Background: Hsp90 binds to functional domains of protein clients, including the ligand-binding domain of the aryl hydrocarbon receptor (AhR).
Results: Hsp90- and ligand-binding sites spatially overlap, and ligand displaces Hsp90 in AhR activation.
Conclusion: Hsp90 binding affects AhR activation and not just protein stability.
Significance: This might be the first mechanism of Hsp90 dissociation in activation of client proteins.
Keywords: Heat Shock Protein, Ligand-binding Protein, Protein Domains, Protein-Protein Interactions, Receptor Structure-Function, PAS Domain, Aryl Hydrocarbon Receptor, Heat Shock Protein 90, Mechanisms of Antagonism
Abstract
Hsp90 (heat shock protein of 90 kDa) is often found associated with functional domains of client proteins, including those for ligand binding, dimerization, DNA binding, and enzymatic activity. Although Hsp90 can maintain the conformation of functionally important domains prior to activation of the client protein, its specific binding site and the mechanism(s) of Hsp90 dissociation during activation are unknown. Here, we have identified and characterized residues involved in Hsp90 binding within the aryl hydrocarbon receptor (AhR) ligand-binding domain and demonstrate that they overlap with those involved in ligand binding. In agreement with this spatial model, ligand binding results in Hsp90 dissociation from the AhR Per-ARNT-Sim B fragment. Interestingly, whereas Hsp90-binding residues within the ligand-binding domain were not involved in Hsp90-dependent AhR protein stability, several of these residues are important for ligand-dependent AhR activation, and their mutation resulted in conversion of two AhR antagonists/partial agonists into full AhR agonists. These studies reveal co-localization of a tentative Hsp90-binding site with that for AhR ligand binding and provide the first molecular mechanism for Hsp90 dissociation in the activation of a client protein.
Introduction
Hsp90 (heat shock protein of 90 kDa) is a molecular chaperone involved in multiple signal transduction pathways and a popular target in cancer therapy. In addition to being responsible for correct conformational folding of client proteins, it is thought to stabilize them against degradation, with exposure to the antibiotic geldanamycin or its derivatives resulting in disruption of Hsp90 protein binding and reductions in cellular levels of Hsp90 client proteins (reviewed in Ref. 1). Hsp90-binding sites on client proteins are generally located within functionally important domains, such as the kinase domains of Akt1, Lck, and Cdk2; the regulatory Polo box domain of Plk1; the DNA-binding domains of p53 and the aryl hydrocarbon receptor (AhR)2; the protein dimerization domains of hypoxia-inducible factor-1α (HIF1α), the AhR, and soluble guanylyl cyclase; and the ligand-binding domains of steroid receptors and the AhR (2–11). Although Hsp90 can maintain the conformation of functionally important domains of client proteins prior to their activation, the mechanism of Hsp90 dissociation during activation events is not known. One of the main limitations in Hsp90 biology has been the lack of a defined consensus Hsp90-binding site on client proteins. Although several studies identified specific amino acids or small regions involved in Hsp90 binding to client proteins (2, 5–7, 10, 12, 13), no actual consensus binding site was identified. It is currently thought that Hsp90 binding may occur through a specific charge pattern present on the surface of client proteins rather than through a specific amino acid consensus sequence (12, 14).
The AhR is an Hsp90-binding ligand-dependent transcription factor that mediates the toxic and biological effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and a wide variety of structurally diverse compounds (15). Hsp90 has been shown to bind to both the AhR basic helix-loop-helix (bHLH) and Per-ARNT-Sim B (PASB) domains of the AhR (8, 9), and these interactions have several functional roles, including protecting the AhR from degradation and maintaining the AhR PASB ligand-binding domain (LBD) in a ligand binding-competent state, although ligand binding to the Hsp90-free AhR has been reported (16). Hsp90 binding to the ligand-free cytosolic AhR maintains it in an inactive state by masking its nuclear localization sequence and ARNT (AhR nuclear translocator) dimerization interface (8, 17–19). During ligand-dependent conversion of the AhR into its high affinity DNA-binding form (referred to as AhR transformation), ligand stimulates AhR nuclear localization through unmasking the nuclear localization sequence. Once in the nucleus, Hsp90 appears to be displaced by ARNT due to their mutually exclusive overlapping binding sites in the AhR (17, 20). Although ligand binding is thought to result in a conformational change(s) in the AhR PASB domain that alters/weakens Hsp90 binding and enhances interaction of ARNT with the AhR Per-ARNT-Sim A (PASA) domain (20), the Hsp90-binding site within the PASB domain and the mechanisms responsible for ligand-dependent effects on Hsp90 and ARNT dimerization remain to be elucidated. In the study described here, we have identified specific amino acid residues involved in Hsp90 binding within the AhR PASB LBD and demonstrate a novel role for these residues in modulating AhR ligand-dependent activity.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Anti-Hsp90 monoclonal antibody 3G3p90 was produced at Antibodies Inc. (Davis, CA) from hybridoma cells kindly provided by Dr. Gary Perdew (Pennsylvania State University). Control mouse IgM was from Rockland Immunochemicals, and AffiniPure goat anti-mouse IgM from Jackson ImmunoResearch Laboratories. TCDD was obtained from Dr. Steven Safe (Texas A&M University). α-Naphthoflavone (ANF) and 3′-methoxy-4′-nitroflavone (MNF) were obtained from Sigma-Aldrich.
Plasmid Constructs
pcDNA3-mβAhR, pcDNA3-mβAhRΔPASB, pcDNA3-GST-PASB, and pcDNA3-mβARNT have been described previously (20, 21). AhR point mutations were generated using the QuikChange system (Agilent).
Co-immunoprecipitation and Western Blot Assays
COS-1 cells were maintained in α-minimal essential medium supplemented with 5% FBS at 5% CO2 and 37 °C. COS-1 cells grown in 100-mm culture plates were transiently transfected with 8 μg of AhR expression vectors and 20 μl of Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were rinsed once with PBS, plates were scraped, and cells were lysed on ice for 30 min using co-immunoprecipitation assay buffer (25 mm MOPS (pH 7.5), 10% (v/v) glycerol, 1 mm EDTA, 1% (v/v) IGEPAL CA-630, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, and 0.5% (v/v) protein inhibitor mixture (Sigma)). Lysates were centrifuged at 14,000 × g for 10 min, and 400-μl aliquots of supernatant (0.8–1.2 mg of protein) were incubated with antibody 3G3 bound to Bio-Rad Affi-Gel 10 substrate (prepared as described previously (20)) for 1 h at 4 °C with shaking. Samples were washed three times with co-immunoprecipitation assay buffer. Proteins were resolved by SDS-PAGE and detected by Western blotting as described (20) using anti-AhR antibody M20 (Santa Cruz Biotechnology) and anti-β-actin antibody (Santa Cruz Biotechnology). For Hsp90 dissociation studies, transiently transfected COS-1 cells were incubated in the presence of 10 nm TCDD (or 0.1% (v/v) solvent control dimethyl sulfoxide (DMSO)) for 3 h prior to cell lysis. Hsp90 co-immunoprecipitation/AhR Western blot analysis was as described above except that instead of antibody M20 (directed against the AhR bHLH domain and used to detect wild-type AhR (wtAhR)), antibody SE-8 (directed against the AhR PASB domain) (22) was used to detect the AhR GST-PASB fragment. In geldanamycin-dependent degradation experiments, transiently transfected COS-1 cells were incubated in the presence of 0.1 μm geldanamycin or 0.1% (v/v) DMSO for the indicated periods of time prior to lysis and Western blot analysis.
Reporter Gene Induction Assays
COS-1 cells were transiently transfected in 24-well plates with (per well) 40 ng of wild-type or mutant AhR expression vector, 200 ng of pGudLuc6.1 (23), 40 ng of pRL-TK (Promega), 520 ng of pcDNA3.1+ (Invitrogen), and 2 μl of Lipofectamine 2000. Twenty-four hours after transfection, cells were incubated with 1 nm TCDD and/or the indicated concentrations of ANF or MNF for 20–24 h and lysed, and aliquots were analyzed for firefly and Renilla luciferase activities using the Dual-Luciferase reporter assay system (Promega) and an Orion microplate luminometer (Berthold Detection Systems).
RESULTS
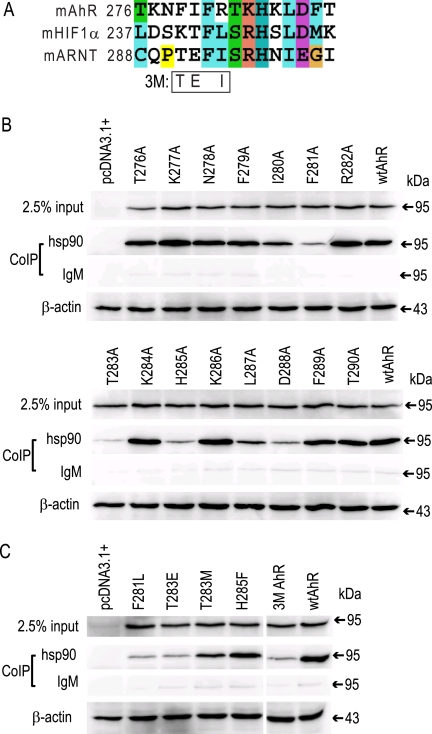
Previous mutational and functional analyses of the AhR revealed that deletion of amino acids 288–418 (which removes the PASB LBD) results in an AhR that is not only deficient in Hsp90 binding and constitutively active (i.e. exhibits ligand-independent transformation/DNA binding) but with an overall level of DNA binding that is much higher than that of the wtAhR (20). These results confirm that Hsp90 maintains the AhR in an inactive state in the absence of ligand and also suggest that Hsp90 binding in the LBD modulates the overall level of AhR transformation/DNA binding. Coincidentally, our preliminary results using in vitro synthesized AhR revealed that a single mutation in the AhR PASB LBD, K284A, dramatically increased the AhR transformation/DNA binding level (to 148% of that obtained with wtAhR (data not shown)), suggesting that an Hsp90-binding site may be located nearby. Intriguingly, the region of the AhR PASB domain encompassing this amino acid is rich in aromatic, basic, and threonine residues (Fig. 1A), a characteristic similar to a region in the Plk1 kinase domain that contains two previously reported natural mutations (P509A and R512W) that eliminate its Hsp90 binding (6). Utilizing these observations as a guide for site-directed mutagenesis and functional analysis, we attempted to identify and characterize amino acid residues involved in Hsp90 binding contained within the PASB LBD of the AhR.
FIGURE 1.
Identification of amino acid residues contributing to Hsp90 binding. A, alignment of a section of the PASB domain of mouse (m) AhR, HIF1α, and ARNT proteins. B and C, lysates from COS-1 cells transiently transfected with the indicated AhR expression constructs were subjected to co-immunoprecipitation (CoIP) assay with anti-Hsp90 antibody 3G3 or control IgM as described under “Experimental Procedures.” Samples were separated by SDS-PAGE, and protein was detected by Western blotting with anti-AhR antibody M20 or anti-β-actin antibody. The 3M AhR contains substitutions F279T, I280E, and R282I. Results are representative of three independent experiments.
Identification of Residues Involved in Hsp90 Binding
The AhR PASB region surrounding Lys-284 was aligned with the homologous regions of the closely related proteins HIF1α, which also binds Hsp90 in its PASB domain, and ARNT, which does not bind Hsp90 (9, 11). Because the alignment did not reveal a clear distinction between Hsp90-binding AhR and HIF1α and non-Hsp90-binding ARNT (Fig. 1A), we attempted to identify key residues by individually mutating each of the AhR residues contained within this region (amino acids 276–290) to alanine. Hsp90 binding analysis of the resulting mutant AhRs was performed using lysates from transiently transfected COS-1 cells. The wild-type and mutant AhR expression constructs produced similar levels of AhR protein (see inputs in Fig. 1B); no protein was expressed from the empty vector. Although several alanine substitutions within this region had no significant effect on Hsp90 binding, others resulted in a dramatic decrease in Hsp90 binding (F281A, T283A, H285A, and D288A) or a moderate decrease in Hsp90 binding (I280A, L287A, and F289A) compared with the wtAhR (Fig. 1B and Table 1). Hsp90 binding to the K284A mutant AhR was not significant different from that to the wtAhR (Table 1), indicating that the relative increase in AhR transformation/DNA binding observed with in vitro synthesized AhR containing the mutation was not simply due to a loss of Hsp90 binding like that proposed for AhRΔPASB. This could also result from functional differences in ligand-dependent activation and/or Hsp90 binding to AhR proteins expressed in transient transfections and compared with those synthesized in vitro.
TABLE 1.
Hsp90 binding to the transiently transfected AhR proteins
Binding to the indicated mutant or wild-type AhR was analyzed in triplicate using the co-immunoprecipitation/Western blot protocol described under “Experimental Procedures.” Values are expressed as a percent of the wtAhR and presented as the means ± S.D. of three replicate reactions adjusted for nonspecific binding reactions (IgM controls). Statistical comparisons were performed using the corresponding wtAhR controls, and only one representative wtAhR value is presented.
| AhR mutation | Hsp90 binding |
|---|---|
| T276A | 111 ± 8 |
| K277A | 105 ± 16 |
| N278A | 105 ± 15 |
| F279A | 105 ± 10 |
| I280A | 65 ± 16 |
| F281A | 5 ± 10a |
| R282A | 83 ± 19 |
| T283A | −1 ± 10a |
| K284A | 90 ± 8 |
| H285A | 9 ± 6a |
| K286A | 89 ± 2 |
| L287A | 29 ± 7a |
| D288A | 13 ± 2a |
| F289A | 63 ± 4a |
| T290A | 73 ± 17 |
| F281L | 17 ± 8a |
| T283E | 20 ± 6a |
| T283M | 64 ± 16a |
| H285F | 123 ± 25 |
| 3M (F279T/I280E/R282I) | 13 ± 6a |
| A375L | 64 ± 17a |
| A375V | 77 ± 18 |
| wtAhR | 100 ± 18 |
a Statistically different from the wtAhR at p < 0.05 as determined by Student's t test.
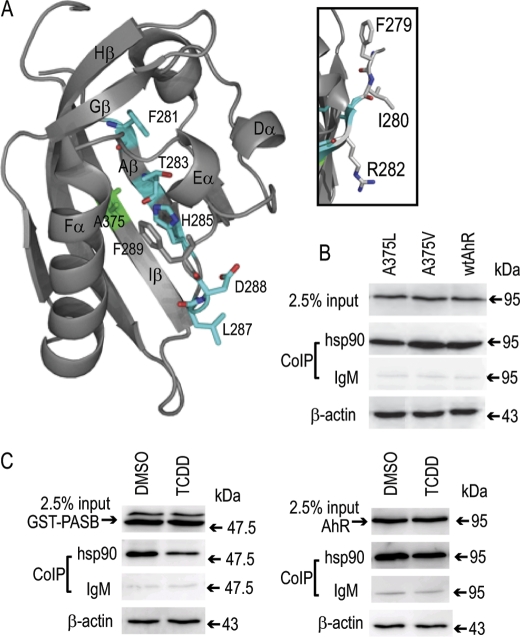
Although the AhR has two Hsp90-binding sites, namely in the bHLH and PASB domains (8, 9), the decrease in Hsp90 binding observed with several point mutations spanning amino acids 280–288 suggests that these residues are part of an Hsp90-binding site within the PASB domain. Additional analysis of the specificity of this tentative Hsp90-binding site using AhR point mutations generated in previous studies (24, 25) revealed that whereas some mutations (T283M and H285F) had little or no loss of Hsp90 binding, compared with an alanine substitution, mutation of Phe-281 to a bulky hydrophobic but non-aromatic leucine residue did not restore Hsp90 binding (Fig. 1C). These findings indicate the importance of Phe-281 in the binding site specificity. Visualizing the position of the key amino acids affecting Hsp90 binding (Phe-281, Thr-283, His-285, Leu-287, and Asp-288) using our homology model of the AhR PASB LBD (25) revealed that the side chains of these residues form a continuous stretch within the ligand-binding cavity (Fig. 2A).
FIGURE 2.
The putative Hsp90-binding site spatially overlaps with the ligand-binding site in the PASB LBD. A, structure and labels are from the AhR PASB homology model (25). Internal Hsp90-interacting resides are shown in cyan, and Ala-375 (implicated in ligand binding) is shown in green. B, Hsp90 binding for mutant AhRs was carried out as described in the legend to Fig. 1B. C, TCDD treatment results in Hsp90 dissociation from the AhR PASB LBD. COS-1 cells transiently transfected with either the GST-PASB (left panel) or wtAhR (right panel) expression plasmid were incubated with TCDD (or solvent control) for 3 h, and cell lysates were subjected to co-immunoprecipitation (CoIP) assay as described in the legend to Fig. 1B. Western blot analysis was carried out using anti-PASB fragment antibody SE-8 for GST-PASB and anti-AhR antibody M20 for the wtAhR. Antibody SE-8 also detected an additional slower migrating nonspecific band (left panel), which did not bind Hsp90 and likely does not represent the functional AhR fragment. Results are representative of three independent experiments.
Although alignment of the AhR with ARNT, which does not bind Hsp90 (9), demonstrated that the specific residues of the putative AhR PASB LBD Hsp90-binding site were relatively conserved (Phe-286, Ser-288, His-290, Ile-292, and Glu-293, respectively, in ARNT (Fig. 1A)), residues surrounding the critical Phe-281 (i.e. Phe-279, Ile-280, and Arg-282) were poorly conserved (Thr, Glu, and Ile, respectively, in ARNT) (Fig. 1A). However, individual mutations of these residues to alanine did not result in significant reduction in Hsp90 binding (Table 1). To test the possible effect of multiple mutations within this region on Hsp90 binding, the combined mutation of all three residues in the AhR to those found in ARNT (referred to as the 3M AhR) was generated and, surprisingly, resulted in a dramatic reduction in Hsp90 binding compared with the wtAhR (Fig. 1C and Table 1). This reduction in Hsp90 binding could result from a change in structure of this region of the AhR LBD, demonstrating that amino acids surrounding Phe-281 can dramatically affect Hsp90 binding and may contribute to the differential ability of Hsp90 to bind to the AhR and not ARNT.
Involvement of the Hsp90-binding Residues in Ligand Binding and AhR Activation
The majority of side chains of the amino acids at positions 279–290 that are involved in Hsp90 binding (judged by our individual mutations) are pointing into the ligand-binding cavity of the PASB LBD (Fig. 2A) and are spatially close to Ala-375, a residue previously implicated in ligand binding (24, 26). To examine the possible overlap of Hsp90- and ligand-binding sites within this region, Hsp90 binding analysis was performed using two AhRs containing single mutations at Ala-375 that either have decreased TCDD-binding affinity (A375V) or lack TCDD binding (A375L) (24). Consistent with our hypothesis that Hsp90 binding occurs within the PASB LBD cavity, we observed a reduction in Hsp90 binding to an AhR containing the A375L substitution (Fig. 2B and Table 1), a mutation that reduces the available space within the cavity and impinges on the side chain of His-285, a residue critical for ligand (24) and Hsp90 (Fig. 1B) binding. In contrast, Hsp90 binding to an AhR containing an A375V substitution (which only lowers ligand-binding affinity) was statistically similar to binding to the wtAhR (Fig. 2B and Table 1). These results suggest that Hsp90- and ligand-binding sites in the AhR PASB LBD spatially overlap but are not identical. The overlapping nature of the ligand- and Hsp90-binding sites suggests that ligand binding in the PASB domain may displace Hsp90, at least to certain extent, from its binding site. However, results in the literature are contradictory with regard to the experimental demonstration of ligand-dependent dissociation of Hsp90 from either the full-length AhR or a GST-tagged AhR PASB fragment (GST-PASB) (8, 20). To examine this, COS-1 cells transiently transfected with full-length AhR or GST-PASB expression vectors were incubated with 10 nm TCDD (or 0.1% (v/v) DMSO) for 3 h, and Hsp90 binding to each protein was examined. These experiments demonstrated a ligand-dependent reduction in Hsp90 binding to the AhR GST-PASB fragment, but no significant change in Hsp90 binding to the full-length AhR was observed; there was no change in inputs (Fig. 2C). Statistical analysis of four replicate reactions revealed that Hsp90 binding to the AhR GST-PASB fragment was significantly decreased (by 45%; p < 0.001), whereas Hsp90 binding to the wtAhR was not significantly changed. Interestingly, we did not previously observe a TCDD-dependent decrease in Hsp90 binding to the same AhR GST-PASB fragment when it was expressed in vitro (20). One possible explanation for the apparent discrepancy is the difference in stringency of the washing buffer used in these two studies (with a more stringent washing buffer used in the current study). Additionally, ligand-dependent Hsp90 dissociation from the AhR appears to require p23 (27), and the limited amount of p23 in this expression system (28) may contribute to the previously observed lack of Hsp90 dissociation from in vitro synthesized GST-PASB.
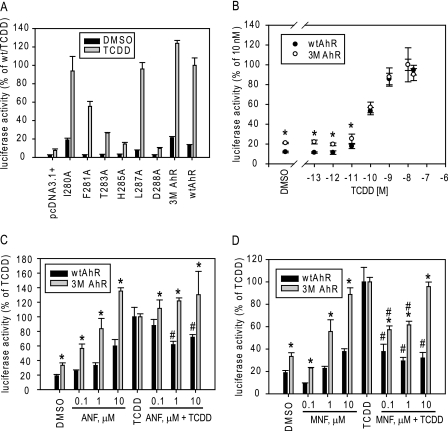
To determine the impact of decreased Hsp90 binding on AhR functionality, TCDD-dependent AhR-responsive reporter gene induction was analyzed in COS-1 cells transiently cotransfected with a mutant or wild-type AhR expression plasmid and an AhR-responsive luciferase reporter plasmid. These experiments revealed that most mutant AhRs that showed dramatically reduced levels of Hsp90 binding (F281A, T283A, H285A, and D288A) (Table 1) also exhibited reduced TCDD-inducible reporter gene activity (Fig. 3A). These decreases in functional activity may be a result of specific mutation-dependent changes in Hsp90-dependent AhR protein stability. This mechanism of Hsp90-dependent AhR protein stability is analyzed below for selected mutations. It is also possible that some of these mutations affect AhR functional activity through Hsp90 binding-independent mechanisms because mutagenesis can cause changes in overall protein conformation. Although some mutant AhRs exhibited moderate reduction in Hsp90 binding (i.e. I280A and L287A) (Table 1), their ability to stimulate TCDD-inducible gene expression was similar to that of the wtAhR (Fig. 3A). Interestingly, although the 3M AhR had even lower levels of Hsp90 binding (only 13 ± 6% of the wtAhR) (Table 1), it exhibited slightly increased reporter gene induction compared with the wtAhR (Fig. 3, A and B). Thus, this AhR construct appears to be fully functional despite its decreased association with Hsp90. These results indicate a limited role of Hsp90 association in TCDD-mediated AhR activation, in agreement with our previous report (16).
FIGURE 3.
The 3M AhR exhibits differences in transformation/DNA binding by partial agonists/antagonists. A–D, transiently transfected COS-1 cells were incubated with the indicated ligands in the absence or presence of the AhR antagonists ANF and MNF, and the luciferase activity of cell lysates was analyzed as described under “Experimental Procedures.” Results are presented as the means ± S.D. of three independent reactions. Values significantly different from the corresponding wtAhR (*) or the TCDD reaction (#) values at p < 0.05 as determined by Student's t test are indicated. Results are representative of three independent experiments.
In contrast to other Hsp90 binding-deficient AhR mutations, the side chains of the residues mutated in the 3M AhR point outside of the ligand-binding cavity (Fig. 2A, inset) and therefore are not likely to interfere with ligand binding directly. Consistent with this observation, the EC50 values for TCDD-induced reporter activity of both the wtAhR and 3M AhR are comparable (Fig. 3B), indicative of similar binding affinities. Because the side chains of the mutated amino acids in the 3M AhR are external to the binding cavity, the decreased Hsp90 binding of the 3M AhR must result from a mutation-dependent change in the structure of the AhR PASB LBD that selectively impacts Hsp90 binding but not ligand (TCDD) binding within the cavity. In addition to modification of specific interaction sites with ligand, Hsp90, and other factors, mutagenesis can also result in nonspecific changes in AhR protein structure conformation that can change the functional properties and protein-protein interactions of the mutant AhR in unexpected ways. Although these nonspecific effects of mutagenesis on protein conformation can never be fully excluded, they are less likely with mutant AhRs that retain full functional activity (i.e. the 3M AhR, which has similar ligand binding and transcriptional activation as the wtAhR yet demonstrated a 87% reduction in Hsp90 binding).
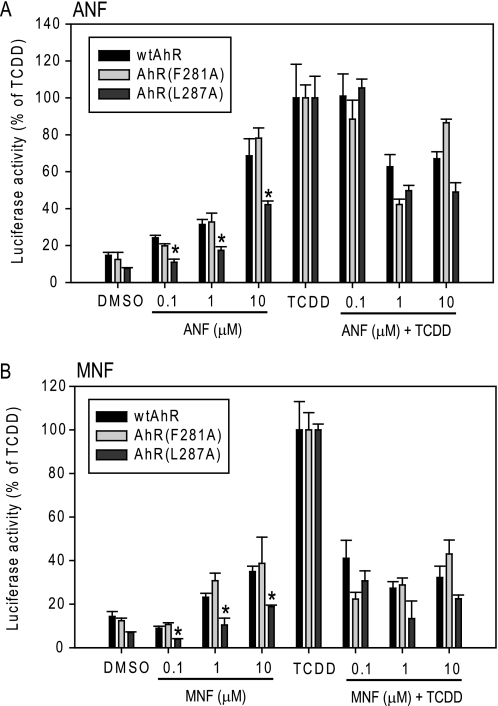
Differences in the ability of ligands to bind to and activate the AhR have been proposed to account for the structural diversity and activity of AhR ligands (agonists and antagonists) (15). Given the alterations in the PASB LBD of the 3M AhR involved in Hsp90 binding, it was of interest to examine AhR activation by ligands that are suggested to interact with the AhR in a distinctly different manner compared with TCDD. The AhR antagonists ANF and MNF, which exhibit agonist activity only at higher concentrations (i.e. they are partial agonists), can bind to the AhR PASB LBD at lower concentrations without initiating transformation (Hsp90 displacement and ARNT dimerization), and as such, they must interact differently from TCDD within the AhR ligand-binding pocket in these conditions (28–31). To test whether the 3M AhR has altered transactivation properties with respect to these partial agonists/antagonists, COS-1 cells were transiently transfected with the wtAhR or 3M AhR and an AhR-responsive luciferase reporter plasmid and then incubated with varying concentrations of ANF and MNF in the absence or presence of TCDD prior to the analysis of reporter gene expression. Not only was the 3M AhR able to activate reporter gene activity to a greater degree with ANF or MNF compared with their effect on the wtAhR (enhanced agonist activity), but their antagonist activity against TCDD was either eliminated (with ANF) or dramatically reduced (with MNF) (Fig. 3, C and D). At 10 μm, both ANF and MNF were full agonists of the 3M AhR but were antagonists of the wtAhR (Fig. 3, C and D). Other AhR constructs that exhibited decreased Hsp90 binding (F281A and L287A) (Fig. 1B) did not demonstrate this dramatic ANF/MNF antagonist-to-agonist switch in AhR transcriptional activation activity (Fig. 4), indicating that this effect with the 3M AhR was not simply the direct consequence of decreased Hsp90 binding but was due to an alteration in ligand-dependent functionality. Moreover, the L287A mutation demonstrated dramatically decreased agonist activity with ANF and MNF relative to the wtAhR (Fig. 4), indicating a possible role of this amino acid residue in ligand-specific AhR transformation mechanisms. Additionally, given the decreased agonist activity and unchanged antagonist activity of the AhR containing this specific mutation (Fig. 4), it is likely that there are distinct mechanisms of agonism and antagonism with ANF (or MNF). Together, the mutagenesis data in this study suggest that distinct amino acid determinants may mediate (i) binding and activation by different AhR ligands, (ii) the mechanism of agonist and antagonist activation by partial agonists/antagonists, and (iii) the switch from antagonist to agonist observed with these compounds. However, nonspecific effects of mutagenesis on the overall protein conformation and functional properties of these mutant AhRs cannot be excluded with certainty.
FIGURE 4.
Transcriptional activation of the wtAhR and mutants F281A and L287A by ANF and MNF. COS-1 cells were transiently transfected and analyzed for activation of AhR-dependent transcription as described in the legend to Fig. 3. L287A mutant AhR agonist activity that was significantly different from the corresponding wtAhR values at p < 0.05 as determined by Student's t test is indicated (*). Results are representative of three independent experiments.
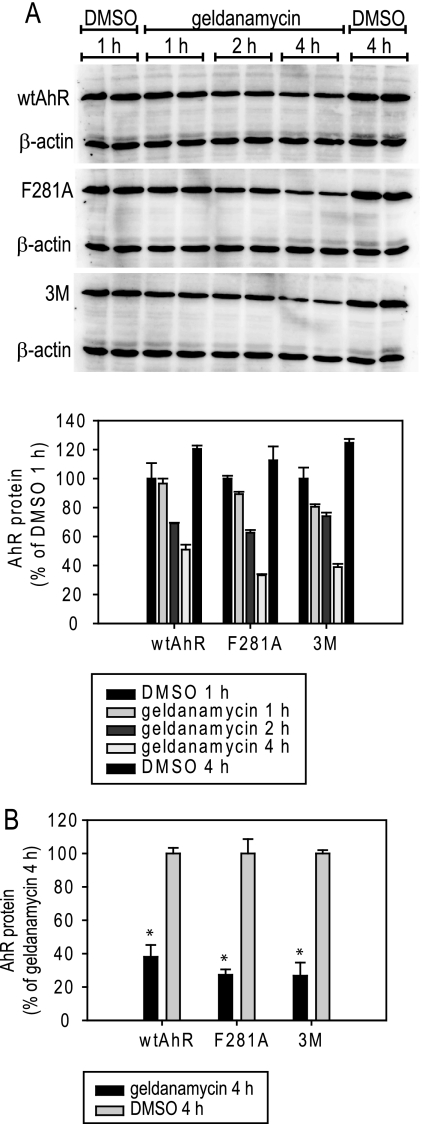
Given the previously reported stabilizing effect of Hsp90 binding on AhR levels (19, 32, 33), it was of interest to analyze whether the decreased Hsp90 binding observed with our mutant AhRs resulted in decreased Hsp90-dependent protein stability as indicated by changes in geldanamycin-dependent degradation. COS-1 cells were transiently transfected with the F281A, 3M, or wild-type AhR and incubated for 1–4 h with 100 nm geldanamycin, a concentration previously shown to affect AhR stability negatively (18). Whole cell lysates were resolved by SDS-PAGE and analyzed by Western blotting, and these studies revealed significant but comparable time- and geldanamycin-dependent decreases in wild-type and mutant AhR protein levels (Fig. 5, A and B). A similar degree of geldanamycin sensitivity of wild-type and mutant AhRs (F281A and 3M) was also observed in COS-1 cells expressing low levels of each AhR (corresponding to the DNA amounts used in reporter assays), indicating that the effects on AhR stability did not depend on AhR expression levels (data not shown). Thus, these two mutant AhRs (F281A and 3M) do not appear to have an alteration in Hsp90-dependent protein stability compared with the wtAhR.
FIGURE 5.
The wtAhR and select AhR mutants demonstrate similar sensitivity to geldanamycin. COS-1 cells were transiently transfected with the indicated AhR constructs (the wtAhR or 3M AhR (F279T/I280E/R282I)) under conditions similar to the co-immunoprecipitation experiments, cells were incubated with 0.1 μm geldanamycin (or 0.1% (v/v) DMSO) for the indicated times, and AhR levels in cell lysates were determined by Western blot assay. A, representative gels are shown, and protein quantitation of duplicate samples is presented in the graph. Values represent the mean of two reactions, and error bars indicate the range of the results. B, geldanamycin-induced decrease in AhR levels is statistically significant and similar among AhR constructs. Geldanamycin-dependent AhR protein degradation after a 4-h incubation was measured as described for A, and the values are expressed as the means ± S.D. of three independent reactions. Values significantly different from the corresponding DMSO reaction values at p < 0.05 as determined by the Student's t test are indicated (*).
DISCUSSION
One of the major obstacles in Hsp90 biology has been the lack of a defined Hsp90-binding site, and although several studies reported selective amino acids or small fragments of diverse client proteins involved in Hsp90 binding (2, 6, 10, 12, 13), no consensus binding site has emerged. It is currently thought that Hsp90 binds to multiple sites in a client protein and may recognize a specific charge pattern on the client protein's surface (12, 13). Unlike previous reports that identified only individual residues involved in Hsp90 binding of client proteins, such as Plk1, ErbB2, and PKC (6, 12, 13), here, we characterized the tentative Hsp90-binding site in more detail (through alanine scanning mutagenesis) and propose its spatial model. In comparison with the previous reports, no obvious similarities were found with the tentative Hsp90-binding sites suggested in ErbB2 and PKC, possibly due to multiple protein segments comprising an Hsp90-binding site or to non-sequence-specific effects of protein structure on Hsp90 binding, as proposed for kinase Hsp90 clients and the glucocorticoid receptor, respectively (2, 34). It is also noteworthy that the abovementioned mutagenesis studies with ErbB2 and PKC did not test the functional activity of the mutant protein, whereas this study and the Plk1 report described below determined Hsp90 binding levels with functional proteins. Because point mutagenesis could produce an incorrectly folded protein that has nonspecific effects on Hsp90 binding, the use of functionally active mutations (which minimize such incorrect folding effects) is desirable for this type of analysis.
In contrast, comparison of our results with the tentative Hsp90-binding site in the Polo box domain of Plk1 provides some interesting novel insights. In Plk1, several naturally occurring mutations that decreased Hsp90 binding were reported (6). When we mapped these specific mutations onto the crystal structure of the Plk1 Polo box domain (35), three of these amino acids (Pro-509, Arg-512, and Asp-457) were located in or near amino acid stretch Phe-515–Thr-513–Arg-512–Tyr-510 (supplemental Fig. 1) similar in charge and polarity distribution to Phe-281–Thr-283–His-285–Phe-289 in the tentative AhR PASB domain Hsp90-binding site identified in this study (Fig. 2A). Although there is little obvious homology in the primary sequences of the AhR and Plk1 that encompass these regions, beyond both being rich in aromatic, basic, and threonine residues, comparisons of the three-dimensional models of each region revealed spatial similarities between these tentative Hsp90-binding sites. Future studies will assess whether these similarities represent an actual consensus Hsp90-binding site (as determined by three-dimensional structural analysis).
Because the client protein binding region of Hsp90 has been proposed to be contained within a flexible loop (plus one tryptophan residue) (36), we envision that this flexible loop protrudes into the AhR ligand-binding pocket and makes key contacts with AhR residues involved in Hsp90 binding. Considering the available internal space in the AhR ligand-binding cavity (25), accessibility of Hsp90 to the modeled binding pocket might initially seem too restrictive. However, given that the current homology model of the AhR LBD most likely represents its ligand-bound, Hsp90-free closed conformation, the binding pocket of the unliganded conformation of the AhR is proposed to be significantly more open and would be more readily accessible to Hsp90 and AhR ligands (24, 25).
Overlap of residues involved in Hsp90 binding and the antagonist-to-agonist switch as demonstrated by decreased Hsp90 binding and altered functional activity of ANF and MNF with the 3M AhR may suggest that Hsp90 binding is involved in the mechanism of antagonism. However, other mutant AhRs with decreased Hsp90 binding (F281A and L387A) (Fig. 4) did not exhibit a similar antagonist-to-agonist switch with ANF or MNF, indicating that simply decreasing Hsp90 binding is not sufficient to allow conversion of an antagonist/partial agonist to a full agonist. The precise role of Hsp90 binding (and ligand-induced dissociation) in this antagonist-to-agonist conversion effect remains to be elucidated. Relative to the mechanism of AhR transformation, these data indicate that in addition to Hsp90 dissociation resulting from ligand displacement as one possible mechanism, additional factors and/or events, such as exposure of the ARNT dimerization interface (20), may contribute to ligand-dependent formation of AhR/ARNT dimers. Interestingly, the existence of partial antagonists/agonists has also been reported for other Hsp90 client proteins such as the steroid hormone receptors (37, 38). In the estrogen and glucocorticoid receptors, Hsp90 binds to the LBD (similar to the AhR); however, whether Hsp90 is directly displaced by bound ligand and how this might affect the properties of partial antagonists/agonists remain to be determined.
ARNT has been previously implicated in the displacement of Hsp90 from their overlapping binding sites within the AhR, with AhR-ARNT binding interactions occurring primarily in the AhR bHLH and PASA domains (8, 17, 18, 20). Although these same mutagenesis and deletion analysis studies revealed little functional requirement of the AhR PASB domain for dimerization with ARNT (e.g. as indicated by constitutive activation of transformation and DNA binding by the AhRΔPASB construct), we have previously observed a small degree of ligand-dependent AhR GST-PASB/ARNT dimerization (20). However, in these experiments, even excess ARNT failed to displace Hsp90 from the GST-PASB fragment, even though it efficiently displaced Hsp90 from the wtAhR or AhR GST-PASA-PASB (20). Thus, ligand-dependent Hsp90 dissociation from GST-PASB observed in this study would likely be independent of any effects of ARNT, which is present at low levels in COS-1 cells. The lack of ligand-dependent Hsp90 dissociation from the wtAhR may further indicate limited effects of ARNT in this system. Although ARNT in COS-1 cells could not be detected by our Western blot assay (data not shown), some must be present as indicated by AhR-dependent induction of reporter gene expression. Together, these data support the mechanism of direct Hsp90 displacement by bound ligand (TCDD) in the PASB LBD. In contrast, ARNT-dependent Hsp90 dissociation effects were found to be important in the context of the full-length AhR and specifically required the presence of the PASA domain (20).
The majority of Hsp90-related studies are focused on its role in the protein stability of client proteins, and they have not addressed its possible involvement in activation of client proteins. Here, we have reported that decreased Hsp90 binding did not affect Hsp90-dependent AhR protein stability but instead dramatically altered the functional properties of AhR activation. Although the second Hsp90-binding site in the AhR (in the bHLH domain) may be responsible for the Hsp90-dependent AhR protein stability, the residues involved in Hsp90 binding in the AhR PASB domain appear to be also involved in the mechanism of ligand-dependent AhR activation. Importantly, several reports suggested that geldanamycin-induced Hsp90 dissociation resulted in temporary activation of diverse client proteins, such as the AhR, HIF1α, and Src1 (39–41), suggesting a common role of Hsp90 in maintaining client proteins in an inactive state. Increased understanding of the mechanisms by which Hsp90 can modulate client protein activity will require detailed knowledge of its specific interactions with client proteins and is an important area for future studies.
Supplementary Material
Acknowledgments
We thank Dr. Steven Safe for the TCDD, Dr. Gary Perdew for the anti-Hsp90 monoclonal antibody and helpful advice on co-immunoprecipitation methods, and Dr. Laura Bonati (University of Milano-Bicocca, Milan, Italy) for the AhR PASB LBD model.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES007685 and R01 ES012498 from NIEHS. This work was also supported by the California Agricultural Experiment Station.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- AhR
- aryl hydrocarbon receptor
- HIF1α
- hypoxia-inducible factor-1α
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- bHLH
- basic helix-loop-helix
- PASB
- Per-ARNT-Sim B
- LBD
- ligand-binding domain
- PASA
- AhR Per-ARNT-Sim A
- ANF
- α-naphthoflavone
- MNF
- 3′-methoxy-4′-nitroflavone
- DMSO
- dimethyl sulfoxide
- wtAhR
- wild-type AhR.
REFERENCES
- 1. Pearl L. H., Prodromou C. (2006) Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 2. Scroggins B. T., Prince T., Shao J., Uma S., Huang W., Guo Y., Yun B. G., Hedman K., Matts R. L., Hartson S. D. (2003) Biochemistry 42, 12550–12561 [DOI] [PubMed] [Google Scholar]
- 3. Sato S., Fujita N., Tsuruo T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10832–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Papapetropoulos A., Zhou Z., Gerassimou C., Yetik G., Venema R. C., Roussos C., Sessa W. C., Catravas J. D. (2005) Mol. Pharmacol. 68, 1133–1141 [DOI] [PubMed] [Google Scholar]
- 5. Prince T., Sun L., Matts R. L. (2005) Biochemistry 44, 15287–15295 [DOI] [PubMed] [Google Scholar]
- 6. Simizu S., Osada H. (2000) Nat. Cell Biol. 2, 852–854 [DOI] [PubMed] [Google Scholar]
- 7. Müller L., Schaupp A., Walerych D., Wegele H., Buchner J. (2004) J. Biol. Chem. 279, 48846–48854 [DOI] [PubMed] [Google Scholar]
- 8. Fukunaga B. N., Probst M. R., Reisz-Porszasz S., Hankinson O. (1995) J. Biol. Chem. 270, 29270–29278 [DOI] [PubMed] [Google Scholar]
- 9. Perdew G. H., Bradfield C. A. (1996) Biochem. Mol. Biol. Int. 39, 589–593 [DOI] [PubMed] [Google Scholar]
- 10. Pratt W. B., Galigniana M. D., Morishima Y., Murphy P. J. (2004) Essays Biochem. 40, 41–58 [DOI] [PubMed] [Google Scholar]
- 11. Katschinski D. M., Le L., Schindler S. G., Thomas T., Voss A. K., Wenger R. H. (2004) Cell. Physiol. Biochem. 14, 351–360 [DOI] [PubMed] [Google Scholar]
- 12. Xu W., Yuan X., Xiang Z., Mimnaugh E., Marcu M., Neckers L. (2005) Nat. Struct. Mol. Biol. 12, 120–126 [DOI] [PubMed] [Google Scholar]
- 13. Gould C. M., Kannan N., Taylor S. S., Newton A. C. (2009) J. Biol. Chem. 284, 4921–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Citri A., Harari D., Shohat G., Ramakrishnan P., Gan J., Lavi S., Eisenstein M., Kimchi A., Wallach D., Pietrokovski S., Yarden Y. (2006) J. Biol. Chem. 281, 14361–14369 [DOI] [PubMed] [Google Scholar]
- 15. Denison M. S., Pandini A., Nagy S. R., Baldwin E. P., Bonati L. (2002) Chem. Biol. Interact. 141, 3–24 [DOI] [PubMed] [Google Scholar]
- 16. Phelan D. M., Brackney W. R., Denison M. S. (1998) Arch. Biochem. Biophys. 353, 47–54 [DOI] [PubMed] [Google Scholar]
- 17. Probst M. R., Reisz-Porszasz S., Agbunag R. V., Ong M. S., Hankinson O. (1993) Mol. Pharmacol. 44, 511–518 [PubMed] [Google Scholar]
- 18. Antonsson C., Whitelaw M. L., McGuire J., Gustafsson J. A., Poellinger L. (1995) Mol. Cell. Biol. 15, 756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H. S., Singh S. S., Perdew G. H. (1997) Arch. Biochem. Biophys. 348, 190–198 [DOI] [PubMed] [Google Scholar]
- 20. Soshilov A., Denison M. S. (2008) J. Biol. Chem. 283, 32995–33005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukunaga B. N., Hankinson O. (1996) J. Biol. Chem. 271, 3743–3749 [DOI] [PubMed] [Google Scholar]
- 22. Brown D. J., Van Beneden R. J., Clark G. C. (1995) Arch Biochem. Biophys. 319, 217–224 [DOI] [PubMed] [Google Scholar]
- 23. Rushing S. R., Denison M. S. (2002) Arch. Biochem. Biophys. 403, 189–201 [DOI] [PubMed] [Google Scholar]
- 24. Pandini A., Denison M. S., Song Y., Soshilov A. A., Bonati L. (2007) Biochemistry 46, 696–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pandini A., Soshilov A. A., Song Y., Zhao J., Bonati L., Denison M. S. (2009) Biochemistry 48, 5972–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poland A., Palen D., Glover E. (1994) Mol. Pharmacol. 46, 915–921 [PubMed] [Google Scholar]
- 27. Kazlauskas A., Poellinger L., Pongratz I. (1999) J. Biol. Chem. 274, 13519–13524 [DOI] [PubMed] [Google Scholar]
- 28. Merchant M., Arellano L., Safe S. (1990) Arch. Biochem. Biophys. 281, 84–89 [DOI] [PubMed] [Google Scholar]
- 29. Lu Y. F., Santostefano M., Cunningham B. D., Threadgill M. D., Safe S. (1995) Arch. Biochem. Biophys. 316, 470–477 [DOI] [PubMed] [Google Scholar]
- 30. Zhou J., Gasiewicz T. A. (2003) Arch. Biochem. Biophys. 416, 68–80 [DOI] [PubMed] [Google Scholar]
- 31. Wilhelmsson A., Whitelaw M. L., Gustafsson J. A., Poellinger L. (1994) J. Biol. Chem. 269, 19028–19033 [PubMed] [Google Scholar]
- 32. Carver L. A., Jackiw V., Bradfield C. A. (1994) J. Biol. Chem. 269, 30109–30112 [PubMed] [Google Scholar]
- 33. Whitelaw M. L., McGuire J., Picard D., Gustafsson J. A., Poellinger L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaul S., Murphy P. J., Chen J., Brown L., Pratt W. B., Simons S. S., Jr. (2002) J. Biol. Chem. 277, 36223–36232 [DOI] [PubMed] [Google Scholar]
- 35. Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N. (2003) EMBO J. 22, 5757–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. (2003) Mol. Cell 11, 647–658 [DOI] [PubMed] [Google Scholar]
- 37. Clark R. D. (2008) Curr. Top. Med. Chem. 8, 813–838 [DOI] [PubMed] [Google Scholar]
- 38. Bai Z., Gust R. (2009) Arch. Pharm. 342, 133–149 [DOI] [PubMed] [Google Scholar]
- 39. Lees M. J., Whitelaw M. L. (1999) Mol. Cell. Biol. 19, 5811–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ibrahim N. O., Hahn T., Franke C., Stiehl D. P., Wirthner R., Wenger R. H., Katschinski D. M. (2005) Cancer Res. 65, 11094–11100 [DOI] [PubMed] [Google Scholar]
- 41. Koga F., Xu W., Karpova T. S., McNally J. G., Baron R., Neckers L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11318–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.