Abstract
Hyperchromicity measurements and quasi-elastic laser light scattering (QELS) have been used to assess the solution structure of the metabolically stable E. coli 4.5S RNA. Results from thermal denaturation measurements revealed the 4.5S species to be markedly more stable than most other RNAs characterized thus far. Optical Tm's range from 79 degrees to 88 degrees with transitions approximately 25 degrees C wide. The Tm values show little dependence on ionic strength, but stability is enhanced considerably by Mg+2. In the QELS experiments the diffusion coefficient does not decrease until T greater than 70 degrees C. Neither the diffusive melting nor the diffusion coefficient at infinite dilution (D0(20,w)) show dependencies on ionic strength but both are influenced by Mg+2. The diffusion behavior is in agreement with that predicted for a rigid cylindrical molecule 125 to 160 A long and 37 to 26 A in diameter. Taken together these results are consistent with the more stable hypothetical secondary structures that can be formed, in which 70-75% of the 114 bases are paired to form a single extended hairpin helix.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
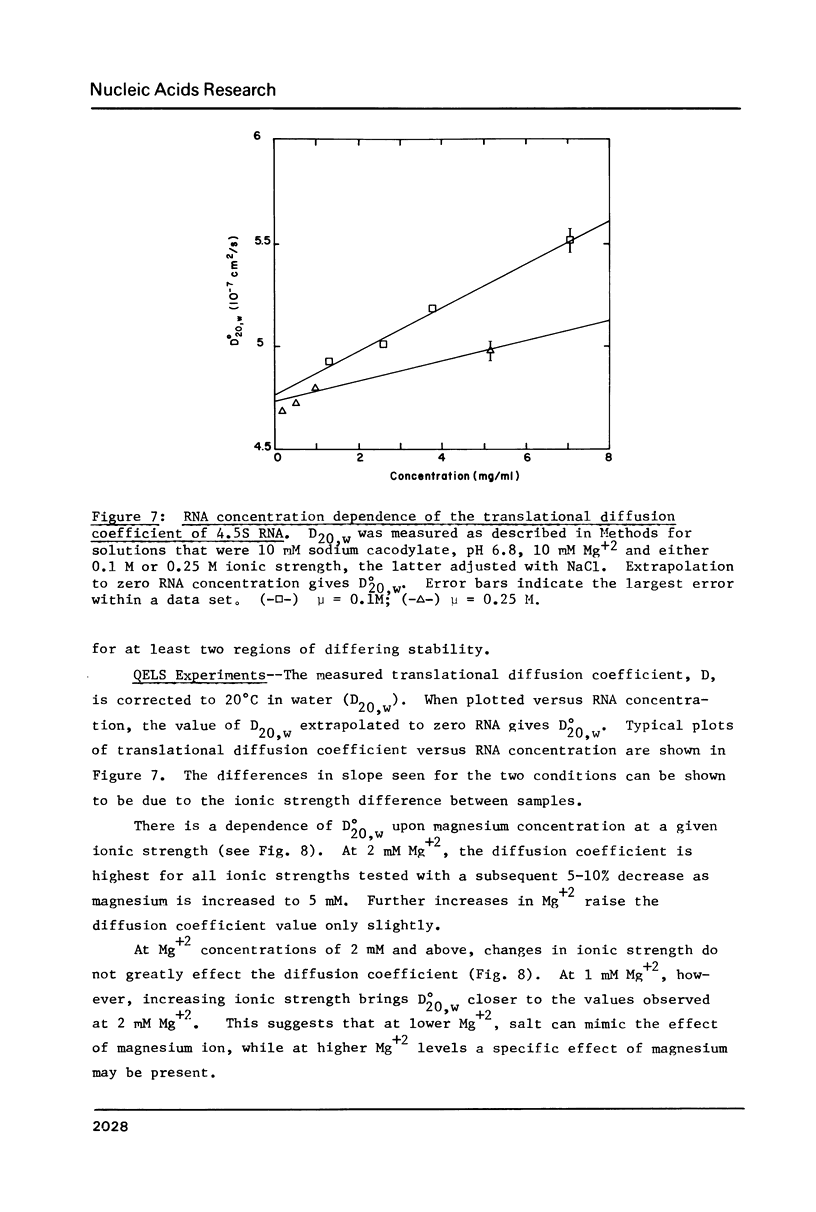
- Bothwell A. L., Garber R. L., Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J Biol Chem. 1976 Dec 10;251(23):7709–7716. [PubMed] [Google Scholar]
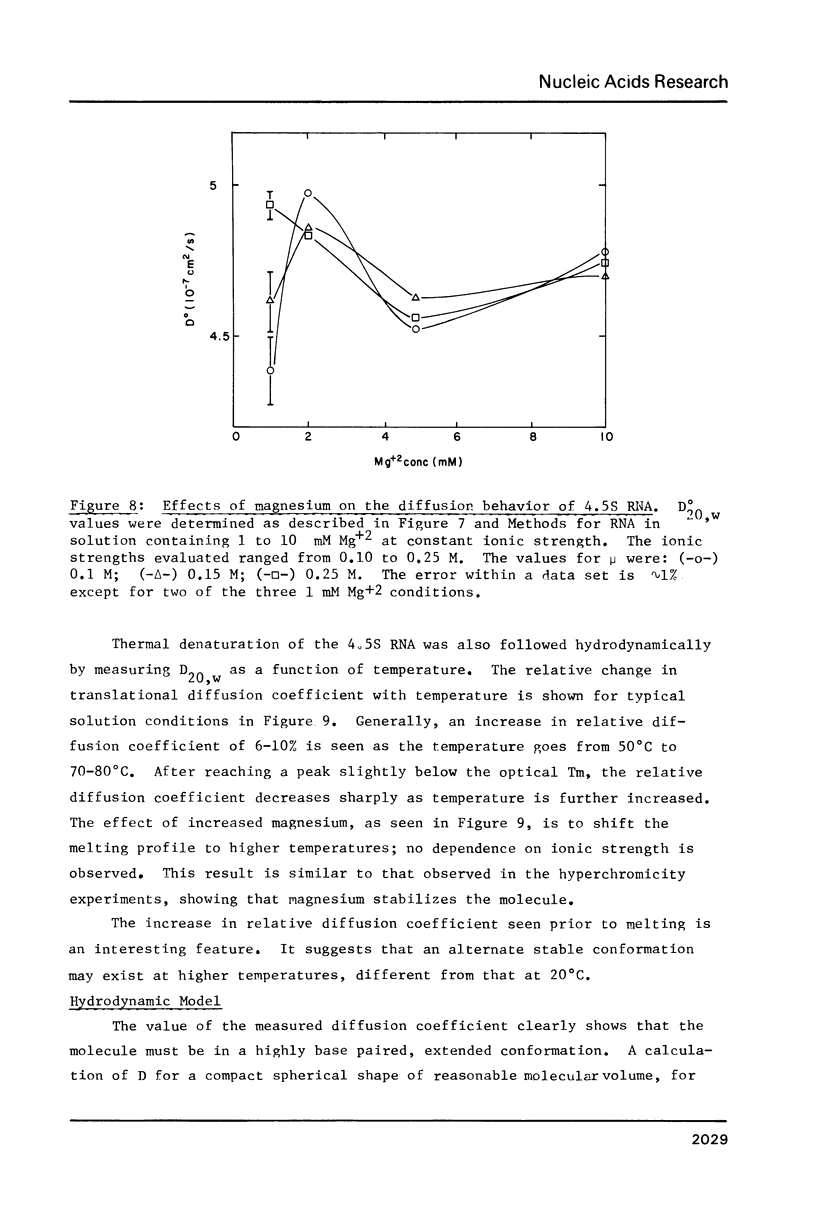
- Fournier M. J., Peterkofsky A. Formation of chromatographically unique species of transfer ribonucleic acid during amino acid starvation of relaxed-control Escherichia coli. J Bacteriol. 1975 May;122(2):538–548. doi: 10.1128/jb.122.2.538-548.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
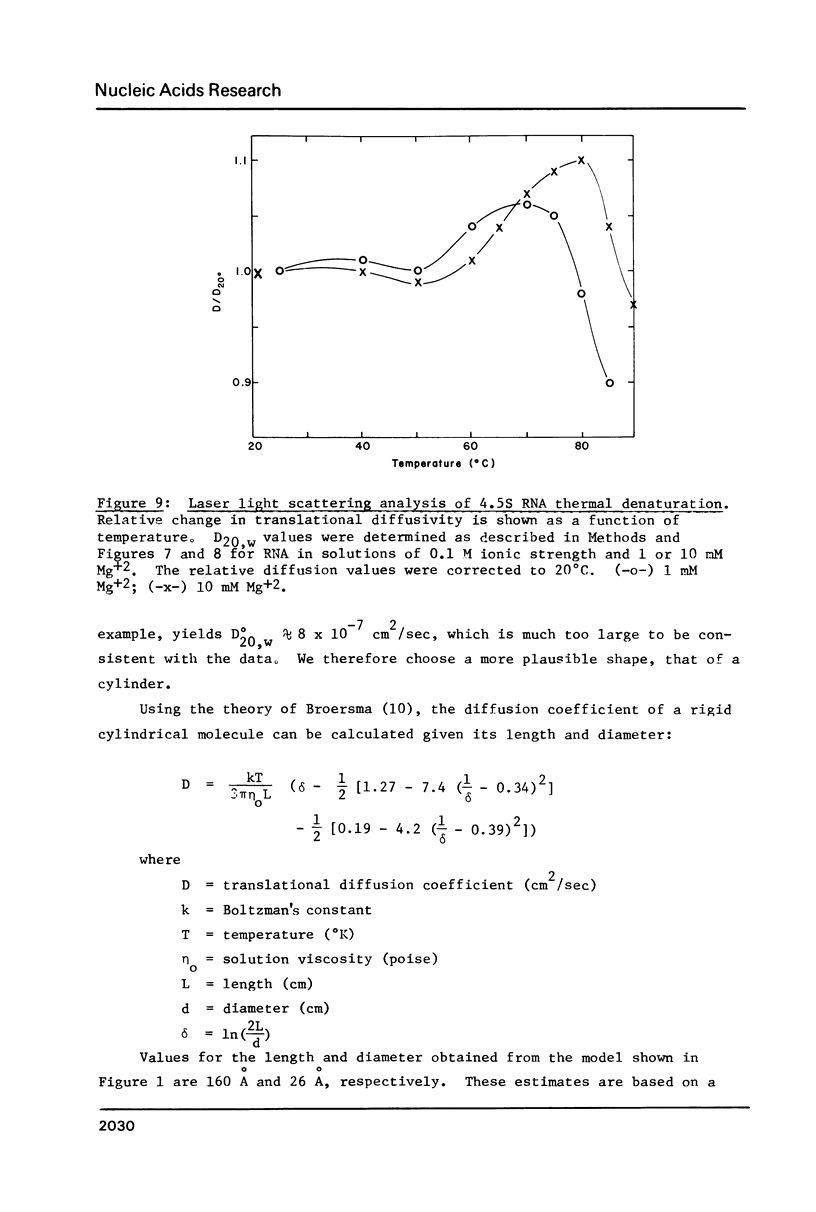
- Griffin B. E. Studies and sequences of Escherichia coli 4.5 S RNA. J Biol Chem. 1975 Jul 25;250(14):5426–5437. [PubMed] [Google Scholar]
- Holmes W. M., Hurd R. E., Reid B. R., Rimerman R. A., Hatfield G. W. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1068–1071. doi: 10.1073/pnas.72.3.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem. 1973 Jul 25;248(14):5033–5041. [PubMed] [Google Scholar]
- Kallenbach N. R., Berman H. M. RNA structure. Q Rev Biophys. 1977 May;10(2):138–236. doi: 10.1017/s0033583500000202. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maruyama S., Sugai S. Folding of yeast 5S ribosomal RNA induced by magnesium binding. J Biochem. 1980 Jul;88(1):151–158. [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson T., Fournier M. J., Langley K. H., Ford N. C., Jr Detection of a major conformational change in transfer ribonucleic acid by laser light scattering. J Mol Biol. 1976 Apr 5;102(2):193–203. doi: 10.1016/s0022-2836(76)80048-4. [DOI] [PubMed] [Google Scholar]
- Pedersen F. S., Haseltine W. A. A micromethod for detailed characterization of high molecular weight RNA. Methods Enzymol. 1980;65(1):680–687. doi: 10.1016/s0076-6879(80)65066-6. [DOI] [PubMed] [Google Scholar]
- Riesner D., Römer R., Maass G. Thermodynamic properties of the three conformational transitions of alanine specific transfer RNA from yeast. Biochem Biophys Res Commun. 1969 May 8;35(3):369–376. doi: 10.1016/0006-291x(69)90508-7. [DOI] [PubMed] [Google Scholar]
- Römer R., Riesner D., Maass G., Wintermeyer W., Thiebe R., Zachau H. G. Cooperative helix-coil transitions in half molecules of phenylalanine specific tRNA from yeast. FEBS Lett. 1969 Sep;5(1):15–19. doi: 10.1016/0014-5793(69)80281-4. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Redfield A. G. Transfer RNA in solution: selected topics. Annu Rev Biophys Bioeng. 1980;9:181–221. doi: 10.1146/annurev.bb.09.060180.001145. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]



