time when polymerase is absent from the promoter remains enigmatic.
Binding of the Swi/Snf and Mediator coactivators to HO URS1 is increased in an ash1 mutant (Fig. 2). Coactivator recruitment to HO URS1 is interdependent in ASH1 cells, as any single coactivator mutation reduces binding of the other coactivators. Swi/Snf and Mediator binding to URS1 is sharply reduced in a gcn5 mutant, but binding is significantly restored in a gcn5 ash1 double mutant. Thus, ash1 suppresses the defect in Swi/Snf and Mediator binding caused by a gcn5 mutation. Ash1 recruits the Rpd3(L) histone deacetylase to HO URS1, whereas GCN5 encodes a histone acetyltransferase, and thus it is reasonable that Ash1 and Gcn5 would act in opposition. This suggests that binding of Swi/Snf and Mediator to HO URS1 is facilitated by histone acetylation.
The Swi2 subunit of Swi/Snf contains a bromodomain, providing a possible mechanism, as bromodomains bind acetylated lysines present on histones (54–56). Mediator interacts directly with nucleosomes (57), although its binding is not known to be affected by histone acetylation. Interestingly, a multicopy plasmid with ASH1 can suppress a temperature-sensitive mutation in the Med21 subunit of Mediator (50), suggesting that Ash1 can modulate Mediator function.
Based on the timing of Ash1 activity at the HO promoter, the effect of Ash1 on coactivator binding appears to be the consequence of an action performed in the previous cell cycle. Swi5 enters the nucleus and binds to promoters such as HO URS1 in late M/early G1, at 20 min following release from a CDC20 arrest (3). Swi/Snf, SAGA, and Mediator are recruited to HO URS concurrently (3). At this time Swi5 also binds to the ASH1 promoter and activates ASH1 expression (14, 27). ChIP experiments show Ash1 binding to HO URS1 at 30 min following release from a CDC20 arrest (Fig. 3A). Thus, the Swi/Snf and Mediator binding that Ash1 affects appears to occur before Ash1 actually binds, and we conclude that the Ash1 effect occurred in the previous cell cycle. Ash1 is an unstable protein (38, 39) and is present only transiently during the cell cycle (Fig. 3D). We suggest that the histone modifications caused by Rpd3(L), recruited by Ash1, persist through the next cell cycle to affect binding of Swi/Snf and Mediator. If correct, this provides a model for studying the epigenetic control of transcription.
HO expression is markedly reduced in strains with a mutation in TAF1, one of the subunits of the TFIID complex that contains TATA-binding protein (Fig. 7). Interestingly, this transcriptional defect caused by a taf1 mutation can be suppressed by an ash1 mutation, despite the fact that Ash1 binds to the URS1 region of the HO promoter more than a kilobase distant from the TATA element. How does Ash1 activity make the HO TATA element TAF-dependent? Although it is possible that the URS1 region where Ash1 binds interacts with the TATA element, the effect can also be explained by our observation that an ash1 mutation results in increased binding of SBF at URS2 (Fig. 4B), as well as increased binding by Swi/Snf and Mediator (Fig. 2B). The TAF dependence of a promoter could therefore be a consequence of the amount of binding by transcription factors and coactivators. Finally, global studies of gene regulation find that most promoters can be categorized as either SAGA-dependent or TFIID-dependent (45). In contrast, transcriptional activation at the HO promoter, at least in an ASH1 strain, requires both SAGA and TFIID. This may reflect the uniquely repressive nature of this promoter, such that both SAGA and TFIID are required to overcome this repression.
Supplementary Material
Acknowledgments
We thank Linda Breeden, David Kirkpatrick, Tetsuro Kokubo, and Kim Nasmyth for plasmids. We thank Tim Formosa and members of the laboratory for comments on the manuscript and for helpful advice throughout the course of this project.
This work was supported, in whole or in part, by National Institutes of Health grants.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Tables S1–S3, and additional references.
- qPCR
- quantitative PCR
- TAF
- TATA-binding protein-associated factor.
REFERENCES
- 1. Cairns B. R. (2009) Nature 461, 193–198 [DOI] [PubMed] [Google Scholar]
- 2. Weake V. M., Workman J. L. (2010) Nat. Rev. Genet. 11, 426–437 [DOI] [PubMed] [Google Scholar]
- 3. Takahata S., Yu Y., Stillman D. J. (2009) Mol. Cell 34, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peterson C. L., Herskowitz I. (1992) Cell 68, 573–583 [DOI] [PubMed] [Google Scholar]
- 5. Stillman D. J., Dorland S., Yu Y. (1994) Genetics 136, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nasmyth K. (1985) Cell 42, 213–223 [DOI] [PubMed] [Google Scholar]
- 7. Cosma M. P., Tanaka T., Nasmyth K. (1999) Cell 97, 299–311 [DOI] [PubMed] [Google Scholar]
- 8. Bhoite L. T., Allen J. M., Garcia E., Thomas L. R., Gregory I. D., Voth W. P., Whelihan K., Rolfes R. J., Stillman D. J. (2002) J. Biol. Chem. 277, 37612–37618 [DOI] [PubMed] [Google Scholar]
- 9. Adkins M. W., Howar S. R., Tyler J. K. (2004) Mol. Cell 14, 657–666 [DOI] [PubMed] [Google Scholar]
- 10. Verdone L., Wu J., van Riper K., Kacherovsky N., Vogelauer M., Young E. T., Grunstein M., Di Mauro E., Caserta M. (2002) EMBO J. 21, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwabish M. A., Struhl K. (2007) Mol. Cell. Biol. 27, 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biddick R. K., Law G. L., Chin K. K., Young E. T. (2008) J. Biol. Chem. 283, 33101–33109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sil A., Herskowitz I. (1996) Cell 84, 711–722 [DOI] [PubMed] [Google Scholar]
- 14. Bobola N., Jansen R. P., Shin T. H., Nasmyth K. (1996) Cell 84, 699–709 [DOI] [PubMed] [Google Scholar]
- 15. Cosma M. P. (2004) EMBO Rep. 5, 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Biochim. Biophys. Acta 1731, 77–87 [DOI] [PubMed] [Google Scholar]
- 17. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 18. Joshi A. A., Struhl K. (2005) Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 19. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 20. Chandarlapaty S., Errede B. (1998) Mol. Cell. Biol. 18, 2884–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan X., Heitman J. (2000) Mol. Cell. Biol. 20, 8364–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Talia S., Wang H., Skotheim J. M., Rosebrock A. P., Futcher B., Cross F. R. (2009) PLoS Biol. 7, e1000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 24. Rothstein R. (1991) Methods Enzymol. 194, 281–301 [DOI] [PubMed] [Google Scholar]
- 25. Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 26. Bhoite L. T., Yu Y., Stillman D. J. (2001) Genes Dev. 15, 2457–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voth W. P., Yu Y., Takahata S., Kretschmann K. L., Lieb J. D., Parker R. L., Milash B., Stillman D. J. (2007) EMBO J. 26, 4324–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eriksson P., Biswas D., Yu Y., Stewart J. M., Stillman D. J. (2004) Mol. Cell. Biol. 24, 6419–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sidorova J., Breeden L. (1999) Genetics 151, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biswas D., Takahata S., Stillman D. J. (2008) Mol. Cell. Biol. 28, 4445–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Breeden L., Nasmyth K. (1987) Cell 48, 389–397 [DOI] [PubMed] [Google Scholar]
- 32. Badis G., Chan E. T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C. D., Gossett A. J., Hasinoff M. J., Warren C. L., Gebbia M., Talukder S., Yang A., Mnaimneh S., Terterov D., Coburn D., Li Yeo A., Yeo Z. X., Clarke N. D., Lieb J. D., Ansari A. Z., Nislow C., Hughes T. R. (2008) Mol. Cell 32, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taba M. R., Muroff I., Lydall D., Tebb G., Nasmyth K. (1991) Genes Dev. 5, 2000–2013 [DOI] [PubMed] [Google Scholar]
- 34. Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G. (2005) Mol. Cell. Biol. 25, 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitra D., Parnell E. J., Landon J. W., Yu Y., Stillman D. J. (2006) Mol. Cell. Biol. 26, 4095–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dorland S., Deegenaars M. L., Stillman D. J. (2000) Genetics 154, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahata S., Yu Y., Stillman D. J. (2009) EMBO J. 28, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McBride H. J., Sil A., Measday V., Yu Y., Moffat J., Maxon M. E., Herskowitz I., Andrews B., Stillman D. J. (2001) Mol. Microbiol. 42, 345–353 [DOI] [PubMed] [Google Scholar]
- 39. Liu Q., Larsen B., Ricicova M., Orlicky S., Tekotte H., Tang X., Craig K., Quiring A., Le Bihan T., Hansen C., Sicheri F., Tyers M. (2011) Mol. Cell. Biol. 31, 584–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costanzo M., Nishikawa J. L., Tang X., Millman J. S., Schub O., Breitkreuz K., Dewar D., Rupes I., Andrews B., Tyers M. (2004) Cell 117, 899–913 [DOI] [PubMed] [Google Scholar]
- 41. de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., 3rd, Wittenberg C. (2004) Cell 117, 887–898 [DOI] [PubMed] [Google Scholar]
- 42. de Bruin R. A., Kalashnikova T. I., Wittenberg C. (2008) Mol. Cell. Biol. 28, 6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wittenberg C., Reed S. I. (2005) Oncogene 24, 2746–2755 [DOI] [PubMed] [Google Scholar]
- 44. Formosa T. (2008) Mol. Biosyst. 4, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 45. Huisinga K. L., Pugh B. F. (2004) Mol. Cell 13, 573–585 [DOI] [PubMed] [Google Scholar]
- 46. Sanders S. L., Jennings J., Canutescu A., Link A. J., Weil P. A. (2002) Mol. Cell. Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cosma M. P., Panizza S., Nasmyth K. (2001) Mol. Cell 7, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 48. Bai L., Ondracka A., Cross F. R. (2011) Mol. Cell 42, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu H., Hu C., Zhang F., Hwang G. J., Swanson M. J., Boonchird C., Hinnebusch A. G. (2005) Mol. Cell. Biol. 25, 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hallberg M., Hu G. Z., Tronnersjö S., Shaikhibrahim Z., Balciunas D., Björklund S., Ronne H. (2006) Mol. Genet. Genomics 276, 197–210 [DOI] [PubMed] [Google Scholar]
- 51. Dohrmann P. R., Butler G., Tamai K., Dorland S., Greene J. R., Thiele D. J., Stillman D. J. (1992) Genes Dev. 6, 93–104 [DOI] [PubMed] [Google Scholar]
- 52. Björklund S., Gustafsson C. M. (2005) Trends Biochem. Sci. 30, 240–244 [DOI] [PubMed] [Google Scholar]
- 53. Kornberg R. D. (2005) Trends Biochem. Sci. 30, 235–239 [DOI] [PubMed] [Google Scholar]
- 54. Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., Zhou M. M. (1999) Nature 399, 491–496 [DOI] [PubMed] [Google Scholar]
- 55. Ornaghi P., Ballario P., Lena A. M., González A., Filetici P. (1999) J. Mol. Biol. 287, 1–7 [DOI] [PubMed] [Google Scholar]
- 56. Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. (2000) EMBO J. 19, 6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lorch Y., Beve J., Gustafsson C. M., Myers L. C., Kornberg R. D. (2000) Mol. Cell 6, 197–201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.