Abstract
Listeria monocytogenes is a Gram-positive intracellular bacterial pathogen that colonizes the cytosol of eukaryotic cells. Recent transcriptomic studies have revealed that intracellular L. monocytogenes alter expression of genes encoding envelope components. However, no comparative global analysis of this cell wall remodeling process is yet known at the protein level. Here, we used high resolution mass spectrometry to define the cell wall proteome of L. monocytogenes growing inside epithelial cells. When compared with extracellular bacteria growing in a nutrient-rich medium, a major difference found in the proteome was the presence of the actin assembly-inducing protein ActA in peptidoglycan purified from intracellular bacteria. ActA was also identified in the peptidoglycan of extracellular bacteria growing in a chemically defined minimal medium. In this condition, ActA maintains its membrane anchoring domain and promotes efficient bacterial entry into nonphagocytic host cells. Unexpectedly, Internalin-A, which mediates entry of extracellular L. monocytogenes into eukaryotic cells, was identified at late infection times (6 h) as an abundant protein in the cell wall of intracellular bacteria. Other surface proteins covalently bound to the peptidoglycan, as Lmo0514 and Lmo2085, were detected exclusively in intracellular and extracellular bacteria, respectively. Altogether, these data provide the first insights into the changes occurring at the protein level in the L. monocytogenes cell wall as the pathogen transits from the extracellular environment to an intracytosolic lifestyle inside eukaryotic cells. Some of these changes include alterations in the relative amount and the mode of association of certain surface proteins.
Keywords: Bacteria, Cell Wall, Membrane Proteins, Pathogen-associated Molecular Pattern, Proteomics
Introduction
The cell wall is the major interface of the bacterial cell with the environment. This macromolecular structure acts as a platform in which processes central for bacterial physiology, such as nutrient transport, protein secretion, assembly of organelles required for motility, and adhesion to biotic and abiotic surfaces, take place (1). The chemistry of the cell wall is based on a giant peptidoglycan macromolecule of repeated N-acetylglucosamine-N-acetylmuramic disaccharide units cross-linked by peptidic side chains bound to the N-acetylmuramic moiety (2). In Gram-positive bacteria, this peptidoglycan scaffold is decorated with proteins, polysaccharides, and teichoic acids, which associate by rather varied mechanisms, including covalent and noncovalent bonds (3, 4). In Gram-positive bacteria, the cell wall reaches a thickness of 20–80 nm and contacts the external milieu (2, 3).
Many surface proteins of Gram-positive bacterial pathogens anchored to the cell wall play essential roles in the infection process (3, 5, 6). Pioneering studies performed in Staphylococcus aureus unraveled the presence of surface proteins covalently bound to the peptidoglycan that allow the pathogen to avoid the host immune response (7). Subsequent data obtained in other Gram-positive bacterial pathogens reinforced the important role of the cell wall in the interaction with the host and the dynamic nature of this structure in terms of the number and type of molecules expressed at a certain time and location (7, 8).
One of the major goals in deciphering the biology of these pathogens is to define the number and type of proteins located in the surface at a certain time or environment. Proteomics based studies have recently identified many novel surface proteins annotated as new bacterial genome sequences became available (9). The combination of gel-free and gel-based proteomic approaches has provided insights on metabolic adaptation and environmental sensing (10). Two-dimensional gel-based proteomics is commonly used to assess changes in proteins levels in response to environmental changes and to analyze post-translational modifications and degradation rates. Instead, gel-free proteomics is mostly applied to identify bacterial proteins located in the envelope, including those inserted in the membrane or directly associated to the peptidoglycan (reviewed in Ref. 9).
Gram-positive bacteria of the genus Listeria contain a large variety of surface proteins that associate with the cell wall (5). Bacteria belonging to this genus carry the largest family of proteins predicted to be covalently anchored to the peptidoglycan upon cleavage of a C-terminal LPXTG sorting motif (3). More than 40 genes encoding LPXTG proteins have been annotated in every Listeria genome sequenced to date (11–13). This feature is shared by nonpathogenic and pathogenic species. Some LPXTG proteins and other surface proteins with predicted noncovalent association to the cell wall are found exclusively in pathogenic species of the genus, as the intracellular pathogen Listeria monocytogenes (14). Many of these proteins are important virulence factors involved in promoting entry of the bacteria into the host cell (reviewed in Refs. 5, 15, 16).
Our previous gel-free proteomics studies, based on nano-liquid chromatography coupled to mass spectrometry, identified a total of 13 LPXTG proteins strongly associated to the peptidoglycan of L. monocytogenes growing in brain-heart infusion (BHI)2 medium (17, 18). Such analyses were possible due to the covalent anchoring of these proteins to the peptidoglycan, which facilitates the co-purification of these molecules upon extensive boiling of envelope material in SDS-containing solutions. Although not yet addressed in L. monocytogenes, it is assumed that changes in environmental conditions should lead to modification in the repertoire of LPXTG proteins anchored to the peptidoglycan. Of interest, differences in the type of virulence-related proteins synthesized by L. monocytogenes have been reported in BHI-rich medium compared with a minimal medium containing distinct fermentable or nonfermentable carbohydrates (19). For instance, the activity of the master virulence regulator of L. monocytogenes PrfA, which controls expression of functions involved in virulence such as the listeriolysin LLO and the phospholipases PlcA and PlcB (15, 20, 21), differs in these distinct growth conditions (19). PrfA also modulates the expression of surface proteins, including proteins bearing an LPXTG-sorting motif, GW-rich modules, and the membrane protein ActA, which induces host actin assembly (15, 20, 21).
The aim of this study was to exploit the most recent proteomic tools to detect changes in the L. monocytogenes cell wall proteome. Highly sensitive gel-free proteomic technology, based on high resolution mass spectrometry (LTQ Orbitrap MS), was used to define the cell wall proteome of intracellular bacteria growing inside eukaryotic cells. This proteome was compared with that of the infecting extracellular bacteria. Besides the identification of novel LPXTG proteins, such as Lmo0514, other surface proteins not previously known to strongly associate with peptidoglycan were detected. This was the case of the actin-binding protein ActA. Such ActA-peptidoglycan association was observed in intracellular bacteria and in bacteria growing in a chemically defined minimal medium. Of note, the association of ActA to the peptidoglycan was found in growth conditions in which the bacteria use this protein to invade cultured nonphagocytic eukaryotic cells. Conversely, a surface protein used by L. monocytogenes for entry into eukaryotic cells as the invasin Internalin-A (InlA) was enriched in the cell wall of intracellular bacteria at late post-infection times. Altogether, these data unravel for the first time the cell wall proteome of a Gram-positive bacterial pathogen growing inside eukaryotic cells and are consistent with major remodeling of the L. monocytogenes cell wall as the bacteria adapt to grow in the eukaryotic cytosol.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The L. monocytogenes serotype 1/2a strains used in this study were EGDe, with genome sequenced (12), and the isogenic mutant derivatives ΔinlA2, ΔinlB2, and ΔactA2 (22, 23). The L. monocytogenes strain P14-A, which carries a gain-of-function variant of PrfA resulting in overexpression of actA and other virulence genes of the PrfA regulon (24), was also used. Bacteria were grown at 37 °C in BHI medium. When indicated, the chemically defined minimal medium IMM (25) supplemented with 17.9 μm ferric citrate (C6H5O7Fe) was used.
Isolation of Intracellular L. monocytogenes for Cell Wall Proteomic Analysis
The human epithelial cell line JEG-3 (ATCC HTB-36), obtained from a choriocarcinoma of placenta, was selected for these studies. JEG-3 cells express InlA and InlB receptors and are efficiently invaded by L. monocytogenes (26). These cells were propagated in Eagle's minimal essential medium (MEM) containing 10% (v/v) fetal bovine serum (FBS), 4 mm l-glutamine, and nonessential amino acids in BioDish-XL plates (BD Biosciences, product 351040) to reach ∼80% confluency (∼5.6 × 107 epithelial cells). At this stage, the epithelial cells contained in five BioDish-XL plates were infected for 50 min with L. monocytogenes EGDe strain grown overnight in BHI medium at 37 °C in static nonshaking conditions. Before infection, bacteria were centrifuged and suspended in tissue culture medium. The multiplicity of infection used in these assays was 10:1 (bacteria/epithelial cell). Noninternalized bacteria were removed by one wash with pre-warmed phosphate-buffered saline (PBS), pH 7.4. Fresh MEM, 10% FBS medium containing 50 μg/ml gentamicin was added. At 2 h post-infection, gentamicin concentration was lowered to 10 μg/ml by appropriate dilution with pre-warmed MEM, 10% FBS medium. At 6 h post-infection, the infected JEG-3 cells were washed three times with PBS, pH 7.4, and lysed in 10 ml of boiling 1% SDS per BioDish-XL plate. The material remaining in the plate was collected with additional 10 ml of boiling 1% SDS. The 20-ml solution was centrifuged at 40,000 × g, 30 min, at 30 °C, and the supernatant was discarded. The pellet was resuspended in 1 ml of PBS, pH 7.4, and washed once at 30,000 × g, 10 min, at room temperature. The pellet for each BioDish-XL plate was finally suspended in 200 μl of PBS. This sample was pooled with those obtained from the remaining four BioDish-XL plates and centrifuged at 30,000 × g, 10 min, 4 °C. The pellet, containing intracellular L. monocytogenes, was kept at −80 °C. JEG-3 cells contained in a total of 100 BioDish-XL plates were infected in 20 independent experiments. These pellets, enriched in intracellular bacteria, were further pooled and resuspended in 20 ml of PBS, pH 7.4, containing a mixture of protease inhibitors (Roche Applied Science) and DNase 100 μg/ml. Bacteria were lysed in three passes through a French press. Unbroken cells were removed by centrifugation (5,000 × g, 5 min, 4 °C) and cell envelopes obtained by centrifugation of supernatant at a high speed (140,000 × g, 45 min, 4 °C). The envelopes contained in the pellet were resuspended in 1.5 ml of PBS, pH 7.4, and gently mixed with 1.5 ml of boiling 8% SDS. The SDS-insoluble material, enriched in peptidoglycan and strongly associated proteins, was collected by centrifugation at high speed (300,000 × g, 20 min, 30 °C) and washed three times with 2.5 ml of warm distilled water. This purified peptidoglycan material was digested with modified trypsin (sequencing grade, Promega, Madison, WI) as described previously (18). The resulting peptide mixture was lyophilized and kept at −20 °C.
Preparation of Peptidoglycan from Bacterial Cultures for Proteomic Analysis
Peptidoglycan was purified from L. monocytogenes EGDe grown in laboratory media as described previously (18). Briefly, 1011 bacteria grown to stationary phase in BHI or IMM media were spun down by centrifugation (10,000 × g, 10 min, 4 °C) and resuspended in 20 ml of PBS, pH 7.4, containing a mixture of protease inhibitors (Roche Applied Science) and 100 μg/ml DNase. Bacterial lysis, preparation of envelope material, peptidoglycan purification, and trypsin digestion were performed as described above for intracellular bacteria. The resulting peptide mixture was lyophilized and kept at −20 °C.
Protein Identification by Liquid Chromatography-Tandem Mass Spectrometry and Data Processing
The tryptic peptide mixtures obtained from peptidoglycan material collected from intracellular or extracellular L. monocytogenes were dissolved in 0.5% acetic acid in water. This solution was cleaned up with cartridges that trap remaining SDS (Michrom BioResources, Auburn, CA) as described previously (18). Peptides were then injected onto a C-18 reversed phase nano-column (100 μm inner diameter × 12 cm, MediterraneaTM Sea, Teknokroma) and analyzed in a continuous acetonitrile gradient consisting of 0–40% B in 90 min and 50–90% B in 1 min (B = 95% acetonitrile, 0.5% acetic acid). A flow rate of ∼300 nl/min was used to elute peptides from the reverse phase nano-column to an emitter nanospray needle for real time ionization and peptide fragmentation on an LTQ-Orbitrap XL ETD mass spectrometer (Thermo Fisher Scientific, San Jose, CA). An enhanced FT resolution spectrum (resolution = 60,000) followed by the MS/MS spectra from the most intense five parent ions were analyzed along the chromatographic run (130 min). Dynamic exclusion was set at 0.5 min. For protein identification, tandem mass spectra were extracted, and charge state was deconvoluted by Proteome Discoverer version 1.0. All MS/MS samples were analyzed using SEQUESTTM (Thermo Fisher Scientific, version 1.0.43.2) and X! Tandem (The GPM, version 2007.01.01.1). Sequest was set up to search an in-house-generated protein database (Listeria_EGDe.fasta, 2846 entries) obtained from the genome sequence entry of L. monocytogenes strain EGDe (accession number NC_003210.1) and assuming the digestion enzyme trypsin. As compilation software, Protein Discoverer version 1.0 was used. X! Tandem was set up to search a subset of the Listeria_EGDe database also assuming trypsin. Sequest and X! Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 15 ppm. Oxidation of methionine was specified in Sequest and X! Tandem as variable modification. Scaffold (version Scaffold_3_00_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (27). Protein identifications were accepted if they could be established at greater than 95% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (28). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The entire set of MS data were compiled using the Scaffold program and is available as supplemental material (CompleteMSdata_BHI_IMM_intracellular).
Selective Reaction Monitoring (SRM) Experiments by Triple-quadrupole Tandem Mass Spectrometry
For these experiments, tryptic peptide mixtures were analyzed with a hybrid triple-quadrupole mass spectrometer (4000 Q-Trap system, Applied Biosystems). The samples were injected onto a C-18 reversed phase nano-column (100 μm inner diameter × 12 cm, MediterraneaTM Sea, Teknokroma) and analyzed in a continuous acetonitrile gradient consisting of 0–40% B in 95 min, 50–90% B in 1 min (B = 95% acetonitrile, 0.5% acetic acid). Multiply charged monoisotopic masses from the Listeria peptides of interest were selected to be monitored in the quadrupole Q1, whereas the corresponding diagnostically derived fragment ions (yielded in the collision-induced dissociation taken place within quadrupole Q2) were targeted in the third quadrupole (Q3). After the SRM analyses of selected pairs (Q1/Q3), an enhanced resolution scan (for charge and mass determination) and an enhanced product ion scan (for sequence collision-induced dissociation fragmentation) were analyzed along the chromatographic run (130 min). The parameters used for the combined analysis were as follows: collision energy for SRM scans 50 V, de-clustering potential 120, entrance potential 10. The ion spray voltage was set to 3000, the interface heater temperature was 130, the curtain gas 20, and the collision gas was set to high. For protein identification, all MS/MS spectra were searched against MSDB database using the Analyst 1.5.1 software (Applied Biosystems).
Subcellular Fractionation and Western Blot Analyses of Peptidoglycan-associated Proteins in Extracellular and Intracellular L. monocytogenes
Fractions containing cytosolic, membrane, and cell wall proteins of L. monocytogenes grown overnight at 37 °C in BHI or IMM media were obtained as described previously (29). Peptidoglycan-associated proteins were obtained by two methods depending on the sample as follows: muramidase (Cellosyl) treatment of previously purified peptidoglycan material (30) or, alternatively, mutanolysin treatment on intact bacteria (17). Cellosyl and mutanolysin are N-acetylmuramidases isolated from different Streptomyces species that cleave the β(1–4)-O-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine moieties. Unless otherwise indicated, Cellosyl treatment was used to characterize proteins associated with peptidoglycan in subcellular fractionation experiments involving mechanical rupture of bacteria (30). To compare levels of peptidoglycan-associated proteins in intracellular and extracellular bacteria and, due the much lower amount of bacteria obtained from inside eukaryotic cells, bacterial fractionation was avoided, and instead mutanolysin was used to digest peptidoglycan in intact cells. Control experiments performed in extracellular bacteria indicated that no difference in the relative amount of representative cell wall-associated proteins, as InlA, was observed when using either Cellosyl or mutanolysin to digest the peptidoglycan (data not shown). For the case of intracellular bacteria, JEG-3 epithelial cells grown in five BioDish-XL plates were infected with L. monocytogenes EGDe strain up to 6 h post-infection as aforementioned. After three washes with 20 ml of PBS, pH 7.4, the infected cells were lysed in a total volume of 20 ml of a 1% SDS boiling solution (31). Upon centrifugation at 40,000 × g, 30 min, room temperature, the pellet containing intracellular bacteria was washed once with PBS, pH 7.4, and further resuspended in 0.5 ml of TS buffer (10 mm MgCl2, 10 mm Tris-HCl, pH 6.9, 0.5 m sacarose). Mutanolysin (Sigma) 40 μg/ml, RNase 250 μg/ml, and a mixture of protease inhibitors (Roche Applied Science) were added. The sample was then incubated overnight at 37 °C under shaking conditions. The next day, the sample was centrifuged (30,000 × g, 15 min, 4 °C), and the supernatant containing peptidoglycan-associated proteins was processed for Western blot analysis as described previously (17). Identical mutanolysin digestion conditions were followed to release peptidoglycan-associated proteins from extracellular bacteria. Antibodies used included the following: mouse monoclonal antibody anti-InlA (clone I4-4); mouse monoclonal antibody anti-InlB (clone B4-6); mouse monoclonal anti-ActA (clone M119); and rabbit polyclonal sera recognizing the LPXTG proteins Lmo0130, Lmo0160, Lmo0610, Lmo0514, Lmo0610, and Lmo0880. These latter antisera were obtained by immunization of rabbits with purified recombinant His6-tagged LPXTG proteins overproduced in Escherichia coli BL21. The details of this immunization will be published elsewhere. Protein separation by SDS-PAGE and Western immunoblotting were performed as described previously (29).
Infection of Eukaryotic Cell Lines and Determination of Invasion Rates
The epithelial cell lines HeLa (ATCC CCL-2) and Vero (ATCC CCL-81) of human and monkey origin, respectively, were used. NRK-49F (ATCC CRL-1570) rat fibroblasts were also included as representative of other nonphagocytic cell type. HeLa and Vero cells were propagated in MEM containing 10% FBS. NRK-49F fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) 5% FBS containing 4 mm l-glutamine. The L. monocytogenes strains were grown overnight at 37 °C in BHI medium or the chemically defined medium IMM in static nonshaking conditions. Under these conditions, the culture reaches an optical density of ∼1.0 (∼109 bacteria/ml) in BHI and ∼0.8 in IMM. Prior to infection, bacteria were spun down at 6,000 × g, 2 min at room temperature, resuspended in the appropriate tissue culture medium, and added at a m.o.i. of 10:1 (bacteria/eukaryotic cell). Co-incubation of bacteria and eukaryotic cells was maintained for 60 min. Noninternalized bacteria were removed by one wash with pre-warmed Hanks' balanced salt solution, pH 7.4. Fresh tissue culture medium containing 100 μg/ml gentamicin was then added to kill extracellular bacteria. Incubation was prolonged for another 60 min, the time in which infected cells were lysed with 100 μl of 1% Triton X-100–0.1% SDS for 5 min at room temperature. Upon addition of 400 μl of PBS, pH 7.4, the mixture was plated onto BHI-agar plates using appropriate dilutions. The invasion rate was calculated as the ratio of viable intracellular bacteria versus the bacteria present in the inoculum used to infect the cultured cells. The infection assays were repeated in a minimum of three independent experiments.
Transmission Electron Microscopy
Samples were prepared from L. monocytogenes wild-type strain EGDe bacteria grown to exponential or stationary growth phases in BHI or minimal medium IMM at 37 °C. The cells were spun down gently and suspended in 1 ml of solution of fixative (2% solution of glutaraldehyde in 0.4 m HEPES buffer, pH 7.2) for 2 h at room temperature. The cells were further spun down, suspended in 0.4 m HEPES buffer, and kept at 4 °C. These cells were further dehydrated, embedded in LR-White resin, sectioned, and contrasted as described previously (32). Samples were visualized at 80 kV in a JEOL1200EX electron microscope.
Immunofluorescence Analysis
To analyze ActA location in the cell surface, bacteria were grown in BHI-rich medium or minimal medium IMM at 37 °C to stationary phase (OD ∼2.0). Bacteria were washed twice in PBS, pH 7.4, and fixed in 3% paraformaldehyde for 10 min at room temperature. Antibody labeling was performed essentially as described previously (33). Primary antibodies used included the aforementioned anti-InlA and anti-ActA antibodies and, in addition, a rabbit polyclonal serum recognizing Listeria proteins that co-purify with peptidoglycan (30). Secondary antibodies included goat anti-mouse Alexa-488 (green fluorescence) and goat anti-rabbit Alexa-594 (red fluorescence). Samples were examined in a Leica fluorescence inverted microscope DMI 6000B.
Statistical Analysis
Data were analyzed with the GraphPad Prism 5.0 software (GraphPad Inc., San Diego) using two-way analysis of variance with Bonferroni's post-test. A p value lower than 0.05 was considered significant.
RESULTS
Identification of Surface Proteins in Cell Wall of Intracellular L. monocytogenes by High Resolution Mass Spectrometry (MS)
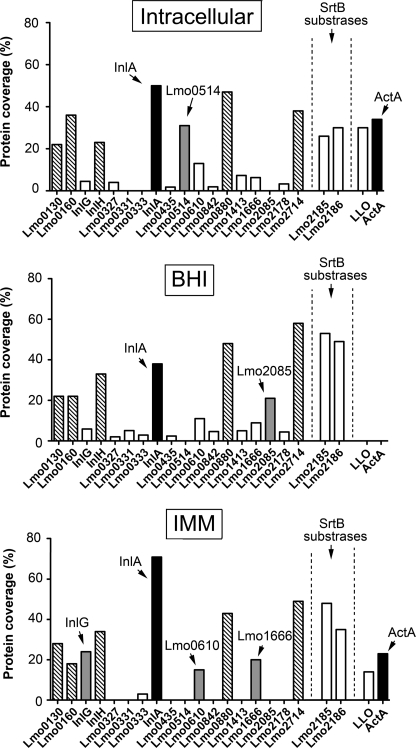
To identify cell wall-associated proteins in intracellular L. monocytogenes, peptidoglycan was isolated at 6 h post-infection from wild-type strain EGDe growing inside human epithelial cells (see “Experimental Procedures”). High resolution MS of the tryptic peptide mixture obtained from this material led to the unequivocal identification of 53 proteins (supplemental Table S1). The most abundant classes identified in this proteome included surface proteins covalently bound to peptidoglycan such as those bearing the LXPTG motif, the two sortase-B substrates Lmo2185 and Lmo2186 (34), and enzymes related to peptidoglycan metabolism (Table 1). A total of 15 LPXTG proteins covalently bound to the peptidoglycan were identified in the cell wall of intracellular bacteria (Table 1). Interestingly, a surface protein not previously reported to be associated with the peptidoglycan, the actin assembly-inducing protein ActA, and the secreted protein listeriolysin-O (LLO) were also identified by proteomics in the peptidoglycan isolated from intracellular bacteria with 19 and 11 unique peptides, respectively (Table 1). These data suggested that these two proteins, known in extracellular bacteria to be anchored to the membrane (ActA) or secreted (LLO), could associate with the cell wall during the intracellular phase of the infection. To test this hypothesis, peptidoglycan-associated proteins were identified by high resolution MS in peptidoglycan isolated from extracellular bacteria growing exponentially in BHI medium. A total of 170 protein species was identified in this case (supplemental Table S1). Similarly to the cell wall proteome obtained in intracellular bacteria, many of the most abundant proteins identified in extracellular bacteria were proteins covalently bound to peptidoglycan or involved in peptidoglycan metabolism (Table 1). Together with the SrtB substrates Lmo2185 and Lmo2186, a total of 17 LPXTG proteins were identified in extracellular bacteria. However, neither ActA nor LLO were identified in the cell wall of extracellular bacteria growing in BHI, despite the fact that in this condition the bacteria produce both surface proteins (see below). The comparison of cell wall proteomes between intracellular and extracellular bacteria revealed other differences, especially in the LPXTG protein profile. For example, the LPXTG protein Lmo0514 was identified exclusively in intracellular bacteria (Table 1). This observation indicated that synthesis of Lmo0514 could respond to signals found by the bacteria in the eukaryotic cytosol. Conversely, the LPXTG proteins Lmo0331, Lmo0333, and Lmo2085 were detected only in extracellular bacteria (Table 1). This comparative proteomic analysis also allowed us to differentiate a subset of “abundant” LPXTG proteins in the cell wall of L. monocytogenes, which were identified with a high coverage irrespective of the growth condition analyzed. These include Lmo0130, Lmo0160, Lmo0263 (InlH), Lmo0433 (InlA), Lmo0880, Lmo1666, and Lmo2714 (Table 1). Given the function of InlA in promoting entry of extracellular bacteria into eukaryotic cells (16), it was rather unexpected to identify this protein in the surface of intracellular bacteria 6 h after entry. Taken together, these proteomic data favored the existence of substantial changes in the cell wall proteome of L. monocytogenes when bacteria grow in the cytosol of epithelial cells.
TABLE 1.
Surface and virulence-related proteins of L. monocytogenes EGDe strain identified by high resolution MS in the cell wall of bacteria growing in the cytosol of JEG-3 epithelial cells (intracellular, 6 h post-infection) or in BHI medium (extracellular)
| Protein | Function | Mass | Intracellular (6 h, epithelial cells) |
Extracellular (BHI medium) |
||
|---|---|---|---|---|---|---|
| Unique peptides | Coverage | Unique peptides | Coverage | |||
| Da | % | % | ||||
| Lmo0130 | Similar to 5-nucleotidase, putative peptidoglycan-bound protein (LPXTG motif) | 82,511 | 12 | 22.0 | 15 | 22.0 |
| Lmo0160 | Putative peptidoglycan-bound protein (LPXTG motif) | 62,374 | 14 | 36.0 | 10 | 22.0 |
| Lmo0262 | Internalin G (InlG) | 53,323 | 2 | 4.5 | 2 | 5.9 |
| Lmo0263 | Internalin H (InlH) | 58,683 | 8 | 23.0 | 13 | 33.0 |
| Lmo0327 | Similar to cell surface proteins (LPXTG motif) | 146,726 | 4 | 4.0 | 2 | 1.3 |
| Lmo0331 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif) | 69,488 | 2 | 5.1 | ||
| Lmo0333 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif). Internalin I (InlI) | 191,652 | 4 | 2.9 | ||
| Lmo0433 | Internalin A (inlA) | 86,497 | 36 | 50.0 | 21 | 38.0 |
| Lmo0435 | Putative peptidoglycan-bound protein (LPXTG motif) | 219,391 | 2 | 1.8 | 2 | 2.4 |
| Lmo0514 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif) | 66,033 | 13 | 31.0 | ||
| Lmo0610 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif) | 62,843 | 5 | 13.0 | 5 | 11.0 |
| Lmo0842 | Putative peptidoglycan bound protein (LPXTG motif) | 222,364 | 3 | 1.9 | 6 | 4.6 |
| Lmo0880 | Similar to wall-associated protein precursor (LPXTG motif) | 49,879 | 16 | 47.0 | 21 | 48.0 |
| Lmo1413 | Similar to internalin, putative peptidoglycan-bound protein (LPXTG motif) | 48,248 | 3 | 7.3 | 2 | 5.0 |
| Lmo1666 | Peptidoglycan linked protein (LPXTG) | 184,529 | 8 | 6.3 | 12 | 8.9 |
| Lmo2085 | Putative peptidoglycan-bound protein (LPXTG motif) | 60,458 | 11 | 21.0 | ||
| Lmo2178 | Putative peptidoglycan-bound protein (LPXTG motif) | 175,077 | 4 | 3.3 | 6 | 5.4 |
| Lmo2714 | Peptidoglycan-anchored protein (LPXTG motif) | 34,669 | 11 | 38.0 | 20 | 58.0 |
| Lmo2185 | Unknown, sortase-B substrate | 63,381 | 9 | 26 | 30 | 53.0 |
| Lmo2186 | Unknown, sortase-B substrate | 22,267 | 4 | 30 | 11 | 49 |
| Lmo0202 | Listeriolysin O (LLO) precursor | 58,689 | 11 | 30.0 | ||
| Lmo0204 | Actin assembly-inducing protein precursor (ActA) | 70,351 | 19 | 34.0 | ||
| Lmo0582 | P60 extracellular protein, invasion-associated protein Iap-containing LysM-modules | 50,340 | 5 | 17.0 | 6 | 17.0 |
| Lmo1941 | Unknown, LysM domain-containing protein | 25,836 | 2 | 10.0 | 1 | 5.9 |
| Lmo2505 | Peptidoglycan lytic protein P45 (Spl) | 42,712 | 9 | 26.0 | 3 | 10.2 |
| Lmo2691 | Similar to autolysin, N-acetylmuramidase-containing LysM domain | 63,574 | 3 | 9.8 | 3 | 11.3 |
InlA Protein Is Up-regulated by Intracellular Bacteria
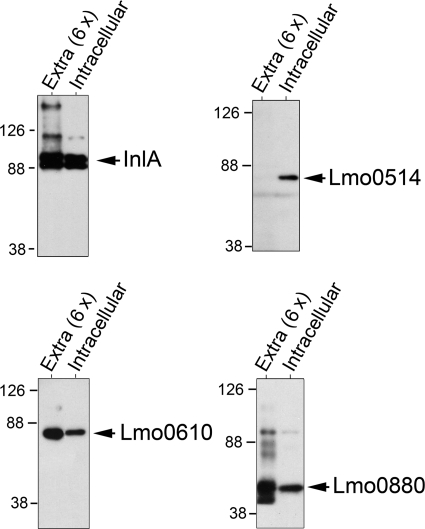
A transcriptomic study performed by Chatterjee et al. (35) showed up-regulation of the inlA gene in bacteria upon infection of murine macrophages. This finding was in line with the known up-regulation of the PrfA regulon occurring in intracellular bacteria, which mediate lysis of the phagosomal membrane and cell-to-cell spread of the pathogen (20, 21). Our proteomic analyses were also in concordance with the transcriptomic data. Thus, InlA was identified in the cell wall of intracellular bacteria with a protein coverage of 50% of the full-length polypeptide, whereas such coverage was of 38% in extracellular bacteria (Table 1). We hypothesized that such difference in the identification rate could reflect distinct protein levels in the cell wall. To test this assumption, extracts containing surface proteins associated with the peptidoglycan were prepared from intracellular and extracellular bacteria using the mutanolysin-digestion procedure (see “Experimental Procedures”). The levels of the LPXTG proteins Lmo0514, InlA, Lmo0610, and Lmo0880 were determined by Western blot. In concordance with the proteomic data, we observed that Lmo0514 was produced exclusively by intracellular bacteria and that the amount of InlA slightly increased in these bacteria (Fig. 1). In contrast, the levels of both Lmo0610 and Lmo0880 remained constant in extracellular and intracellular bacteria. These data confirmed that intracellular bacteria alter the relative amounts of some LPXTG proteins covalently bound to the peptidoglycan with respect to extracellular bacteria growing in BHI medium.
FIGURE 1.
L. monocytogenes located in the cytosol of eukaryotic cells contain increased levels of the LPXTG proteins InlA and Lmo0514 associated with the cell wall. L. monocytogenes wild-type strain EGDe was used to infect JEG-3 epithelial cells for 6 h. A fraction enriched in proteins strongly bound to peptidoglycan in intracellular bacteria was prepared upon digestion of peptidoglycan with mutanolysin (see “Experimental Procedures”). Peptidoglycan-associated proteins were also obtained following the same digestion procedure from bacteria grown in BHI medium to the stationary phase (extracellular). Shown are the Western assays performed to detect the LPXTG proteins InlA, Lmo0514, Lmo0610, and Lmo0880. The cell wall extracts loaded in the different cases corresponded to 8 × 108 and 1.3 × 108 bacteria in extracellular and intracellular samples, respectively. Note that, considering this ∼6-fold difference in total bacterial protein, the cell wall of intracellular bacteria contains higher amounts of InlA and Lmo0514.
Characterization of Cell Wall Proteome of L. monocytogenes Grown in a Chemically Defined Minimal Medium
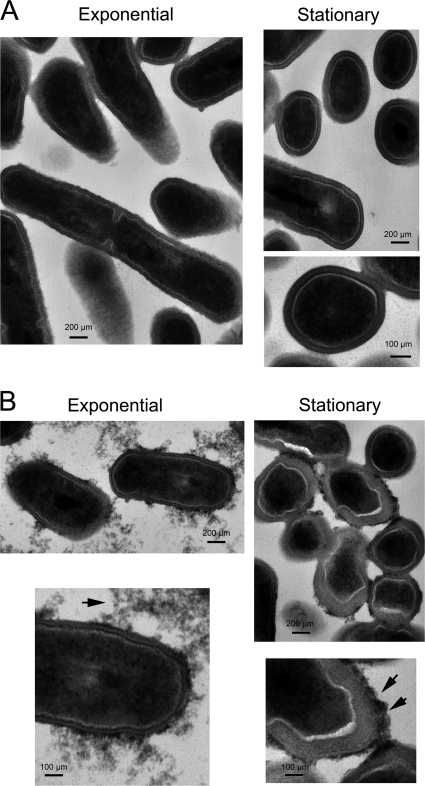
As part of the proteomic studies performed in intracellular L. monocytogenes, we sought to find extracellular conditions mimicking the intracellular infection in terms of a roughly similar composition of peptidoglycan-associated proteins. This was of significance because it could allow us to perform biochemical studies requiring large amounts of peptidoglycan, technically difficult to obtain from intracellular bacteria. For this purpose, bacteria were grown at 37 °C in the chemically defined medium IMM described to be optimal for growing Listeria (25). Examination at the electron microscope of bacteria grown in BHI and IMM media denoted drastic morphological differences in the cell wall. Thus, bacteria in BHI medium exhibited a uniform cell wall type irrespective of the growth phase (Fig. 2A), whereas those bacteria grown in minimal medium IMM contained an electro-dense material associated with the cell wall in exponential phase and a thick cell wall decorated with electro-dense material in stationary phase (Fig. 2B). These differences suggested that the structure of the cell wall could differ in these two growth conditions and, as a result, the type and relative abundance of surface proteins associated with it. To test this hypothesis, peptidoglycan was purified from bacteria growing in minimal medium IMM and the cell wall proteome defined by high resolution MS. A total of 249 proteins was identified in this growth condition. As expected, many of the most abundant proteins corresponded to LPXTG proteins, sortase-B substrates, and peptidoglycan hydrolases (supplemental Table S1). Despite identifying more proteins than in the peptidoglycan of bacteria growing in BHI medium or intracellular bacteria, the LPXTG protein repertoire in bacteria growing in IMM medium was less varied as only 10 protein species of this family were identified (Fig. 3). Interestingly, InlA predominated in the cell wall of bacteria growing in minimal medium IMM because it was the protein identified by MS with the highest coverage. This accounted for the identification of 569 amino acids of the 800 amino acids that constitute the full-length of the InlA protein (73% coverage). Considering the processing of this protein at the signal peptide (N terminus) and the cleavage of the LPXTG motif (C terminus), the coverage of the mature protein form covalently bound to the peptidoglycan reached 77%. This extremely high coverage of InlA in bacteria growing in minimal medium IMM denoted the abundance of InlA in this particular growth condition (Fig. 3).
FIGURE 2.
Different growth conditions result in morphological changes in L. monocytogenes cell wall. The wild-type strain EGDe was grown in BHI medium (A) or chemically defined minimal medium IMM (B). Bacteria were collected at an OD = 0.25 (exponential phase) or after a 16-h cessation of growth (stationary phase). Samples were processed for transmission electron microscopy as described under “Experimental Procedures.” Arrows indicate electron dense material located in the outer surface of the cell wall that is evident only in bacteria growing in IMM medium. Scale bars, 100 and 200 μm as indicated.
FIGURE 3.
Profile of L. monocytogenes surface proteins identified by proteomics in the cell wall of intracellular and extracellular bacteria. Cell wall extracts were prepared from intracellular bacteria after 6 h post-infection of JEG-3 epithelial cells or from extracellular bacteria grown in BHI or minimal medium IMM to stationary phase. These extracts were processed for high resolution MS to identify proteins associated with the peptidoglycan. Indicated is the coverage obtained in the three conditions (intracellular, BHI, and IMM) for surface proteins covalently bound to the peptidoglycan, including members of the LPXTG family and the two sortase-B (SrtB) substrates Lmo2185 and Lmo2186. The PrfA-regulated proteins ActA and LLO are also shown. LPXTG proteins with differential expression in some of the conditions tested are shown as gray bars and those identified with a high coverage in the three experimental conditions as hatched bars. InlA and ActA are highlighted as black bars.
Interestingly, ActA and LLO were also identified in the cell wall of bacteria growing in minimal medium IMM. In this case, protein coverages of 23% (ActA) and 14% (LLO) were obtained by MS (Fig. 3). These data suggested that the peptidoglycan of bacteria growing in minimal medium IMM might differ in structural terms to that synthesized by bacteria growing in BHI. Such changes could facilitate a stronger association of LLO and ActA to the peptidoglycan, as it was observed in intracellular bacteria (Table 1). However, differences must also exist in the environment found by L. monocytogenes in the minimal medium IMM and the cytosol of eukaryotic cells. Thus, Lmo0514, an LPXTG protein produced by intracellular bacteria (Table 1, Fig. 1), was not detected by proteomics in the peptidoglycan of extracellular bacteria growing in minimal medium IMM (Fig. 3). Another example was Lmo0842, an LPXTG protein identified in intracellular bacteria and bacteria grown in BHI but not in minimal medium IMM (Fig. 3). Overall, these data indicated that the cell wall of L. monocytogenes growing in minimal medium IMM might share only a few structural features with that of intracellular bacteria. These common features could support the abundance of InlA and the association to the cell wall of LLO and ActA.
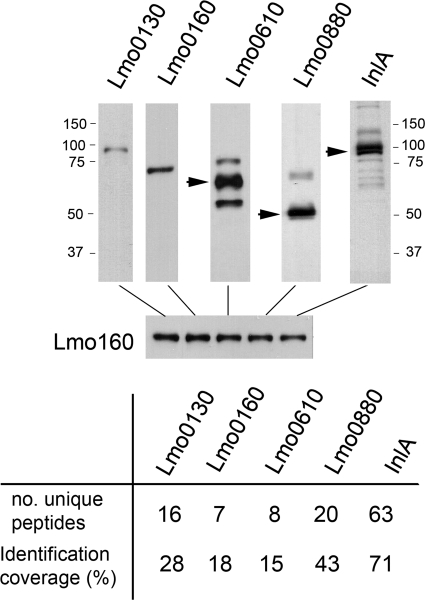
The coverage at which high resolution MS identifies a concrete polypeptide is considered proportional to the relative abundance of the protein. To assess this, we were interested in comparing this type of estimation using alternative methods. A total of five LPXTG proteins identified with higher coverage by MS (range 15–70%) were analyzed by immunological methods using specific sera (Fig. 4). To this aim, we used antibodies against the LPXTG proteins Lmo0130, Lmo0160, Lmo0610, Lmo0880, and InlA. Western analysis demonstrated that although all the proteins were detected in the cell wall extracts, the respective signals slightly varied depending on the protein species analyzed (Fig. 4). A representative example was that of Lmo0610, easily detected by Western blot but exhibiting the lowest identification rate among the set of proteins tested (Fig. 4). Differences in the affinity of the antisera used may in part account for the diverse immunological signal found. Other factors include the specific location of the LPXTG protein in the peptidoglycan, which could affect the rate at which the protein is released upon enzymatic digestion of this macromolecule. It is worth noting that LPXTG proteins of L. monocytogenes released from the peptidoglycan upon digestion with muramidases are not visible by Coomassie or silver staining (data not shown). This fact hampers the evaluation of the relative abundance of these proteins by their direct visualization in gels. Overall, these observations indicate that, although not totally overlapping, high resolution MS and immunological methods can provide semi-quantitative estimation of the relative abundance of LPXTG proteins covalently anchored to the peptidoglycan.
FIGURE 4.
Comparison of relative abundance estimated by high resolution MS and individual Western data for five LPXTG proteins identified with high coverage in proteomics: Lmo0130, Lmo0160, Lmo0610, Lmo0880, and InlA. Cell wall extracts were prepared from L. monocytogenes EGDe strain grown in minimal medium IMM to stationary phase. Note that for an equal volume of sample loaded in the gel (internal control represented by levels of Lmo0160 protein), distinct intensities are observed in the Western assays performed with the corresponding anti-Lmo0130, anti-Lmo0160, anti-Lmo0610, anti-Lmo0880, and anti-InlA antisera. Note the differences observed in comparative terms with the rest of the proteins for the case of Lmo0610, detected as a very abundant protein in the Western assay but with a lower coverage in proteomics. Sizes of the molecular weight markers used are also indicated.
ActA Associates with Peptidoglycan of Bacteria Growing in Minimal Medium IMM Maintaining Its Membrane Anchor
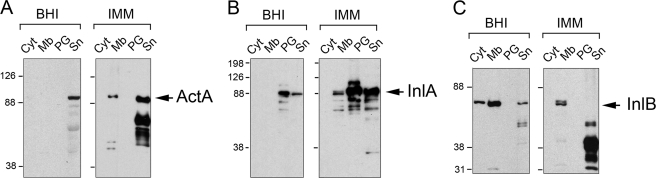
ActA is a surface protein of 639 amino acids with a membrane spanning region close to its C terminus that encompasses residues 613–635. To our knowledge, association of ActA to the peptidoglycan has not been previously reported. Based on this, we investigated whether this putative ActA-peptidoglycan association was specific. Increased production of the protein could potentially lead to unspecific retention into the cell wall during the purification procedure. However, we also reasoned that varied interactions of ActA with cell wall components could take place when bacteria grow in markedly different environmental conditions such as the cytosol of eukaryotic cells and laboratory media. Of note, several authors have reported the production in laboratory media of multiple ActA-derived breakdown products resulting from cleavage at defined sites (36, 37). Considering this fact, we assessed whether the ActA polypeptide identified by proteomics could display a different envelope association compared with ActA molecules present either in the extracellular milieu or in the membrane but not bound to the peptidoglycan. The level of ActA was examined in subcellular extracts containing proteins present in cytosol, membrane, cell wall (peptidoglycan), or culture supernatant. These extracts were prepared from extracellular bacteria growing in BHI or minimal medium IMM. Western assays shown in Fig. 5A revealed that ActA is produced at higher levels in minimal medium IMM and that the protein is present mostly in the membrane and in the extracellular medium. Interestingly, most of the ActA produced by the bacteria in BHI medium appeared to be secreted. This latter result suggested that some proteolytic activity could act on ActA to release the protein from its membrane anchor. Proteolytic activity acting on ActA was also evident in minimal medium IMM, in which several breakdown products were easily detected in the supernatant fraction (Fig. 5A).
FIGURE 5.
Subcellular distribution of distinct surface proteins of L. monocytogenes in bacteria grown in BHI or IMM media. Shown are cytosol (Cyt), membrane (Mb), peptidoglycan (PG), and culture supernatant (Sn) extracts of wild-type strain EGDe grown in BHI or minimal medium IMM to stationary phase (overnight culture). The panels indicate the distribution of the following: A, ActA (a membrane protein having a stretch of hydrophobic residues in its C terminus); B, InlA (an LPXTG protein covalently bound to peptidoglycan); and C, InlB (a protein that associates with the lipotheicoic acids via its GW modules). Extracts were adjusted to the same number of bacteria (5 × 108 cfu) with the following exceptions: cytosolic fractions, which were 5-fold diluted, and peptidoglycan fractions, 20-fold concentrated for the cases of ActA and InlB. Note the increased production of ActA, InlA, and InlB in bacteria growing in minimal medium IMM. These three proteins are positively regulated by PrfA. Indicated are the sizes of the molecular weight markers used.
In concordance to the proteomic data (Fig. 3 and supplemental Table S1), these Western assays also showed increased levels of InlA in the peptidoglycan fraction in IMM compared with BHI medium (Fig. 5B). InlA was also detected in higher relative amounts in the supernatant of IMM cultures (Fig. 5B). Remarkably, a surface protein such as InlB, which lacks a membrane anchor domain and associates weakly to the cell wall via interaction of its GW modules to lipoteichoic acids (38), was also produced at higher levels in minimal medium IMM. In this case, InlB was mainly present in the extracellular milieu as breakdown products (Fig. 5C). Of note, InlB is positively regulated by PrfA, as are InlA and ActA. However, both the proteomic data (supplemental Table S1) and these Western assays indicated that InlB does not strongly associate with the peptidoglycan in any of the conditions tested. These biochemical assays discarded an unspecific retention of ActA into the peptidoglycan lattice. Thus, InlB was not retained in the peptidoglycan despite being also produced at high levels in the minimal medium IMM.
To definitively discard the possibility of an effect in the identification of ActA in the peptidoglycan linked to the high levels at which the protein is produced in medium IMM, we prepared envelope material from L. monocytogenes strain P14-A, known to overexpress the PrfA regulon in BHI medium, including the actA gene (24). The levels of ActA were determined in membrane fractions obtained from EGDe strain grown in BHI or IMM media and from the P14-A strain grown in BHI medium. As expected, Western assays revealed a higher amount of full-length ActA in the membrane of P14-A compared with EGDe in minimal medium IMM (Fig. 6A). No ActA was visible in EGDe grown in BHI (Fig. 6A). Interestingly, ActA was identified by proteomics at a much larger extent in the peptidoglycan of EGDe strain grown in minimal medium IMM (Fig. 6B). This was evident considering the high number of peptides and independent identifications of each peptide that were obtained by high resolution MS in the respective peptidoglycan extracts (Fig. 6B). Taken together, these data demonstrate that the identification of ActA in the cell wall is not a consequence of an increase of the protein content present in the envelope. On the contrary, such identification may reflect a new mode of association to peptidoglycan adopted by a fraction of the population of ActA molecules only under certain growth conditions.
FIGURE 6.
Association of ActA with peptidoglycan is not dictated by the relative amount of ActA present in the membrane. Membrane protein extracts were prepared from L. monocytogenes strain EGDe grown in BHI or IMM media to stationary phase and from strain P14-A, which overproduces ActA, in BHI medium. In parallel, peptidoglycan material was isolated and processed for high resolution MS proteomics as described under “Experimental Procedures.” A, relative levels of ActA detected in the membrane fraction by Western assay; B, ActA peptides identified by high resolution MS in peptidoglycan purified from bacteria grown in the distinct conditions. Indicated are the peptide sequence and the number of independent identifications for each peptide. Note that despite the higher content of ActA detected in the membrane of strain P14-A, the protein is identified with a lower rate than in peptidoglycan prepared from EGDe bacteria grown in minimal medium IMM.
ActA Polypeptide That Associates with Peptidoglycan Is Anchored to the Membrane
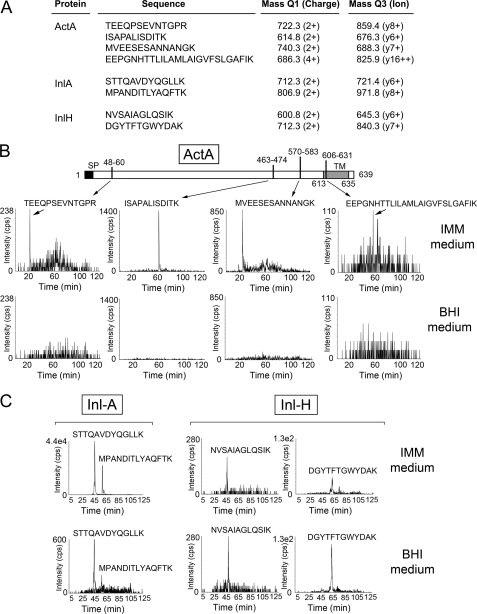
Although ActA was unequivocally identified with 21 unique peptides and 46% sequence coverage in the peptidoglycan of bacteria growing in IMM medium (Fig. 6B), none of the tryptic peptides detected overlapped with the stretch of hydrophobic residues at positions 613–635; TLILAMLAIGVFSLGAFIKIIQL. Targeted proteomics by SRM of ActA-derived tryptic peptides was used to gain insights on the topology and size of the ActA form that associates with the peptidoglycan. Samples were prepared from EGDe bacteria growing in IMM and BHI media. Four distinct ActA peptides mapping at different regions of the protein were monitored by SRM (Fig. 7A) as follows: (i) one peptide was positioned close to the N terminus of the protein (residues 48–60); (ii) two peptides covered residues 463–474 and 570–583; and (iii) one peptide of residues 606–631 overlapped part of the hydrophobic stretch (EEPGNHTTLILAMLAIGVFSLGAFIK) (Fig. 7B). As controls, four additional tryptic peptides of two LPXTG proteins (two from InlA and InlH, each) were also monitored (Fig. 7C). MS/MS spectra from all the peptides monitored by SRM are shown as supplemental material (supplemental Fig. S1). In agreement with the data obtained by Western analysis (Fig. 6A), the SRM analysis demonstrated that none of the four ActA peptides monitored were detected in peptidoglycan purified in BHI medium, although all of them were identified in peptidoglycan samples prepared from bacteria growing in minimal medium IMM (Fig. 7B). This latter result supported the idea that the ActA form that associates with peptidoglycan is tethered to the membrane. InlA and InlH tryptic peptides were identified in peptidoglycan prepared from bacteria grown in BHI and IMM media (Fig. 7C). Altogether, the SRM data confirmed that the ActA form that associates with the peptidoglycan is anchored to the membrane.
FIGURE 7.
SRM analyses reveal in peptidoglycan material the presence of an ActA-derived tryptic peptide corresponding to the membrane hydrophobic region. A, ion pair transitions for tryptic peptides of diverse L. monocytogenes surface proteins monitored in the SRM experiments. Shown is the identification by SRM of peptides corresponding to the following: B, ActA; and C, LPXTG proteins InlA and InlH. The SRM study was designed to identify four different tryptic peptides of ActA (positions 48–60, 463–474, 570–583, and 606–631). The peptide encompassing residues 606–631 (EEPGNHTTLILAMLAIGVFSLGAFIK) contains part of the residues 613–635 of the transmembrane region of ActA (TLILAMLAIGVFSLGAFIKIIQL). None the four ActA tryptic peptides was identified in peptidoglycan samples prepared from bacteria grown in BHI medium. The data correspond to peptidoglycan prepared from wild-type strain EGDe grown to stationary phase in the indicated media.
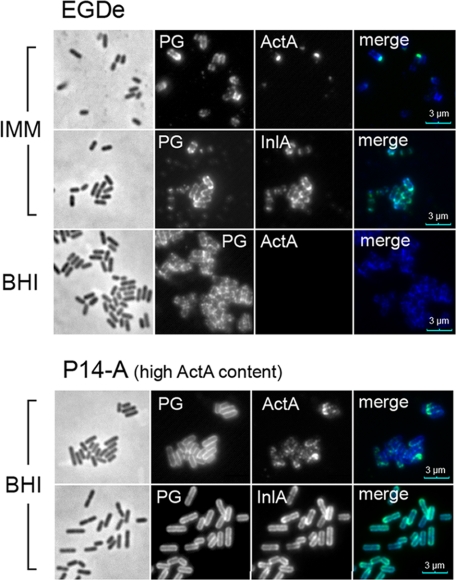
ActA Exposure in the Cell Surface Correlates to the Amount of Protein Produced by L. monocytogenes
Previous studies have shown that ActA is not detected on the bacterial surface of L. monocytogenes growing in BHI medium (39). Our proteomic data suggested that ActA may adopt a distinct envelope location in minimal medium IMM, which could translate in different exposure on the bacterial surface. To test this hypothesis, ActA distribution was examined by immunofluorescence microscopy in intact EGDe bacteria. For this analysis, we also include the P14-A strain grown in BHI, which overproduces ActA (Fig. 6A) but has not much ActA associated with the peptidoglycan (Fig. 6B). ActA was accessible to antibody labeling in EGDe strain grown in minimal medium IMM and P14-A strain grown in BHI, being barely detected in EGDe collected from BHI medium (Fig. 8). The signal observed for ActA was more intense in the case of the P14-A strain, which correlates with a higher level of ActA in the envelope (Fig. 6A). A similar observation was found for InlA and also visualized with a stronger signal in P14-A (Fig. 8). This difference may account for the overexpression of PrfA-regulated genes reported to occur in the P14-A strain (24). Altogether, these data suggest that exposure of ActA on the bacterial surface may be facilitated by an increase of the relative levels of this surface protein and not by a concrete mode of association to the cell wall.
FIGURE 8.
ActA exposure in bacterial cell surface to the amount of ActA protein present in the membrane. L. monocytogenes P14-A (ActA-overproducing strain) and EGDe (wild-type) were grown to stationary phase in BHI or minimal medium IMM, fixed in 3% paraformaldehyde, and processed for immunofluorescence microscopy. Monoclonal mouse antibodies anti-InlA, anti-ActA, and a polyclonal rabbit antibody raised against proteins strongly bound to peptidoglycan (PG) were used. As secondary antibodies, goat anti-mouse Alexa-488 (green) and goat anti-rabbit Alexa-594 (red) were used. The red fluorescence was pseudo-colored to blue to better differentiate the two labels in the overlay image. Note that ActA is clearly visible on the surface of the overproducing strain P14-A, although it is visible at the poles of EGDe bacteria grown in minimal medium IMM. No ActA was detected in EGDe bacteria grown in BHI. The surface distribution of the LPXTG protein InlA is shown for comparison. Scale bar, 3 μm.
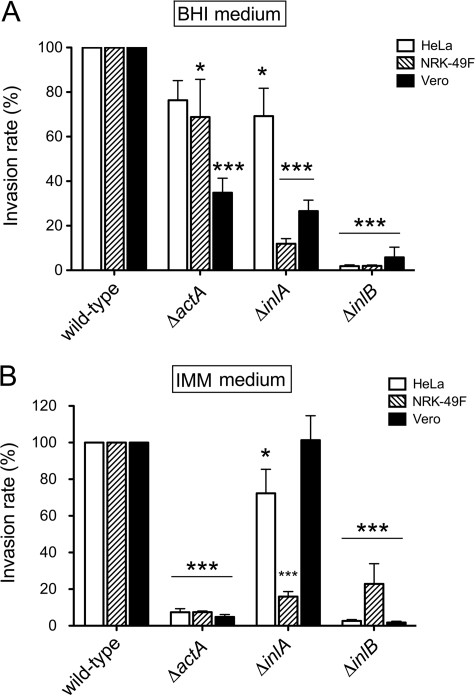
ActA Promotes Efficient Entry into Eukaryotic Cells Only under Conditions in Which It Associates with Peptidoglycan
A previous study reported the capacity of ActA to promote entry of L. monocytogenes into eukaryotic cells (40). This study however compared wild-type bacteria with mutants overproducing the PrfA regulon and derivative strains lacking ActA. No comparison of wild-type and ΔactA strains was performed. Because our proteomic analyses suggested that a fraction of ActA molecules might adopt distinct conformations in the cell wall, we tested the role of ActA in bacterial invasion using different growth media (BHI and IMM) and several nonphagocytic cell lines, including fibroblasts NRK-49F and the epithelial cells HeLa and Vero. Mutants defective in the two major invasins of L. monocytogenes, InlA and InlB, were also used for comparison. The lack of ActA impacted minimally the invasion rate of bacteria growing in BHI medium, with a significant difference detected only for the case of Vero cells. In this case, ΔactA mutant bacteria invaded with an efficiency of ∼40% compared with that shown by the wild-type strain (Fig. 9A). In contrast, ΔinlA and ΔinlB mutants exhibited a drastic reduction of their invasion rates when growing in BHI medium, and the only exception was the behavior of the ΔinlA mutant in HeLa epithelial cells (Fig. 9A). Strikingly, the invasion rate of the ΔactA mutant decreased significantly when bacteria were grown in minimal medium IMM. In this condition, lack of ActA resulted in invasion rates of ∼5% compared with those shown by the wild type in the three eukaryotic cell lines tested (Fig. 9B). Growth in the medium IMM also resulted in a low invasion rate for the ΔinlB mutant, whereas ΔinlA bacteria displayed major changes in the invasion rate only in NRK-49F fibroblasts (Fig. 9B). Altogether, these assays demonstrate a strong requirement of ActA for L. monocytogenes invasion when bacteria are grown under conditions favoring association of the protein to the peptidoglycan.
FIGURE 9.
ActA is required for entry of L. monocytogenes EGDe into epithelial and fibroblast cells when bacteria are grown in minimal medium IMM. Infection of epithelial (HeLa and Vero) and fibroblast (NRK-49F) cells was performed as described under “Experimental Procedures.” Shown are the relative invasion values obtained at 2 h post-infection using bacteria grown in the following: A, nutrient-rich BHI medium or B, minimal medium IMM. Strains used included EGDe (wild-type) and isogenic ΔinlA, ΔinlB, and ΔactA mutants. The data are the means ± S.D. from three independent experiments. *, p = 0.01–0.05; ***, p < 0.001, by two-way analysis of variance with Bonferroni's post-test. For the inoculum in BHI medium, the average number of viable intracellular bacteria counted in the three independent experiments was (per 105 eukaryotic cells) as follows: 3.4 × 103 cfu (HeLa epithelial cells), 5.2 × 103 cfu (NRK-49F fibroblasts), and 1.5 × 104 cfu (Vero epithelial cells). Values obtained with bacteria grown in minimal medium IMM were as follows: 1.15 × 106 cfu (HeLa epithelial cells), 9.6 × 104 cfu (NRK-49F fibroblasts), and 6.15 × 105 cfu (Vero epithelial cells). Note the drastic reduction in invasiveness of the ΔactA mutant in bacteria growing in minimal medium IMM.
DISCUSSION
To our knowledge, the work described here with L. monocytogenes represents the first analysis of a bacterial cell wall proteome using high resolution mass spectrometry based on an LTQ-Orbitrap MS system. This study also reports for the first time the cell wall proteome of an intracellular bacterial pathogen growing in the cytosol of eukaryotic cells. Of note, a very recent report describes protein expression profiles of three L. monocytogenes strains growing inside macrophages using standard multidimensional protein identification technology (41). This study, however, focuses only on total protein extracts and provides rather low identification rates, in the range of 45–80 protein species for total extracts prepared from intracellular bacteria. In our case, the high resolution proteomic approach used led to a much higher identification rate. Thus, 53 proteins were identified in peptidoglycan purified from intracellular bacteria upon extensive boiling of the envelope material in SDS. Such a subcellular fraction contains mostly surface proteins strongly associated with the cell wall. A high percentage of the proteins identified in this material corresponds with surface proteins expected to be associated with peptidoglycan, i.e. sortase substrates (covalently bound to the peptidoglycan) and enzymes related to peptidoglycan metabolism. However, most of the rest of identified proteins were predicted cytosolic proteins that have been largely documented as “moonlighting” proteins also displaying subcellular localization at the cell wall (42). Examples are chaperons (DnaK), elongation factors (EF-Tu), and diverse metabolic enzymes as enolase and glyceraldehyde-3-phosphate dehydrogenase, among others. Indeed, most of the moonlighting proteins we detected using this high resolution MS approach are coincident with the subproteome of L. monocytogenes reported by Schaumburg et al. (42), which extracted proteins from the cell wall of bacteria grown in BHI medium.
The “intracellular cell wall proteome” defined here for L. monocytogenes at 6 h post-invasion was compared with those of extracellular bacteria growing in two distinct laboratory media. A major conclusion of this comparative study was that L. monocytogenes produces inside of the eukaryotic most of the LPXTG proteins covalently bound to the peptidoglycan that are synthesized by extracellular bacteria. This finding was at some extent unexpected given the metabolic changes predicted to occur during the intracytosolic growth compared with the growth in BHI medium. For instance, only intracellular L. monocytogenes utilize glycerol or glucose 6-phosphate instead of glucose as the main carbon sources and produce oxalacetate predominantly via carboxylation of pyruvate (43, 44). Despite this, a few differences were also observed in the proteome, especially concerning the up-regulation of the LPXTG proteins InlA and Lmo0514 in intracellular bacteria. The function of Lmo0514, a protein carrying features of internalin proteins such as the leucine-rich repeat (LRR) domains, is at present unknown. Lmo0514 carries also two PKD repeats conferring immunoglobulin-like folding properties (5, 45). Such characteristics suggest that Lmo0514 could interact with other bacterial or eukaryotic proteins once the bacteria colonize the eukaryotic cytosol. In agreement with our proteomic data, previous transcriptomic analyses revealed that bacteria growing inside macrophages up-regulate the expression of the lmo0514 gene (35). However, the same study reported in intracellular bacteria up-regulation of other genes encoding LPXTG proteins, such as lmo2085, for which we identified the protein only in the cell wall of extracellular bacteria growing in BHI (Fig. 3). Up-regulation of Lmo0514 in intracellular L. monocytogenes upon infection of JEG-3 epithelial cells also contrasts with other studies monitoring global expression in L. monocytogenes during mouse infection (46) or in Caco-2 epithelial cells (47). None of these two studies reported changes in expression of the specific gene lmo0514. Different infection conditions, the distinct source of the epithelial cells used (JEG-3 from placenta and Caco-2 from intestine), or even post-transcriptional regulatory mechanisms acting differently in bacteria depending on the type of cell colonized could account for these discrepancies. However, our proteomic data were in concordance with the up-regulation of inlA and other PrfA-regulated genes as actA was reported in this same series of transcriptomic studies (35, 46, 47). As a feature distinctive to our work, none of these former transcriptomic studies anticipated the abundance of InlA with respect to most of the other LPXTG proteins in the cell wall of intracellular bacteria. Our analysis was performed at 6 h post-infection, so it is unlikely that the InlA identified by proteomics corresponds to that present in the infecting bacteria. For other intracellular bacterial pathogens, such as Salmonella enterica, it has been shown that invasion-related functions are activated in intracellular bacteria at late infection times to facilitate infection of neighbor cells upon extrusion from the infected cell (48, 49). In contrast, the dissemination of intracellular L. monocytogenes to neighbor cells proceeds with no “extracellular phase,” which makes difficult to reconcile with such model. Thus, the role of the InlA synthesized de novo by intracellular bacteria remains totally unknown. Further work is clearly needed to elucidate whether the up-regulation of InlA in intracellular bacteria is a phenomenon related (or not) to the bacterial entry process.
Together with InlA, other LPXTG proteins easily identified in the L. monocytogenes cell wall included Lmo0130, Lmo0160, Lmo263 (InlH), Lmo0880, Lmo1666, and Lmo2714. With the exception of Lmo1666, which has recently been shown to be a new L. monocytogenes adhesion protein required for virulence (50), and Lmo0263 (InlH), which also contributes to virulence (51), the rest of these LPXTG proteins have orthologs in nonpathogenic Listeria species such as Listeria innocua (12). This fact suggests that the role of some abundant LPXTG proteins in Listeria could be related to the maintenance of cell wall integrity or other conserved physiological processes, such as nutrient transport. In other Gram-positive bacteria such as S. aureus and Bacillus anthracis, several proteins anchored covalently to the peptidoglycan are known to form a heme transport system for iron acquisition (52, 53). Further work could certainly focus on these abundant LPXTG proteins of L. monocytogenes of unknown function.
At present, we are uncertain as the reason why a relatively small fraction of the 41 LPXTG proteins predicted to be encoded in the genome of the L. monocytogenes strain EGD-e (12) are detected with the most sensitive high resolution proteomic technology known to date. Our analyses in intracellular and extracellular bacteria identified a total of 18 distinct LPXTG protein species (Fig. 3 and supplemental Table S1). In some cases, additional LPXTG proteins were also identified with a single peptide and fragmentation spectra of high quality (data not shown). These proteins could be present in extremely low amounts in the cell wall, close to the current threshold of these sensitive proteomic technologies. A paradigmatic example supporting this postulate is that of Lmo0320. This LPXTG protein, also known as Vip, acts as an invasin promoting bacterial entry (54). The mutant defective in this protein exhibits a strong defect for entry in several cell lines and is attenuated in virulence animal models (54). Strikingly, we have been unable to detect this protein by high resolution MS in any of the conditions tested, which favors the idea of Vip being an LPXTG protein present in scarce amounts in the cell wall of L. monocytogenes. Attempts to detect this protein with anti-Lmo0320 antibody in peptidoglycan of wild-type bacteria have also been unsuccessful (data not shown). Nonetheless, the proteomic analysis reported here provides a considerable increase in the knowledge of this large family of LPXTG proteins of Listeria, for which previous studies had identified a maximum of 13 distinct members (17, 18).
A major unexpected finding of our study was the identification of surface and secreted proteins regulated positively by PrfA, such as listeriolysin-O and ActA, in the peptidoglycan of intracellular L. monocytogenes. Peptidoglycan purification involves extensively boiling in 4% SDS, so the identification of a certain protein in this material presumes a strong association (covalent or not) that is not disrupted by this harsh detergent treatment. Proper controls with other surface proteins of known association with the cell wall, such as InlB, discarded the possibility of unspecific retention of either listeriolysin-O or ActA in the cell wall. Interestingly, these new protein-peptidoglycan associations were reproduced in extracellular bacteria growing in the chemically defined minimal medium IMM. ActA contains a stretch of hydrophobic residues at its C terminus, which is thought to tether the protein to the membrane (5). The SRM data indicated that the ActA form that associates with the peptidoglycan still maintains its membrane anchorage. ActA does not contain “cell wall association domains” as LysM, GW, or WXL (5), so it is tempting to postulate that other factors, such as an interaction with yet unknown cell wall components, are responsible for ActA association with the peptidoglycan. ActA is known to display discrete positioning at distinct sites on the surface early after synthesis, which is followed by redistribution in helices and further polarization (39). It is possible that during these changes in surface distribution, a small population of ActA molecules establishes an association with the peptidoglycan strong enough to allow co-purification in boiling conditions. Differences in the relative amount of the protein located in the membrane, as observed in minimal medium IMM with respect to BHI, might influence this association and therefore affect the capacity of the bacteria to perform new activities. However, the experiments performed with the ActA-overproducing strain P14-A discarded a direct correlation between the amount of ActA present in the envelope and the association of the protein to the peptidoglycan. Our data are therefore consistent with a flexibility of ActA for adopting distinct conformations in the surface depending on the growth conditions. It is also worth noting that our subcellular fractionation analyses based on Western assays were unable to detect ActA in purified peptidoglycan fractions. Given the strong evidence obtained by high resolution MS of the presence of ActA in this fraction, we hypothesize that at steady state only a small fraction of the ActA molecules present in the envelope of L. monocytogenes growing either in medium IMM or inside epithelial cells might be associated with peptidoglycan. However, this small fraction could accomplish distinct roles during infection or outside host cells. A precedent of this situation is found for the S. enterica membrane protein InvH, required for proper function of a specialized type III secretion machinery and for which it was estimated that only ∼2% of the molecules were associated with peptidoglycan (55). The disruption of such interaction following changes in peptidoglycan structure compromised protein secretion (55).
An intriguing observation was the apparent lack of ActA in the membrane or peptidoglycan fractions of the wild-type EGDe strain growing in BHI (Figs. 5A and 6A). The ΔactA mutant exhibited a slight reduction in the invasion rate when growing in this nutrient-rich BHI medium (Fig. 9A). It is therefore possible that a low number of ActA molecules, hardly detectable by Western analysis, may exist in the membrane of bacteria growing in BHI medium. This postulate agrees with observations reported by other authors (37). Likewise, the ΔactA mutant was highly impaired in the ability to promote invasion of epithelial and fibroblast cells when bacteria were grown in IMM, a condition in which a fraction of ActA molecules associates with peptidoglycan. Under identical growth conditions, mutants defective in InlA or InlB also displayed a strong defect in invasion (Fig. 9B). At present, we have no further data supporting why the lack of different surface proteins led to a defect in entry into a specific eukaryotic cell line. A plausible explanation is that the absence of any of these proteins has a deleterious effect on the amount or distribution of the other invasin(s). In the case of the ΔactA mutant, we have, however, not found evidence supporting this fact for the case of either InlA or InlB. This was tested by proteomic analysis of the cell wall of the ΔactA mutant, which identified high amounts of InlA (data not shown). Further microscopy and Western analyses revealed no difference in the exposure and surface distribution of InlA in the ΔactA mutant. Similar findings were obtained when determining the levels of InlB in membrane fractions of the ΔactA mutant (data not shown). Based on these observations, our interpretation for the requirement of ActA to invade cultured cells is that under certain growth conditions, such as the minimal medium IMM, the ActA molecules that associate with the peptidoglycan may modulate the topology in the cell wall of invasins as InlA or InlB by establishing direct or indirect protein-protein interactions. We plan to test this postulate by searching for protein complexes involving these surface proteins. Our findings are also consistent with studies demonstrating the requirement of other surface proteins besides InlA and InlB for efficient entry of L. monocytogenes into eukaryotic cells (56).
In summary, our study provides the first insights into cell wall proteome changes occurring during the intracellular infection of eukaryotic cells by L. monocytogenes. Our data also account for novel protein-peptidoglycan associations not suspected by analysis of the primary sequence, as it was the case of ActA.
Supplementary Material
Acknowledgments
We thank Prof. Pascale Cossart for providing us with anti-InlA and anti-InlB antibodies; Prof. Trinad Chakraborty for the anti-ActA antibody and EGDe isogenic mutant strains defective in InlA, InlB, or ActA; and Prof. J.A. Vázquez-Boland for the P14-A strain. The technical assistance of M. Laura Navarro and Diana Barroso is greatly acknowledged. The expert assistance of the Electron Microscope Unit of the National Centre of Biotechnology (CNB-CSIC) is also acknowledged.
This work was funded by Grants PIM2010EPA-00714 from the ERA-NET Pathogenomics LISTRESS Consortium (to F. G.-P.) and BIO2010-18962 (to M. G. P.) from the Spanish Ministry of Science and Innovation.
This paper is dedicated to the memory of Professor Jürgen Wehland, expert in the field of Listeria-host cell interactions and former Scientific Director of the Helmholtz Centre for Infection Research in Braunschweig, Germany, who unexpectedly passed away on August 16, 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- BHI
- brain-heart infusion medium
- IMM
- chemically defined minimal medium
- SRM
- selective reaction monitoring
- MEM
- Eagle's minimal essential medium
- LLO
- listeriolysin-O.
REFERENCES
- 1. Silhavy T. J., Kahne D., Walker S. (2010) Cold Spring Harb. Perspect. Biol. 2, a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vollmer W., Blanot D., de Pedro M. A. (2008) FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 3. Marraffini L. A., Dedent A. C., Schneewind O. (2006) Microbiol. Mol. Biol. Rev. 70, 192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pucciarelli M. G., Bierne H., García-Del Portillo F. (2007) in Listeria monocytogenes: Pathogenesis and Host Response (Goldfine H., Sghen H. eds) pp. 81–110, Springer Science & Business Media LLC., New York [Google Scholar]
- 5. Bierne H., Cossart P. (2007) Microbiol. Mol. Biol. Rev. 71, 377–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hendrickx A. P., Willems R. J., Bonten M. J., van Schaik W. (2009) Trends Microbiol. 17, 423–430 [DOI] [PubMed] [Google Scholar]
- 7. Fedtke I., Götz F., Peschel A. (2004) Int. J. Med. Microbiol. 294, 189–194 [DOI] [PubMed] [Google Scholar]
- 8. Boneca I. G., Dussurget O., Cabanes D., Nahori M. A., Sousa S., Lecuit M., Psylinakis E., Bouriotis V., Hugot J. P., Giovannini M., Coyle A., Bertin J., Namane A., Rousselle J. C., Cayet N., Prévost M. C., Balloy V., Chignard M., Philpott D. J., Cossart P., Girardin S. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poetsch A., Wolters D. (2008) Proteomics 8, 4100–4122 [DOI] [PubMed] [Google Scholar]
- 10. Hecker M., Antelmann H., Büttner K., Bernhardt J. (2008) Proteomics 8, 4958–4975 [DOI] [PubMed] [Google Scholar]
- 11. Nelson K. E., Fouts D. E., Mongodin E. F., Ravel J., DeBoy R. T., Kolonay J. F., Rasko D. A., Angiuoli S. V., Gill S. R., Paulsen I. T., Peterson J., White O., Nelson W. C., Nierman W., Beanan M. J., Brinkac L. M., Daugherty S. C., Dodson R. J., Durkin A. S., Madupu R., Haft D. H., Selengut J., Van Aken S., Khouri H., Fedorova N., Forberger H., Tran B., Kathariou S., Wonderling L. D., Uhlich G. A., Bayles D. O., Luchansky J. B., Fraser C. M. (2004) Nucleic Acids Res. 32, 2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Charbit A., Chetouani F., Couvé E., de Daruvar A., Dehoux P., Domann E., Domínguez-Bernal G., Duchaud E., Durant L., Dussurget O., Entian K. D., Fsihi H., García-del Portillo F., Garrido P., Gautier L., Goebel W., Gómez-López N., Hain T., Hauf J., Jackson D., Jones L. M., Kaerst U., Kreft J., Kuhn M., Kunst F., Kurapkat G., Madueno E., Maitournam A., Vicente J. M., Ng E., Nedjari H., Nordsiek G., Novella S., de Pablos B., Pérez-Diaz J. C., Purcell R., Remmel B., Rose M., Schlueter T., Simoes N., Tierrez A., Vázquez-Boland J. A., Voss H., Wehland J., Cossart P. (2001) Science 294, 849–852 [DOI] [PubMed] [Google Scholar]
- 13. Hain T., Chatterjee S. S., Ghai R., Kuenne C. T., Billion A., Steinweg C., Domann E., Kärst U., Jänsch L., Wehland J., Eisenreich W., Bacher A., Joseph B., Schär J., Kreft J., Klumpp J., Loessner M. J., Dorscht J., Neuhaus K., Fuchs T. M., Scherer S., Doumith M., Jacquet C., Martin P., Cossart P., Rusniock C., Glaser P., Buchrieser C., Goebel W., Chakraborty T. (2007) Int. J. Med. Microbiol. 297, 541–557 [DOI] [PubMed] [Google Scholar]
- 14. Doumith M., Cazalet C., Simoes N., Frangeul L., Jacquet C., Kunst F., Martin P., Cossart P., Glaser P., Buchrieser C. (2004) Infect. Immun. 72, 1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freitag N. E., Port G. C., Miner M. D. (2009) Nat. Rev. Microbiol. 7, 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonazzi M., Lecuit M., Cossart P. (2009) Cell. Microbiol. 11, 693–702 [DOI] [PubMed] [Google Scholar]
- 17. Pucciarelli M. G., Calvo E., Sabet C., Bierne H., Cossart P., García-del Portillo F. (2005) Proteomics 5, 4808–4817 [DOI] [PubMed] [Google Scholar]
- 18. Calvo E., Pucciarelli M. G., Bierne H., Cossart P., Albar J. P., García-Del Portillo F. (2005) Proteomics 5, 433–443 [DOI] [PubMed] [Google Scholar]
- 19. Stoll R., Mertins S., Joseph B., Müller-Altrock S., Goebel W. (2008) Microbiology 154, 3856–3876 [DOI] [PubMed] [Google Scholar]
- 20. Scortti M., Monzó H. J., Lacharme-Lora L., Lewis D. A., Vázquez-Boland J. A. (2007) Microbes Infect. 9, 1196–1207 [DOI] [PubMed] [Google Scholar]
- 21. Cossart P., Toledo-Arana A. (2008) Microbes Infect. 10, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 22. Guzman C. A., Rohde M., Chakraborty T., Domann E., Hudel M., Wehland J., Timmis K. N. (1995) Infect. Immun. 63, 3665–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chakraborty T., Ebel F., Domann E., Niebuhr K., Gerstel B., Pistor S., Temm-Grove C. J., Jockusch B. M., Reinhard M., Walter U., et al. (1995) EMBO J. 14, 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ripio M. T., Domínguez-Bernal G., Lara M., Suárez M., Vazquez-Boland J. A. (1997) J. Bacteriol. 179, 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phan-Thanh L., Gormon T. (1997) Int. J. Food Microbiol. 35, 91–95 [DOI] [PubMed] [Google Scholar]
- 26. Mostowy S., Nam Tham T., Danckaert A., Guadagnini S., Boisson-Dupuis S., Pizarro-Cerdá J., Cossart P. (2009) PLoS One 4, e4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 28. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 29. Bierne H., Garandeau C., Pucciarelli M. G., Sabet C., Newton S., Garcia-del Portillo F., Cossart P., Charbit A. (2004) J. Bacteriol. 186, 1972–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bierne H., Mazmanian S. K., Trost M., Pucciarelli M. G., Liu G., Dehoux P., Jänsch L., Garcia-del Portillo F., Schneewind O., Cossart P. (2002) Mol. Microbiol. 43, 869–881 [DOI] [PubMed] [Google Scholar]
- 31. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. (2003) Mol. Microbiol. 47, 103–118 [DOI] [PubMed] [Google Scholar]
- 32. Pucciarelli M. G., Prieto A. I., Casadesús J., García-del Portillo F. (2002) Microbiology 148, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 33. Lebrun M., Mengaud J., Ohayon H., Nato F., Cossart P. (1996) Mol. Microbiol. 21, 579–592 [DOI] [PubMed] [Google Scholar]
- 34. Mariscotti J. F., García-del Portillo F., Pucciarelli M. G. (2009) J. Biol. Chem. 284, 6140–6146 [DOI] [PubMed] [Google Scholar]
- 35. Chatterjee S. S., Hossain H., Otten S., Kuenne C., Kuchmina K., Machata S., Domann E., Chakraborty T., Hain T. (2006) Infect. Immun. 74, 1323–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niebuhr K., Chakraborty T., Rohde M., Gazlig T., Jansen B., Köllner P., Wehland J. (1993) Infect. Immun. 61, 2793–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lathrop A. A., Banada P. P., Bhunia A. K. (2008) J. Appl. Microbiol. 104, 627–639 [DOI] [PubMed] [Google Scholar]
- 38. Jonquières R., Bierne H., Fiedler F., Gounon P., Cossart P. (1999) Mol. Microbiol. 34, 902–914 [DOI] [PubMed] [Google Scholar]
- 39. Rafelski S. M., Theriot J. A. (2006) Mol. Microbiol. 59, 1262–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suárez M., González-Zorn B., Vega Y., Chico-Calero I., Vázquez-Boland J. A. (2001) Cell. Microbiol. 3, 853–864 [DOI] [PubMed] [Google Scholar]
- 41. Donaldson J. R., Nanduri B., Pittman J. R., Givaruangsawat S., Burgess S. C., Lawrence M. L. (2011) J. Proteomics 74, 1906–1917 [DOI] [PubMed] [Google Scholar]
- 42. Schaumburg J., Diekmann O., Hagendorff P., Bergmann S., Rohde M., Hammerschmidt S., Jänsch L., Wehland J., Kärst U. (2004) Proteomics 4, 2991–3006 [DOI] [PubMed] [Google Scholar]
- 43. Eylert E., Schär J., Mertins S., Stoll R., Bacher A., Goebel W., Eisenreich W. (2008) Mol. Microbiol. 69, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 44. Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010) Nat. Rev. Microbiol. 8, 401–412 [DOI] [PubMed] [Google Scholar]
- 45. Cabanes D., Dehoux P., Dussurget O., Frangeul L., Cossart P. (2002) Trends Microbiol. 10, 238–245 [DOI] [PubMed] [Google Scholar]
- 46. Camejo A., Buchrieser C., Couvé E., Carvalho F., Reis O., Ferreira P., Sousa S., Cossart P., Cabanes D. (2009) PLoS Pathog. 5, e1000449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Joseph B., Przybilla K., Stühler C., Schauer K., Slaghuis J., Fuchs T. M., Goebel W. (2006) J. Bacteriol. 188, 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knodler L. A., Vallance B. A., Celli J., Winfree S., Hansen B., Montero M., Steele-Mortimer O. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17733–17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hautefort I., Thompson A., Eriksson-Ygberg S., Parker M. L., Lucchini S., Danino V., Bongaerts R. J., Ahmad N., Rhen M., Hinton J. C. (2008) Cell. Microbiol. 10, 958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reis O., Sousa S., Camejo A., Villiers V., Gouin E., Cossart P., Cabanes D. (2010) J. Infect. Dis. 202, 551–562 [DOI] [PubMed] [Google Scholar]
- 51. Personnic N., Bruck S., Nahori M. A., Toledo-Arana A., Nikitas G., Lecuit M., Dussurget O., Cossart P., Bierne H. (2010) Infect. Immun. 78, 1979–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres V. J., Pishchany G., Humayun M., Schneewind O., Skaar E. P. (2006) J. Bacteriol. 188, 8421–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maresso A. W., Chapa T. J., Schneewind O. (2006) J. Bacteriol. 188, 8145–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cabanes D., Sousa S., Cebriá A., Lecuit M., García-del Portillo F., Cossart P. (2005) EMBO J. 24, 2827–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pucciarelli M. G., García-del Portillo F. (2003) Mol. Microbiol. 48, 573–585 [DOI] [PubMed] [Google Scholar]
- 56. Chatterjee S. S., Otten S., Hain T., Lingnau A., Carl U. D., Wehland J., Domann E., Chakraborty T. (2006) Int. J. Med. Microbiol. 296, 277–286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.