Background: Mef2c gene expression is significantly diminished in the retinas of NRL (neural retina leucine zipper) knock-out mice.
Results: NRL binding, RNA polymerase II association, and acetylation of histone H3, revealed a novel alternate Mef2c promoter.
Conclusions: Activation of the retinal Mef2c promoter is NRL-dependent and specific to rod photoreceptor cells.
Significance: Mef2c represents a novel regulatory node downstream of NRL in adult rod photoreceptor cells.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Expression, Histone Modification, Photoreceptors, Retinal Degeneration, Transcription Promoter, Retinal Development
Abstract
Neural retina leucine zipper (NRL) is an essential transcription factor for cell fate specification and functional maintenance of rod photoreceptors in the mammalian retina. In the Nrl−/− mouse retina, photoreceptor precursors fail to produce rods and generate functional cone photoreceptors that predominantly express S-opsin. Previous global expression analysis using microarrays revealed dramatically reduced expression of myocyte enhancer factor Mef2c in the adult Nrl−/− retina. We undertook this study to examine the biological relevance of Mef2c expression in retinal rod photoreceptors. Bioinformatics analysis, rapid analysis of cDNA ends (5′-RACE), and reverse transcription coupled with qPCR using splice site-specific oligonucleotides suggested that Mef2c is expressed in the mature retina from an alternative promoter. Chromatin immunoprecipitation (ChIP) studies showed the association of active RNA polymerase II and acetylated histone H3 just upstream of Mef2c exon 4, providing additional evidence for the utilization of an alternative promoter in the retina. In concordance, we observed the binding of NRL to a putative NRL-response element (NRE) at this location by ChIP-seq and electrophoretic mobility shift assays. NRL also activated the Mef2c alternative promoter in vitro and in vivo. Notably, MEF2C could support Rhodopsin promoter activity in rod photoreceptors. We conclude that Mef2c expression from an alternative promoter in the retina is regulated by NRL. Our studies also implicate MEF2C as a transcriptional regulator of homeostasis in rod photoreceptor cells.
Introduction
Distinct gene expression patterns dictate cellular identity and function (1–3). Spatiotemporal control of gene expression is achieved by integrating multiple regulatory mechanisms, including transient or stable interactions between cis-regulatory DNA elements and trans-regulatory factors, epigenetic code, and signaling molecules (4). The neural retina provides an excellent paradigm to elucidate qualitative and quantitative control of gene expression in the developing and adult central nervous system (5). In the retina, six types of neuronal cells and one type of glial cell (Muller) differentiate from multipotent progenitor cells in a conserved order of birth, with each key step controlled by transcriptional regulatory proteins (6, 7). Rod and cone photoreceptors in the retina are light sensors that convert photons into electrical signals (phototransduction) under dim and bright light conditions, respectively (8). Cell type-specific genes are activated during the differentiation and maturation of photoreceptors (5). Precise control of gene expression is also essential for photoreceptor survival as both under- or overexpression of Rhodopsin and other genes can lead to photoreceptor degeneration (9–11). Dysfunction and death of rod photoreceptors is a common hallmark of many inherited retinal dystrophies in humans (5, 12). Pathological mutations involve genes required for phototransduction, outer segment morphogenesis, intracellular transport, and/or transcription factors that control the expression of genes involved in photoreceptor homeostasis (12).
Photoreceptor development and homeostasis are tightly regulated by key transcription factors that include orthodenticle homeobox 2 (13), cone rod homeobox (CRX)2 (14–17), photoreceptor-specific orphan nuclear receptor (NR2E3) (18–21), thyroid hormone receptor β2 (TRβ2) (22), retinoid-related orphan receptor β (RORβ) (23, 24), neural retina leucine zipper (NRL) (17, 25, 26), and estrogen-related receptor β (27). Although several of these transcriptional regulators are individually essential to generate a normal compliment of rod and cone photoreceptors during development, they also function cooperatively to regulate photoreceptor-specific genes in the mature retina (5).
NRL is the primary regulator of rod versus cone photoreceptor cell fate choice (25). Abrogation of Nrl expression in the mouse leads to a complete loss of rod photoreceptors and an increase in the fraction of cones expressing S-opsin (25). Ectopic expression of Nrl results in a rod-only mouse retina devoid of cone photoreceptors (28). NRL and its interacting protein CRX activate many rod photoreceptor-specific genes, whereas NRL and its downstream transcriptional target NR2E3 repress cone genes (18, 20, 21, 26, 29). Mutations of human NRL cause retinal degenerative diseases (30–32).
Nrl expression in mice is detected as early as embryonic day 12.5; however, the expression of phototransduction genes including Rhodopsin, are not detected until age P3 (5, 33). This “delay” in expression of rod maturation genes suggests the involvement of additional regulatory signals. We previously showed that NR2E3 is a direct transcriptional target of NRL (26). The primary role of NR2E3 appears to be suppression of cone gene expression, although it can contribute to the induction of rod-specific genes (18, 20, 21, 34). Estrogen-related receptor β, another potential target of NRL, participates in rod survival by regulating the expression of many genes (27). To identify additional transcription factors that control rod-specific gene expression, we took advantage of gene profiles generated from wild type and Nrl−/− retina and from their respective purified photoreceptors (33, 35). A survey of genes that are down-regulated in the absence of Nrl identified Mef2c (myocyte enhancer factor 2c) as a potential novel target of NRL.
MEF2C belongs to the MADS (MCM1-agamous-deficiens serum response factor) family of transcription factors and is essential for muscle, cardiovascular, and bone development (36–38). Among the four vertebrate MEF2 proteins, postnatal expression of MEF2C is restricted to muscle, brain, and spleen, whereas others (MEF2A, MEF2B, and MEF2D) are expressed ubiquitously (39). MEF2C and two other transcription factors (GATA4 and TBX5) are sufficient to reprogram mouse fibroblasts into functional cardiomyocytes (40). Conditional knock-out of Mef2c in mouse brain has revealed its essential role in early neurogenesis, neuronal migration, and differentiation (41, 42). MEF2C activity is modulated by post-translational modifications in response to cytoplasmic signals including calcium (37). Mutations in human MEF2C are associated with neurological disorders including mental retardation and seizures (43, 44).
To examine the expression and function of Mef2c in the retina, we have performed a comprehensive analysis using in silico, in vitro, and in vivo methods. Here we show that Mef2c transcripts in the retina originate from an alternative promoter upstream of exon 4. We also demonstrate that the Mef2c promoter is a direct transcriptional target of NRL, and MEF2C contributes to the regulation of Rhodopsin promoter activity in vivo.
EXPERIMENTAL PROCEDURES
Animal Care and Use
Animal Care and Use committees of Oakland University and the National Eye Institute approved all animal care and tissue collection procedures. C57BL/6J, rd1, and CD1 mice were obtained from Charles River Laboratories (Wilmington, MA) or the Jackson Laboratory (Bar Harbor, ME).
Antibodies
The following ChIP grade antibodies were used: anti-Pol II, a polyclonal antibody to total RNA polymerase II (Santa Cruz Biotechnology, Santa Cruz, CA); anti-Pol-II-S2, an antibody against transcriptionally active Pol II, phosphorylated on serine 2 of the C-terminal repeat domain (Abcam Inc., Cambridge, MA); anti-H3K9-Ac, a polyclonal antibody to histone H3 acetylated on lysine 9 (Abcam Inc.); and anti-NRL polyclonal antibody (45).
Cloning of the Mouse Mef2c Retinal Proximal Promoter and shRNA Plasmids
A 540-bp genomic fragment around Mef2c exon 4 was amplified from C57BL/6 mouse genomic DNA using Pfu Ultra (Agilent Technologies, Santa Clara, CA) and cloned into pGemTeasy (Promega, Madison, WI) to produce pGemTeasy-Mef2cP-Ret. The PCR primers including restriction sites for KpnI and XhoI are listed here with the genomic sequences underlined: forward KpnI, 5′-CGCAGGTACCTTTCATGTGTGTGTTCATATTTGCAC-3′, reverse XhoI, 5′-TGCACTCGAGATACCCCAATGAGACAAGAAGGC-3′. The Mef2c promoter was subcloned from pGemTEasy-Mef2cP-Ret into the KpnI/XhoI sites of pGL3-basic (Promega) to produce the luciferase-reporter construct: pGL3-Mef2cP-Luc. The same Mef2c promoter was released from pGL3-Mef2cP-Luc using KpnI/XhoI, blunt ended, and ligated into a GFP vector to produce the Mef2cP-GFP construct. Sequencing validated all constructs. The Nrl shRNA plasmid was generated to knockdown Nrl expression (46). Mef2c and Gapdh shRNA plasmids were purchased from Open Biosystems (Huntsville, AL).
5′ Rapid Analysis of cDNA Ends Derived from Full-length RNA (RACE)
Total RNA was isolated from mouse skeletal muscle, brain, and retina, respectively, using TRIzol reagent (Invitrogen). 5′-RACE was performed using the GeneRacerTM kit (Invitrogen) according to the manufacturer's instructions (47, 48). Briefly, 2 μg of RNA was treated with calf intestinal phosphatase to eliminate truncated mRNA and non-mRNA. End caps were removed from full-length mRNAs with tobacco acid pyrophosphatase, leaving a 5′-phosphate for ligation to the GeneRacer RNA oligo. Reverse transcription with random hexamers was used to synthesize cDNA. To obtain 5′ ends, the first strand cDNA was amplified using a Mef2c-specific reverse primer (5′-ATCTCACAGTCGCACAGCAC-3′) and the GeneRacerTM 5′ primer. The RACE PCR product was visualized on a 1.5% agarose gel, purified, and cloned into the pCR 4-TOPO vector using the TOPO TA cloning kit (Invitrogen) for sequencing. At least 15 clones were sequenced for each reaction.
RNA Polymerase II (Pol II) Chromatin Immunoprecipitation (ChIP) Hybridized to Promoter Tiling Arrays (ChIP-on-chip)
Data for the Mef2c gene were derived from the complete Pol II ChIP-on-Chip data set, which is available under GEO accession number GSE 19999. ChIP-on-chip was performed as previously detailed (49). Briefly, Pol II ChIP was carried out using retinas from CL57BL/6 mice at age P2 and P25. The ChIP DNA was amplified in a linear fashion using random priming amplification, fragmented, and labeled using the GeneChip Double-stranded DNA Terminal Labeling Kit (Affymetrix, Santa Clara, CA). The resulting biotinylated DNA probe was hybridized to GeneChip Mouse Promoter 1.0R arrays (Affymetrix). Biotinylated probe prepared from total genomic DNA (non-ChIP) was hybridized to a separate GeneChip for paired normalization. Tiling arrays were scanned with an Affymetrix GeneChip scanner. Fluorescence data for the ChIP samples (P2 and P25) were normalized to the total genomic DNA control using Affymetrix Tiling Analysis software. Signal values were exported in bar-file format for interval determination with Affymetrix Tiling Analysis software and visualization with Integrated Genome Browser software. Track intervals parameters were: minimum length 300, gap 180, and intermediate threshold of 4.
ChIP Quantitative Real-time PCR (qPCR)
ChIP assays using an antibody recognizing the active form of Pol II (50), or H3K9-Ac were performed with P2 and P25 mouse retinas as previously described (51). The ChIP DNA was quantified in triplicate by real time qPCR using SYBR Green Super mixture (Bio-Rad). The copy number of ChIP DNA for each test region was obtained by comparing the Ct values from ChIP DNA with a standard curve composed of known copy numbers of genomic DNA and their corresponding Ct values. The ChIP-qPCR signal was normalized to the qPCR signal from input DNA, and calculated as copies of DNA detected per 1000 genome equivalents of input DNA. An untranslated region on Chromosome-6 (Untr) served as a negative control (49). Primer sequences used were: Mef2c exon 4 promoter (forward, 5′-TGCAGAAAAGATTCCCACTTG-3′, reverse, 5′-AGACACTCACAAGGCAAAGAC-3′), Rhodopsin promoter (forward, 5′-CCCCTCTGCAAGCCAATT-3′, reverse, 5′-GCAACTCCAGGCACTGAC-3′), Recoverin (Rcvrn) promoter (forward, 5′-CTCCTCCCTCCAAGGACTG-3′, reverse, 5′-CAAGGCTGTGTGCTGCTATG-3′), Untr (forward, 5′-TCAGGCATGAACCACCATAC-3′, reverse, 5′-AACATCCACACGTCCAGTGA-3′).
NRL ChIP-sequencing (Seq) and ChIP Assays
ChIP using NRL antibody or normal IgG control was performed as described (26). ChIP DNA from retinas of C57BL/6J mice (age P28), was used to construct NRL and IgG (control) ChIP-seq libraries, according to the manufacturer's protocol (Illumina, San Diego, CA). Briefly, ChIP DNA was end-repaired, ligated to universal adaptors, and amplified in a linear fashion. The amplified DNA was purified and used for cluster generation and sequencing analysis using the Illumina 1G Genome Analyzer. The ChIP-seq reads were mapped to the mouse genome (mm8) using the Solexa Analysis Pipeline and NRL-binding regions were identified by Model-based Analysis of ChIP-seq (MACS) with a false discovery rate <10−6 (52).
For ChIP assays, separate experiments were performed using retinas from C57BL/6J mice and Nrl−/− mice (on C57BL/6J background) at P2 and P28 using NRL antibody. Normal rabbit IgG was used as a negative control. The ChIP DNA and input DNA control (without immunoprecipitation) were analyzed by PCR using the following primers: Mef2c exon 4 promoter (forward, 5′-TGCAGAAAAGATTCCCACTTG-3′, reverse, 5′-AGACACTCACAAGGCAAAGAC-3′), Rhodopsin promoter (forward, 5′-CCCCTCTGCAAGCCAATT-3′, reverse, 5′-GCAACTCCAGGCACTGAC-3′), Rhodopsin intron negative control (forward, 5′-TGTGGTCTTCACCTGGATCATG-3′, reverse, 5′-TACCTGGACCAGCCAACGA-3′).
Gene Expression Assays
Total RNA was isolated from C57BL/6 or Rd1 mouse retinas using the Absolute RNA Miniprep kit, and cDNA was synthesized using the AffinityScript qPCR cDNA Synthesis kit (Agilent Technologies, La Jolla, CA). Gene expression was detected by qPCR using TaqMan probes and 2× Gene Expression Master mix (Applied Biosystems, Foster City, CA) on an MX3000P real time PCR unit (Agilent).
Cell Culture, Transient Transfection, and Dual Luciferase Assay
HEK293 cells (American Type Culture Collection, Manassas, VA) were cultured in minimal essential medium-α containing 10% FBS and 100 units/ml of penicillin/streptomycin at 37 °C with 5% CO2. The cells were seeded into 24-well plates (150,000 cells/well), 24 h before transfection with Lipofectamine 2000 (Invitrogen) as described previously (17, 53). Triplicate wells were co-transfected with the Firefly luciferase reporter construct, pGL3-Mef2cP-Luc (667 ng/ml); the transfection control plasmid, Renilla luciferase reporter pRL-CMV (11 ng/ml); and expression plasmid for human NRL (pED-NRL at 55 or 278 ng/ml) and/or expression plasmid for human CRX (pcDNA3.1/HisC-hCRX at 278 ng/ml). The corresponding empty vectors (pED, pcDNA3.1/HisC) were used to adjust the total amount of transfected DNA. Cells were harvested 48 h after transfection for assay of Firefly and Renilla luciferase activities using the Dual Reporter Luminescence Reagent and a Turner dual-injector luminometer (Promega). Renilla luciferase activity was used to normalize for transfection efficiency. Fold-activation was calculated relative to transfection with empty expression vectors. All experiments were repeated at least three times. Statistical comparisons used analysis of variance with Tukey-HSD post-analysis using VassarStats.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as previously described (26). Briefly, nuclear extracts from transfected HEK 293T cells were prepared using a kit (Active Motif, Carlsbad, CA). The DNA oligonucleotide (NRE, forward, 5′-TAGACAGTGACCTCCTCCCTGCTGAGCCACTATGCTCT-3′, NRE reverse, 5′-AGAGCATAGTGGCTCAGCAGGGAGGAGGTCACTGTCTA-3′) contain a putative NRL-response element (NRE) (underlined) predicted by Genomatix programs in the mouse Mef2c promoter region. 32P-Labeled double-stranded oligonucleotide (40,000 cpm) was incubated with nuclear extracts at 4 °C for 20 min. In competition studies, nuclear extracts were preincubated with a 50-fold excess of unlabeled oligonucleotide for 30 min at room temperature and incubated with labeled oligonucleotide for 20 min. A mutant oligonucleotide (forward, 5′-TAGACAGTGACCTCCTCCCTGCCGGGTCGCTATGCTCT-3′, reverse, 5′-AGAGCATAGCGACCCGGCAGGGAGGAGGTCACTGTCTA-3′) with four nucleotides changed in the NRE site was also used to compete for the protein binding to the oligonucleotide. To test the presence of NRL in the protein-DNA complexes, nuclear extracts were incubated with 2 μg of the anti-NRL antibody or normal rabbit IgG for 30 min at room temperature, followed by the addition of labeled oligonucleotide and a further incubation for 20 min at room temperature. The reaction mixtures were electrophoresed on 8% polyacrylamide gels at 100 V for 1.5 h and subjected to autoradiography.
Western Blot
HEK293 cells (300,000 cells) were co-transfected with 0.5 μg of mouse Mef2c expression plasmid (Mef2c cDNA), and 0.5 μg of Gapdh shRNA or Mef2c shRNA 1 or Mef2c shRNA 2. Cells were harvested 48 h after transfection and lysed by sonication in radioimmunoprecipitation buffer supplemented with 20 mm N-ethylmaleimide and protease inhibitor mixture (Roche Applied Science). The protein concentration of the supernatant was measured by the bicinchoninic acid assay (Thermo Scientific, Waltham, MA) and an equal amount of lysate was boiled in 2× SDS-PAGE loading buffer (Invitrogen). The lysate was resolved by SDS-PAGE and transferred to nitrocellulose membrane (Invitrogen). The membrane was probed sequentially with MEF2C and tubulin antibodies and visualized by enhanced chemiluminescence (Thermo Scientific).
In Vivo Electroporation
Retinas of CD1, C57BL/6J, or Nrl−/− mouse pups at P0 were electroporated in vivo as previously described (46, 54, 55). Briefly, equal amounts of plasmids, Mef2cP-GFP and CAG-mCherry, were mixed with either Nrl shRNA or Gapdh shRNA control and injected into the subretinal space of CD1 pups at P0. Equal amounts of Mef2cP-GFP and CAG-mCherry were injected into the subretinal space of C57BL/6J and Nrl−/− P0 pups. Equal amounts of Rho-TdT (Td tomato) and Ub-GFP were mixed with either Mef2c shRNA or Gapdh shRNA and injected into the subretinal space of CD1 pups at P0. The injection volume was 0.2 μl. The DNA concentration was 300 nm for all constructs: Mef2cP-GFP, CAG-mCherry, Rho-TdT, Ub-GFP, Nrl shRNA, Mef2c shRNA, and Gapdh shRNA. Voltage pulses (80 V, 1 Hz, 5 pulses) were applied across the heads of pups using an ECM830 squarewave electroporator and 10-mm diameter BTX Tweezertrode electrodes (Holliston, MA). Retinas were harvested at P20 and P28. Tissues were fixed in 4% paraformaldehyde, and cryoprotected in 30% sucrose. Cryosections (10 μm) were counterstained with DAPI, and then imaged with an Olympus FluoView FV1000 confocal laser scanner.
RESULTS
Prediction of an Alternative Promoter Regulating Mef2c Expression in the Retina
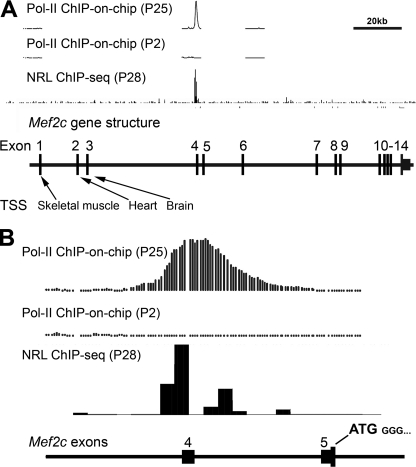
Global expression profiling of adult Nrl−/− retina or purified photoreceptors demonstrated massive down-regulation of Mef2c transcripts, leading to the hypothesis that Mef2c is a transcriptional target of NRL and participates in rod photoreceptor homeostasis (33, 35). The analysis of expressed sequence tags (GenBankTM) and previous studies (56) demonstrated that the mouse Mef2c gene consists of 14 exons with alternative transcription start sites and alternate splicing in specific tissues. Exons 1, 2, or 3 of Mef2c are spliced directly to exon 5 in skeletal muscle, heart, or brain, respectively. These tissue-specific transcripts all include exon 5, which contains the translation start site (56). Our retinal ChIP-on-chip analysis detected a clear association of RNA Pol II immediately upstream of exon 4, but not upstream of exon 1 (Fig. 1, A and B), suggesting that most adult retinal transcripts of Mef2c start at exon 4. ChIP-seq analysis also detected a strong association of NRL with the genomic region immediately upstream of exon 4, providing further evidence for an alternative promoter that dictates Mef2c expression in the neural retina (Fig. 1, A and B).
FIGURE 1.
Identification of a novel retina-specific promoter of Mef2c by ChIP-on-chip and ChIP-seq analysis. A, RNA Pol II ChIP-on-chip peaks at P25 and P2, and NRL ChIP-seq peaks at P28 were mapped to the mouse genome. The Mef2c gene structure is aligned in scale with the exons (vertical black bars) numbered. Tissue-specific Mef2c transcription start sites (TSS) previously reported for skeletal muscle, heart, and brain are indicated. Scale bar, 20 kb. B, Pol II ChIP-on-chip peaks (P25 and P2) and NRL ChIP-seq peaks (P28) at the Mef2c locus plotted at higher resolution. The Mef2c start codon (ATG) is located in exon 5, and is common to all known tissue-specific transcripts.
5′-RACE Confirms a Retinal Transcription Start Site at Exon 4
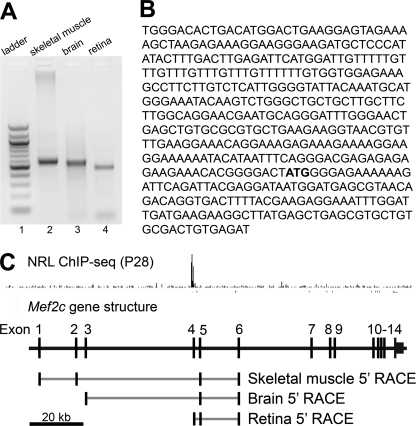
5′-RACE PCR products derived from full-length RNA of different tissues (retina, brain, and skeletal muscle) were of different size, demonstrating alternative transcription start sites of Mef2c in retina, brain, and skeletal muscle (Fig. 2A). Sequence analyses of 5′-RACE products from the neural retina were comprised of sequences aligning to exons 4, 5, and 6 (Fig. 2B). Alignment of the tissue-specific 5′-RACE sequences revealed the following preferred transcription start sites: exon 1 for skeletal muscle, exon 3 for brain, and exon 4 for neural retina (Fig. 2C). All transcripts included exon 5, which contains the translation start site for the MEF2C protein.
FIGURE 2.
Mapping of retinal Mef2c promoter by 5′-RACE. The 5′-untranslated regions of the Mef2c gene in mouse skeletal muscle, brain, and retina were defined by 5′-RACE. The 5′-RACE products were visualized on a 1.5% agarose gel (A), purified, and cloned for sequencing (B). The 5′ UTR sequences were aligned to the Mef2c gene structure (C). At least 15 clones were sequenced for each reaction. A, DNA ladder (lane 1) and tissue-specific 5′-RACE products using RNA from mouse skeletal muscle (lane 2), brain (lane 3), and retina (lane 4) were visualized on a 1.5% agarose gel. B, the retina-specific 5′ UTR sequence in which the Mef2c start codon (ATG) is shown in bold. C, alignment of the 5′-RACE sequences to the Mef2c gene structure. Exons are shown as black bars and the spliced regions in gray lines. NRL ChIP-seq peaks at P28 were mapped to the mouse genome. The Mef2c structure is aligned and shown in scale under the peaks. Scale bar, 20 kb.
Enhanced Pol II and H3K9-Ac Association with the Novel Mef2c Promoter in Mature Retina Compared with Developing Retina
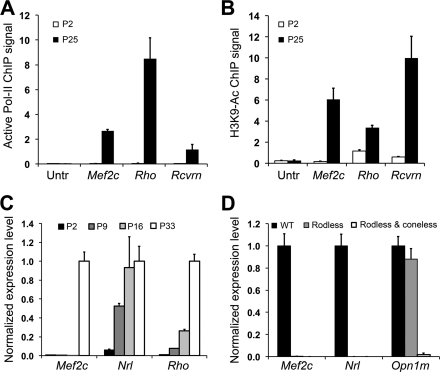
It should be noted that Pol II ChIP-on-chip only detected significant Pol II association around exon 4 in P25 but not in P2 retina (Fig. 1A). We next observed that the transcriptionally active form of Pol II, phosphorylated on serine 2 (Pol-II-S2), associated with the retinal Mef2c promoter (exon 4) at age P25 but not at age P2 (Fig. 3A). In concordance, ChIP-qPCR analysis also detected Pol-II-S2 association with the promoters of two rod-specific genes, Rhodopsin (Rho) and Recoverin (Rcvrn), when the genes were active at P25 but not at P2 (Fig. 3A). An intergenic untranslated region (Untr), downstream of Rho, was negative for Pol-II-S2 binding at both P2 and P25 (Fig. 3A).
FIGURE 3.
Activation of an alternative Mef2c promoter during photoreceptor maturation. Binding of RNA Pol II and acetylation of histones at the retinal Mef2c promoter were measured by ChIP-qPCR using a Pol-II-S2 antibody recognizing the active form of Pol II (A) or H3K9-Ac antibody (B) in P2 (immature) and P25 (mature) mouse retina. The ChIP DNA was quantified in triplicate by qPCR. Results are presented as copy number detected per 1000 genome equivalents of input DNA. An untranslated region on chromosome-6 (untranslated region, Untr) served as a negative control region. Rhodopsin (Rho) and Recoverin (Rcvrn) promoter regions served as positive controls. C, expression of Mef2c, Nrl, and Rhodopsin were measured by qRT-PCR during retina development (at P2, P9, P16, and P33) and normalized to β-Actin (Actb). TaqMan probes targeting Mef2c exons 5 and 6 were used to detect Mef2c expression. D, expression of Mef2c, Nrl, and M-Opsin (Opn1m) in P33 normal retina (WT), P33 rod-less Rd1 retinas (rodless), and P117 Rd1 retinas lacking both rods and cone (rodless and coneless) were measured by RT-qPCR and normalized to β-Actin (Actb). Bars indicate S.D. (n = 3).
Acetylation of lysine 9 on histone H3 favors the formation of an open chromatin architecture and marks actively transcribed genes (57). To examine the epigenetic state of the Mef2c promoter, we performed H3K9-Ac ChIP-qPCR and detected increased acetylation of H3K9 corresponding to increased Pol-II-S2 binding at P25 compared with P2 (Fig. 3B). We identified a similar increase in H3K9-Ac near the transcription start sites of the two NRL-activated rod-specific genes: Rho and Rcvrn. H3K9-Ac levels remained at background in the untranslated control region (UTR) at both P25 and P2 (Fig. 3B).
Mef2c Is Expressed in Mature But Not in Developing Retinas
We next examined the temporal expression profile of Mef2c using a qPCR assay (TaqMan) targeting the exon 5–6 splicing junction in wild type (WT) retina at critical developmental stages: P2, P9, P16, and P33 (Fig. 3C). Mef2c transcripts were only detected in mature retinas (P33) but not in developing retinas (P2, P9, and P16) (Fig. 3C). TaqMan probes specifically targeting Mef2c exons 1 and 2 detected relatively little or no transcript in the retina at any developmental stage, compared with exon 5–6 transcripts (data not shown). As predicted, Nrl expression preceded Rhodopsin (Rho) and Mef2c transcripts (Fig. 3C).
Retinal Cell Type-specific Expression of Mef2c
To examine whether Mef2c is expressed specifically in photoreceptors, we examined its expression in rd1 mice. This mouse strain is homozygous for a mutation in Pde6b and they lose almost all rod photoreceptors by age P33, but retain most of their cones and other non-photoreceptor neurons (58). By age P99, rd1 retinas also lose cones and are photoreceptor-less. Gene expression assays for Mef2c transcripts (exons 5–6), Nrl (rod-specific marker), and M-opsin (Opn1mw, cone-specific marker) were performed for WT (P33), rod-less (rd1, P33), and photoreceptor-less retinas (rd1, P117) (Fig. 3D). Similar to Nrl, Mef2c transcripts were detected in mature WT retina but not in rod-less or photoreceptor-less retinas. Additionally, we could not detect Mef2c transcripts (exons 5–6) or Nrl transcripts in the Nrl−/− retina at age P28, whereas both were abundant in WT retina at P28 (data not shown). The rd1 and Nrl−/− data suggest that Mef2c transcripts starting from exon 4 are specific to rod, and not cone, photoreceptors.
In Vivo and in Vitro Association of NRL with the Retinal Mef2c Promoter
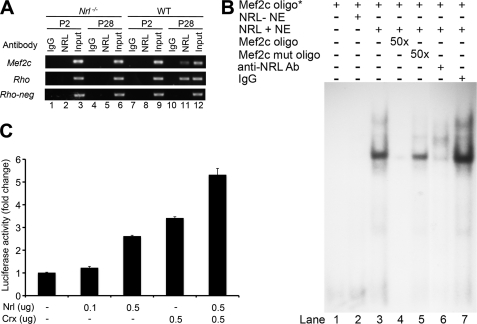
A number of observations implicate NRL as an excellent candidate for controlling the activity of an alternative retinal Mef2c promoter; these include the timing of Mef2c expression specifically in rod photoreceptors and in vivo association of NRL immediately upstream of exon 4. ChIP-seq analysis detected NRL association overlapping with the location of Pol II binding, upstream of exon 4, in the adult mouse retina (see Fig. 1B). Further examination of this promoter region by ChIP assays detected NRL binding at P28 but not at P2 (Fig. 4A). NRL did not bind to a negative control region (Rho-neg) in wild type retina. NRL binding was not detected in Nrl−/− retina at either P28 or P2, confirming the specificity of the ChIP assay (Fig. 4A).
FIGURE 4.
NRL binds to and activates the retinal Mef2c promoter. A, in vivo binding of NRL with the retinal Mef2c promoter was detected by ChIP assays. ChIP was performed using NRL-deficient (Nrl−/−) retina (lanes 1–6) and wild type (WT) retina (lanes 7–12), at P2 (lanes 1–3 and 7–9) and P28 (lanes 4–6 and 10–12) with antibody to NRL (lanes 2, 5, 8, and 11). Input (chromatin samples without IP) served as positive control (lanes 3, 6, 9, and 12) and IP with normal IgG (IgG) served as negative control (lanes 1, 4, 7, and 10). The Rhodopsin (Rho) promoter was a positive control for NRL binding, whereas a region in Rho intron (Rho-neg) served as a negative control. B, NRL directly binds to NRE in the Mef2c promoter in vitro. Components present in each binding reaction are indicated above each lane in the autoradiograph. Mef2c oligo*, representing a 32P-labeled NRE containing oligonucleotide, was included in all reactions (lanes 1–7). Reactions included 10 μg of nuclear extract from HEK293T cells transfected with either empty vector (NRL-NE, lane 2) or Nrl cDNA expression plasmid (NRL+NE, lanes 3–7). Lanes 4 and 5 included 50-fold molar excess of unlabeled NRE oligonucleotide and unlabeled mutant NRE oligonucleotide, respectively. Incubation with 2 μg of anti-NRL antibody (lane 6) or 2 μg of normal rabbit IgG (lane 7) were used to detect the presence of NRL in the shifted protein complex. Experiments were repeated three times, and similar results were obtained. C, activation of the retinal Mef2c promoter in transfected HEK293 cells by NRL alone, CRX alone, and in combination. HEK293 cells were co-transfected with the Mef2c promoter driving luciferase-reporter construct (pGL3-Mef2cP-Luc), Renilla luciferase construct (pRL-CMV) for normalization, and expression plasmids for human NRL and/or human CRX. Cells were harvested 48 h after transfection for luciferase assays. Fold-activation was calculated relative to empty expression vectors. All experiments were repeated at least three times. Statistical comparisons by analysis of variance with Tukey-HSD post-analysis (p < 0.01 for all paired comparisons) are shown. Bars indicate S.D.
In silico analysis of this retinal Mef2c promoter region revealed the presence of a putative NRE: TGGCTCAG. To test whether NRL can directly bind to the region of interest, EMSA were carried out using an oligonucleotide probe (Mef2c oligo) spanning the NRE in the retinal Mef2c promoter (Fig. 4B). Nuclear extracts from HEK293T cells transfected with Nrl cDNA (NRL+ NE) shifted the 32P-labeled Mef2c oligo, whereas nuclear extracts from cells lacking NRL (NRL− NE) did not, suggesting that NRL could bind to the Mef2c oligo (Fig. 4B, lanes 1–3). Specificity of the binding was validated by competition with an excess of unlabeled Mef2c oligo. An excess of the NRE mutant oligo did not compete for the binding (Fig. 4B, lanes 4 and 5). Consistent with these results, an anti-NRL antibody abolished the gel shift, whereas normal IgG did not (Fig. 4B, lanes 6 and 7), further confirming the direct binding of NRL to the retinal Mef2c promoter.
NRL Activates the Retinal Mef2c Promoter in Vitro and in Vivo
As a functional test of the ability of NRL to activate transcription from the retinal Mef2c promoter, we cloned a 540-bp region including a part of exon 4 into the pGL3 Firefly luciferase reporter vector. This construct (pGL3-Mef2cP-Luc) was co-transfected with or without an NRL expression plasmid into HEK293 cells. The Mef2c promoter was able to drive luciferase expression, and its activity was significantly enhanced by NRL (p < 0.01, analysis of variance) in a dose-dependent manner (Fig. 4C). NRL and CRX often cooperate to induce gene expression in mature rod photoreceptors (17, 59, 60), so we also tested the ability of CRX to activate the retinal Mef2c promoter (14, 58). CRX could activate the promoter alone and produced additional activation with NRL (Fig. 4C).
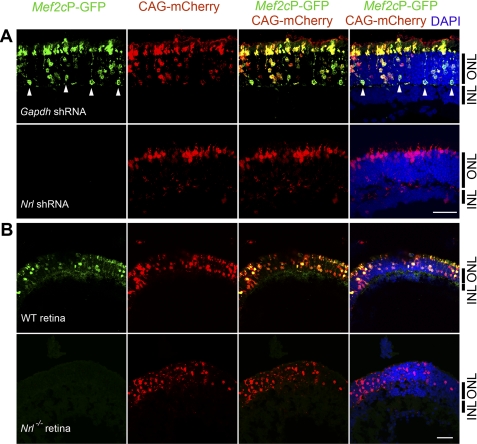
To examine whether NRL is able to activate the Mef2c promoter in vivo, we subcloned the same retinal Mef2c promoter into a GFP vector. This Mef2cP-GFP construct was transfected into mouse retinas at P0 by in vivo electroporation, and the promoter activity was monitored at P20 and P28 (Fig. 5). We observed similar results at P20 and P28. To confirm successful electroporation and normalize for transfection efficiency, a plasmid expressing mCherry driven by the CAG promoter (CAG-mCherry) was electroporated with Mef2cP-GFP. Nrl shRNA was co-electroporated into CD1 WT mouse pups to knockdown Nrl expression (46). Confocal imaging of retinal sections revealed that only photoreceptor cells, comprising the outer nuclear layer (ONL), expressed GFP (Fig. 5A). Expression of GFP was abrogated by co-electroporation with Nrl shRNA, but not with Gapdh shRNA, demonstrating that NRL is a key activator of the Mef2c promoter in vivo (Fig. 5A). To further evaluate whether the Mef2c promoter is expressed only in rod photoreceptors and is dependent on NRL, in vivo electroporation was carried out in cone-only Nrl−/− mice (Fig. 5B). We observed GFP expression only in sections of P20 WT retina, but not in P20 Nrl−/− retina (Fig. 5B).
FIGURE 5.
Induction of retinal Mef2C promoter activity by NRL in mature rod photoreceptors. A, representative confocal images of P28 CD1 wild type mouse retinas that were electroporated in vivo at P0 with Mef2cP-GFP, CAG-mCherry (electroporation control), and either Gapdh shRNA (top row) or Nrl shRNA (bottom row). Photoreceptor cells reside in the ONL but not in the inner nuclear layer (INL). B, representative confocal images of P20 C57BL/6J mouse retina (top row) and P20 Nrl−/− mouse (on C57BL/6J background) retina (bottom row) that were electroporated in vivo at P0 with Mef2cP-GFP and CAG-mCherry (electroporation control). Mef2cP-GFP expression is shown in green and CAG-mCherry in red. DAPI staining is in blue. Activation of the Mef2cP-GFP reporter in rod-photoreceptor cell bodies is indicated (white arrowheads). At least 3 biological replicates were collected for each electroporation condition and similar phenotypes were observed. Scale bar, 20 μm.
Mef2c Activates the Rhodopsin Promoter in Vivo
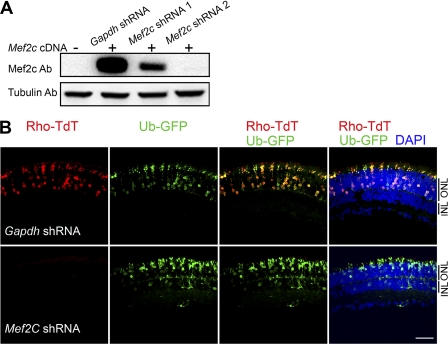
To examine whether Mef2c can regulate rod photoreceptor-specific genes, we knocked down endogenous Mef2c expression in the retina by in vivo shRNA electroporation. To test the efficacy of Mef2c shRNA constructs, we first cotransfected HEK293 cells with a Mef2c expression construct (Mef2c cDNA) and Gapdh shRNA (control) or Mef2c shRNA. Knockdown efficiency was evaluated by immunoblotting (Fig. 6A), and the more efficient Mef2c shRNA construct (shRNA-2) was used for in vivo co-electroporation of the retina with a Rhodopsin-promoter fluorescence reporter construct (Rho-TdT). Electroporation of Rho-TdT resulted in photoreceptor-specific expression of TdT that was restricted to the ONL (Fig. 6B). In vivo co-electroporation of Mef2c shRNA reduced the TdT expression driven from the Rhodopsin promoter, whereas Gapdh shRNA (control) had no effect on the TdT expression (Fig. 6B).
FIGURE 6.
Reduced Rhodopsin promoter activity by Mef2c knockdown in adult retina. A, efficacy of Mef2c shRNA was validated in transfected HEK293 cells. HEK293 cells were co-transfected with the mouse Mef2c expression plasmid (Mef2c cDNA), and Gapdh shRNA or Mef2c shRNA 1 or Mef2c shRNA 2. Cells were harvested 48 h after transfection and lysed for immunoblot analysis with Mef2c antibody. Anti-tubulin antibody was used as loading control. B, knockdown of Mef2c by shRNA decreased Rhodopsin promoter activity in adult mouse retina. Representative confocal images of P20 CD1 wild type mouse retinas that were electroporated in vivo at P0 with the Rhodopsin promoter-Td tomato construct (Rho-TdT), ubiquitin promoter-GFP construct (Ub-GFP) (electroporation control), and either Gapdh shRNA (top) or Mef2c shRNA (bottom). Ub-GFP is shown in green. Rho-TdT is shown in red. DAPI is shown in blue. Photoreceptor cells reside in the ONL. At least 3 biological replicates were collected for each electroporation condition and a similar phenotype was observed. Scale bar, 20 μm.
DISCUSSION
Spatiotemporal control of gene expression dictates development and homeostasis (1). Transcriptional promoters and enhancers are key determinants in the transcription of any gene by serving to integrate cis-regulatory DNA elements, transcription factor binding, epigenetic regulation, and signal transduction events (4). Alternative promoter usage is an important contributor to tissue and cell-specific gene expression (61, 62). In this study, we demonstrate that Mef2c expression in photoreceptors is regulated by NRL through a novel alternative promoter upstream of a 4th exon. The major lines of evidence are: 1) correlation of the expression profile of Mef2c with Pol II binding and increased H3K9 acetylation at the alternate Mef2c promoter in WT retina, 2) decreased Mef2c expression in Nrl−/− retina, 3) in vivo association of NRL with the Mef2c promoter and direct in vitro binding of NRL with the NRE within this promoter, and 4) NRL-mediated activation of the Mef2c alternate promoter in HEK293 cells and in the neural retina in vivo.
Our in vivo electroporation data show that the activation of the retinal-specific Mef2c promoter was restricted to the ONL of the mature retina, and included rod photoreceptors. The ONL is comprised of rod and cone photoreceptor cell bodies, with the cones restricted to the outermost (scleral) side (63, 64). Failure to detect GFP expression in the cone-only Nrl−/− retina strongly suggests that the retina-specific alternative Mef2c promoter is activated by NRL and only in rod photoreceptors.
Expression of Mef2c from the alternative promoter in the retina after age P16 correlated with the association of NRL and Pol II, as indicated by ChIP-qPCR in the mature retina (P28 or P25) but not in the developing retina (P2). A similar Mef2c promoter construct was reportedly inactive at P8 in electroporated mouse retinal explants (29). These observations support the hypothesis that the retina-specific Mef2c promoter is activated relatively late, in mature rod photoreceptors. We have previously shown that a minimal transactivation domain of NRL interacts with TATA-binding protein (65). It is possible that NRL activates Mef2c transcription by recruiting or stabilizing the TATA-binding protein and consequently the general transcription machinery at the Mef2c promoter. The mechanism underlying “late” induction of the retinal Mef2c promoter, despite the presence of NRL and CRX in rod progenitor cells, remain to be elucidated.
CRX activates the transcription of photoreceptor-specific genes (including all Opsins) by recruiting co-activators possessing histone acetyltransferase activity (65). In Crx−/− mice, the association of histone acetyltransferase-containing co-activators and acetylated histone H3 at Opsin promoters are decreased significantly, which correlates with diminished Opsin expression (65). Increased acetylation of histone H3, on Lys-9 or Lys-14, has been associated with enhanced transcription, whereas histone hypoacetylation is correlated with transcription silencing (66). CRX association at the retina-specific Mef2c promoter region was recently reported in the mature mouse retina (29). Our H3K9-Ac ChIP-qPCR analysis demonstrates that acetylation of H3K9 increases substantially at the Mef2c alternative promoter, suggesting the transition of this chromatin region from a closed architecture to an open architecture within mature photoreceptors. Thus, it is possible that CRX recruits histone acetyltransferase-containing co-activators to the retinal-specific Mef2c promoter, promoting an open chromatin architecture that facilitates NRL binding and recruitment of the basal transcription machinery. This is consistent with the observation that CRX alone was not sufficient to activate the Mef2c promoter as shown by our in vivo electroporation results of Nrl−/− mice or shRNA knockdown of Nrl expression in WT mice.
Elucidation of the NRL-centered gene regulatory network, which dictates photoreceptor cell fate and function, requires identification of the transcriptional targets of NRL. Most importantly, transcription factor targets represent critical secondary nodes that regulate the expression of distinct or shared downstream target genes. Gene expression profiles from Nrl−/− mice continue to accelerate this discovery process (33, 35, 67).
The transcription factor Mef2c, reported here as a novel NRL target in the rod photoreceptor, was previously shown to regulate cell specification and survival in multiple tissues. Mef2c knockdown by shRNA, using in vivo electroporation, significantly decreased Rhodopsin promoter activity within rod photoreceptors. Hence, Mef2c might play an important role in regulating rod homeostasis by participating in the regulation of specific genes. Further investigations are required to identify the downstream targets of MEF2C in rod photoreceptors. Our studies, however, implicate MEF2C as an important regulator of rod gene expression downstream of NRL.
Acknowledgments
We thank Shiming Chen (Washington University) for CRX expression construct, Danielle Hine (Oakland University) and Matthew Brooks (National Eye Institute) for technical assistance, and Paul Labhart (Genpathway) for programming assistance. We acknowledge Robert Fariss (National Eye Institute) for confocal training.
This work was supported, in whole or in part, by National Institutes of Health Grants EY14626 (to K. P. M.), EY14803 (Oakland University Eye Research Institute), and the intramural program of the NEI, National Institutes of Health.
- CRX
- cone rod homeobox
- Nrl
- neural retina leucine zipper
- qPCR
- quantitative polymerase chain reaction
- Mef2c
- myocyte enhancer factor-2c
- RACE
- rapid analysis of cDNA ends
- ChIP-seq
- ChIP followed by high throughput sequencing
- ChIP-on-chip
- ChIP followed by tiling array analysis
- NRE
- NRL response element
- P
- postnatal day
- Pol II
- polymerase II
- TdT
- Td tomato.
REFERENCES
- 1. Levine M., Tjian R. (2003) Nature 424, 147–151 [DOI] [PubMed] [Google Scholar]
- 2. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chin M. C., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H. W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K. R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Yuan X. F., Zhang B., Zwingman T. A., Jones A. R. (2007) Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- 3. Ooi L., Wood I. C. (2008) Biochem. J. 414, 327–341 [DOI] [PubMed] [Google Scholar]
- 4. Heintzman N. D., Ren B. (2007) Cell Mol. Life Sci. 64, 386–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swaroop A., Kim D., Forrest D. (2010) Nat. Rev. Neurosci. 11, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marquardt T., Gruss P. (2002) Trends Neurosci. 25, 32–38 [DOI] [PubMed] [Google Scholar]
- 7. Livesey F. J., Cepko C. L. (2001) Nat. Rev. Neurosci. 2, 109–118 [DOI] [PubMed] [Google Scholar]
- 8. Luo D. G., Xue T., Yau K. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9855–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lem J., Krasnoperova N. V., Calvert P. D., Kosaras B., Cameron D. A., Nicolò M., Makino C. L., Sidman R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Humphries M. M., Rancourt D., Farrar G. J., Kenna P., Hazel M., Bush R. A., Sieving P. A., Sheils D. M., McNally N., Creighton P., Erven A., Boros A., Gulya K., Capecchi M. R., Humphries P. (1997) Nat. Genet. 15, 216–219 [DOI] [PubMed] [Google Scholar]
- 11. Tan E., Wang Q., Quiambao A. B., Xu X., Qtaishat N. M., Peachey N. S., Lem J., Fliesler S. J., Pepperberg D. R., Naash M. I., Al-Ubaidi M. R. (2001) Invest. Ophthalmol. Vis. Sci. 42, 589–600 [PubMed] [Google Scholar]
- 12. Wright A. F., Chakarova C. F., Abd El-Aziz M. M., Bhattacharya S. S. (2010) Nat. Rev. Genet. 11, 273–284 [DOI] [PubMed] [Google Scholar]
- 13. Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T. (2003) Nat. Neurosci. 6, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 14. Chen S., Wang Q. L., Nie Z., Sun H., Lennon G., Copeland N. G., Gilbert D. J., Jenkins N. A., Zack D. J. (1997) Neuron 19, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 15. Hennig A. K., Peng G. H., Chen S. (2008) Brain Res. 1192, 114–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Furukawa T., Morrow E. M., Li T., Davis F. C., Cepko C. L. (1999) Nat. Genet. 23, 466–470 [DOI] [PubMed] [Google Scholar]
- 17. Mitton K. P., Swain P. K., Chen S., Xu S., Zack D. J., Swaroop A. (2000) J. Biol. Chem. 275, 29794–29799 [DOI] [PubMed] [Google Scholar]
- 18. Chen J., Rattner A., Nathans J. (2005) J. Neurosci. 25, 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haider N. B., Mollema N., Gaule M., Yuan Y., Sachs A. J., Nystuen A. M., Naggert J. K., Nishina P. M. (2009) Exp. Eye Res. 89, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng G. H., Ahmad O., Ahmad F., Liu J., Chen S. (2005) Hum. Mol. Genet. 14, 747–764 [DOI] [PubMed] [Google Scholar]
- 21. Cheng H., Aleman T. S., Cideciyan A. V., Khanna R., Jacobson S. G., Swaroop A. (2006) Hum. Mol. Genet. 15, 2588–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng L., Hurley J. B., Dierks B., Srinivas M., Saltó C., Vennström B., Reh T. A., Forrest D. (2001) Nat. Genet. 27, 94–98 [DOI] [PubMed] [Google Scholar]
- 23. Jia L., Oh E. C., Ng L., Srinivas M., Brooks M., Swaroop A., Forrest D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17534–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srinivas M., Ng L., Liu H., Jia L., Forrest D. (2006) Mol. Endocrinol. 20, 1728–1741 [DOI] [PubMed] [Google Scholar]
- 25. Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001) Nat. Genet. 29, 447–452 [DOI] [PubMed] [Google Scholar]
- 26. Oh E. C., Cheng H., Hao H., Jia L., Khan N. W., Swaroop A. (2008) Brain Res. 1236, 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Onishi A., Peng G. H., Poth E. M., Lee D. A., Chen J., Alexis U., de Melo J., Chen S., Blackshaw S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11579–11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oh E. C., Khan N., Novelli E., Khanna H., Strettoi E., Swaroop A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Corbo J. C., Lawrence K. A., Karlstetter M., Myers C. A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L. G., Weber B. H., Langmann T. (2010) Genome Res. 20, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeAngelis M. M., Grimsby J. L., Sandberg M. A., Berson E. L., Dryja T. P. (2002) Arch. Ophthalmol. 120, 369–375 [DOI] [PubMed] [Google Scholar]
- 31. Bessant D. A., Payne A. M., Mitton K. P., Wang Q. L., Swain P. K., Plant C., Bird A. C., Zack D. J., Swaroop A., Bhattacharya S. S. (1999) Nat. Genet. 21, 355–356 [DOI] [PubMed] [Google Scholar]
- 32. Kanda A., Friedman J. S., Nishiguchi K. M., Swaroop A. (2007) Hum. Mutat. 28, 589–598 [DOI] [PubMed] [Google Scholar]
- 33. Akimoto M., Cheng H., Zhu D., Brzezinski J. A., Khanna R., Filippova E., Oh E. C., Jing Y., Linares J. L., Brooks M., Zareparsi S., Mears A. J., Hero A., Glaser T., Swaroop A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng H., Khanna H., Oh E. C., Hicks D., Mitton K. P., Swaroop A. (2004) Hum. Mol. Genet. 13, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 35. Yoshida S., Mears A. J., Friedman J. S., Carter T., He S., Oh E., Jing Y., Farjo R., Fleury G., Barlow C., Hero A. O., Swaroop A. (2004) Hum. Mol. Genet. 13, 1487–1503 [DOI] [PubMed] [Google Scholar]
- 36. McKinsey T. A., Zhang C. L., Olson E. N. (2002) Trends Biochem. Sci. 27, 40–47 [DOI] [PubMed] [Google Scholar]
- 37. Black B. L., Molkentin J. D., Olson E. N. (1998) Mol. Cell. Biol. 18, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Potthoff M. J., Olson E. N. (2007) Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 39. McDermott J. C., Cardoso M. C., Yu Y. T., Andres V., Leifer D., Krainc D., Lipton S. A., Nadal-Ginard B. (1993) Mol. Cell. Biol. 13, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010) Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H., Radford J. C., Ragusa M. J., Shea K. L., McKercher S. R., Zaremba J. D., Soussou W., Nie Z., Kang Y. J., Nakanishi N., Okamoto S., Roberts A. J., Schwarz J. J., Lipton S. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9397–9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barbosa A. C., Kim M. S., Ertunc M., Adachi M., Nelson E. D., McAnally J., Richardson J. A., Kavalali E. T., Monteggia L. M., Bassel-Duby R., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le Meur N., Holder-Espinasse M., Jaillard S., Goldenberg A., Joriot S., Amati-Bonneau P., Guichet A., Barth M., Charollais A., Journel H., Auvin S., Boucher C., Kerckaert J. P., David V., Manouvrier-Hanu S., Saugier-Veber P., Frébourg T., Dubourg C., Andrieux J., Bonneau D. (2010) J. Med. Genet. 47, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nowakowska B. A., Obersztyn E., Szymańska K., Bekiesińska-Figatowska M., Xia Z., Ricks C. B., Bocian E., Stockton D. W., Szczałuba K., Nawara M., Patel A., Scott D. A., Cheung S. W., Bohan T. P., Stankiewicz P. (2010) Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 1042–1051 [DOI] [PubMed] [Google Scholar]
- 45. Swain P. K., Hicks D., Mears A. J., Apel I. J., Smith J. E., John S. K., Hendrickson A., Milam A. H., Swaroop A. (2001) J. Biol. Chem. 276, 36824–36830 [DOI] [PubMed] [Google Scholar]
- 46. Matsuda T., Cepko C. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schaefer B. C. (1995) Anal. Biochem. 227, 255–273 [DOI] [PubMed] [Google Scholar]
- 48. Maruyama K., Sugano S. (1994) Gene 138, 171–174 [DOI] [PubMed] [Google Scholar]
- 49. Tummala P., Mali R. S., Guzman E., Zhang X., Mitton K. P. (2010) Mol. Vis. 16, 252–271 [PMC free article] [PubMed] [Google Scholar]
- 50. Labhart P., Karmakar S., Salicru E. M., Egan B. S., Alexiadis V., O'Malley B. W., Smith C. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mali R. S., Peng G. H., Zhang X., Dang L., Chen S., Mitton K. P. (2008) BMC Mol. Biol. 9, 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Feng J., Liu T., Zhang Y. (2011) Curr. Protoc. Bioinformatics 34, 2.14.1–2.14.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitton K. P., Swain P. K., Khanna H., Dowd M., Apel I. J., Swaroop A. (2003) Hum. Mol. Genet. 12, 365–373 [DOI] [PubMed] [Google Scholar]
- 54. Kautzmann M. A., Kim D. S., Felder-Schmittbuhl M. P., Swaroop A. (2011) J. Biol. Chem. 286, 28247–28255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsuda T., Cepko C. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang D. Z., Valdez M. R., McAnally J., Richardson J., Olson E. N. (2001) Development 128, 4623–4633 [DOI] [PubMed] [Google Scholar]
- 57. Liang G., Lin J. C., Wei V., Yoo C., Cheng J. C., Nguyen C. T., Weisenberger D. J., Egger G., Takai D., Gonzales F. A., Jones P. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bowes C., Li T., Danciger M., Baxter L. C., Applebury M. L., Farber D. B. (1990) Nature 347, 677–680 [DOI] [PubMed] [Google Scholar]
- 59. Swaroop A., Wang Q. L., Wu W., Cook J., Coats C., Xu S., Chen S., Zack D. J., Sieving P. A. (1999) Hum. Mol. Genet. 8, 299–305 [DOI] [PubMed] [Google Scholar]
- 60. Rehemtulla A., Warwar R., Kumar R., Ji X., Zack D. J., Swaroop A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 191–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V. B., Brenner S. E., Batalov S., Forrest A. R., Zavolan M., Davis M. J., Wilming L. G., Aidinis V., Allen J. E., Ambesi-Impiombato A., Apweiler R., Aturaliya R. N., Bailey T. L., Bansal M., Baxter L., Beisel K. W., Bersano T., Bono H., Chalk A. M., Chiu K. P., Choudhary V., Christoffels A., Clutterbuck D. R., Crowe M. L., Dalla E., Dalrymple B. P., de Bono B., Della Gatta G., di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C. F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T. R., Gojobori T., Green R. E., Gustincich S., Harbers M., Hayashi Y., Hensch T. K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S. P., Kruger A., Kummerfeld S. K., Kurochkin I. V., Lareau L. F., Lazarevic D., Lipovich L., Liu J., Liuni S., McWilliam S., Madan Babu M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K. C., Pavan W. J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J. F., Ring B. Z., Ringwald M., Rost B., Ruan Y., Salzberg S. L., Sandelin A., Schneider C., Schönbach C., Sekiguchi K., Semple C. A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S. L., Tang S., Taylor M. S., Tegner J., Teichmann S. A., Ueda H. R., van Nimwegen E., Verardo R., Wei C. L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S. M., Teasdale R. D., Liu E. T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J. S., Hume D. A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y. (2005) Science 309, 1559–1563 16141072 [Google Scholar]
- 62. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corbo J. C., Cepko C. L. (2005) PLoS Genet. 1, e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carter-Dawson L. D., LaVail M. M. (1979) J. Comp. Neurol. 188, 245–262 [DOI] [PubMed] [Google Scholar]
- 65. Peng G. H., Chen S. (2007) Hum. Mol. Genet. 16, 2433–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kurdistani S. K., Tavazoie S., Grunstein M. (2004) Cell 117, 721–733 [DOI] [PubMed] [Google Scholar]
- 67. Mustafi D., Kevany B. M., Genoud C., Okano K., Cideciyan A. V., Sumaroka A., Roman A. J., Jacobson S. G., Engel A., Adams M. D., Palczewski K. (June 9, 2011) Faseb J. 25, 3157–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]