Background: Synucleinopathies are a group of neurodegenerative disorders associated with the formation of amyloid inclusions composed of the normally soluble presynaptic protein α-synuclein.
Results: We describe transgenic mice expressing E46K human α-synuclein.
Conclusion: These E46K α-synuclein transgenic mice accumulate age-dependent intracytoplasmic neuronal α-synuclein inclusions and motor impairments.
Significance: Our findings provide novel insights into the mechanisms of formation and consequence of α-synuclein inclusions.
Keywords: Neurodegeneration, Neuroscience, Parkinson Disease, Protein Folding, Synuclein, Transgenic
Abstract
Synucleinopathies are a group of neurodegenerative disorders associated with the formation of aberrant amyloid inclusions composed of the normally soluble presynaptic protein α-synuclein (α-syn). Parkinson disease is the most well known of these disorders because it bears α-syn pathological inclusions known as Lewy bodies (LBs). Mutations in the gene for α-syn, including the E46K missense mutation, are sufficient to cause Parkinson disease as well as other synucleinopathies like dementia with LBs. Herein, we describe transgenic mice expressing E46K human α-syn in CNS neurons that develop detrimental age-dependent motor impairments. These animals accumulate age-dependent intracytoplasmic neuronal α-syn inclusions that parallel disease and recapitulate the biochemical, histological, and morphological properties of LBs. Surprisingly, the morphology of α-syn inclusions in E46K human α-syn transgenic mice more closely resemble LBs than the previously described transgenic mice (line M83) that express neuronal A53T human α-syn. E46K human α-syn mice also develop abundant neuronal tau inclusions that resemble neurofibrillary tangles. Subsequent studies on the ability of E46K α-syn to induce tau inclusions in cellular models suggest that both direct and indirect mechanisms of protein aggregation are probably involved in the formation of the tau inclusions observed here in vivo. Re-evaluation of presymptomatic transgenic mice expressing A53T human α-syn reveals that the formation of α-syn inclusions in mice must be synchronized; however, inclusion formation is diffuse within affected areas of the neuroaxis such that there was no clustering of inclusions. Collectively, these findings provide insights in the mechanisms of formation of these aberrant proteinaceous inclusions and support the notion that α-syn aggregates are involved in the pathogenesis of human diseases.
Introduction
Synucleinopathies are a group of neurodegenerative diseases associated with neuronal, and in some cases oligodendritic, amyloid inclusions composed of the presynaptic protein α-synuclein (α-syn)3 (1–4). Parkinson disease (PD), the most common known synucleinopathy, is a progressive movement disorder (5, 6) associated with the loss of dopaminergic neurons in the substantia nigra (SN) pars compacta and the formation of α-syn inclusions, known as Lewy bodies (LBs), in some of the remaining dopaminergic neurons (3, 4, 7–9). However, PD is also associated with a range of nonmotor symptoms (10, 11), and there is increasing awareness that many other neuronal populations are also affected in PD (2, 9, 12, 13).
α-Syn is a small, highly charged 140-amino acid residue protein characterized by three major regions: 1) an N-terminal region containing several imperfect KTKEGV repeats, 2) a hydrophobic center domain, and 3) a highly negatively charged C-terminal region (3, 4, 14). α-Syn is predominantly expressed in neurons of the central nervous system (CNS), where it is localized at presynaptic terminals in close proximity to synaptic vesicles (14–17), and several studies suggest that it is involved in modulating synaptic transmission (16–21). The molecular interactions, protein folding, and many of the steps involved in the conversion of normally soluble α-syn into the formation of amyloid inclusions have been studied extensively using in vitro α-syn (4, 22, 23). For example, it is well established that the hydrophobic region of α-syn is necessary for amyloid formation, but both the N- and C-terminal regions and modifications therein influence this process (4, 22, 23). The importance of these findings notwithstanding, the biological events that lead to the aggregation of α-syn into pathological inclusions in vivo are still poorly understood (4, 22, 23).
The most direct and compelling evidence for a fundamental role of α-syn in the pathogenesis of synucleinopathies is the causal relationship between genetic mutations and disease. The missense mutation (c.G209A) in the α-syn gene (SNCA) resulting in the amino acid substitution A53T was first identified in a large Italian family (Contursi) and three small Greek families with autosomal dominant PD (24) and soon thereafter in at least eight additional PD kindreds (25–28). Another autosomal dominant missense mutation (A30P) was identified in a German kindred (29). Short chromosomal duplications or trisomies containing the α-syn gene plus relatively short flanking regions on chromosome 4 were also discovered in patients with PD and the related disease dementia with LBs (30–32). The E46K mutation in α-syn was identified in a kindred manifesting classical PD or dementia with LB (33). Transgenic mice generated to express wild-type (WT) human α-syn as well as those that express the A53T or A30P mutation have provided important insights in the pathogenic role of these mutations and the pathogenic mechanisms of α-syn inclusions (4, 34–37); however, to date, no E46K human α-syn transgenic mouse modeling human disease has been reported. Here we describe two α-syn transgenic lines that express E46K human α-syn and that develop a detrimental motor phenotype associated with the formation of Lewy-like pathology and tau pathological inclusions.
EXPERIMENTAL PROCEDURES
Antibodies
pSer129 is a mouse monoclonal antibody specific to α-syn phosphorylated at Ser-129 (38). Syn211 is a mouse monoclonal antibody specific for human α-syn (39). SNL-1 is a rabbit polyclonal antibody raised against a synthetic peptide corresponding to amino acids 104–119 of α-syn and specifically reacts with both murine and human α-syn (39). SNL-4 is a rabbit polyclonal antibody raised against a synthetic peptide corresponding to amino acids 2–12 of α-syn (39). Syn505 and Syn506 are conformational anti-α-syn mouse monoclonal antibodies that preferentially detect α-syn in pathological inclusions (40, 41). 17025 (generously provided by Dr. Virgnia Lee, University of Pennsylvania, Philadelphia, PA) is a rabbit polyclonal antibody for tau. Mouse monoclonal antibody AT8 (Thermo-Fisher) is specific toward phosphorylation sites Ser-202 and Thr-205 in tau (42). PHF1 (generously provided by Dr. Peter Davies, Albert Einstein University, New York) is specific toward phosphorylation sites Ser-396 and Ser-404 in tau (43). AT100 (Thermo-Fisher) is specific toward phosphorylation sites Ser-212 and Thr-214 in tau (44). Anti-actin (clone C4) (Millipore) is an affinity-purified monoclonal antibody that reacts with all vertebrate isoforms of actin. Anti-tyrosine hydroxylase (TH) antibody (Millipore) is an affinity-purified rabbit polyclonal antibody. Anti-Iba1, a marker for activated microglial cells, is a rabbit polyclonal antibody raised against ionized calcium-binding adaptor molecule 1 (Iba1) (Wako Chemicals USA, Inc., Richmond, VA). Anti-glial fibrillary acidic protein (GFAP) is a rabbit polyclonal antibody against glial fibrillary acidic protein, a specific marker for astrocytes (Promega Corp., Madison, WI).
α-Syn Transgenic Mice
Transgenic mice expressing WT human α-syn (line M20) or A53T human α-syn (line M83) were described previously (35). For the generation of transgenic mice expressing E46K human α-syn, the cDNA harboring the E46K mutation was cloned into the MoPrP.Xho expression vector (45) at the XhoI restriction site. The 14-kb NotI linear fragments containing the α-syn cDNA and the mouse prion protein (PrP) gene promoter together with its 5′-untranslated region (UTR) containing an intron and its 3′-untranslated sequences (Fig. 1A) were used as the transgene to create mice expressing E46K α-syn on a C57Bl/C3H background. The transgenic DNA was microinjected into C57Bl/C3H mouse eggs as a service provided by the Transgenic and Chimeric Mouse Facility of the University of Pennsylvania. Genomic DNA samples were isolated from mouse tails with the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). Potential founders were identified by Southern blot analysis with a 32P-labeled oligonucleotide-primed α-syn DNA probe. Stable transgenic lines carrying the mutant E46K human α-syn constructs (lines M31 and M47) were established, and transgenic offspring were identified by Southern blot analysis of tail DNA. Homozygous transgenic lineages were identified by quantitative Southern blot analysis and verified by backcrossing. Mice were generated, maintained, and sacrificed by CO2 euthanization as approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
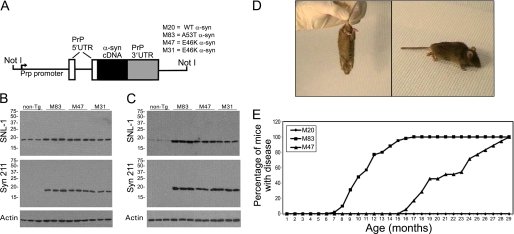
FIGURE 1.
Transgenic lines expressing human α-syn driven by the murine PrP promoter and motor impairment due to the E46K α-syn pathological mutant. A, schematic of the constructs used to generate transgenic α-syn mice. All constructs were identical except for the presence of missense mutations corresponding to either the E46K (lines M31 and M47) or A53T (line M83) amino acid substitution as indicated. WT, E46K, and A53T human α-syn cDNAs were cloned into the XhoI site of MoPrP.Xho mammalian expression vector. The resulting constructs are composed of the murine PrP promoter, the 5′-UTR of the PrP gene containing an intron, the human α-syn cDNA, and the 3′-UTR of the PrP gene (not drawn to scale). Transgenic mouse lines expressing WT or A53T human α-syn were previously generated and described (35). Transgenic lines expressing E46K human α-syn (M31 and M47) were generated here as described under “Experimental Procedures.” Western blot analysis of total protein extracts from the brain (B) or spinal cord (C) of 3-month-old non-transgenic mice and transgenic mouse lines M83, M47, and M31 with antibody SNL-1 (specific for mouse and human α-syn), antibody Syn211 (specific for human α-syn), or anti-actin antibody (clone C4). Samples from three independent mice from each genotype were loaded on the polyacrylamide gels for each analysis. The positions of molecular mass markers are depicted on the left. D, photographs of a 21-month-old homozygous M47 transgenic mouse displaying quadriparesis, arched back, and impaired axial rotation to upright posture, without retraction of its hind limbs. E, comparison of the age of onset of motor phenotype in M20 (n = 52), M83 (n = 102), and M47 (n = 42) α-syn transgenic mice.
Mouse Footprint Analysis
Motor coordination was assessed using footprint analysis testing as described previously (46). Briefly, the fore and hind paws were dipped in non-toxic water-based paints of different colors, and mice were allowed to walk on strips of white paper through a narrow walkway (5 cm wide, 70 cm long). 3–4-month-old mice (WT, n = 6; M47, n = 6), 5–9-month-old mice (WT, n = 6; M47, n = 8), and 15–19-month-old mice (WT, n = 6; M47, n = 4) were subjected to 3 days of testing with three trials per session (with an additional day for non-compliant subjects). The footprint patterns were analyzed for stride length and fore/hind paw overlap as an assessment of gait during ambulation. Stride length was measured as the distance between footprints of each stride. Fore/hind paw overlap was measured as the distance between fore and hind footprints of overlapping steps. Trials were averaged for each mouse, and individual days of testing were analyzed by one-way analysis of variance with Bonferroni post hoc analysis (p was set at <0.05 as level of significance) to assess reproducibility. Data for age cohorts were calculated from the average performance of each mouse, and statistical significance was determined using a one-tailed t test.
Wire Hang Test
To evaluate motor performance, a wire hang test was performed as described previously with slight modifications (47). Briefly, mice were placed on an elevated wire cage lid that was waved lightly to ensure mice gripped the wires before the lid was inverted, and the latency to fall was recorded. A 60-s cut-off was used. 2–4-month-old mice (WT, n = 8; M47, n = 24), 6–8-month-old mice (WT, n = 8; M47, n = 6), and 15–19-month-old mice (WT, n = 7; M47, n = 5) were subjected to 3 days of testing with three trials per session with no prior training. The ability to hang from the wire cage lid is used as an assessment of neuromuscular function and grip strength. Trials were averaged for each mouse, and individual days of testing were analyzed by one-way analysis of variance with Bonferroni post hoc analysis (p was set at <0.05 as level of significance) to assess reproducibility. Data for age cohorts were calculated from the average performance of each mouse, and statistical significance was determined using a two-tailed t test.
Immunohistochemical Analysis
Mice were sacrificed with CO2 euthanization and perfused with phosphate-buffered saline (PBS)/heparin, followed by perfusion with either 70% ethanol plus 150 mm NaCl or PBS-buffered formalin. The brain and spinal cord were then removed and fixed for 24 h in the respective fixatives used for perfusion. As described previously, tissues were dehydrated at room temperature through a series of ethanol solutions, followed by xylene, and then were infiltrated with paraffin at 60 °C (48). The tissues were then embedded into paraffin blocks, which were cut into 7-μm sections. Immunostaining of the sections was performed using methods described previously (48). Briefly, paraffin-embedded tissue sections were rehydrated and labeled with primary antibodies in 0.1 m Tris, pH 7.6, 2% horse serum overnight at 4 °C. Following washing, sections were sequentially incubated with biotinylated goat anti-mouse or anti-rabbit antibodies and avidin-biotin complex system (Vectastain ABC Elite Kit, Vector Laboratories, Burlingame, CA) for 1 h at room temperature. Immunocomplexes were visualized with the chromogen 3,3′-diaminobenzidine, and the sections were counterstained with hematoxylin.
Double-labeling Immunofluorescence Analysis of Mouse Brain Tissue
Paraffin-embedded tissue sections were deparaffinized and hydrated through a series of graded ethanol solutions followed by 0.1 m Tris, pH 7.6. The sections were blocked with 5% dry milk, 0.1 m Tris, pH 7.6, and were incubated simultaneously with combinations of primary antibodies diluted in 5% dry milk, 0.1 m Tris, pH 7.6. After extensive washing, sections were incubated with goat anti-mouse secondary antibody conjugated to Alexa 594 and goat anti-rabbit secondary antibody conjugated to Alexa 488 (Invitrogen). Sections were postfixed with formalin, incubated with Sudan Black, and stained with 5 μg/ml Hoechst 33342. The sections were coverslipped with Fluoromount-G (SouthernBiotech, Birmingham, AL) and visualized using an Olympus BX51 microscope mounted with a DP71 Olympus digital camera to capture images.
Sequential Biochemical Fractionation of Mouse Brain and Spinal Cord Tissue
The cortex or brainstem/spinal cord was dissected from mice. For each mouse, the brainstem and spinal cord were pooled together, and then all tissues were weighed and homogenized with a pellet pestle motor in 3 tissue volumes of high salt (HS) buffer (50 mm Tris, pH 7.5, 750 mm NaCl, 20 mm NaF, 5 mm EDTA) with a mixture of protease inhibitor (1 mm phenylmethylsulfonyl fluoride and 1 mg/ml each of pepstatin, leupeptin, N-tosyl-l-phenylalanyl chloromethyl ketone, N-tosyl-lysine chloromethyl ketone, and soybean trypsin inhibitor) followed by sedimentation at 100,000 × g for 20 min. Supernatants were saved as the HS fraction. Pellets were rehomogenized in 3 tissue volumes of HS buffer and sedimented at 100,000 × g for 20 min. Pellets were homogenized in 3 tissue volumes of HS buffer with 1% Triton X-100 (HS-TX buffer) and sedimented at 100,000 × g for 20 min. These supernatants were saved as the HS-TX fraction. The pellets were subjected to a sucrose myelin float by homogenizing in 3 pellet volumes of sucrose buffer (HS buffer plus 1 m sucrose), and after centrifugation, the myelin-rich supernatants were discarded. Pellets were then homogenized in 2 tissue volumes of radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS with the protease inhibitors), sedimented at 100,000 × g for 20 min, and the supernatants were saved as the RIPA fraction. Pellets were then sonicated in 1 pellet volume of 2% SDS, 4 m urea and were designated to be the SDS/urea fractions. The fractions were then quantified using the bicinchoninic acid (BCA) assay (Pierce). SDS sample buffer was added to each sample, and all samples, except for the SDS-urea fractions, were heated to 100 °C for 5 min prior to Western blot analysis. Equal amounts of protein extracts (10 μg) were resolved by SDS-PAGE and analyzed by Western blot.
Western Blot Analysis
Protein samples were separated by electrophoresis onto 15% SDS-polyacrylamide gels for α-syn analysis or 10% SDS-polyacrylamide gels for tau analysis and transferred by electrophoresis onto nitrocellulose membranes. Membranes were blocked in Tris-buffered saline (TBS) with 5% dry milk and incubated overnight at 4 °C with Syn211, SNL-1, SNL-4, or anti-actin in TBS with 5% dry milk or with PHF1, AT8, or pSer129 in TBS with 5% bovine serum albumin (BSA). Each incubation was followed by goat anti-mouse-conjugated horseradish peroxidase (HRP) (Amersham Biosciences) or goat anti-rabbit conjugated HRP (Cell Signaling Technology, Danvers, MA), and immunoreactivity was detected using chemiluminescent reagent (PerkinElmer Life Sciences) followed by exposure on x-ray film.
For total protein lysate of mouse tissues, the specified tissues were lysed in 3% SDS, 50 mm Tris, pH 6.8, by sonication and heating to 100 °C for 10 min. Total protein extracts were then quantified using the BCA assay using BSA as the standard. Equal amounts of protein extracts (3 μg) were resolved by SDS-PAGE and analyzed by Western blot.
Immunoelectron Microscopic Analysis of E46K α-Syn Transgenic Mice
Mice were sacrificed with CO2 euthanization and perfused with PBS/heparin, followed by 2% paraformaldehyde plus 0.5% glutaraldehyde in 0.1 m cacodylate, pH 7.4. The tissue was further fixed for 12 h, washed with PBS, cut into 50 μm sections with a vibratome, and reacted with 0.1% sodium borohydrate in PBS for 10 min. Following washing and blocking with 5% horse serum, 1% bovine serum albumin, 0.2% cold water fish skin gelatin in PBS for 24 h, the sections were incubated with antibody SNL-4 or pSer129 in blocking solution for 24 h. After extensive washing, the sections were sequentially incubated with a biotinylated goat anti-mouse antibody or a biotinylated goat anti-rabbit antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and developed with 3,3′- diaminobenzidine. The chemical product was enhanced using a modification of the Rodriguez silver/gold enhancement method (49, 50). After extensive washes, sections were postfixed with 2% paraformaldehyde, 2.5% glutaraldehyde, 0.1 m sodium cacodylate buffer, pH 7.4, followed by 2% osmium tetraoxide. Sections were dehydration in a graded ethanol series prior to infiltration and embedding in EMbed-812 (Electron Microscopy Sciences, Fort Washington, PA). Epone blocks were cut, and sections were stained with uranyl acetate and lead citrate. Sections were examined with a JEOL 1010 transmission electron microscope (Peabody, MA) fitted with a Hamamatsu digital camera (Bridgewater, MA) and AMT Advantage image capture software (Danvers, MA).
Cell Culture and Transfection
QBI293 cells were maintained using Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. The mammalian expression vector pcDNA3.1 cloned with WT human α-syn cDNA was described previously (51). The same mammalian expression vector expressing human α-syn with the E46K mutation was created with oligonucleotides corresponding to the amino acid substitution by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). The pcDNA3.1 expression plasmid containing the WT (2N/4R) human tau cDNA was generously provided by Dr. Virginia Lee (University of Pennsylvania, Philadelphia, PA).
Cells were plated onto poly-d-lysine-coated 6-well plates and transfected at ∼30% confluence, using calcium phosphate precipitation, as described previously (52). Cells were transfected with an equal proportion of tau-containing expression plasmid and α-syn-containing expression plasmid or pcDNA3.1 (empty vector). 4 h after transfection, 1 μm (final concentration) water bath-sonicated α-syn fibrils was added dropwise to medium, after which cells were incubated overnight at 37 °C, 5% CO2.
Approximately 16 h after transfection, calcium phosphate precipitation was washed twice with PBS for removal, and medium was replaced with warm DMEM containing 3% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (reduced serum). Cells were harvested for biochemical cellular fractionation or fixed 72 h after transfection.
Generation of Recombinant Human α-Syn and Fibril Preparation of Recombinant α-Syn Fibrils for in Situ Experimentation
The bacterial expression vector pRK172 expressing WT and E46K human α-syn was described previously (53, 54). The pRK172 DNA construct expressing N-terminal truncated 21–140 α-syn (with a Met codon added before amino acid 21) was generously provided by Dr. Virginia Lee. α-Syn proteins were expressed in Escherichia coli BL21 (DE3) and purified as described previously (53, 54). α-Syn proteins were assembled into filaments by incubation at 37 °C at concentrations greater than 5 mg/ml in sterile PBS (Invitrogen) with continuous shaking at 1050 rpm (Thermomixer R, Eppendorf, Westbury, NY). Experimentation was planned so that α-syn would be visibly assembled (by filamentous clusters observed in the solution) by the day of cellular experimentation. α-Syn fibrils were diluted to a concentration of 3 mg/ml in sterile PBS and treated by water bath sonication for a minimum of 2 h.
Biochemical Cellular Fractionation
Cells were washed one time in ice-cold PBS, and samples were harvested in 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 20 mm NaF, and a mixture of protease inhibitors described above. Samples were sedimented at 100,000 × g for 30 min at 4 °C. Supernatants were removed, and pellets were sonicated in 1.5× Laemmli sample buffer (75 mm Tris-HCl, pH 6.8, 3% SDS, 15% glycerol, 3.75 mm EDTA, pH 7.4). SDS sample buffer was added, and samples were heated to 100 °C for 5 min prior to Western blot analysis. Equal proportions of supernatant (Triton-soluble protein) and pellet (Triton-insoluble protein) were loaded for Western blot analysis.
Cell Culture Double Immunofluorescence
Double immunofluorescence of transfected cells was completed as described previously (52, 55). Cells were fixed at −20 °C with 100% MeOH for 20 min. Following washes with PBS, coverslips were blocked with PBS containing 3% BSA, 1% dry milk, and 1% fish gelatin, and incubated with primary antibodies diluted into blocking solution for 1–2 h at room temperature. Following PBS washes, coverslips were incubated in secondary antibodies conjugated to Alexa488 or Alexa594 for 1 h. For double immunofluorescence between pSer129 and PHF1, secondary antibodies goat anti-mouse IgG1 conjugated to Alexa488 (for PHF1) and IgG2A conjugated to Alexa594 (for pSer129) were used. For double immunofluorescence between AT8 and 17025, secondary antibodies goat anti-mouse IgG conjugated to Alexa594 and goat anti-rabbit IgG conjugated to Alexa488 were used. Nuclei were counterstained with Hoechst trihydrochloride trihydrate 33342 (Invitrogen), and coverslips were mounted using Fluoromount-G (SouthernBiotech, Birmingham, AL). Confocal microscopy images were captured on a Zeiss Axiovert 200 M inverted confocal microscope mounted with a Zeiss LSM510 META NLO digital camera utilizing Zeiss LSM510 META version 3.2 confocal microscope software (Zeiss, Thornwood, NY).
Evaluation of the Abundance of α-Syn Pathology in Asymptomatic M83 Transgenic Mice
The whole brain and spinal cord of asymptomatic 11–12-month-old M83 transgenic mice were analyzed by immunocytochemistry as described above with antibodies Syn505, Syn506, and pSer129. The abundance of α-syn inclusions was assessed using a semiquantitative assessment strategy, as described previously (56). The regional density of immunoreactive inclusions was graded in each region as follows: 3, abundant; 2, moderate; 1, sparse; 0, none.
Statistical Analysis
All of the mouse footprint analysis and wire hang test data are presented as mean ± S.E. For behavioral experiments, individual measurements for each mouse were calculated as mean ± S.E. for each group. The means were analyzed by one-way analysis of variance. A post hoc Bonferroni test was used to compare means after analysis of variance (GraphPad Prism version 5.0, San Diego, CA). A probability value of p < 0.05 was considered statistically significant.
RESULTS
Generation of Novel Transgenic Mice Expressing E46K Human α-Syn
Two transgenic mouse lines (M47 and M31) expressing E46K human α-syn were generated to study the consequences of this mutation in vivo (Fig. 1). These lines were generated using the human α-syn cDNA carrying the E46K mutation cloned into the MoPrP.Xho expression plasmid (Fig. 1A), which drives high expression of the transgene in most CNS neurons (45). For comparison, the previously described transgenic mouse lines that express WT (line M20) or A53T (line M83) human α-syn (35) were included in these studies because all transgenic mice possess identical transgenic constructs except for the absence or presence of disease-causing missense mutations (Fig. 1A). It was previously described that the M20 transgenic line expresses higher levels (∼1.5-fold) of human α-syn than the M83 transgenic line (35) (also see Fig. 6). The levels of transgene expression of the new mouse lines expressing E46K α-syn were slightly lower than that of the M83 transgenic mice (75% ± 10 for M47 and 72% ± 15 for the M31 expression relative to M83) (Fig. 1, B and C). The anti-α-syn-specific antibody SNL-1, which reacts equally with murine and human α-syn (39), was used to detect total α-syn, and Syn211, which is specific for human α-syn (39), was used to specifically detect transgene expression. The expression levels of the transgenes were similar in the cortex and spinal cord, although their relative levels of overexpression compared with native mouse α-syn were much higher in the spinal cord than in the cortex (Fig. 1, B and C). This is due to the much lower expression levels of endogenous α-syn in the spinal cord than in the cerebral cortex (35, 57).
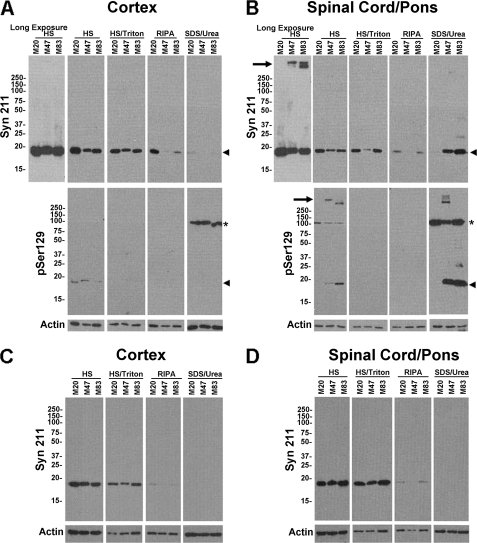
FIGURE 6.
Accumulation of detergent-insoluble and hyperphosphorylated α-syn in affected area of symptomatic transgenic mice expressing E46K and A53T mutant α-syn. Cortex (A) or spinal cord/brain stem (B) from a 12-month-old M20 transgenic mouse, a 12-month-old symptomatic M83 transgenic mouse, and a 20-month-old symptomatic M47 transgenic mouse were sequentially extracted, as described under “Experimental Procedures.” HS, HS/T, RIPA, and SDS/urea sequentially extracted fractions were analyzed by Western blotting using antibody Syn211 (human α-syn-specific), antibody pSer129 (specific for α-syn phosphorylated at Ser-129), and anti-actin antibody C3. The position of monomeric α-syn is indicated with an arrowhead, whereas the positions of molecular mass markers are depicted on the left. A longer exposure for the HS fractions with antibody Syn211 is shown to demonstrate the specific presence of soluble α-syn oligomers that do not enter the resolving gel in the spinal cord/brain stem of affected M83 and M47 mice (depicted with an arrow). The asterisk indicates a non-specific protein band. C and D, for comparison, similar analysis was performed on non-affected 3-month-old M20, M47, and M83 transgenic mice.
It was previously reported that the transgenic mice of line M83 that express A53T human α-syn remain healthy without any overt phenotype up to the age of 7 months. By 8 months of age, a few of these mice begin to develop a dramatic motor phenotype (35) (also see Fig. 1E). The initial changes include neglect of grooming, weight loss, and reduced ambulation. These changes are followed by severe movement impairments with resistance to passive movement and partial paralysis of limbs, which typically affects all four limbs within 2–3 days. The animals are eventually unable to stand up and support their own body weight.
Mice expressing E46K human α-syn developed a similar phenotype (Fig. 1D) with several notable differences (also see supplemental Video 1). Most A53T human α-syn transgenic mice are affected by 16 months of age; however, this was the earliest age of onset for E46K human α-syn transgenic mice (Fig. 1E). In addition, the disease progression was slower in E46K human α-syn transgenic mice such that once a single hind leg exhibited paralysis, there was typically a 4–5-day lag before the other hind leg was affected. Forelimbs were typically affected several days after this, but mice were usually sacrificed before this occurred to prevent suffering. At late stages, E46K human α-syn transgenic mice were unable to right themselves when placed on their sides, and they developed hunched backs (Fig. 1D). Notably, in contrast to many other mouse models of human disease (58–60), E46K human α-syn transgenic mice did not show retraction of their hind limbs when held inverted by the tail (Fig. 1D). To date, all M47 (n = 48) and M31 (n = 6) E46K human α-syn transgenic mice have developed the phenotype described above within 29 months of age. These effects are unique to the transgenic mice carrying pathological mutations in human α-syn because the WT human α-syn transgenic mice (line M20) exhibit no phenotype (Fig. 1E) (35).
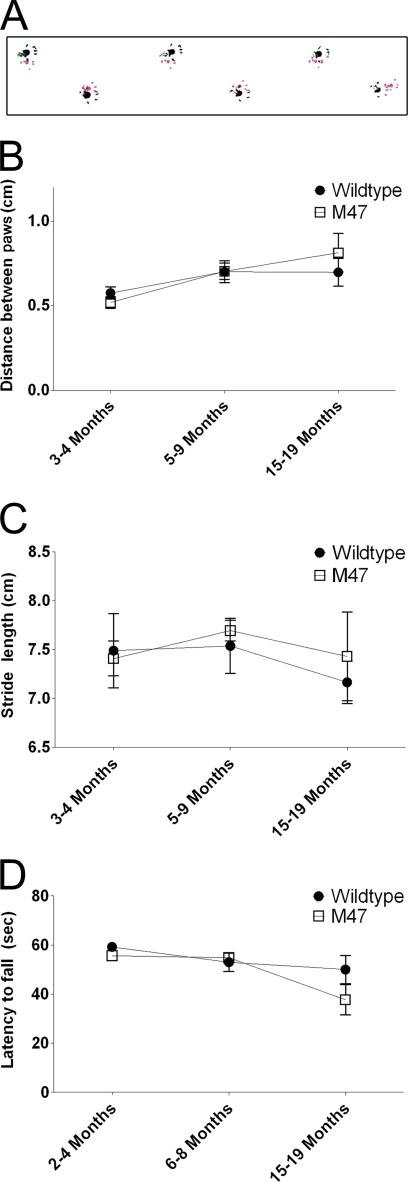
To further study the presentation of more subtle motor impairment in E46K human α-syn transgenic mice, cohorts of mice at different ages (2–4 months, 6–8 months, and 15–19 months) were subjected to footprint analysis to asses gait during ambulation and wire hang testing to assess neuromuscular function and grip strength (Fig. 2). No statistically significant differences were found between presymptomatic E46K human α-syn transgenic and non-transgenic mice for any of the age cohorts examined. It is important to note that some transgenic mice began to exhibit severe motor impairment within the 15–19-month range (Fig. 1E); however, only mice not yet exhibiting any overt phenotype were used for behavioral testing because severely affected mice could not execute these assays. The degree of variability in the E46K human α-syn transgenic mice at 15–19 months of age is noteworthy and probably reflects the high variability in the age at which these transgenic mice can develop an overt motor phenotype (Fig. 1E). Mice at this age will eventually display severe motor impairment at different ages thereafter, and they are probably at different stages of disease progression.
FIGURE 2.
Motor behavioral analyses of M47 E46K human α-syn transgenic mice. A, representative track of fore (red) and hind (black) paws of a 9-month-old M47 E46K α-syn transgenic mouse. Footprint analysis of walking performance for distance between fore and hind footprints of overlapping steps (B) and stride length (C) showed no significant difference between presymptomatic E46K α-syn transgenic and nontransgenic mice at various ages as indicated. No significance was also observed in assessing motor performance by the wire hang test (D). Data are presented as group means ± S.E. (error bars) and were analyzed by Student's t test; p was set at <0.05 as level of significance.
Histological Characterization of α-Syn Pathology in E46K Human α-Syn Transgenic Mice
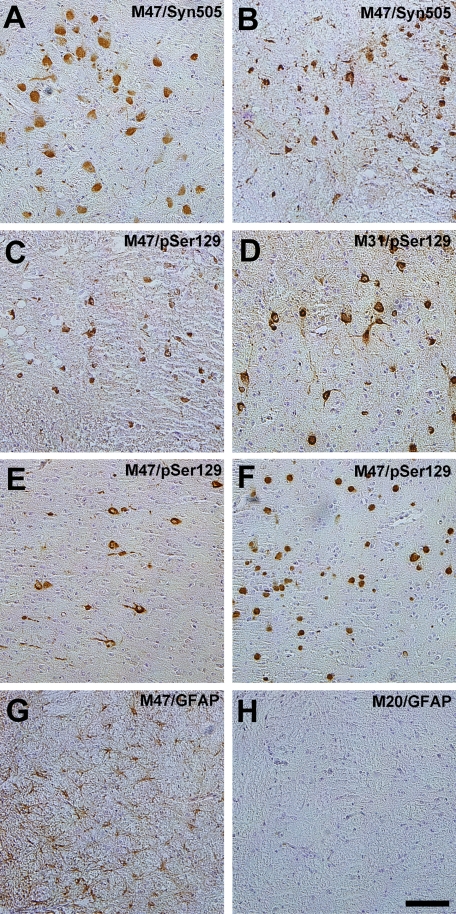
Previous immunostaining analysis of the nervous tissue of M20 and M83 transgenic mice has shown the normal neuropil staining pattern expected for α-syn (14, 17, 57). Immunostaining analyses of the nervous tissue of presymptomatic E46K α-syn transgenic mice demonstrated similar normal staining patterns (data not shown). However, affected E46K human α-syn transgenic mice demonstrated abundant and widespread accumulation of α-syn inclusions in the spinal cord, throughout the brainstem, the deep cerebellar nuclei, most of the thalamus, and the motor cortex (Fig. 3) (data not shown). Other regions of the cerebral cortex, the olfactory bulb, and the hippocampus were devoid of α-syn pathology. The α-syn inclusions were highly reactive with conformation-specific anti-α-syn antibodies Syn505 and Syn506 (Figs. 3 (A and B) and 5 (C and D)) and with anti-α-syn phosphospecific antibody pSer129 (Figs. 3 (C–F) and 5A), consistent with findings in human brains and other transgenic mice that show inclusions containing α-syn hyperphosphorylated at Ser-129 (38, 40, 41, 61). In addition, consistent with neuronal injury associated with inclusion formation, staining for GFAP revealed significant astrocytic gliosis in regions with α-syn inclusions (Fig. 3, G and H). Similarly, analysis with the antibody Iba-1 also revealed elevated reactive microglia specifically in affected areas (data not shown).
FIGURE 3.
Representative images of α-Syn pathology in symptomatic E46K α-syn transgenic mice. Immunocytochemistry was performed as described under “Experimental Procedures.” Shown are abundant perikaryal α-syn pathological inclusion in the pons of a sick 22-month-old M47 transgenic mouse stained with anti-α-syn antibody Syn505 (A). Perikaryal and neuritic α-syn pathological inclusions in the spinal cord of a sick M47 transgenic mouse were stained with anti-α-syn antibody Syn505 (B) or pSer129 (C). α-Syn pathological inclusions were labeled with pSer129 antibody in the periaqueductal gray area of a sick 18-month-old M31 transgenic mouse (D), in the motor cortex of a sick 22-month-old M47 transgenic mouse (E), and in the hypothalamus of a sick 22-month-old M47 transgenic mouse (F). Staining with anti-GFAP antibody depicts astrogliosis in the pons of a symptomatic 22-month old M47 transgenic mouse (G) versus the comparative normal staining in a non-affected 20-month-old M20 transgenic mouse (H). Sections were counterstained with hematoxylin. Bar, 100 μm.
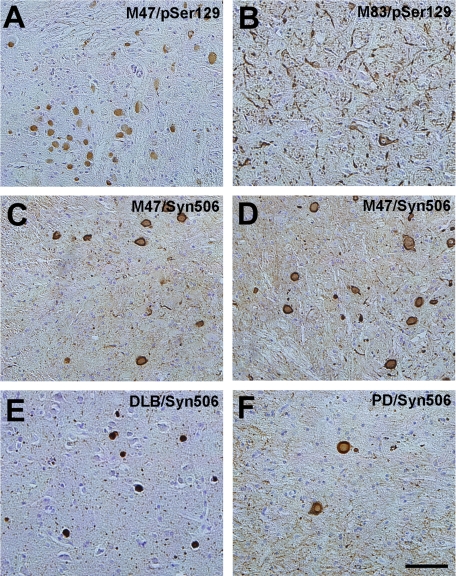
FIGURE 5.
α-Syn inclusions in E46K α-syn transgenic mice bear close morphological resemblance to Lewy bodies in human brains. A, uniformly labeled round α-syn inclusions stained with pSer129 antibody in the pons of a symptomatic 22-month-old M47 transgenic mouse. B, labeling of somatodendritic and neuritic α-syn inclusions stained with pSer129 antibody in the pons of a symptomatic 12-month-old M83 transgenic mouse. Staining of α-syn inclusions with antibody Syn506 in the cerebellum (C) or pons (D) of a symptomatic 22-month-old M47 transgenic mouse showing a clear increased labeling of the outer rim of the inclusions yielding a core and halo profile. Shown are staining with antibody Syn506 of cortical LBs in a human patient with dementia with LB (DLB) (E) and nigral LBs in the SN of a patient with PD (F). Note that cortical LBs usually have a solid round profile, whereas nigral LBs typically have a core and halo profile, where the core is not as intensely labeled. Sections were counterstained with hematoxylin. Bar, 100 μm.
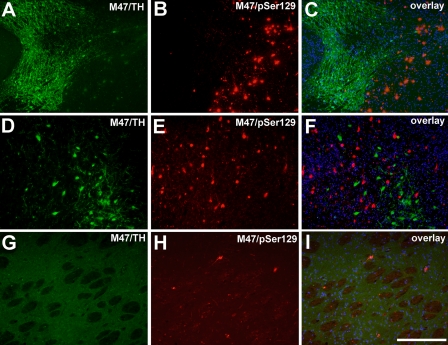
Certain cell populations were completely spared of α-syn inclusions including the Purkinje cells and granular cells in the cerebellum (data not shown) and TH-positive neurons of the SN (Fig. 4). The paucity of α-syn inclusions in TH-positive neurons was also demonstrated in neuronal processes in the striatum (Fig. 4). This selective vulnerability (or protection) for α-syn inclusion formation in the E46K human α-syn mice is similar to that previously described for the mice expressing A53T human α-syn (35); however, one important difference is the morphology of the α-syn inclusions in E46K human α-syn transgenic mice. Although most of the inclusions in the A53T human α-syn transgenic mice fill the somatodendritic compartment, the inclusions in the E46K human α-syn mice are much more compact and round, closely reminiscent of LBs in human brains (Fig. 5). In addition, many α-syn inclusions in E46K human α-syn transgenic mice demonstrate “core and halo” antibody labeling profiles similar to classical LBs in the human brain (Fig. 5).
FIGURE 4.
Paucity of α-syn inclusions in TH-positive neurons of the substantia nigra and projection in the striatum in E46K α-syn transgenic mice. A, D, and G, staining with anti-TH antibody (green); B, E, and H, pSer129 antibody (red) in the midbrain (A–F) or striatum (G–I) of a 22-month-old symptomatic M47 transgenic mouse. C, F, and I, overlay of the immune staining with Hoechst labeling (blue). The images demonstrate a paucity of α-syn inclusions in these TH-positive neurons and processes. Bar, 400 μm (A–C) and 200 μm (D–I).
Accumulation of Insoluble and Aggregated Hyperphosphorylated α-Syn in E46K Human α-Syn Transgenic Mice
The cortex and spinal cord of transgenic mice expressing WT (M20), A53T (M83), and E46K (M47) human α-syn were sequentially extracted with buffers with increasing strength of protein solubilization. Western analyses were performed using antibodies specific for human α-syn (Syn211), α-syn phosphorylated at Ser-129 (pSer129), and an anti-actin antibody. In transgenic mice expressing WT α-syn, the majority of α-syn was recovered in the HS and HS/TX fractions, but a small amount could also be observed in the RIPA fraction (Fig. 6). In affected A53T and E46K human α-syn transgenic mice, there was a significant accumulation of RIPA-insoluble (i.e. SDS/urea-soluble) α-syn hyperphosphorylated at Ser-129 in the spinal cord but not in the cortex (Fig. 6, A and B), coinciding with the presence of α-syn inclusions observed by light microscopy in these brain regions. Quantitative assessment determined that there is a relative 5-fold increase in the amount of pSer129 in monomeric α-syn in the SDS/urea-soluble fractions of spinal cord compared with the HS soluble fractions. The presence of hyperphosphorylated HS-soluble α-syn aggregates that do not enter the resolving SDS-polyacrylamide gel was also observed in affected A53T and E46K α-syn transgenic mice (Fig. 6B, arrow). In contrast, aggregated and insoluble forms of human α-syn were not observed in unaffected younger A53T and E46K α-syn transgenic mice (Fig. 6, C and D).
Immunoelectron Microscopy Analysis of Inclusions in E46K α-Syn Transgenic Mice
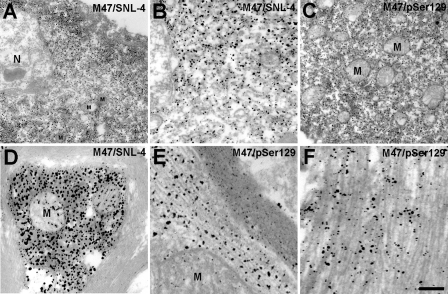
In order to better characterize the structure and consequences of α-syn inclusions in E46K α-syn transgenic mice, immunoelectron microscopy analysis was performed. α-Syn inclusions with filamentous profiles that filled most of the neuronal cell body were observed (Fig. 7, A and B). These inclusions typically contained numerous organelles, including mitochondria (Fig. 7, A and C). Similarly, intensely labeled α-syn inclusions with “trapped” mitochondria were also observed in axons (Fig. 7D). The ultrastructure of α-syn inclusions in intensely labeled structures was obscured by the silver labeling; however, higher magnification of more lightly labeled inclusions with antibody SNL-4 or pSer129 demonstrated the fibrillar structure of most α-syn inclusions (Fig. 7, E and F).
FIGURE 7.
Ultrastructural analyses of α-syn inclusions in E46K α-syn transgenic mouse. Immmunoelectron microscopy analysis was performed in the pons of a 16-month-old symptomatic E46K α-syn transgenic mouse with antibodies SNL-4 (A, B, and D) or pSer129 (C, D, and F). A and B, progressively increased magnification of an α-syn inclusion within a neuronal cell body. The inclusion demonstrates trapped mitochondria and the fibrillar profile of α-syn inclusions. C and D, intensely labeled α-syn inclusion in a cell body (C) or axon with numerous trapped mitochondria. E and F, higher magnification of more lightly labeled anti-α-syn inclusions, allowing observation of the clear fibrillar profile of α-syn inclusions. N, nucleus; M, mitochondria. Scale bar, 660 nm (A and C), 266 nm (B), 200 nm (D), and 133 nm (E and F).
Accumulation of Tau Inclusions in E46K α-Syn Transgenic Mice
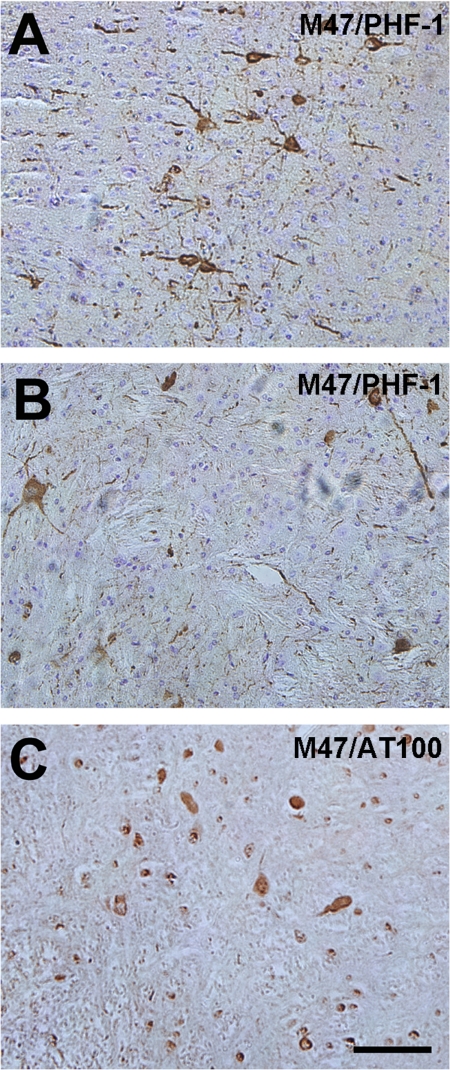
Because α-syn has the ability to induce the aggregation of tau protein, which is the major component of neurofibrillary tangles in Alzheimer disease (62, 63) and this process was previously observed in M83 A53T human α-syn transgenic mice (62), E46K human α-syn transgenic mice were examined for tau pathology. Staining with several tau antibodies, including PHF1, which recognizes a tau phosphoepitope elevated in pathological inclusions (43), revealed tau inclusions in brain areas where α-syn inclusions were present (Fig. 8, A and B). Staining with AT100, which recognizes aggregated tau phosphorylated at Thr-212 and Ser-214 in a unique disease-associated conformation state (44), also revealed tau inclusions in affected E46K α-syn transgenic mice (Fig. 8C). The abundance of tau inclusions in E46K transgenic mice was greater than observed previously in M83 A53T human α-syn transgenic mice (62). Double immunofluorescence analysis revealed that α-syn and tau inclusions in E46K α-syn transgenic mice were typically not co-localized (data not shown), similar to observations in diseased human brains (63).
FIGURE 8.
Tau pathology in E46K α-syn transgenic mice. Tau inclusions in the motor cortex (A) and pons (B) of a symptomatic 22-month old M47 transgenic mouse stained with anti-phospho-tau antibody PHF-1. C, tau inclusions are also intensely labeled with anti-phospho-tau antibody AT100. Sections were counterstained with hematoxylin. Bar, 100 μm.
Assessment of the Ability of E46K α-Syn to Induce Tau Inclusion Formation
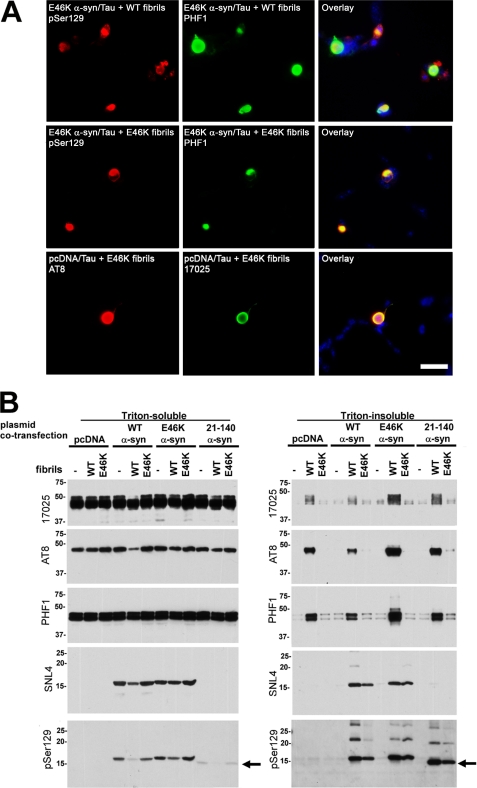
Because E46K α-syn transgenic mice demonstrate the formation of inclusions composed of endogenous tau, we wanted to test whether this mutation enhanced the ability of α-syn to induce tau aggregate formation. To assess this phenomenon, we used a cell culture model of tau inclusion formation, where the addition of extracellular α-syn fibrils to cells via calcium phosphate precipitation seeds intracellular amyloid fibrillar tau inclusion formation with or without intracellular co-expression of α-syn (64). Using double immunofluorescence, we first determined that treatment with either extracellular WT or E46K α-syn recombinant preformed fibrils induced the formation of tau intracellular inclusions. These inclusions developed in the presence (Fig. 9, A and B) or absence (Fig. 9C) of intracellular E46K α-syn co-expression, as observed with tau antibody 17025 and phosphospecific tau antibodies AT8 or PHF1. To directly assess the relative ability of WT and E46K α-syn fibrils to induce tau inclusion formation, biochemical cellular fractionation was performed. QBI293 cells were co-transfected with plasmids for the expression of tau and either pcDNA (empty vector), WT α-syn, E46K α-syn, or 21–140 (N-terminally truncated) α-syn. Each condition received either no treatment, WT α-syn fibril treatment, or E46K α-syn fibril treatment. Under all transfection conditions, cells treated with recombinant WT α-syn fibrils developed a significant amount of Triton-insoluble tau (Fig. 9B). The appearance of Triton-insoluble tau was further confirmed with the phospho-tau-specific antibody AT8 or PHF1. Comparatively, treatment with E46K α-syn fibrils produced much lower levels of Triton-insoluble tau. Interestingly, WT and E46K α-syn fibrils were similarly capable of producing intracellular α-syn aggregates, as observed by SNL-4 and pSer129 immunoreactivity. Therefore, although WT and E46K α-syn recombinant fibrils had a similar effect in seeding cellular α-syn aggregates, WT α-syn recombinant fibrils were more effective at seeding cellular tau aggregates.
FIGURE 9.
E46K α-syn is less efficient than WT α-syn at promoting tau inclusions in cultured cells. A, representative double immunofluorescence of QBI293 cells co-transfected with expression plasmids for human E46K α-syn and tau (2N/4R) or empty pcDNA vector and tau (2N/4R) with the addition of WT or E46K recombinant, prefibrillized α-syn protein as indicated in each panel described under “Experimental Procedures.” Co-immunofluorescence was performed with anti-phospho-α-syn antibody pSer129 (red) and anti-phospho-tau antibody PHF1 (green) or anti-phospho-tau antibody AT8 (red) and anti-pan tau antibody 17025 (green) as indicated. These images show the formation of α-syn or tau intracellular inclusions. The images in the right column show the overlay with Hoechst 33342 counterstain (blue). Bar, 50 μm. B, biochemical cellular fractionation assessing the formation of tau and α-syn aggregates. QBI293 cells were co-transfected with expression plasmids for WT tau and pcDNA3.1 (pcDNA) empty vector, WT α-syn, E46K α-syn, or N-truncated 21–140 α-syn. Parallel samples either received no treatment or were treated with recombinant prefibrillized WT or E46K α-syn protein. Cells were harvested 72 h after transfection, and biochemical cellular fractionation was performed, as described under “Experimental Procedures.” Shown are representative immunoblots of biochemical fractionation with anti-tau antibody 17025, with phospho-specific tau antibodies PHF1 or AT8, with anti-α-syn antibody SNL-4, or with phosphospecific anti-α-syn antibody pSer129. Triton-insoluble tau and α-syn only accumulated after treatment with recombinant, prefibrillized α-syn treatment. Minimal recombinant α-syn was determined to be pSer129-immunoreactive, as determined by the predominant shift of α-syn noted in cells co-transfected with the 21–140 α-syn expression plasmid (indicated with an arrow). Treatment with recombinant E46K α-syn prefibrils produced Triton-insoluble tau, but to a much lesser extent, whereas both WT and E46K α-syn recombinant fibrils were similarly capable of producing intracellular α-syn aggregates, as observed by SNL-4 and pSer129 immunoreactivity.
Assessment of the Onset and Distribution of α-Syn Pathology in A53T and E46K Human α-Syn Transgenic Mice
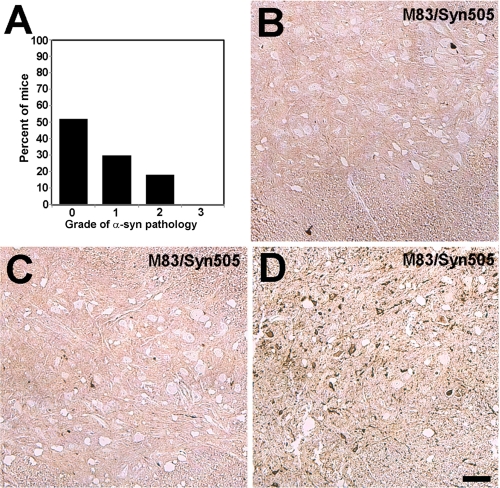
Given several recent reports suggesting that α-syn inclusion formation may proceed by a cell-to-cell transmission and seeding mechanisms (65, 66), we assessed the distribution of α-syn pathology in presymptomatic transgenic mice to provide insights into this possible mode of disease progression. Due to the limited cohort of E46K α-syn transgenic mice available, we performed the majority of analyses on M83 A53T human α-syn transgenic mice. Twenty-seven M83 transgenic mice were analyzed at 11–12 months of age, approximately the age when half the population of mice develops symptoms. Of the M83 mice studied, 52% (14 of 27) did not display any α-syn pathological inclusions. 30% (8 of 27) of the M83 mice demonstrated the presence of sparse α-syn pathological inclusions, whereas 18% (5 of 27) reveal moderate levels of α-syn inclusions (Fig. 10A). None of the asymptomatic mice demonstrated abundant α-syn inclusions, compared with the symptomatic mice, which all presented with abundant α-syn inclusions. Furthermore, in mice with sparse or moderate levels of α-syn inclusions, there was no region that was affected first, and the distribution of aggregates was diffuse within affected areas in the neuroaxis such that there was no clustering of inclusions (Fig. 10B). Using the mice available, several presymptomatic E46K α-syn transgenic mice (line M47) were analyzed for α-syn pathologic inclusions. Ten mice between the ages of 6 and 10 months were studied and revealed no α-syn pathologic inclusions. Four presymptomatic mice between the ages of 18 and 20 months were also analyzed. One of these mice (20 months old) demonstrated no α-syn inclusions, and one 18-month-old mouse revealed rare inclusions without clustering. However, two M47 mice at 18 and 19 months of age demonstrated moderate levels of inclusions with morphologies and distribution similar to those of symptomatic M47 mice.
FIGURE 10.
Presentation of α-syn pathological inclusions in asymptomatic M83 A53T human α-syn transgenic mice. A, semiquantitative estimate of α-syn inclusions as described under “Experimental Procedures” in the affected neuroaxis for 11–12-month-old asympomatic M83 A53T human α-syn transgenic mice. The density of α-syn inclusions was graded as follows: 0, none; 1, sparse; 2, moderate; 3, abundant. B and C, representative images of spinal cord sections of asymptomatic 12-month-old M83 A53T human α-syn transgenic mice stained with antibody Syn505. D, representative image of a spinal cord section from a symptomatic 14-month-old M83 A53T human α-syn mouse stained with antibody Syn505. Sections were counterstained with hematoxylin. Bar, 100 μm.
DISCUSSION
We describe the first E46K human α-syn transgenic mice associated with a pathological phenotype. These mice develop an age-dependent severe motor phenotype associated with the formation of α-syn inclusions that closely resemble properties of authentic human pathological inclusions as assessed immunologically, histologically, and biochemically. These inclusions are strongly labeled with antibodies that preferentially mark human pathological inclusions (i.e. Syn505 and Syn506) (40, 41), and, like human inclusions, they contain α-syn highly phosphorylated at Ser-129 (38, 61). In addition, similar to human inclusions (48, 67–70), the inclusions in the E46K human α-syn transgenic mice are detergent-insoluble and composed of α-syn fibrils. Furthermore, many of the inclusions in the E46K human α-syn transgenic mice are round and display a core and halo antibody labeling profile similar to those seen in human brains.
Compared with the previously described A53T human α-syn transgenic mice (line M83), the mice expressing E46K human α-syn have a delayed age of onset and a slower overall progression of disease; however, this may in part be due to the lower expression of the E46K human α-syn transgene. In addition, the age of onset of the overt motor impairment in E46K transgenic mice presents within a significant large age period (16–29 months), and the reason for this variability is unclear. Footprint analysis and wire hang testings were performed in several younger cohorts of E46K human α-syn transgenic mice to try to investigate more subtle phenotypic changes; however, no significant difference was observed between these mice and non-transgenic mice of the several age-matched groups. Importantly, for the 15–19 month mice cohort, transgenic mice that began to exhibit severe motor impairments could not execute these tests and were excluded from analyses. Therefore, only presymptomatic mice were used for behavioral testing. The greater degree of variability within this older cohort of transgenic mice may be attributed to subtle developments of behavioral changes that may present at different ages in different mice, consistent with the variability seen in the overt motor phenotype. This variability probably contributes to the lack of detectable significant changes in the behavioral tasks assayed. These findings are also consistent with a relatively rapid progression of disease. Limited histological assessment of α-syn pathology in presymptomatic E46K human α-syn transgenic mice also supports the variability in the presentation of pathological changes.
Both the E46K and A53T α-syn mutations have been shown to promote the propensity of α-syn to polymerize into amyloid in vitro (54, 71), which is consistent with these two mutations resulting in inclusion formation associated with disease in mice, whereas inclusions and disease do not present in similar mice expressing even higher levels of WT human α-syn (Fig. 1) (35). It is fascinating that the α-syn inclusions in the E46K human α-syn transgenic mice more closely resemble the morphology of Lewy pathology in human brains, and it is possible that the additional effects of the E46K mutation on the lipid binding property of α-syn (71) may be important in altering this process. Many studies suggest that α-syn lipid binding can regulate and alter α-syn inclusion formation (72, 73). Alternatively, other age-dependent changes associated with the later disease onset in E46K human α-syn transgenic mice may also be involved.
The distribution of α-syn inclusions throughout the neuroaxis for both the A53T and E46K human α-syn transgenic mice is very similar, and for reason(s) that are still not clear, there is a divergent selectivity for the formation of α-syn inclusions in mice and human brains (2, 4, 9, 13). For example, in these transgenic mice, neurons in the hippocampus and dopaminergic neurons of the SN are selectively spared, whereas these regions are selectively vulnerable in human brains (2, 13). This unexpected selective protection against α-syn inclusion formation in mice is not unique to these transgenic mice (37, 74), and even in some transgenic mice designed with a specific promoter to drive high expression of WT, A30P, or A53T α-syn in dopaminergic neurons, there is a paucity of α-syn inclusions (75, 76). It has been proposed (and shown in some experimental paradigms) that the ability of dopamine byproducts to inhibit α-syn fibril formation may be involved in the paucity of pathology in murine nigral dopaminergic neurons (55, 77, 78), but other mechanisms must be involved to selectively protect other murine neuronal populations.
The E46K human α-syn transgenic mice also demonstrate tau inclusions, consistent with the ability of α-syn to induce amyloid tau fibril formation (62). Although A53T human α-syn transgenic mice demonstrate some tau pathology (62), the abundance of tau inclusions observed in the E46K human α-syn transgenic mouse model is comparatively profound. This is especially impressive considering the relative paucity of tau pathology observed in many transgenic mouse models of Alzheimer disease, including mice expressing amyloid precursor protein with Aβ deposits (79, 80) and even in mice overexpressing WT tau (81, 82). To assess whether this finding was solely due to an increased ability of the E46K α-syn mutant to induce tau polymerization, we compared the ability of WT and E46K α-syn to seed and induce tau inclusion formation using a novel cellular model of α-syn-induced tau inclusion formation (64). Although E46K α-syn was capable of inducing cellular tau inclusion in this system, this mutant was much less efficient than WT α-syn. This finding is in contrast with the ability of extracellular α-syn fibril treatment to induce cellular α-syn inclusions, where both WT and E46K α-syn demonstrated similar abilities. Therefore, the reason for the accumulation of tau inclusions in E46K α-syn transgenic mice is not completely clear, and other pathological events in addition to α-syn seeding must be involved. One explanation may be the older age of onset of disease in E46K human α-syn transgenic mice, thereby exposing subjects to additional age-related factors that could contribute to tau inclusion formation.
All of the E46K and A53T human α-syn transgenic mice that developed a motor phenotype that underwent histological analysis demonstrated abundant α-syn pathological inclusions, suggesting that the aggregation of α-syn is directly involved in the disease phenotype. Recent studies have demonstrated the ability of α-syn to be transported in or out across cell membranes (66, 83–85), to be transferred cell-to-cell leading to aggregation, and to be transferred between host and graphed neurons in mouse brains (65, 66). These studies suggest a possible prion-like spreading disease mechanism for α-syn inclusions (86, 87). This notion is consistent with the Braak staging of disease that appears to follow neuroanatomical pathways (13) and the presence of LBs in grafted embryonic dopaminergic neurons in the brains of PD patients (88, 89). Furthermore, cellular studies have shown that the entry of a small amount of preformed α-syn polymers can induce the formation of large intracellular amyloid inclusions (52, 90). Because α-syn transgenic mice develop disease within a defined age range, we wanted to assess whether pathological findings in presymptomatic mice may provide evidence for the prion-like spreading model of synucleinopathies. Due to animal availability, we primarily performed this analysis on A53T human α-syn transgenic mice. A cohort of asymptomatic mice was sacrificed at 11–12 months, the age when about half the mice would typically be affected. We found that 52% of these mice did not display any α-syn pathological inclusions, whereas 30 and 18% of the mice demonstrated sparse or moderate α-syn pathological inclusions, respectively. None of the presymptomatic mice demonstrated abundant α-syn inclusions, similar to the levels found in symptomatic mice. Furthermore, in mice with sparse or moderate levels of α-syn inclusions, no region showed selective initiation of aggregation, and the distribution of aggregates was diffuse within affected areas of the neuroaxis such that there was no clustering of inclusions. Interestingly, using a limited cohort of E46K human α-syn transgenic mice, only one of four of the presymptomatic mice was without α-syn pathological inclusions, suggesting that these mice may be more likely to bear pathological aggregates prior to symptoms. Future studies may identify additional differences between A53T and E46K human α-syn transgenic mice.
Based on the pathological data generated, the process by which widespread α-syn inclusions occur is unknown. Although the lack of clustering of neurons with inclusions does not appear to support a seeding mechanism, the long processes of neurons could allow for transmission over long distances. The lack of α-syn inclusions in specific neuronal populations that express high levels of α-syn and are adjacent to regions that form a high density of inclusions also does not support a seeding mechanism. Nevertheless, because most of the asymptomatic M83 mice analyzed would have abundant α-syn inclusions within 5 months or less from the time they were studied, these findings indicate that the process of α-syn inclusion formation must be relatively rapid and probably synchronized. Other mechanisms that could result in this phenomenon of widespread pathological inclusions include oxidative stress, excitotoxicity, and neuroinflammation (91).
We report on the first E46K human α-syn transgenic mice that develop a pathological phenotype associated with formation of α-syn inclusions that closely resemble the morphology and properties of LBs in human brains. These findings further support the notion that α-syn aggregates are involved in the pathogenesis of human disease known as synucleinopathies, but further studies are needed to better understand the mechanisms that promote the formation of these inclusions.
Acknowledgment
We thank Michael Heenan for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NIA, Grant AG09215 and NIH, NINDS, Grant NS053488.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Video 1.
- α-syn
- α-synuclein
- GFAP
- glial fibrillary acidic protein
- LB
- Lewy body
- PD
- Parkinson disease
- RIPA
- radioimmunoprecipitation assay
- SN
- substantia nigra
- TH
- tyrosine hydroxylase
- PrP
- prion protein
- HS
- high salt.
REFERENCES
- 1. Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 2. Forman M. S., Lee V. M., Trojanowski J. Q. (2005) Neuron 47, 479–482 [DOI] [PubMed] [Google Scholar]
- 3. Goedert M. (2001) Nat. Rev. Neurosci. 2, 492–501 [DOI] [PubMed] [Google Scholar]
- 4. Waxman E. A., Giasson B. I. (2009) Biochim. Biophys. Acta 1792, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simuni T., Hurtig H. I. (2000) in Neurodegenerative Dementias (Clark C. M., Trojanoswki J. Q. eds) pp. 193–203, McGraw-Hill, New York [Google Scholar]
- 6. Gelb D. J., Oliver E., Gilman S. (1999) Arch. Neurol. 56, 33–39 [DOI] [PubMed] [Google Scholar]
- 7. Pakkenberg B., Møller A., Gundersen H. J., Mouritzen Dam A., Pakkenberg H. (1991) J. Neurol. Neurosurg. Psychiatry 54, 30–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Damier P., Hirsch E. C., Agid Y., Graybiel A. M. (1999) Brain 122, 1437–1448 [DOI] [PubMed] [Google Scholar]
- 9. Forno L. S. (1996) J. Neuropathol. Exp. Neurol. 55, 259–272 [DOI] [PubMed] [Google Scholar]
- 10. Chaudhuri K. R., Healy D. G., Schapira A. H. (2006) Lancet Neurol. 5, 235–245 [DOI] [PubMed] [Google Scholar]
- 11. Poewe W. (2008) Eur. J. Neurol. 15, Suppl. 1, 14–20 [DOI] [PubMed] [Google Scholar]
- 12. Cornford M. E., Chang L., Miller B. L. (1995) Brain Cogn. 28, 321–341 [DOI] [PubMed] [Google Scholar]
- 13. Braak H., Bohl J. R., Müller C. M., Rüb U., de Vos R. A., Del Tredici K. (2006) Mov. Disord. 21, 2042–2051 [DOI] [PubMed] [Google Scholar]
- 14. Jakes R., Spillantini M. G., Goedert M. (1994) FEBS Lett. 345, 27–32 [DOI] [PubMed] [Google Scholar]
- 15. George J. M., Jin H., Woods W. S., Clayton D. F. (1995) Neuron 15, 361–372 [DOI] [PubMed] [Google Scholar]
- 16. Withers G. S., George J. M., Banker G. A., Clayton D. F. (1997) Brain Res. Dev. Brain Res. 99, 87–94 [DOI] [PubMed] [Google Scholar]
- 17. Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., de Silva H. A., Kittel A., Saitoh T. (1995) Neuron 14, 467–475 [DOI] [PubMed] [Google Scholar]
- 18. Murphy D. D., Rueter S. M., Trojanowski J. Q., Lee V. M. (2000) J. Neurosci. 20, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., Orrison B., Chen A., Ellis C. E., Paylor R., Lu B., Nussbaum R. L. (2002) J. Neurosci. 22, 8797–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W. H., Castillo P. E., Shinsky N., Verdugo J. M., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A. (2000) Neuron 25, 239–252 [DOI] [PubMed] [Google Scholar]
- 21. Burré J., Sharma M., Tsetsenis T., Buchman V., Etherton M. R., Südhof T. C. (2010) Science 329, 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uversky V. N. (2007) J. Neurochem. 103, 17–37 [DOI] [PubMed] [Google Scholar]
- 23. Cookson M. R. (2005) Annu. Rev. Biochem. 74, 29–52 [DOI] [PubMed] [Google Scholar]
- 24. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 25. Athanassiadou A., Voutsinas G., Psiouri L., Leroy E., Polymeropoulos M. H., Ilias A., Maniatis G. M., Papapetropoulos T. (1999) Am. J. Hum. Genet. 65, 555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Markopoulou K., Wszolek Z. K., Pfeiffer R. F., Chase B. A. (1999) Ann. Neurol. 46, 374–381 [DOI] [PubMed] [Google Scholar]
- 27. Papadimitriou A., Veletza V., Hadjigeorgiou G. M., Patrikiou A., Hirano M., Anastasopoulos I. (1999) Neurology 52, 651–654 [DOI] [PubMed] [Google Scholar]
- 28. Spira P. J., Sharpe D. M., Halliday G., Cavanagh J., Nicholson G. A. (2001) Ann. Neurol. 49, 313–319 [PubMed] [Google Scholar]
- 29. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 30. Chartier-Harlin M. C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., Waucquier N., Defebvre L., Amouyel P., Farrer M., Destée A. (2004) Lancet 364, 1167–1169 [DOI] [PubMed] [Google Scholar]
- 31. Farrer M., Kachergus J., Forno L., Lincoln S., Wang D. S., Hulihan M., Maraganore D., Gwinn-Hardy K., Wszolek Z., Dickson D., Langston J. W. (2004) Ann. Neurol. 55, 174–179 [DOI] [PubMed] [Google Scholar]
- 32. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) Science 302, 841 [DOI] [PubMed] [Google Scholar]
- 33. Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Gomez Tortosa E., del Ser T., Muñoz D. G., de Yebenes J. G. (2004) Ann. Neurol. 55, 164–173 [DOI] [PubMed] [Google Scholar]
- 34. van der Putten H., Wiederhold K. H., Probst A., Barbieri S., Mistl C., Danner S., Kauffmann S., Hofele K., Spooren W. P., Ruegg M. A., Lin S., Caroni P., Sommer B., Tolnay M., Bilbe G. (2000) J. Neurosci. 20, 6021–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giasson B. I., Duda J. E., Quinn S. M., Zhang B., Trojanowski J. Q., Lee V. M. (2002) Neuron 34, 521–533 [DOI] [PubMed] [Google Scholar]
- 36. Lee M. K., Stirling W., Xu Y., Xu X., Qui D., Mandir A. S., Dawson T. M., Copeland N. G., Jenkins N. A., Price D. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8968–8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Magen I., Chesselet M. F. (2010) Prog. Brain Res. 184, 53–87 [DOI] [PubMed] [Google Scholar]
- 38. Waxman E. A., Giasson B. I. (2008) J. Neuropathol. Exp. Neurol. 67, 402–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giasson B. I., Jakes R., Goedert M., Duda J. E., Leight S., Trojanowski J. Q., Lee V. M. (2000) J. Neurosci. Res. 59, 528–533 [DOI] [PubMed] [Google Scholar]
- 40. Duda J. E., Giasson B. I., Mabon M. E., Lee V. M., Trojanowski J. Q. (2002) Ann. Neurol. 52, 205–210 [DOI] [PubMed] [Google Scholar]
- 41. Waxman E. A., Duda J. E., Giasson B. I. (2008) Acta Neuropathol. 116, 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goedert M., Jakes R., Vanmechelen E. (1995) Neurosci. Lett. 189, 167–169 [DOI] [PubMed] [Google Scholar]
- 43. Otvos L., Jr., Feiner L., Lang E., Szendrei G. I., Goedert M., Lee V. M. (1994) J. Neurosci. Res. 39, 669–673 [DOI] [PubMed] [Google Scholar]
- 44. Zheng-Fischhöfer Q., Biernat J., Mandelkow E. M., Illenberger S., Godemann R., Mandelkow E. (1998) Eur. J. Biochem. 252, 542–552 [DOI] [PubMed] [Google Scholar]
- 45. Borchelt D. R., Davis J., Fischer M., Lee M. K., Slunt H. H., Ratovitsky T., Regard J., Copeland N. G., Jenkins N. A., Sisodia S. S., Price D. L. (1996) Genet. Anal. 13, 159–163 [DOI] [PubMed] [Google Scholar]
- 46. Heck D. H., Zhao Y., Roy S., LeDoux M. S., Reiter L. T. (2008) Hum. Mol. Genet. 17, 2181–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sango K., Yamanaka S., Hoffmann A., Okuda Y., Grinberg A., Westphal H., McDonald M. P., Crawley J. N., Sandhoff K., Suzuki K., Proia R. L. (1995) Nat. Genet. 11, 170–176 [DOI] [PubMed] [Google Scholar]
- 48. Duda J. E., Giasson B. I., Gur T. L., Montine T. J., Robertson D., Biaggioni I., Hurtig H. I., Stern M. B., Gollomp S. M., Grossman M., Lee V. M., Trojanowski J. Q. (2000) J. Neuropathol. Exp. Neurol. 59, 830–841 [DOI] [PubMed] [Google Scholar]
- 49. Rodríguez E. M., Yulis R., Peruzzo B., Alvial G., Andrade R. (1984) Histochemistry 81, 253–263 [DOI] [PubMed] [Google Scholar]
- 50. Teclemariam-Mesbah R., Wortel J., Romijn H. J., Buijs R. M. (1997) J. Histochem. Cytochem. 45, 619–621 [DOI] [PubMed] [Google Scholar]
- 51. Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. (2001) J. Neurosci. 21, 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waxman E. A., Giasson B. I. (2010) J. Neurochem. 113, 374–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giasson B. I., Murray I. V., Trojanowski J. Q., Lee V. M. (2001) J. Biol. Chem. 276, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 54. Greenbaum E. A., Graves C. L., Mishizen-Eberz A. J., Lupoli M. A., Lynch D. R., Englander S. W., Axelsen P. H., Giasson B. I. (2005) J. Biol. Chem. 280, 7800–7807 [DOI] [PubMed] [Google Scholar]
- 55. Mazzulli J. R., Mishizen A. J., Giasson B. I., Lynch D. R., Thomas S. A., Nakashima A., Nagatsu T., Ota A., Ischiropoulos H. (2006) J. Neurosci. 26, 10068–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giasson B. I., Mabon M. E., Duda J. E., Montine T. J., Robertson D., Hurtig H. I., Lee V. M., Trojanowski J. Q. (2003) Acta Neuropathol. 106, 243–250 [DOI] [PubMed] [Google Scholar]
- 57. Giasson B. I., Duda J. E., Forman M. S., Lee V. M., Trojanowski J. Q. (2001) Exp. Neurol. 172, 354–362 [DOI] [PubMed] [Google Scholar]
- 58. Ishihara T., Hong M., Zhang B., Nakagawa Y., Lee M. K., Trojanowski J. Q., Lee V. M. (1999) Neuron 24, 751–762 [DOI] [PubMed] [Google Scholar]
- 59. Côté F., Collard J. F., Julien J. P. (1993) Cell 73, 35–46 [DOI] [PubMed] [Google Scholar]
- 60. Lee M. K., Marszalek J. R., Cleveland D. W. (1994) Neuron 13, 975–988 [DOI] [PubMed] [Google Scholar]
- 61. Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T. (2002) Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 62. Giasson B. I., Forman M. S., Higuchi M., Golbe L. I., Graves C. L., Kotzbauer P. T., Trojanowski J. Q., Lee V. M. (2003) Science 300, 636–640 [DOI] [PubMed] [Google Scholar]
- 63. Lee V. M., Giasson B. I., Trojanowski J. Q. (2004) Trends Neurosci. 27, 129–134 [DOI] [PubMed] [Google Scholar]
- 64. Waxman E. A., Giasson B. I. (2011) J. Neurosci. 31, 7604–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hansen C., Angot E., Bergström A. L., Steiner J. A., Pieri L., Paul G., Outeiro T. F., Melki R., Kallunki P., Fog K., Li J. Y., Brundin P. (2011) J. Clin. Invest. 121, 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Desplats P., Lee H. J., Bae E. J., Patrick C., Rockenstein E., Crews L., Spencer B., Masliah E., Lee S. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dickson D. W., Liu W., Hardy J., Farrer M., Mehta N., Uitti R., Mark M., Zimmerman T., Golbe L., Sage J., Sima A., D'Amato C., Albin R., Gilman S., Yen S. H. (1999) Am. J. Pathol. 155, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., Trojanowski J. Q., Iwatsubo T. (1998) Am. J. Pathol. 152, 879–884 [PMC free article] [PubMed] [Google Scholar]
- 69. Galvin J. E., Giasson B., Hurtig H. I., Lee V. M., Trojanowski J. Q. (2000) Am. J. Pathol. 157, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lippa C. F., Fujiwara H., Mann D. M., Giasson B., Baba M., Schmidt M. L., Nee L. E., O'Connell B., Pollen D. A., St George-Hyslop P., Ghetti B., Nochlin D., Bird T. D., Cairns N. J., Lee V. M., Iwatsubo T., Trojanowski J. Q. (1998) Am. J. Pathol. 153, 1365–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi W., Zibaee S., Jakes R., Serpell L. C., Davletov B., Crowther R. A., Goedert M. (2004) FEBS Lett. 576, 363–368 [DOI] [PubMed] [Google Scholar]
- 72. Auluck P. K., Caraveo G., Lindquist S. (2010) Annu. Rev. Cell Dev. Biol. 26, 211–233 [DOI] [PubMed] [Google Scholar]
- 73. Ruipérez V., Darios F., Davletov B. (2010) Prog. Lipid Res. 49, 420–428 [DOI] [PubMed] [Google Scholar]
- 74. Dawson T. M., Ko H. S., Dawson V. L. (2010) Neuron 66, 646–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Matsuoka Y., Vila M., Lincoln S., McCormack A., Picciano M., LaFrancois J., Yu X., Dickson D., Langston W. J., McGowan E., Farrer M., Hardy J., Duff K., Przedborski S., Di Monte D. A. (2001) Neurobiol. Dis. 8, 535–539 [DOI] [PubMed] [Google Scholar]
- 76. Richfield E. K., Thiruchelvam M. J., Cory-Slechta D. A., Wuertzer C., Gainetdinov R. R., Caron M. G., Di Monte D. A., Federoff H. J. (2002) Exp. Neurol. 175, 35–48 [DOI] [PubMed] [Google Scholar]
- 77. Norris E. H., Giasson B. I., Hodara R., Xu S., Trojanowski J. Q., Ischiropoulos H., Lee V. M. (2005) J. Biol. Chem. 280, 21212–21219 [DOI] [PubMed] [Google Scholar]
- 78. Conway K. A., Rochet J. C., Bieganski R. M., Lansbury P. T., Jr. (2001) Science 294, 1346–1349 [DOI] [PubMed] [Google Scholar]
- 79. Janus C., Chishti M. A., Westaway D. (2000) Biochim. Biophys. Acta 1502, 63–75 [DOI] [PubMed] [Google Scholar]
- 80. Götz J., Streffer J. R., David D., Schild A., Hoerndli F., Pennanen L., Kurosinski P., Chen F. (2004) Mol. Psychiatry 9, 664–683 [DOI] [PubMed] [Google Scholar]
- 81. Götz J., Deters N., Doldissen A., Bokhari L., Ke Y., Wiesner A., Schonrock N., Ittner L. M. (2007) Brain Pathol. 17, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hutton M., Lewis J., Dickson D., Yen S. H., McGowan E. (2001) Trends Mol. Med. 7, 467–470 [DOI] [PubMed] [Google Scholar]
- 83. Ahn K. J., Paik S. R., Chung K. C., Kim J. (2006) J. Neurochem. 97, 265–279 [DOI] [PubMed] [Google Scholar]
- 84. Lee H. J., Suk J. E., Bae E. J., Lee J. H., Paik S. R., Lee S. J. (2008) Int. J. Biochem. Cell Biol. 40, 1835–1849 [DOI] [PubMed] [Google Scholar]
- 85. Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S. D., Ntzouni M., Margaritis L. H., Stefanis L., Vekrellis K. (2010) J. Neurosci. 30, 6838–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Goedert M., Clavaguera F., Tolnay M. (2010) Trends Neurosci. 33, 317–325 [DOI] [PubMed] [Google Scholar]
- 87. Lee S. J., Desplats P., Sigurdson C., Tsigelny I., Masliah E. (2010) Nat. Rev. Neurol. 6, 702–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kordower J. H., Chu Y., Hauser R. A., Freeman T. B., Olanow C. W. (2008) Nat. Med. 14, 504–506 [DOI] [PubMed] [Google Scholar]
- 89. Li J. Y., Englund E., Holton J. L., Soulet D., Hagell P., Lees A. J., Lashley T., Quinn N. P., Rehncrona S., Björklund A., Widner H., Revesz T., Lindvall O., Brundin P. (2008) Nat. Med. 14, 501–503 [DOI] [PubMed] [Google Scholar]
- 90. Luk K. C., Song C., O'Brien P., Stieber A., Branch J. R., Brunden K. R., Trojanowski J. Q., Lee V. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brundin P., Li J. Y., Holton J. L., Lindvall O., Revesz T. (2008) Nat. Rev. Neurosci. 9, 741–745 [DOI] [PubMed] [Google Scholar]