Background: DJ-1 is a protein in association with Parkinson disease and tumorigenesis.
Results: DJ-1 increases its mitochondrial distribution in response to ultraviolet B (UVB) irradiation and binds to Bcl-XL in an oxidation-dependent manner to prevent Bcl-XL degradation.
Conclusion: Bcl-XL is a new partner of DJ-1 in mitochondria and DJ-1 stabilizes Bcl-XL in response to UVB irradiation.
Significance: DJ-1 plays important roles in mitochondria by regulating Bcl-XL functions.
Keywords: Mitochondria, Parkinson's Disease, Protein Degradation, Protein Stability, Ubiquitination, Bcl-XL, DJ-1
Abstract
Parkinson disease (PD)- and cancer-associated protein, DJ-1, mediates cellular protection via many signaling pathways. Deletions or mutations in the DJ-1 gene are directly linked to autosomal recessive early-onset PD. DJ-1 has potential roles in mitochondria. Here, we show that DJ-1 increases its mitochondrial distribution in response to ultraviolet B (UVB) irradiation and binds to Bcl-XL. The interactions between DJ-1 and Bcl-XL are oxidation-dependent. DJ-1(C106A), a mutant form of DJ-1 that is unable to be oxidized, binds Bcl-XL much less than DJ-1 does. Moreover, DJ-1 stabilizes Bcl-XL protein level by inhibiting its ubiquitination and degradation through ubiquitin proteasome system (UPS) in response to UVB irradiation. Furthermore, under UVB irradiation, knockdown of DJ-1 leads to increases of Bcl-XL ubiquitination and degradation upon UVB irradiation, thereby increasing mitochondrial Bax, caspase-3 activation and PARP cleavage. These data suggest that DJ-1 protects cells against UVB-induced cell death dependent on its oxidation and its association with mitochondrial Bcl-XL.
Introduction
DJ-1, the product of the PARK7 gene, is associated with Parkinson disease (PD) and cancers. Homozygous deletion or mutations in the DJ-1 gene including L166P, A104T, M26I, D149A, and E64D are associated with forms of autosomal recessive early-onset PD (1–2). Mutations in the DJ-1 gene account for almost 1–2% of all early-onset PD (3). DJ-1 was identified as an oncoprotein (4) involved in tumorigenesis because high expression of DJ-1 is a causative risk factor for prostate cancer, ovarian carcinoma, breast cancer, and lung carcinoma (5–8).
DJ-1 is a multifunctional protein that protects cells via several molecular processes including regulating gene transcription, stabilizing protective proteins, mediating cell signal transduction pathways and scavenging excessive reactive oxygen species (ROS)2 (9). Overexpression of DJ-1 protects neurons and somatic cells against various oxidative agents including UV irradiation, H2O2, paraquat and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Moreover, deletion or down-regulation of the DJ-1 gene sensitizes cells to oxidative stress (10–14). The antioxidant activity of DJ-1 is dependent on oxidization of Cys-106 to cysteine sulfinic acid to scavenge ROS, and its C106A artificial mutant form abolishes its anti-apoptotic properties (12–13, 15–16).
Mitochondrial dysfunction is one of the key pathological processes involved in the pathogenesis of sporadic PD and familial PD (17–18). Several lines of evidence suggest that DJ-1 affects mitochondrial functions (19–24). Many studies have shown that DJ-1 is localized in both the cytoplasm and nucleus, and partially localized to mitochondria (2, 13, 25–26). Interestingly, DJ-1 was found to be increased in the mitochondrial fraction under death stimuli (13, 16, 26–28). Mitochondrial DJ-1 exhibits stronger protective effects than cytosolic or nuclear DJ-1 (28). In addition, the cytosolic and mitochondrial pools of DJ-1 are mediated by Cys106 oxidation (16). The oxidation-induced mitochondrial translocation and protection could be abolished by an artificial point mutation C106A (13, 16). Recently, it was reported that DJ-1 maintains mitochondrial functions under oxidative stress and loss of DJ-1 leads to mitochondrial phenotypes such as reduction of membrane potential and increases of fragmentation (22–23). These data suggest that DJ-1 may have roles in association with mitochondria in response to oxidative stress. However, the mechanisms by which DJ-1 functions in mitochondria are largely unknown.
In our present study, we show that Bcl-XL, a protein primarily localized in the outer mitochondrial membrane (OMM), is a novel partner of DJ-1 under UVB irradiation. DJ-1 stabilizes Bcl-XL protein to protect cells under UVB irradiation. Our results show that the regulation of Bcl-XL by DJ-1 may play roles in the control of the mitochondrial apoptotic pathway.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmid Transfection
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Hyclone). H1299 cells were cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum (Hyclone). Cells were transfected with expression plasmids using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. H1299 cells stably expressing EGFP-Bcl-XL were obtained by a limiting dilution and a selection with 200 μg/ml Geneticin (Invitrogen) after transfection.
UVB Irradiation Treatment
The culture medium was removed just prior to irradiation and replaced with a thin layer of phosphate buffer solution (PBS, pH 7.4) to cover the cells. Then, HEK293 or H1299 cells were exposed to UVB irradiation (312 nm) with a UV crosslinker (SCIENTZ03-II, Ningbo, China). After UVB irradiation, the cells were fed fresh culture medium and returned to culture for 16 h. After culturing, the treated cells were subjected to additional experiments.
siRNA Knockdown
si-DJ-1328 was described previously (29). siRNA against human Bcl-XL mRNA was synthesized by GenePharma (GenePharma, Shanghai, China) with the following sequences: sense: 5′-GAGAUGCAGGUAUUGGUGATT-3′, antisense: 5′-UCACCAAUACCUGCAUCUCTT-3′. The oligonucleotides were transfected with Oligofectamine reagent (Invitrogen) following the manufacturer's instructions. Briefly, cultured cells were washed with Opti-MEM medium (Invitrogen) and then transfected with siRNA using Oligofectamine reagent in Opti-MEM medium without serum. Six hours after transfection, the culture medium was replaced with fresh complete medium. The cells were subjected to further experiments 72 h after transfection.
Plasmid Constructs
Full-length DJ-1 in p3×Flag-myc-cmv-24, pET-15b and pGEX-5x-1 were described previously (30). pDsRed-N1-DJ-1 was generated by subcloning the PCR product amplified with the primers 5′-CGGGATCCCCATGGCTTCCAAAAGAAG-3′ and 5′-CGCTCGAGCTGCTGGAGTCTTTAAGAAC-3′ into pDsRed-N1 at its BglII/SalI sites. DJ-1(C106A) was obtained by site-directed mutagenesis with the following primers: 5′-GCTGCAGGTCCTACTGCT-3′ and 5′-GATGGCGGCTATCAG-3′. Full length Bcl2 cDNA was amplified from a human fetal brain cDNA library (Clonetech) with the primers 5′-CGGAATTCAATATGGCGCACGCTG-3′ and 5′-GCGTCGACTCACTTGTGGCCCAG-3′ and then inserted into pET-21a at its EcoRI/SalI sites. A full-length Bax cDNA was amplified from a human fetal brain cDNA library (Clonetech) with the primers 5′-CGGAATTCATGGACGGGTCCGGGGA-3′ and 5′-CCCTCGAGCAGCCCATCTTCTTCCAGA-3′ and then inserted into pET-21a at its EcoRI/XhoI sites. Full-length Bcl-XL cDNA was amplified from a human fetal brain cDNA library (Clontech) with the primers 5′-CGGAATTCATGTCTCAGAGCAAC-3′ and 5′-GCGTCGACTCATTTCCGACTGAAG-3′ and inserted into pEGFP-C2, pGEX-5x-1, or pET-21a at EcoRI/SalI sites. p3×Flag-myc-cmv-24-Bcl-XL was generated by subcloning the PCR product, amplified with the primers 5′-CGGAATTCCATGTCTCAGAGCAAC-3′ and 5′-GCGTCGACTCATTTCCGACTGAAG-3′ into p3×Flag-myc-cmv-24 at its EcoRI/SalI sites. Deletion mutants of Bcl-XL encoding amino acids 1–85, 86–195, or 196–233 were created by subcloning PCR products into pGEX-5x-1 at its EcoRI/SalI sites. The PCR products were amplified with the following primers: 5′-CGGAATTCATGTCTCAGAGCAAC-3′ and 5′-GCGTCGACTCATGCTGCCATGGGGAT-3′ for 1–85aa; 5′-CGGAATTCGTAAAGCAAGCGCTG-3′ and 5′-GCGTCGACTCAATAGAGTTCCACAAA-3′ for 86–195aa; 5′-CGGAATTCGGGAACAATGCAGCA-3′ and 5′-GCGTCGACTCATTTCCGACTGAAG-3′ for 196–233aa.
GST Pull-down Assay
An aliquot containing 20 μg of GST or a recombinant GST-fused protein expressed by Escherichia coli strain JM109 was incubated with 20 μl of glutathione-agarose beads (Pharmacia) for 30 min at 4 °C. After washing three times with ice-cold PBS, the beads were incubated with 50 μg of a protein expressed by E. coli strain BL21 for 2 h at 4 °C. After incubation, the beads were washed five times with ice-cold HNTG buffer (20 mm Hepes-KOH, pH 7.5, 100 mm NaCl, 0.1% Triton X-100 and 10% glycerol). Bound proteins were eluted from the beads and subjected to immunoblot analysis. The input represents 10% of the protein incubated with GST or a GST-fusion protein. The inputs of purified GST and GST-fusion proteins are stained with Coomassie Brilliant Blue (CBB) or anti-GST antibody.
Immunoprecipitation Assay
Cells were lysed in cell lysis buffer (50 mm Tris-HCl pH 7.5 buffer containing 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and protease inhibitor mixture (Roche) for 30 min at 4 °C. Cellular debris was removed by centrifugation at 12,000 × g for 15 min at 4 °C. The supernatant was incubated with polyclonal anti-GFP antibodies (Santa Cruz Biotechnology), or monoclonal anti-Bcl-XL antibody (Santa Cruz Biotechnology) coupled to protein G-Sepharose (Roche). The immunoprecipitated proteins were then washed five times with cell lysis buffer. Bound proteins and cell lysates were subjected to immunoblot analysis. The input represents 10% of the cell lysate used in the co-immunoprecipitation experiment.
Immunoblot Analysis
Proteins were separated by 12% or 15% SDS-PAGE followed by transfer onto polyvinylidene difluoride membranes (Millipore). Monoclonal anti-Bcl2, anti-Bcl-XL, anti-GFP, anti-GST, anti-Tom20, and anti-Ub antibodies were from Santa Cruz Biotechnology, as were rabbit polyclonal anti-Bax antibodies. Polyclonal anti-Bcl-XL, anti-cleaved caspase-3 and anti-PARP antibodies were from Cell Signaling. Polyclonal anti-DJ-1 antibody was from Chemicon. Monoclonal anti-Flag, anti-Flag-HRP and anti-α-Tubulin antibodies were from Sigma. Monoclonal anti-GAPDH antibody was from Millipore. The secondary antibodies, sheep anti-mouse IgG-HRP and anti-rabbit IgG-HRP, were from Amersham Biosciences. The proteins were visualized using an ECL detection kit (Amersham Biosciences).
Quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted using the RNA extraction kit (TaKaRa, Japan) and used for reverse transcription with an oligo (dT)18 primer and TransScript RT/RI Enzyme (TaKaRa, Japan). qRT-PCR was performed using the SYBR Green PCR Kit (TaKaRa, Japan) and a StepOne Real-Time PCR System (Applied Biosystems) in 20 μl for 40 cycles (95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min). The following specific primers were used, as described elsewhere (31): 5′-GTAAACTGGGGTCGCATTGT-3′ and 5′-TGGATCCAAGGCTCTAGGTG-3′ for Bcl-XL; 5′-GATGAGTATGCCTGCCGTGTG-3′ and 5′-CAATCCAAATGCGGCATCT-3′ for the house-keeping gene B2MG. The relative amount of the target gene was calculated using the 2−ΔΔCt method (32).
Subcellular Fractionation Assay
Isolation of cytosolic and mitochondrial fractions from transfected cells was performed according to instructions from the Mitochondria Isolation kit for Cultured Cells (Beyotime, China). The isolated fractions were subjected to immunoblot analysis. Tom20 was used as the mitochondrial maker and α-tubulin was used as the cytosolic maker.
MTT Assay
Cells were washed with DMEM without phenol red and incubated with MTT (3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-di-phenytetrazoliumromide, Sigma). After 3 h of incubation, the formazan crystals were dissolved in DMSO (dimethyl sulfoxide). The optical density (OD) was measured by a photometer at 570 nm and background at 630 nm was subtracted. The data were normalized to a control and the ratios are presented as means ± S.E. from three independent experiments.
Pulse Chase Analysis
H1299 cells stably expressing EGFP-Bcl-XL were starved in methionine-free medium and incubated for 30 min. The cells were then labeled with 200 μCi/ml [35S]methionine (PerkinElmer) for 2 h. Pulse-labeling was terminated by incubating cells in fresh medium containing an excess of unlabeled methionine (300 mg/liter), and the cells were harvested at 0, 7, and 14 h. Cells lysates were immunoprecipitated with anti-GFP antibodies (Santa Cruz Biotechnology) and analyzed by SDS-PAGE and autoradiography.
Statistical Analysis
Western blot densitometric analysis of three independent experiments was performed using Photoshop 7.0 (Adobe). The data were analyzed by one-way analysis of variance (ANOVA) using origin 6.0 software (Originlab, Northampton, MA).
RESULTS
The Interactions between DJ-1 and Bcl-XL under UVB Irradiation
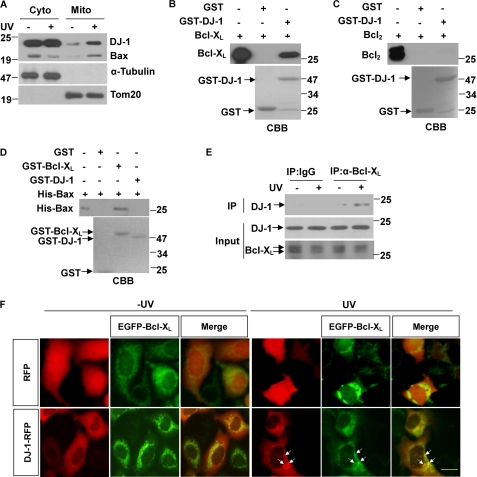
The mitochondrial localization of DJ-1 increases under oxidative stresses such as paraquat treatment, H2O2 and UV irradiation (13, 26–28). Consistent with those findings, we observed that UVB irradiation increased the mitochondrial relocalization of DJ-1 (Fig. 1A). Bax was shown as positive control which is the well-known protein translocation to mitochondria under UV irradiation (33). The mitochondrial fraction of DJ-1 has been reported to localize to the OMM (13, 28), we hypothesized that DJ-1 may interact with Bcl-2 family proteins. The Bcl-2 family, which includes both pro-apoptotic (Bax, Bid, Bad) and anti-apoptotic (Bcl2, Bcl-XL, Mcl-1) members, acts as an apoptotic checkpoint upstream of caspases and mitochondria. Many Bcl-2 family proteins function and localize in the OMM (33–34). We therefore performed GST pull-down assays to test the interactions between DJ-1 and three typical Bcl-2 family proteins, Bcl-XL, Bcl2, and Bax. Interestingly, DJ-1 interacted with Bcl-XL but not with Bcl2 and Bax (Fig. 1, B–D), although they share sequence homology and structural similarity (33). Bcl-XL can form homodimers by itself or heterodimers with other Bcl-2 family proteins, such as Bax (35–36). In our observations, GST-Bcl-XL interacted with His-Bax (Fig. 1D). To further confirm these interactions, we performed co-immunoprecipitation assays using anti-Bcl-XL antibody in H1299 cells. Interestingly, DJ-1 was precipitated by anti-Bcl-XL antibody under UVB irradiation, but much less DJ-1 was precipitated under normal culture conditions (Fig. 1E). In addition, neither Bax nor Bcl2 interacted with DJ-1 in cells (data not shown).
FIGURE 1.
Interactions between DJ-1 and Bcl-XL under UVB irradiation. A, H1299 cells were treated with or without 80 mJ/cm2 UVB irradiation, and the cells were then subjected to the subcellular fractionation assay. The cytosolic and mitochondrial fractions were subjected to immunoblot analysis. B–D, supernatant of E. coli crude extract containing 50 μg of recombinant Bcl-XL, Bcl2 or His-Bax protein was incubated with 20 μg of GST, GST-DJ-1 or GST-Bcl-XL coupled to glutathione-agarose beads. The input and bound proteins were detected by immunoblot analysis. (E) The supernatant of H1299 cells treated with or without 80 mJ/cm2 UVB irradiation was subjected to immunoprecipitation with normal mouse serum or anti-Bcl-XL antibody. F, HEK293 cells transiently transfected with RFP or DJ-1-RFP along with EGFP-Bcl-XL were treated with or without 80 mJ/cm2 UVB irradiation, and then the cells were observed using an inverted fluorescence microscope (Olympus, IX71). Co-localization of EGFP-Bcl-XL with DJ-1-RFP in punctuate structures is indicated with arrows. Bar, 10 μm.
Consistent with the data from the immunoprecipitation assays, UVB irradiation resulted in small DJ-1-RFP punctate formations, and these dots were partially co-localized with EGFP-Bcl-XL signals (Fig. 1F). These results suggest that UVB irradiation increases the translocation of DJ-1 to OMM and promotes its binding to Bcl-XL.
Cys-106 of DJ-1 Is Required for Its Binding to Bcl-XL
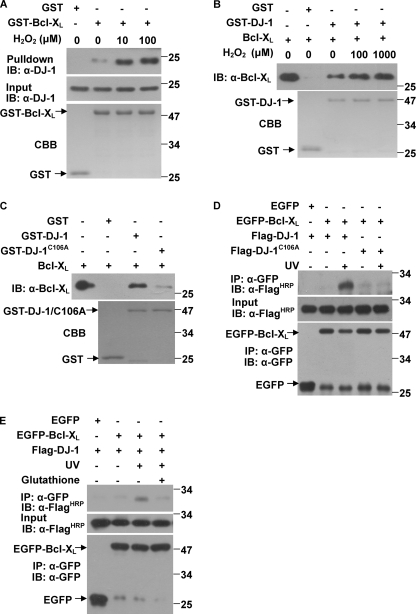
The ability of DJ-1 to protect against oxidative stress is dependent on C106 oxidation and oxidation-driven mitochondrial localization (12–13). To examine if the interaction between DJ-1 and Bcl-XL is mediated by the oxidative state of DJ-1, we treated H1299 cells with various concentrations of H2O2 and then collected cells for pull-down assays. The results showed that more DJ-1 was pulled down by GST-Bcl-XL when more H2O2 was used (Fig. 2A). Using recombinant proteins in vitro, we also observed that an equivalent amount of GST-DJ-1 treated with more H2O2 pulled down more Bcl-XL (Fig. 2B). We next created a C106A mutant to examine whether C106 is required for DJ-1 to bind to Bcl-XL. GST-DJ-1(C106A), which cannot be oxidized, bound to Bcl-XL much less than GST-DJ-1 did in GST pulldown assays (Fig. 2C) and could not bind to EGFP-Bcl-XL with or without UVB irradiation in cells (Fig. 2D). Moreover, interactions between DJ-1 and Bcl-XL induced by UVB irradiation could be blocked by reduced glutathione (Fig. 2E). These results suggest that the interactions between DJ-1 and Bcl-XL are oxidation-dependent.
FIGURE 2.
Cys-106 is required for DJ-1 binding to Bcl-XL. A, supernatants of H1299 cells treated with various concentrations of H2O2 for 12 h were incubated with 20 μg of GST or GST-Bcl-XL coupled to glutathione-agarose beads. B, GST-DJ-1 was incubated with various concentrations of H2O2 for 2 h at 30 °C in PBS. Then, the supernatant of E. coli crude extract containing 50 μg of recombinant Bcl-XL was incubated with 20 μg of GST or oxidized GST-DJ-1. The bound proteins were detected using immunoblot analysis. C, supernatant of E. coli crude extract containing 50 μg of recombinant Bcl-XL was incubated with 20 μg of GST, GST-DJ-1 or GST-DJ-1(C106A). The bound proteins were detected using immunoblot analysis. D, H1299 cells transiently transfected with Flag-DJ-1 or Flag-DJ-1(C106A) along with EGFP or EGFP-Bcl-XL were treated with or without 80 mJ/cm2 UVB irradiation, and then the supernatants of the cell lysates were subjected to immunoprecipitation with anti-GFP antibodies. E, H1299 cells transfected with Flag-DJ-1 along with EGFP or EGFP-Bcl-XL, were treated with or without 80 mJ/cm2 UVB irradiation and treated with or without 5 mm reduced glutathione for 16 h. The cells were then collected and subjected to immunoprecipitation using anti-GFP antibodies.
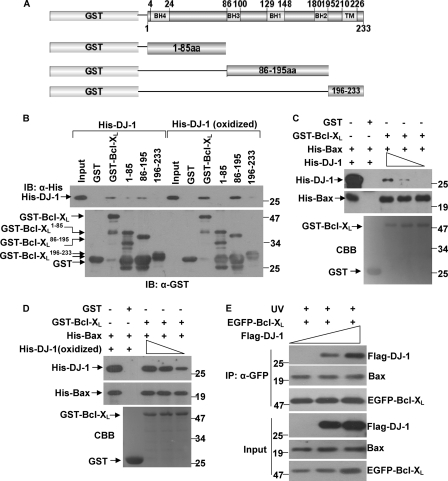
DJ-1 Interacts with BH1–3 Domains of Bcl-XL
Bcl-XL protein contains four conserved Bcl-2 homology domains (BHs), BH1–4 (34). To identify the regions of Bcl-XL that are required for its binding to DJ-1, we generated three deletion mutants of Bcl-XL fused to GST: amino acids 1–85, which contain the BH4 domain; amino acids 86–195, which contain domains BH1–3; and amino acids 196–233, which contain a transmembrane domain (Fig. 3A). GST-pull down assays showed that DJ-1 mainly bound to the middle fragment of Bcl-XL containing amino acids 86–195, and it bound significantly less to the N-terminal and C-terminal fragments (Fig. 3B). However, oxidized DJ-1 also bound to these domains with more strong affinity (Fig. 3B).
FIGURE 3.
DJ-1 interacts with BH1–3 domains of Bcl-XL. A, schematic structure of human Bcl-XL and a diagram of the Bcl-XL deletion mutants used in the GST pulldown assay. B, supernatant of E. coli crude extract containing recombinant His-DJ-1 or 100 μm H2O2 pretreated His-DJ-1 (oxidized DJ-1) was incubated with GST, GST-Bcl-XL, GST-Bcl-XL(1–85), GST-Bcl-XL(86–195) or GST-Bcl-XL(196–233) coupled to glutathione-agarose beads. Both the input and the bound proteins were subjected to immunoblot analysis. C–D, supernatant containing 50 μg of recombinant His-Bax was incubated with 20 μg of GST or GST-Bcl-XL coupled to glutathione-agarose beads. After being washed to remove unbound proteins, the glutathione-agarose beads were incubated with various amounts supernatant of His-DJ-1 or 100 μm H2O2 pretreated His-DJ-1 (oxidized DJ-1). The bound proteins were subjected to immunoblot analysis. E, H1299 cells stably expressing EGFP-Bcl-XL were transiently transfected with various amounts of Flag-DJ-1 and treated with 80 mJ/cm2 UVB irradiation. The supernatants of cell lysates were subjected to immunoprecipitation with anti-GFP antibodies.
The BH1, BH2, and BH3 domains in Bcl-XL (aa 86–195) and the C terminus of Bcl-XL are required for Bcl-XL/Bax heterodimer formation (36–38). We therefore investigated if DJ-1 affects the interactions between Bcl-XL and Bax. Using in vitro competitive binding experiments, we found neither non-oxidized DJ-1 nor oxidized DJ-1 affected the heterodimerization between Bcl-XL and Bax (Fig. 3, C and D). To further confirm this findings, we transfected various amounts of Flag-DJ-1 into H1299 cells that stably expressed EGFP-Bcl-XL and performed immunoprecipitation assays after UVB irradiation. DJ-1 did not affect the interactions between Bcl-XL and Bax (Fig. 3E). Interestingly, Flag-DJ-1 increased EGFP-Bcl-XL protein levels in a dose-dependent manner under UVB irradiation (Fig. 3E, lowest panel).
Stabilization of Bcl-XL by DJ-1 under UVB Irradiation
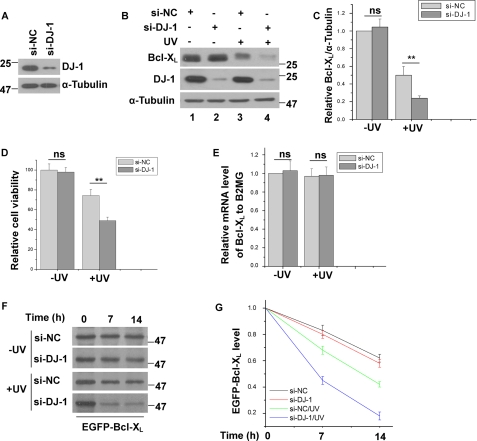
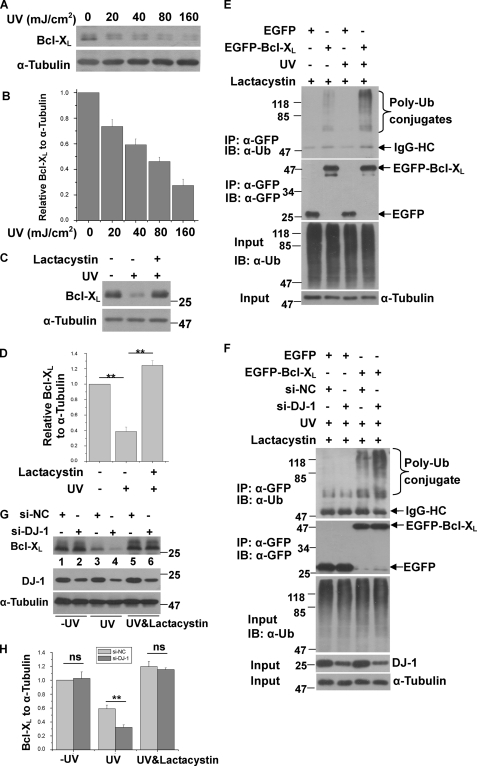
To further confirm the effects of DJ-1 on Bcl-XL protein level, we silenced DJ-1 using small interfering RNA (si-DJ-1) and examined its effects on Bcl-XL. The knockdown efficiency of si-DJ-1 is shown in Fig. 4A. It has been reported that UVB irradiation promotes Bcl-XL degradation via the UPS (39–40). Consistent with these findings, Bcl-XL protein levels decreased after UVB irradiation (Fig. 4B, lanes 1 and 3). Without UVB irradiation, knockdown of DJ-1 did not significantly change Bcl-XL protein levels (Figs. 4, B and C, lanes 1 and 2). However, with UVB irradiation, Bcl-XL protein levels were significantly lower in DJ-1 knockdown cells than in control cells (Fig. 4, B and C, lanes 3 and 4). We next examined the effects of DJ-1 knockdown on cell viability. Although DJ-1 knockdown alone did not affect cell death, under UVB irradiation, DJ-1 knockdown significantly increased cell death (Fig. 4D). Knockdown of DJ-1 did not change Bcl-XL mRNA levels with or without UVB irradiation as detected by qRT-PCR (Fig. 4E), suggesting that DJ-1 may influence Bcl-XL levels post-translationally. We therefore performed pulse-chase assays to assess the effects of DJ-1 on Bcl-XL stability. Without UVB irradiation, DJ-1 knockdown did not affect the degradation rate of Bcl-XL. And consist with previous findings (39–40), the degradation rate of Bcl-XL protein was accelerated by UVB irradiation (Fig. 4, F and G). Moreover, under UVB irradiation the degradation rate of Bcl-XL was further accelerated in DJ-1 knockdown cells as compared with cells transfected with control siRNA (Fig. 4, F and G). These results further suggest that DJ-1 stabilizes Bcl-XL protein to protect against UVB irradiation-induced cell death.
FIGURE 4.
Stabilization of Bcl-XL by DJ-1 under UVB irradiation. A, total cell lysates of H1299 cells transfected with si-NC or si-DJ-1 was subjected to immunoblot analysis. B–E, H1299 cells transfected with si-NC or si-DJ-1 were treated with or without 80 mJ/cm2 UVB irradiation. The samples were subjected to immunoblot analysis (B and C), MTT assays (D) or qRT-PCR experiments (E) (mean ± S.E., n = 3; **, p < 0.01; ns, not statistical significance, by one-way ANOVA). The relative ratios of Bcl-XL to α-tubulin in B were analyzed by densitometry (mean ± S.E., n = 3; **, p < 0.01; ns, not statistical significance, by one-way ANOVA) (C). F, H1299 cells stably expressing EGFP-Bcl-XL transfected with si-NC or si-DJ-1 were treated with or without 80 mJ/cm2 UVB irradiation, and then the cells were subjected to pulse-chase assays. G, relative levels of EGFP-Bcl-XL in F were analyzed using densitometry (mean ± S.E., n = 3; by one-way ANOVA).
Decreased Ubiquitination and Degradation of Bcl-XL by DJ-1 under UVB Irradiation
Bcl-XL protein is markedly degraded through the proteasome in UVB-irradiated cells (39–40). In our observations, Bcl-XL protein levels decreased in a UV dose-dependent manner (Fig. 5, A and B). We also found that in the presence of Lactacystin, an inhibitor of the proteasome, UVB irradiation failed to decrease Bcl-XL protein levels (Fig. 5, C and D), suggesting that Bcl-XL is rapidly degraded through the proteasome after UVB irradiation. We next performed immunoprecipitation assays to examine the ubiquitination of Bcl-XL with or without UVB irradiation. In cells treated with Lactacystin, more ubiquitination of EGFP-Bcl-XL was observed in cells treated with UVB irradiation than in those without UVB irradiation (Fig. 5E), suggesting that UVB irradiation increases Bcl-XL ubiquitination. We therefore asked if the ubiquitination of Bcl-XL upon UVB irradiation is influenced by DJ-1. Knockdown of DJ-1 significantly increased EGFP-Bcl-XL ubiquitination after UVB irradiation (Fig. 5F, lanes 3 and 4). Furthermore, the decreased Bcl-XL levels induced by knockdown of DJ-1 plus UVB irradiation (Fig. 5, G and H, lanes 3 and 4) were restored by Lactacystin treatment (Fig. 5, G and H, lanes 5 and 6). Our data demonstrate that under UVB irradiation, DJ-1 inhibits UVB-induced Bcl-XL degradation by the proteasome.
FIGURE 5.
DJ-1 decreases the ubiquitination and degradation of Bcl-XL after UVB irradiation. A, H1299 cells were exposed to various UVB doses and then cultured for 16 h. The total cell lysates were subjected to immunoblot analysis. B, relative ratios of Bcl-XL to α-tubulin in A were analyzed using densitometric analysis (mean ± S.E., n = 3; by one-way ANOVA). C, H1299 cells were treated with or without 80 mJ/cm2 UVB and 5 μm lactacystin for 16 h, and then total cell lysates were subjected to immunoblot analysis. D, relative ratios of Bcl-XL to α-tubulin were analyzed from densitometric analysis (mean ± S.E., n = 3; **, p < 0.01; by one-way ANOVA). E, H1299 cells transfected with EGFP or EGFP-Bcl-XL were treated with or without 80 mJ/cm2 UVB and 5 μm lactacystin for 16 h. The supernatants of the cell lysates were subjected to immunoprecipitation with anti-GFP antibodies. F, H1299 cells expressing EGFP or EGFP-Bcl-XL were transfected with si-NC or si-DJ-1 and treated with 80 mJ/cm2 UVB irradiation and 5 μm lactacystin for 16 h. The supernatants of the cell lysates were subjected to immunoprecipitation with anti-GFP antibodies. The immunoprecipitants were detected using anti-GFP antibody or anti-ubiquitin antibodies. G, H1299 cells were transfected with si-NC or si-DJ-1 for 60 h. After being pre-treated with or without 10 μm lactacystin for 2 h, the cells were then treated with or without 80 mJ/cm2 UVB irradiation and total cell lysates were then subjected to immunoblot analysis. H, relative ratios of Bcl-XL to α-tubulin was analyzed using densitometry (mean ± S.E., n = 3; **, p < 0.01; ns, no statistical significance by one-way ANOVA).
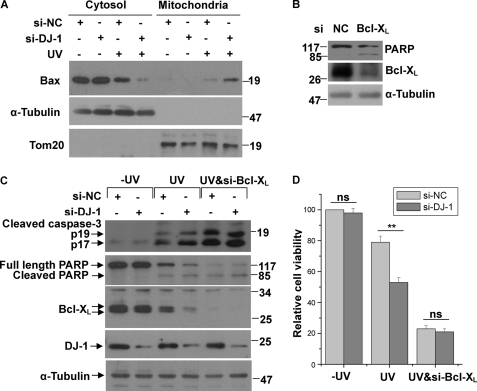
Decreased Mitochondrial Bax Levels and Cell Death by DJ-1 under UVB Irradiation
The mitochondrial localization of Bax is important for its ability to induce cell death (34). Because DJ-1 re-distributes to mitochondria upon UVB irradiation and influence Bcl-XL protein levels, and Bcl-XL protects mitochondria intact partially by heterodimerizating with Bax to inhibit it translocation to OMM (36, 41), we performed cytosolic and mitochondrial fractionation assays to examine the effects of DJ-1 on mitochondrial Bax levels. It has been reported that DJ-1 inhibits Bax transcription by binding to p53 (30, 42–43). In order to eliminate the influence of p53 on Bax protein levels, we performed the experiments in H1299 cells, a p53 null cell line. In the absence of UVB irradiation, knockdown of DJ-1 had no significant effects on the translocation of Bax from the cytosol to the mitochondria (Fig. 6A). However, under UVB irradiation, less Bax was present in the cytosolic fraction but more in the mitochondrial fraction in cells transfected with si-DJ-1 (Fig. 6A). These results suggest that DJ-1 inhibits Bax re-localization to the mitochondria. To further examine whether the protective effects of DJ-1 is Bcl-XL-dependent, we examined the effects of DJ-1 on cell death or cell viability in Bcl-XL silencing cells. Knockdown of Bcl-XL spontaneously induced apoptosis characterized by PARP cleavage (Fig. 6B). Knockdown of DJ-1 alone had no significant effects on Bcl-XL levels, or cleavages of caspase-3 and PARP (Fig. 6C), or cell viability (Fig. 6D). However, with UVB irradiation, knockdown of DJ-1 decreased Bcl-XL levels and accordingly increased the cleavages of caspase-3 and PARP (Fig. 6C), leading to a decrease of cell viability (Fig. 6D). Moreover, the effects of knockdown DJ-1 on cell death under UVB irradiation were abrogated by Bcl-XL silence (Fig. 6, C and D). These results suggest that the ability of DJ-1 to protect cells under UVB irradiation are Bcl-XL-dependent.
FIGURE 6.
Decreased mitochondrial Bax levels and cell death by DJ-1 under UVB irradiation. A, H1299 cells transfected with si-NC or si-DJ-1 were treated with or without 80 mJ/cm2 UVB irradiation, and the cells were then subjected to the subcellular fractionation assay. The cytosolic and mitochondrial fractions were subjected to immunoblot analysis. B, cell lysates of H1299 cells transfected with si-NC or si-Bcl-XL were subjected to immunoblot analysis. C and D, H1299 cells transfected with si-NC, si-DJ-1, or along with si-Bcl-XL were treated with or without 80 mJ/cm2 UVB irradiation. Cell lysates were then subjected to immunoblot analysis (C) or the cells were analyzed with the MTT assay (D) (mean ± S.E., n = 3; *, p < 0.05; ns, no statistical significance by one-way ANOVA).
DISCUSSION
PD is one of the most common progressive neurodegenerative movement disorders, and its most common feature is selective loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNc) (44). Lines of evidence have suggested that mitochondrial dysfunction plays essential roles in the pathogenesis of PD (17–18). PD-associated proteins such as DJ-1, α-synuclein, Parkin and PINK1 are all linked to mitochondria and affect mitochondrial functions (17). DJ-1 has potential functions in maintaining mitochondrial dynamics, integrity and fusion (19–23). A proportion of DJ-1 localizes to mitochondria, and its mitochondrial localization is enhanced under death stimuli such as UVB irradiation, H2O2 or paraquat treatment (13, 26–28). These data suggest that DJ-1 has functions in mitochondria in response to oxidative stress. In the present study, we showed that DJ-1 translocates to mitochondria under UVB irradiation and binds to Bcl-XL. The precise localization of DJ-1 in mitochondria has been confirmed by several groups to be in the OMM (13, 28), although it has also been reported to be inserted into the inter-membrane space or in the mitochondrial matrix (25). In our observations, under UVB irradiation DJ-1 binds to Bcl-XL, a typical Bcl-2 family protein that primarily localizes in the OMM (34), supporting the idea that mitochondrial DJ-1 localizes to the OMM.
DJ-1's translocation to mitochondria under oxidative stress is dependent on oxidation of its Cys106 site (13, 16). However, DJ-1(C106A), a loss of oxidized form of DJ-1, does not translocate to the mitochondria and fails to perform its protective function under oxidative stress (13, 16). We found that UVB irradiation promotes the translocation of DJ-1 to the mitochondria (Fig. 1F) and its binding to Bcl-XL (Fig. 1E), and the binding of DJ-1 and Bcl-XL is dependent on the oxidative state of DJ-1 (Fig. 2, A–E). However, DJ-1(C106A) exhibits very low binding affinity to Bcl-XL in co-immunoprecipitation and GST pull-down assays (Fig. 2, C and D). Oxidation of DJ-1 at Cys-106 is important for its protective effects. For instance, the oxidation of Cys-106 is required for DJ-1 protection against mitochondrial fragmentation and cell death, and DJ-1 fails to protect cells when Cys-106 is in reduced state (16). Taken together with our findings, we propose that the protective effects of oxidized DJ-1 on mitochondria are mediated by the enhanced interactions between DJ-1 and Bcl-XL and by its stabilizing Bcl-XL.
The Bcl-2 family proteins are involved in PD pathogenesis (45). For instance, Bax levels are up-regulated in PD patients and animal PD models (43, 46–47). Bcl2 inhibits cellular toxicity induced by dopamine (48), and Bcl-XL is required for mouse substantial nigra development (49) and has a protective role in PD that inhibits dopaminergic neuronal death induced by MPTP in the SNc (50). Bcl-2 family proteins play important roles in the control of mitochondrial cell death. Bcl-2 family proteins include both anti-apoptotic (Bcl2, Bcl-XL, Bcl-w) and pro-apoptotic (Bax, Bak, Bad, Bid) molecules (34). Most of Bax is normally localized in the cytosol and a small part is loosely attached to the OMM, but under lethal stimuli, the cytosolic Bax (as monomers) is re-distributed to mitochondria and oligomerizes to form channels that result in cytochrome c release and apoptosis. The anti-apoptotic proteins Bcl-XL (primarily localized to the OMM) and Bcl2 (which translocates to the OMM during apoptosis) play roles in keeping mitochondria intact to inhibit cytochrome c release (33, 51–52). The protective effects of Bcl-XL and Bcl2 on mitochondria are partially caused by their formation of heterodimers with Bax and subsequent inhibition of Bax activity (34, 36, 38, 53).
In our present study, we found that DJ-1 binds to amino acids 86–195 of Bcl-XL (Fig. 3B), which contain the BH1, BH2 and BH3 domains that form a hydrophobic core and are essential for Bcl-XL's structural stability (54–55). DJ-1 binds to these regions and represses the rapid degradation of Bcl-XL that is induced by UVB irradiation. Upon UVB irradiation, the interaction between DJ-1 and Bcl-XL is enhanced, and the ubiquitination of Bcl-XL is decreased by DJ-1 (Fig. 5F). It has been reported that DJ-1 inhibits the ubiquitination of Nrf2 (NF-E2-related factor 2) by preventing its association with its E3 ligase Keap1 (56). It is therefore possible that the interaction of DJ-1 with Bcl-XL blocks the relevant E3 ligase(s) from interacting with and ubiquitinating Bcl-XL. Interestingly, even though Bcl-XL, Bcl2, and Bax share sequence homology and structural similarity, DJ-1 and DJ-1(L166P) bind only to Bcl-XL and not Bcl2 or Bax, suggesting that DJ-1 specifically acts on Bcl-XL.
In conclusion, our results provide evidence that Bcl-XL is a new partner of DJ-1 in mitochondria. DJ-1 stabilizes Bcl-XL to prevent its UVB irradiation-induced degradation, thereby maintains mitochondria intact and survives cells. Our study provides evidence that DJ-1 plays important roles in mitochondria by regulating Bcl-XL functions.
This work was supported in part by the National Natural Sciences Foundation of China (No. 30970921), the National High-tech Research and Development program of China 973-Projects (2011CB5004102) and the Chinese Academy of Sciences Knowledge Innovation Project (KSCX2-YW-R138).
- ROS
- reactive oxygen species
- ANOVA
- analysis of variance
- CBB
- Coomassie Brilliant Blue
- EGFP
- enhanced green fluorescent proteins
- HRP
- horseradish peroxidase
- OMM
- outer mitochondrial membrane
- PD
- Parkinson disease
- qRT-PCR
- quantitative reverse transcription PCR
- UPS
- ubiquitin proteasome system
- UVB
- ultraviolet B.
REFERENCES
- 1. Abou-Sleiman P. M., Healy D. G., Quinn N., Lees A. J., Wood N. W. (2003) Ann. Neurol 54, 283–286 [DOI] [PubMed] [Google Scholar]
- 2. Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., van Dongen J. W., Vanacore N., van Swieten J. C., Brice A., Meco G., van Duijn C. M., Oostra B. A., Heutink P. (2003) Science 299, 256–259 [DOI] [PubMed] [Google Scholar]
- 3. Hedrich K., Djarmati A., Schäfer N., Hering R., Wellenbrock C., Weiss P. H., Hilker R., Vieregge P., Ozelius L. J., Heutink P., Bonifati V., Schwinger E., Lang A. E., Noth J., Bressman S. B., Pramstaller P. P., Riess O., Klein C. (2004) Neurology 62, 389–394 [DOI] [PubMed] [Google Scholar]
- 4. Nagakubo D., Taira T., Kitaura H., Ikeda M., Tamai K., Iguchi-Ariga S. M., Ariga H. (1997) Biochem. Biophys. Res. Commun. 231, 509–513 [DOI] [PubMed] [Google Scholar]
- 5. Davidson B., Hadar R., Schlossberg A., Sternlicht T., Slipicevic A., Skrede M., Risberg B., Flørenes V. A., Kopolovic J., Reich R. (2008) Hum. Pathol. 39, 87–95 [DOI] [PubMed] [Google Scholar]
- 6. Lev N., Roncevic D., Ickowicz D., Melamed E., Offen D. (2006) J. Mol. Neurosci 29, 215–225 [DOI] [PubMed] [Google Scholar]
- 7. Hod Y. (2004) J. Cell Biochem. 92, 1221–1233 [DOI] [PubMed] [Google Scholar]
- 8. MacKeigan J. P., Clements C. M., Lich J. D., Pope R. M., Hod Y., Ting J. P. (2003) Cancer Res. 63, 6928–6934 [PubMed] [Google Scholar]
- 9. da Costa C. A. (2007) Curr. Mol. Med. 7, 650–657 [DOI] [PubMed] [Google Scholar]
- 10. Mo J. S., Kim M. Y., Ann E. J., Hong J. A., Park H. S. (2008) Cell Death Differ. 15, 1030–1041 [DOI] [PubMed] [Google Scholar]
- 11. Inden M., Taira T., Kitamura Y., Yanagida T., Tsuchiya D., Takata K., Yanagisawa D., Nishimura K., Taniguchi T., Kiso Y., Yoshimoto K., Agatsuma T., Koide-Yoshida S., Iguchi-Ariga S. M., Shimohama S., Ariga H. (2006) Neurobiol Dis. 24, 144–158 [DOI] [PubMed] [Google Scholar]
- 12. Taira T., Saito Y., Niki T., Iguchi-Ariga S. M., Takahashi K., Ariga H. (2004) EMBO Rep 5, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canet-Avilés R. M., Wilson M. A., Miller D. W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M. J., Ringe D., Petsko G. A., Cookson M. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokota T., Sugawara K., Ito K., Takahashi R., Ariga H., Mizusawa H. (2003) Biochem. Biophys. Res. Commun. 312, 1342–1348 [DOI] [PubMed] [Google Scholar]
- 15. Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T. M., Thomas B., Ko H. S., Sasaki M., Ischiropoulos H., Przedborski S., Dawson T. M., Dawson V. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blackinton J., Lakshminarasimhan M., Thomas K. J., Ahmad R., Greggio E., Raza A. S., Cookson M. R., Wilson M. A. (2009) J. Biol. Chem. 284, 6476–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henchcliffe C., Beal M. F. (2008) Nat. Clin. Pract Neurol. 4, 600–609 [DOI] [PubMed] [Google Scholar]
- 18. Moore D. J., West A. B., Dawson V. L., Dawson T. M. (2005) Annu. Rev. Neurosci. 28, 57–87 [DOI] [PubMed] [Google Scholar]
- 19. Kamp F., Exner N., Lutz A. K., Wender N., Hegermann J., Brunner B., Nuscher B., Bartels T., Giese A., Beyer K., Eimer S., Winklhofer K. F., Haass C. (2010) EMBO J. 29, 3571–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Irrcher I., Aleyasin H., Seifert E. L., Hewitt S. J., Chhabra S., Phillips M., Lutz A. K., Rousseaux M. W., Bevilacqua L., Jahani-Asl A., Callaghan S., MacLaurin J. G., Winklhofer K. F., Rizzu P., Rippstein P., Kim R. H., Chen C. X., Fon E. A., Slack R. S., Harper M. E., McBride H. M., Mak T. W., Park D. S. (2010) Hum. Mol. Genet. 19, 3734–3746 [DOI] [PubMed] [Google Scholar]
- 21. Hao L. Y., Giasson B. I., Bonini N. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9747–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomas K. J., McCoy M. K., Blackinton J., Beilina A., van der Brug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M. R. (2011) Hum. Mol. Genet. 20, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoy M. K., Cookson M. R. (2011) Autophagy 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cookson M. R. (2010) Mov. Disord. 25, Suppl. 1, S44–S48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L., Shimoji M., Thomas B., Moore D. J., Yu S. W., Marupudi N. I., Torp R., Torgner I. A., Ottersen O. P., Dawson T. M., Dawson V. L. (2005) Hum. Mol. Genet. 14, 2063–2073 [DOI] [PubMed] [Google Scholar]
- 26. Shinbo Y., Niki T., Taira T., Ooe H., Takahashi-Niki K., Maita C., Seino C., Iguchi-Ariga S. M., Ariga H. (2006) Cell Death Differ. 13, 96–108 [DOI] [PubMed] [Google Scholar]
- 27. Blackinton J., Ahmad R., Miller D. W., van der Brug M. P., Canet-Avilés R. M., Hague S. M., Kaleem M., Cookson M. R. (2005) Brain Res. Mol. Brain Res. 134, 76–83 [DOI] [PubMed] [Google Scholar]
- 28. Junn E., Jang W. H., Zhao X., Jeong B. S., Mouradian M. M. (2009) J. Neurosci. Res. 87, 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren H., Fu K., Mu C., Li B., Wang D., Wang G. (2010) Cancer Lett. 297, 101–108 [DOI] [PubMed] [Google Scholar]
- 30. Fan J., Ren H., Jia N., Fei E., Zhou T., Jiang P., Wu M., Wang G. (2008) J. Biol. Chem. 283, 4022–4030 [DOI] [PubMed] [Google Scholar]
- 31. Kanli A., Savli H. (2007) Exp. Oncol. 29, 314–316 [PubMed] [Google Scholar]
- 32. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 33. Chao D. T., Korsmeyer S. J. (1998) Annu. Rev. Immunol. 16, 395–419 [DOI] [PubMed] [Google Scholar]
- 34. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 35. Cheng E. H., Levine B., Boise L. H., Thompson C. B., Hardwick J. M. (1996) Nature 379, 554–556 [DOI] [PubMed] [Google Scholar]
- 36. Yin X. M., Oltvai Z. N., Korsmeyer S. J. (1994) Nature 369, 321–323 [DOI] [PubMed] [Google Scholar]
- 37. Jeong S. Y., Gaume B., Lee Y. J., Hsu Y. T., Ryu S. W., Yoon S. H., Youle R. J. (2004) EMBO J. 23, 2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 39. Zhang H., Rosdahl I. (2006) Int. J. Oncol. 28, 661–666 [PubMed] [Google Scholar]
- 40. Park K., Lee J. H. (2009) Oncol. Rep. 21, 689–692 [PubMed] [Google Scholar]
- 41. Burlacu A. (2003) J. Cell Mol. Med. 7, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giaime E., Sunyach C., Druon C., Scarzello S., Robert G., Grosso S., Auberger P., Goldberg M. S., Shen J., Heutink P., Pouysségur J., Pagès G., Checler F., Alves da Costa C. (2010) Cell Death Differ 17, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bretaud S., Allen C., Ingham P. W., Bandmann O. (2007) J. Neurochem 100, 1626–1635 [DOI] [PubMed] [Google Scholar]
- 44. Cookson M. R. (2005) Annu. Rev. Biochem. 74, 29–52 [DOI] [PubMed] [Google Scholar]
- 45. Ethell D. W., Fei Q. (2009) Antioxid. Redox Signal. 11, 529–540 [DOI] [PubMed] [Google Scholar]
- 46. Vila M., Jackson-Lewis V., Vukosavic S., Djaldetti R., Liberatore G., Offen D., Korsmeyer S. J., Przedborski S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tatton N. A. (2000) Exp. Neurol. 166, 29–43 [DOI] [PubMed] [Google Scholar]
- 48. Ziv I., Offen D., Haviv R., Stein R., Panet H., Zilkha-Falb R., Shirvan A., Barzilai A., Melamed E. (1997) Apoptosis 2, 149–155 [DOI] [PubMed] [Google Scholar]
- 49. Savitt J. M., Jang S. S., Mu W., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 6721–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dietz G. P., Stockhausen K. V., Dietz B., Falkenburger B. H., Valbuena P., Opazo F., Lingor P., Meuer K., Weishaupt J. H., Schulz J. B., Bähr M. (2008) J. Neurochem. 104, 757–765 [DOI] [PubMed] [Google Scholar]
- 51. Gross A., Jockel J., Wei M. C., Korsmeyer S. J. (1998) EMBO J. 17, 3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolter K. G., Hsu Y. T., Smith C. L., Nechushtan A., Xi X. G., Youle R. J. (1997) J. Cell Biol. 139, 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., Wang X. (1997) Science 275, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 54. Feng Y., Zhang L., Hu T., Shen X., Ding J., Chen K., Jiang H., Liu D. (2009) Arch Biochem. Biophys. 484, 46–54 [DOI] [PubMed] [Google Scholar]
- 55. Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 56. Clements C. M., McNally R. S., Conti B. J., Mak T. W., Ting J. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15091–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]