Background: Eukaryotic Cys-loop receptors contain three domains, whereas prokaryotic ones contain two because they lack the intracellular domain (ICD).
Results: Functional chimeras were engineered by adding the serotonin receptor ICD to prokaryotic GLIC channels.
Conclusion: The ICD is portable between eukaryotic and prokaryotic Cys-loop receptors.
Significance: This portability will allow functional and structural studies of the ICD of diverse eukaryotic Cys-loop receptors.
Keywords: Cys-loop Receptors, GABA Receptors, Ion Channels, Membrane Proteins, Nicotinic Acetylcholine Receptors
Abstract
Prokaryotic members of the Cys-loop receptor ligand-gated ion channel superfamily were recently identified. Previously, Cys-loop receptors were only known from multicellular organisms (metazoans). Contrary to the metazoan Cys-loop receptors, the prokaryotic ones consist of an extracellular (ECD) and a transmembrane domain (TMD), lacking the large intracellular domain (ICD) present in metazoa (between transmembrane segments M3 and M4). Using a chimera approach, we added the 115-amino acid ICD from mammalian serotonin type 3A receptors (5-HT3A) to the prokaryotic proton-activated Gloeobacter violaceus ligand-gated ion channel (GLIC). We created 12 GLIC-5-HT3A-ICD chimeras by replacing a variable number of amino acids in the short GLIC M3M4 linker with the entire 5-HT3A-ICD. Two-electrode voltage clamp recordings after expression in Xenopus laevis oocytes showed that only two chimeras were functional and produced currents upon acidification. The pH50 was comparable with wild-type GLIC. 5-HT3A receptor expression can be inhibited by the chaperone protein RIC-3. We have shown previously that the 5-HT3A-ICD is required for the attenuation of 5-HT-induced currents when RIC-3 is co-expressed with 5-HT3A receptors in X. laevis oocytes. Expression of both functional 5-HT3A chimeras was inhibited by RIC-3 co-expression, indicating appropriate folding of the 5-HT3A-ICD in the chimeras. Our results indicate that the ICD can be considered a separate domain that can be removed from or added to the ECD and TMD while maintaining the overall structure and function of the ECD and TMD.
Introduction
The Cys-loop receptor gene superfamily consists of homologous ligand-gated ion channels that play crucial roles in human health. Diseases linked to Cys-loop receptor family members include: for γ-amino butyric acid receptors, epilepsy and anxiety; for nicotinic acetylcholine receptors (nAChR),2 Alzheimer and Parkinson diseases, schizophrenia, depression, attention deficit hyperactivity disorder, and myasthenia gravis; and for serotonin R (5-HT3), depression and psychosis. Cys-loop receptor subunits assemble to form pentameric receptors. The subunits share the same three-domain structure: a large N-terminal extracellular domain (ECD), a transmembrane domain (TMD) consisting of four α-helical segments (M1–M4), and an intracellular domain (ICD). Two antiparallel β-sheets in each subunit contribute to the ECD, which harbors the agonist-binding site at subunit interfaces. Conformational changes in the ECD upon agonist binding trigger a conformational wave that is transmitted to the TMD to initiate channel opening. The ICD plays a critical role in the localization of receptors at synapses and in receptor trafficking to modulate synaptic strength (1–6). The ICD is by far the most divergent domain among metazoan Cys-loop receptors with respect to both length (∼50–270 amino acids) and amino acid sequence.
In 2005, iterative sequence profile searches identified several Cys-loop receptor homologues in prokaryotes (7). Alignments showed that the overall hydrophobic core structure of the ECD β-sheet and a transmembrane domain topology with four α-helical segments were also conserved for the prokaryotic homologues. Interestingly, however, the eponymous disulfide-linked loop (Cys-loop) in the ECD is not present in prokaryotic members. In addition, all prokaryotic members lack an extensive ICD. The computed length of the M3M4 loop in prokaryotes is 3–14 amino acids (7). Subsequently, it was confirmed that the homologue from a cyanobacterium, the Gloeobacter violaceus ligand-gated ion channel (GLIC), is a homopentameric, proton-gated cation channel (8).
High-resolution crystal structures of the closed and open states of bacterial homologues, the GLIC (open) and Erwinia chrysanthemi LGIC (ELIC, closed), have been published (9–11). Whether the conformation of GLIC obtained by crystallization at acidic pH represents an open or a desensitized conformation is highly controversial. Initially, it was published that GLIC does not desensitize at acidic pH (8, 9); however, several studies have recently shown that it does desensitize (12, 13). The prokaryotic structures have demonstrated a conserved core subunit architecture of metazoan and prokaryotic homologues: an ECD with two antiparallel β-sheets and a TMD with four α-helical segments. The same secondary and tertiary motifs of ECD and TMD had previously been observed in the electron microscopy-derived Torpedo nAChR structural model, as well as in the high-resolution x-ray structures of acetylcholine-binding proteins, which are homologous to the ECD, and of the ECD of α1 nAChR (14–17). The most recent x-ray structure of a truncated (ICD replaced by tripeptide) eukaryotic family member from Caenorhabditis elegans, a glutamate-gated chloride channel (GluCl), shows essentially the same overall fold (18). As compared with the Torpedo nAChR structure, the GluCl structure showed a shift of one helical turn for the M2 and M3 segments. The earlier start of M3 made the M3 segment longer than previously anticipated. M4 is longer as well, albeit it is unclear whether this is the result of the engineering that was required to obtain a crystallizable construct; the M3M4 loop was removed and replaced by a tripeptide. Importantly, the functionality of the GluCl construct was severely impaired. The most significant divergence between prokaryotic and eukaryotic ligand-gated ion channels is the absence of an ICD in the former. The M3M4 loop in prokaryotes is barely longer than what is required to link the two transmembrane segments (3–14 amino acids).
Previously we showed that the large intracellular domain in 5-HT3A receptors (115 amino acids) and in GABAρ receptors (82 amino acids) can be replaced by a short linker and that the modified receptors fold, assemble, and traffic to the membrane and function as ion channels (19). As the linker, we chose a heptapeptide that alignment studies suggested was the linker between the α-helical transmembrane segments M3 and M4 in GLIC (SQPARAA)(7). However, the GLIC x-ray structure revealed that the linker is shifted by several amino acids (9, 10).
In the present study, we engineered a prokaryotic Cys-loop receptor to be more metazoan-like. The major domains of the chimeras stem from the bacterial homologue GLIC, whereas the ICD, in general not present in prokaryotes, was added from eukaryotes, namely the 5-HT3A-ICD (see Fig. 1, A and B). To date, there have been no reports as to whether functional ICDs can be added to prokaryotic Cys-loop receptors. Only fusion proteins have been generated by adding GFP either to complete three-domain eukaryotic Cys-loop receptors (20–23) or to partially ICD-deleted eukaryotic Cys-loop receptors (24).
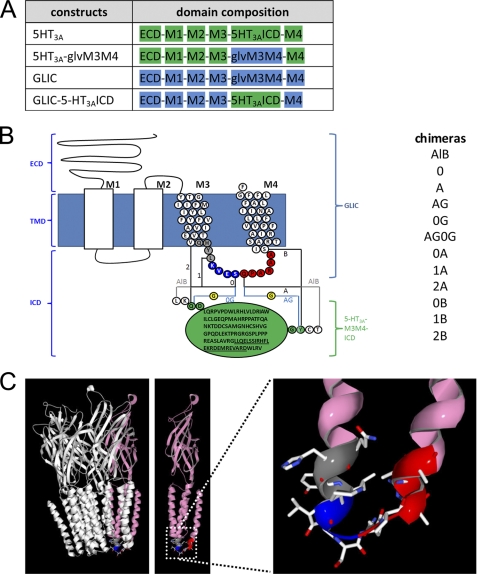
FIGURE 1.
Design and optimization of the GLIC-5-HT3A-ICD chimeras. A, domain composition of the chimeras: ECD, transmembrane domain consisting of M1–M4, and ICD. Domains contributed by 5-HT3A subunits are in green, and domains contributed by GLIC are in blue. B, schematic representation of a GLIC subunit and the different insertion points of the 5-HT3A-ICD (green circles, MA helix residues underlined). Transmembrane α-helix numbering is given on top of each helix (M1–M4). The AlB construct contained the complete GLIC sequence and the complete 5-HT3A-ICD sequence, in addition to the amino acids LK and CT N- and C-terminal to the insertion, respectively. From this construct, we gradually removed residues, as indicated by the color-coded circles. Insertion sites for the ICD are coded by numbers for the N-terminal side (0–2) and by letters for the C-terminal side (A and B). An additional Gly (yellow) was inserted for the 0G and AG constructs. All combinations of N- and C-terminal ICD insertion points generated and tested in this study are indicated in the list on the right. C, left, the GLIC channel structure from Protein Data Bank (PDB) number 3EAM coordinates, with one subunit colored in pink. Middle, unobstructed view of the pink subunit. Right, zoomed in view of the M3M4 linker with the color coding for the junction fragments in the chimeras as in B, M1, and M2 removed for clarity.
We expressed the chimeras heterologously in Xenopus laevis oocytes and investigated the ion channel function by two-electrode voltage clamp experiments. Out of 12 chimeras, two were functional proton-gated ion channels. To investigate whether the ICD in the functional chimeras was properly folded, we investigated the known interaction of the protein resistance to inhibitors of cholinesterase (RIC-3) with the 5-HT3A-ICD. RIC-3 co-expression decreased the expression of the chimeras on the plasma membrane, indicating that the engineered ICD is at least partly folded.
Our study thus provides further evidence for the modular design theory for Cys-loop receptors that we put forth previously (19). Other studies have shown that functional chimeras can be obtained by exchanging the ECD between Cys-loop receptors and thus provided evidence for two modules (25–29). The identification of acetylcholine-binding protein also corroborated the view of the ECD as an independent module. Our results show that the ICDs can be removed from three-domain Cys-loop receptors and added to two-domain receptors while retaining their overall functionality as ion channels. However, the modules are not absolutely interchangeable because when the ECD was exchanged between subunits, certain electrostatic interactions between modules had to be preserved (25–29), or when the ICD was added and removed, linker lengths between modules had to be optimized. Overall the various chimera studies, including the present one, indicate the presence of three separate domains that are exchangeable and thus modular for Cys-loop receptors.
EXPERIMENTAL PROCEDURES
Materials
Horse serum and primers were obtained from Sigma. Antibiotic-antimycotic (100×) liquid (10,000 IU/ml penicillin, 10,000 μg/ml streptomycin, and 25 μg/ml amphotericin B) was from Invitrogen.
Plasmids
A G. violaceus culture (ATCC 29082) was heated to 92 °C for 10 min and then used as a template in a PCR utilizing the primers 5′-GTA TAG GAT CCA CCA TGT TCC CGA CCG GCT GGC GGC CCA AAC-3′ and 3′-CTC GAT GCG GCC GCC TAA AAT CCA AAG AAA AGA AAT GCC AG-5′ containing a BamHI and NotI site (underlined), respectively. Segments aligning with the gene of interest to be subcloned are in bold. The PCR fragment and pXOON vector (30) were cut with BamHI (all enzymes from New England Biolabs, Ipswich, MA) and NotI, and the cut fragment was ligated into the cut vector to obtain GLIC. A C-terminal FLAG epitope tag was introduced in GLIC utilizing PCR with the primer 5′-CTG GCA TTT CTT TTC TTT GGA TTT GAC TAT AAG GAC GAT GAT GAC AAG TAG GCG GCC GCC TAG AAA TAG CTT GAT CTG-3′ and its reverse complement. Sequence coding for the FLAG tag (DYKDDDDK) is in bold (31). Inside the GLIC M3 M4 linker region (HYLKVES-insert-QPARAA), we added sequence for unique AflII and BsrGI (sites underlined) sites by PCR using the primer 5′-CTA AAA GTT GAG AGC CTT AAG AAC GTA TGT ACA CAG CCC GCC AGG-3′ and its reverse complement (31). These two restriction sites were utilized to ligate an appropriately obtained mouse 5-HT3A-ICD fragment (QDL QRP VP···RDW LRV GY) to obtain GLIC-AlB (see Fig. 1B). The loop was inserted at the apex of the GLIC M3M4 loop between KVES and QPAR (see Fig. 1B) where the two α-helical segments are linked by a short loop in GLIC (see Fig. 1C). Subsequently, to optimize the insertion points of the ICD, the flanking regions of GLIC both N-terminal and C-terminal to the insert were cut back stepwise, yielding 11 additional chimeric GLIC-5-HT3A-ICD constructs (stepwise removal color-coded similarly in Fig. 1, B and C). Similarly, by first creating an AlB construct and then polishing the N- and C-terminal ends of GLIC, a comparable set of 12 MBP-GLIC (11) chimeras in the bacterial expression vector pET26b (Novagen) was obtained. The identity of all constructs was verified by DNA sequencing (Genewiz, South Plainfield, NJ) of the complete coding region.
Oocyte Expression
Plasmids were linearized with XbaI for in vitro transcription using T7 RNA polymerase (mMESSAGE mMACHINE kit, Applied Biosystems/Ambion, Austin, TX). Capped mRNA was purified (MEGAclear kit, Applied Biosystems/Ambion) and precipitated with ammonium acetate. mRNA was dissolved in nuclease-free water and stored at −80 °C. The integrity of the mRNA was tested by agarose gel electrophoresis. X. laevis oocytes were harvested and defolliculated as described previously (32). Unless otherwise noted, oocytes were injected 24 h after isolation with 10 ng of mRNA (0.2 μg/μl) and were kept in standard oocyte saline (100 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2, 5 mm HEPES, pH 7.5) supplemented with 1% antibiotic-antimycotic (100×) and 5% horse serum for 4–7 days at 17 °C.
Oocyte Two-electrode Voltage Clamp (TEVC) Experiments
Electrophysiological recordings were conducted 3–7 days after mRNA injection at room temperature in an ∼250-μl chamber continuously perfused at a rate of 5–6 ml/min with GLIC oocyte recording buffer (GORB, 100 mm NaCl, 20 mm NaOH, 2.5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm HEPES, 5 mm citric acid, pH adjusted to 7.5 or as indicated with HCl). Currents were recorded from individual oocytes under two-electrode voltage clamp conditions at a holding potential of −60 mV. The ground electrode was connected to the bath via a 3 m KCl/agar bridge. Glass microelectrodes had a resistance of <2 megaohms when filled with 3 m KCl. Data were acquired and analyzed using a TEV-200 amplifier (Dagan Instruments, Minneapolis, MN), a Digidata 1440A data interface (Molecular Devices, Sunnyvale, CA), and pClamp 10.2 software (Molecular Devices). Currents (IpH) elicited by lowering the pH of GORB to <pH 7.5 were separated by GORB pH 7.5 wash.
pH50 Determination
Proton concentration-response curves were obtained by exposing the oocytes stepwise to GORB of successively lower pH until a steady-state current was reached for each respective pH. After GORB pH 4 application, perfusion was switched back to GORB pH 7.5. Currents were normalized to the maximum current obtained for each oocyte. Most oocytes tolerated pH 4, and currents returned rapidly to the initial baseline upon switching back to pH 7.5. Oocytes that became unstable upon exposure to acidic pH (5–10%) were eliminated from data analysis. No significant currents were obtained in mock oocytes (water-injected or uninjected). At pH 4, the current in mock oocytes was less than 100 nA, which was 10% or less of what was recorded in functional GLIC constructs at the same pH. Data were fit with the equation
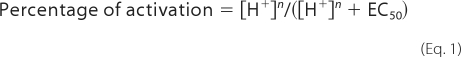 |
where H+ is the proton concentration, n is the Hill coefficient, and EC50 is the H+ concentration for half-maximal activation. pH50 = −logEC50.
RIC-3 Inhibition of Expression of 5-HT3A Receptors and GLIC-5-HT3A-ICD Chimeras
We co-injected X. laevis oocytes with mRNAs for rat 5-HT3A (10 ng) and human RIC-3 (5 ng). We investigated the time course of inhibition by measuring serotonin-induced (10 μm) current amplitudes at different time points after mRNA injection with two-electrode voltage clamp recordings. Subsequently, we investigated RIC-3 inhibition of currents produced by switching from pH 7.5 to pH 5.0 buffer for GLIC and GLIC chimeras by co-injecting mRNAs in a molar ratio comparable with wild-type GLIC (10 ng) to RIC-3 (5 ng) in TEVC recordings performed 72 h after injection or later.
Bacterial Expression
Escherichia coli BL21-CodonPlus (DE3)-RIPL Competent Cells (Agilent) were transformed with the MBP-GLIC-pET26b plasmids and grown in LB medium containing kanamycin and chloramphenicol (both 50 μg/ml) at 37 °C to an A600 of ∼0.4. Protein expression was induced overnight at 20 °C with 0.2 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation at 4,000 × g at 4 °C for 5 min and washed twice with PBS. After resuspension in lysis buffer (100 mm sodium phosphate buffer, pH 8.0, 400 mm NaCl, 1 mm phenylmethanesulfonyl fluoride), cells were disrupted by sonication. Unbroken cells were removed from the cell lysate by centrifugation (20 min at 10,000 × g). Subsequently, membranes were isolated by ultracentrifugation (60 min at 100,000 × g). After the addition of sample buffer, proteins were separated by SDS-PAGE (4–15% TGX gel) (Bio-Rad Laboratories), blotted to PVDF membranes (Bio-Rad Laboratories), and probed with HRP-conjugated anti-MBP antibody (New England Biolabs) and ECL (Thermo Fisher Pierce).
RESULTS
GLIC-5-HT3A-ICD Chimera Construction and Functional Testing
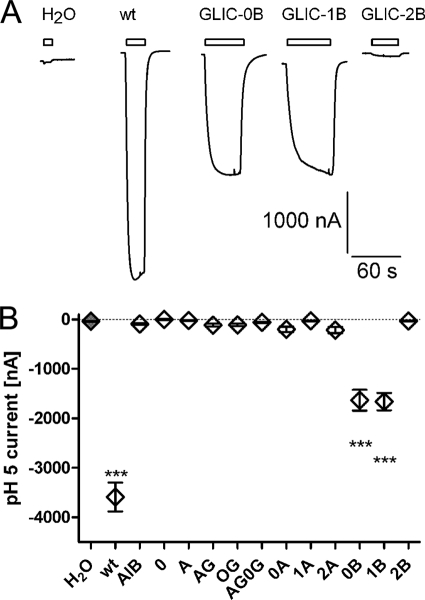
Twelve GLIC-5-HT3A-ICD chimeras were constructed (Fig. 1). All chimeras contain the complete 5-HT3A-ICD but differ in the number of amino acids contributed by the GLIC M3M4 linker and its flanking regions both N-terminal and C-terminal to the 5-HT3A-ICD insertion (Fig. 1, B and C). We expressed the constructs in Xenopus oocytes and tested the macroscopic functionality of the chimeras in TEVC experiments by measuring the current amplitude induced by changing the extracellular pH from 7.5 to pH 5. Two chimeras, GLIC-0B and GLIC-1B, were gated by switching from pH 7.5 to pH 5 (Fig. 2). For these two chimeras, the currents induced by pH 5 were significantly different from water-injected oocytes (Fig. 2). The other 10 chimeras did not show currents significantly different from water-injected oocytes. To test the possibility of functional chimeras with lower pH50 than wild-type GLIC, we repeated the experiment, utilizing a shift from pH 7.5 to pH 4.5 instead of 5. Again, only the two chimeras GLIC-0B and GLIC-1B showed currents significantly different from those of water-injected oocytes (data not shown).
FIGURE 2.
Functional characterization of GLIC-5-HT3A-ICD chimeras. A, currents obtained by TEVC in X. laevis oocytes injected with water, wild-type (wt) GLIC, GLIC-0B, GLIC-1B, or GLIC-2B mRNA, recorded by TEVC by switching perfusion buffer from pH 7.5 to pH 5. Holding potential was −60 mV. B, average current amplitudes recorded for all constructs by switching from pH 7.5 to pH 5. All current amplitudes were compared with water-injected oocytes (gray filled symbol) by one-way analysis of variance with Dunnett's multiple comparison post test. Results significantly different from water-injected oocytes are indicated by *** (p < 0.05). All others were not significantly different. Error bars indicate S.E.
pH50 Determination
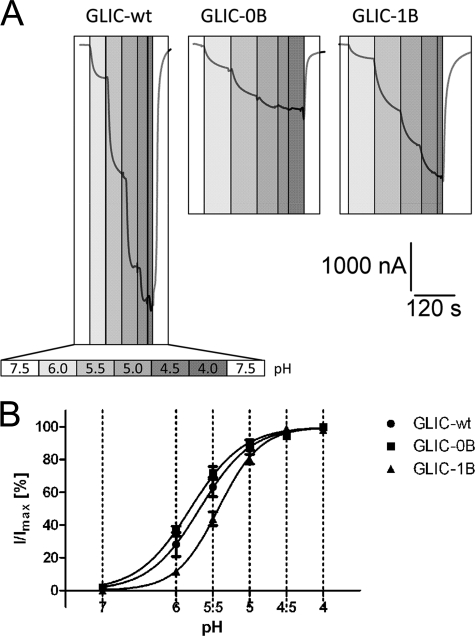
GLIC is activated by external protons (8) (Fig. 3A). For wild-type GLIC and the functional chimeras GLIC-0B and GLIC-1B, we determined the pH that induces half-maximal activation (pH50) (Fig. 3B). The pH50 values for wild-type and GLIC-0B were similar, with 5.70 ± 0.3 (n = 9) and 5.78 ± 0.13 (n = 7), respectively. The pH50 for GLIC-1B was shifted to a slightly more acidic pH of 5.42 ± 0.13 (n = 9), which is significantly different from wild-type GLIC (one-way analysis of variance with Dunnett's post hoc test). Positive cooperativity was observed for all constructs, wild-type (n = 1.47 ± 0.20), GLIC-0B (n = 1.29 ± 0.28), and GLIC-1B (n = 1.62 ± 0.28), with no significant difference between constructs.
FIGURE 3.
Dose-response relationships for functional GLIC-5-HT3A-ICD chimeras. A, currents recorded by TEVC from X. laevis oocytes injected with wild-type GLIC (GLIC-wt), GLIC-0B, or GLIC-1B mRNA, by graded reductions of perfusion buffer pH. Increasing proton concentrations are indicated by increasingly darker gray boxes. pH scale legend is given for wild-type GLIC. Holding potential was −60 mV. B, pH-response curves obtained by TEVC in oocytes by plotting the current amplitudes obtained for the respective pH normalized against the maximum current amplitude obtained at pH 4. Holding potential was −60 mV. Error bars indicate S.E.
RIC-3 Co-expression Decreases Current Amplitudes in 5-HT3A at 24 h or Later
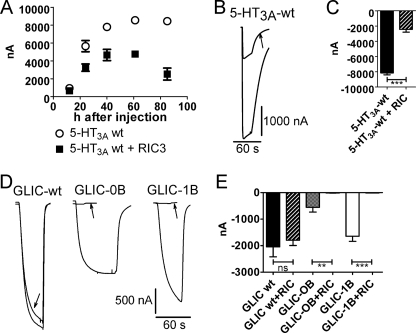
The chaperone protein RIC-3 influences the plasma membrane expression level of various Cys-loop receptors such as α7 and α4β2 nAChR and 5-HT3A receptors by modulating maturation (folding and assembly) of these receptors (33–35). We have shown previously that expression of wild-type 5-HT3AR on oocyte plasma membranes is inhibited by co-expression of RIC-3 by measuring the maximum 5-HT-inducible current with TEVC (19). Deletion of the ICD in 5-HT3A-glvM3M4 abolished RIC-3 inhibition, indicating that the ICD is required for the RIC-3 effect (19). The 5-HT3A-glvM3M4 chimera has the 115-amino acid 5-HT3A-ICD replaced by the G. violaceus M3M4 linker SQPARAA. For the current studies, we investigated the time course of this inhibition to optimize the time point at which to detect a possible modulation (Fig. 4A). Our investigations revealed that recordings at 24 h after injection or later yielded significant inhibition by RIC-3. RIC-3 co-expression with 5-HT3A significantly decreased 5-HT (10 μm)-induced currents by 70 ± 4% (n = 8) as compared with oocytes expressing 5-HT3A alone (Fig. 4, B and C)(19). We tested for modulation of GLIC-5-HT3A chimeras by RIC-3 co-expression 72 h or more after injection of mRNAs.
FIGURE 4.
RIC-3 current inhibition. A, time course of 5-HT3A-ICD-mediated RIC-3 inhibition on 10 μm 5-HT-induced currents in 5-HT3AR. B, current traces for RIC-3 inhibition of currents induced by 10 μm 5-HT for wild-type 5-HT3A. The arrow indicates current trace from RIC-3 co-expressing oocyte. C, averaged RIC-3 inhibition. Currents were obtained by 10 μm 5-HT for wild-type 5-HT3A with and without RIC-3 co-expression. D, current traces for RIC-3 inhibition of currents induced by switching from pH 7.5 to pH 5 for wild-type GLIC, GLIC-0B, and GLIC-1B. The arrows indicate current trace from RIC-3 co-expressing oocytes. E, averaged RIC-3 inhibition. Currents were obtained by switching from pH 7.5 to pH 5 for wild-type GLIC, GLIC-0B, and GLIC-1B with and without RIC-3 co-expression. ns, not significant. Error bars in A, C, and D indicate S.E.
RIC-3 Decreases Current Amplitudes in the Functional Chimeras GLIC-0B and GLIC-1B
To test for correct folding of the 5-HT3A-ICD in the chimeras, we utilized its interaction with the chaperone protein RIC-3 (19). We co-expressed GLIC and the functional GLIC-5-HT3A-ICD chimeras with and without RIC-3 in Xenopus oocytes and measured the current amplitudes induced by changing the pH from pH 7.5 to pH 5 (Fig. 4, D and E). RIC-3 co-expression with GLIC did not significantly change current amplitudes as compared with expression of GLIC alone. However, co-expression of the functional chimeric GLIC-5-HT3A-ICD constructs (GLIC-0B and GLIC-1B) together with RIC-3 significantly decreased current amplitudes as compared with expression of the GLIC chimeras alone (94 ± 2.2% inhibition for GLIC-0B and 99 ± 0.6% inhibition for GLIC-1B).
GLIC Chimeras GLIC-0B and GLIC-1B Are Expressed on E. coli Membranes
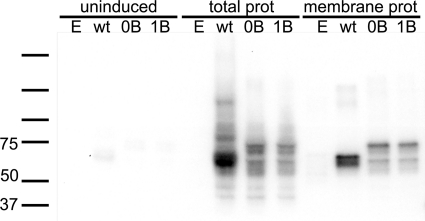
We expressed wild-type GLIC and the two functional chimeras in E. coli as N-terminal fusion proteins with maltose-binding protein (MBP) and investigated their presence on the membrane. All three constructs were found in total bacterial lysates as well as in isolated bacterial membranes (Fig. 5).
FIGURE 5.
GLIC chimeras express in E. coli membranes. Wild-type GLIC and GLIC-0B and GLIC-1B were expressed in E. coli. Fractions shown represent uninduced total protein (uninduced), induced total protein (total prot), and membrane protein (membrane prot). Bacteria transformed with empty plasmid (E) were used as a negative control.
DISCUSSION
The recent crystal structures of the prokaryotic Cys-loop receptors ELIC and GLIC and the most recent one of the eukaryotic GluCl have provided insights into these ion channels at ∼3 Å resolution (9–11, 18). Additional co-crystal structures of Cys-loop superfamily ligand-gated ion channels with a variety of ligands have been obtained; GLIC was crystallized at acidic pH without and with allosteric modulators such as general anesthetics (36) and with channel blockers such as lidocaine (37). GluCl was crystallized in the presence of ivermectin together with and without glutamate or picrotoxin (18). The location of the ivermectin site in the GluCl x-ray structures closely coincides with the binding site for the intravenous anesthetics propofol and etomidate at the TMD lipid interface previously identified (38–40). The picrotoxin-binding site location inside the channel had also been previously identified by functional studies (41).
It is important to emphasize that all Cys-loop receptors for which a crystal structure has been solved consist of only two domains, the ECD and the TMD. This has greatly aided our insights into these two domains that are the targets for all drugs in current clinical use that target Cys-loop receptors. However, these domains harbor significant sequence identities between subtypes belonging to one Cys-loop family but also between members of other families within the Cys-loop superfamily. This hampers the design of subtype-selective compounds that would be devoid of side effects caused by the interference with off-target subunits. While the ECD and TMD bear significant similarities, the ICD is by far the most diverse domain with respect to both length and amino acid composition. Together with the involvement of the ICD in functional but also regulatory aspects of Cys-loop receptors, this diversity makes the ICD an intriguing target for future drug design. The electron microscopy-derived structure of Torpedo nAChR is the only structure that resolved part of the ICD. However, only one-third, the so-called membrane-associated (MA) helix that precedes M4, was resolved. It is controversial whether the MA helix is present in only cationic or also anionic Cys-loop receptors. When they were cloned, the anionic receptors were suspected to lack an MA helix (42–44), but some recent studies assumed that anionic receptors do contain an MA helix (45, 46). However, this has not been definitely investigated. To date, the MA helix has been implicated in gating kinetics, assembly, and unitary conductance (24, 45, 47–53). At present, knowledge about the structure and function of the ICD is limited.
We generated a systematic set of chimeras consisting of the ECD and TMD of GLIC and the ICD of 5-HT3A receptors. Both receptors can form functional homopentamers (8, 54, 55). The initial chimera contained the entire GLIC sequence including the loop connecting the M3 and M4 transmembrane segments in addition to the complete 5-HT3A-ICD and 2 additional amino acids both N-terminal and C-terminal to the inserted loop because of our cloning procedure. Subsequently, we removed the amino acids from cloning and varied the peptide fragments from the GLIC M3M4 loop at both the N terminus and the C terminus of the inserted 5-HT3A-ICD in various combinations. Out of 12 GLIC-5-HT3A-ICD chimeras, two were functional, as evidenced by electrophysiological studies that show that the GLIC-0B and GLIC-1B chimeras are proton-gated channels similar to the parent GLIC. Our experimental data further indicate that the insertion of a complete additional domain did not interfere with the overall macroscopic function, as evidenced by similar pH50 values and Hill coefficients.
We can only speculate why two chimeras were functionally comparable with wild-type GLIC, whereas 10 chimeras were not. All chimeras that left the parent GLIC M3 and M4 helices completely intact were not functional: GLIC-AlB, GLIC-0, GLIC-A, GLIC-AG, GLIC-0G, GLIC-AG0G, and GLIC-0A. The linker between the helical segments M3 and M4 in the GLIC x-ray structure is 3 amino acids long (SQP). For the optimization of the C-terminal side of the insertion point of the 5-HT3A-ICD, among the chimeras tested, it was absolutely required to cut back to position B (Fig. 1B) in M4 of GLIC to gain function; GLIC-0A and -1A were not functional, whereas GLIC-0B and -1B were functional. The difference between the A and B constructs is the absence of the QPARAA fragment that contains the start of M4 (underlined) from the B constructs. Notably, the peptide contains a proline at the boundary of the M4 helix with the M3M4 loop. Due to the cyclic nature of the proline side chain, the conformational flexibility of this amino acid is severely reduced, resulting in a rigid component. The inserted 5-HT3A-ICD is predicted, like all other cationic Cys-loop receptors, to contain an α-helical segment preceding M4, the so-called MA helix. Together with the presence of the proline contributed by the GLIC linker of the chimeras between the inserted 5-HT3A-ICD and the GLIC-M4 segment, this might have posed a higher degree of conformational strain on the chimeras and thus may have interfered with correct folding/functionality. With regard to the optimization on the N-terminal side of the insertion, the 0 or 1 sites lead to functional chimeras. The difference between these is the peptide KVES including the end of M3 (underlined). Removing additional amino acids (QHYL) as in the 2 constructs leads to a loss of function, as seen in GLIC-1B that is functional and GLIC-2B that is not, likely due to removal of too many residues (total of 7 residues, 3 helical ones from the 1 insertion point plus 4 more from the 2 insertion point) from M3.
Our goal was to generate chimeras with wild-type-like functionality, which we obtained with GLIC-0B and GLIC-1B. Therefore, we optimized the insertion points in steps of several amino acids instead of further amino acid-based optimization. Functional chimeras between GLIC and the 5-HT3A-ICD will be critical for further studies to investigate the structure and function of the ICD. Purification of membrane proteins for structural studies is often difficult. One strategy that has been used successfully is to generate fusion proteins combining a soluble protein to the target protein of interest. The soluble protein facilitates the purification, and in some cases, the crystallization process. MBP, glutathione S-transferase (GST), thioredoxin, and lysozyme have been utilized as the soluble partner for the fusion protein. Several crystal structures of membrane proteins have been solved by creating such fusion proteins. Recently, several G-protein-coupled receptor (dopamine R, chemokine R, and adenosine R) structures have been solved with T4-lysozyme (56–60), and a transmembrane domain from T cell leukemia virus type 1 gp21 ectodomain has been solved with MBP (61). Chimeras obtained by exchanging domains or segments between two homologous proteins have also greatly facilitated membrane-protein studies, for example, with potassium channels (62, 63), glutamate receptors (64), and also Cys-loop receptors (25–29). Here we designed and optimized a chimera combining a transmembrane protein with a known crystal structure, GLIC, as a carrier and the intracellular loop of 5-HT3A receptors as the target. The ICD of Cys-loop receptors is predicted to contain a high degree of disorder by bioinformatics approaches and experimental methods (15, 19, 65, 66). This might preclude the ICD from x-ray structural determination. However, spectroscopic methods such as Fourier transform infrared and circular dichroism spectroscopy (CD) are well suited to investigate the secondary structure content of the ICD. Our chimeric approach is applicable to other homopentameric Cys-loop receptors as well.
To assess the folding of the engineered third domain in our chimeras, we utilized the modulation of expression of 5-HT3A-ICD-containing receptors by the chaperone protein RIC-3 (Fig. 4). Under our experimental conditions, the expression of 5-HT3A receptors and functional GLIC-5-HT3A-ICD chimeras, but not of 5-HT3A-glvM3M4 or wild-type GLIC, was reduced by co-expression of RIC-3, as determined by attenuation of maximum inducible currents in TEVC experiments in X. laevis oocytes. This supports the conclusion that the 5-HT3A-ICD in the chimeras is folded in a native conformation or at least in a conformation that is amenable to interaction with RIC-3. We infer that at least the portion of the 5-HT3A-ICD inside the GLIC-5-HT3A-ICD chimeras that interacts with RIC-3 can adapt a conformation suitable for RIC-3 interaction.
We have demonstrated previously by removing the ICD in 5-HT3AR that the inhibitory effect of RIC-3 requires the presence of the ICD (19). This was in contrast to a previous study suggesting that a single amino acid in the ECD close to the first transmembrane segment M1 in a chimera consisting of the ECD from α7 nAChR and the TMD and ICD from 5-HT3AR was responsible for the RIC-3 inhibition (67). Later the same laboratory had shown involvement of parts of the ICD, although of α7 nAChR, in RIC-3 modulation (68). The present study supports our previous conclusion that the 5-HT3A-ICD is required for RIC-3 inhibition by using the converse approach to the one we used previously. By adding the 5-HT3A-ICD to RIC-3-insensitive GLIC subunits, the resulting chimeric channels were now RIC-3-sensitive, additionally supporting our hypothesis that RIC-3 interacts with the 5-HT3A-ICD.
Our results further support the hypothesis that the ICD is an independent module that can be removed from and added to Cys-loop receptors while retaining the overall ability of the receptor to fold, assemble, and function as an ion channel. Contrary to the ECD and TMD, however, the ICD is extremely divergent with regard to length and amino acid composition between different Cys-loop receptor subunits. The presently created chimeras will allow structural studies of the ICD after expression in E. coli. These functional chimeras will allow structural studies of the ICD in a “native-like” environment, tethered and thus constrained by M3 and M4 and at the lipid bilayer. Similar chimeras will be created for several homopentameric anionic (glycine receptor (GlyR) and GABAρ) and cationic (α7) Cys-loop family members to experimentally investigate the contribution of their respective ICDs to the structure and function of GLIC chimeras. This will allow us to investigate and compare the secondary structure content between cationic and anionic receptors.
Acknowledgments
We thank Phaneendra Kumar Duddempudi, Juan Pedro Ortega, Jonathan Pauwels, Robin Rajan, and Valentina Snetkova for helpful discussions. We thank Professor Millet Treinin, Hebrew University of Jerusalem, Israel, for providing the human RIC-3 construct in the pGEMH19 expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grant NS059841 (to M. J.). This work was also supported by grants from the South Plains Foundation (SPF) (to M. J.) and the Texas Tech University Health Sciences Center (TTUHSC) School of Medicine Student Summer Research Program (to A. A. S.).
- nAChR
- nicotinic acetylcholine receptor
- ECD
- extracellular domain
- ICD
- intracellular domain
- TMD
- transmembrane domain
- ELIC
- E. chrysanthemi ligand-gated ion channel
- GLIC
- G. violaceus ligand-gated ion channel
- GORB
- GLIC oocyte recording buffer
- 5-HT3
- 5-hydroxy tryptamine (serotonin) type 3
- MBP
- maltose-binding protein
- RIC-3
- resistance to inhibitors of cholinesterase type 3
- TEVC
- two-electrode voltage clamp
- GluCl
- glutamate-gated chloride channel
- MA
- membrane-associated.
REFERENCES
- 1. Wang H., Bedford F. K., Brandon N. J., Moss S. J., Olsen R. W. (1999) Nature 397, 69–72 [DOI] [PubMed] [Google Scholar]
- 2. Meyer G., Kirsch J., Betz H., Langosch D. (1995) Neuron 15, 563–572 [DOI] [PubMed] [Google Scholar]
- 3. Tretter V., Jacob T. C., Mukherjee J., Fritschy J. M., Pangalos M. N., Moss S. J. (2008) J. Neurosci. 28, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charych E. I., Yu W., Miralles C. P., Serwanski D. R., Li X., Rubio M., De Blas A. L. (2004) J. Neurochem. 90, 173–189 [DOI] [PubMed] [Google Scholar]
- 5. Keller S. H., Lindstrom J., Ellisman M., Taylor P. (2001) J. Biol. Chem. 276, 18384–18391 [DOI] [PubMed] [Google Scholar]
- 6. Ren X. Q., Cheng S. B., Treuil M. W., Mukherjee J., Rao J., Braunewell K. H., Lindstrom J. M., Anand R. (2005) J. Neurosci. 25, 6676–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tasneem A., Iyer L. M., Jakobsson E., Aravind L. (2005) Genome Biol. 6, R4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J. P., Corringer P. J. (2007) Nature 445, 116–119 [DOI] [PubMed] [Google Scholar]
- 9. Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 10. Hilf R. J., Dutzler R. (2009) Nature 457, 115–118 [DOI] [PubMed] [Google Scholar]
- 11. Hilf R. J., Dutzler R. (2008) Nature 452, 375–379 [DOI] [PubMed] [Google Scholar]
- 12. Parikh R. B., Bali M., Akabas M. H. (2011) J. Biol. Chem. 286, 14098–14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez-Gutierrez G., Grosman C. (2010) J. Mol. Biol. 403, 693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Celie P. H., van Rossum-Fikkert S. E., van Dijk W. J., Brejc K., Smit A. B., Sixma T. K. (2004) Neuron 41, 907–914 [DOI] [PubMed] [Google Scholar]
- 15. Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 16. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 17. Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 18. Hibbs R. E., Gouaux E. (2011) Nature 474, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jansen M., Bali M., Akabas M. H. (2008) J. Gen. Physiol. 131, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gensler S., Sander A., Korngreen A., Traina G., Giese G., Witzemann V. (2001) Eur. J. Biochem. 268, 2209–2217 [DOI] [PubMed] [Google Scholar]
- 21. Li M., Lester H. A. (2002) Biophys. J. 83, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ilegems E., Pick H., Deluz C., Kellenberger S., Vogel H. (2005) Chembiochem 6, 2180–2185 [DOI] [PubMed] [Google Scholar]
- 23. Ilegems E., Pick H. M., Deluz C., Kellenberger S., Vogel H. (2004) J. Biol. Chem. 279, 53346–53352 [DOI] [PubMed] [Google Scholar]
- 24. Valor L. M., Mulet J., Sala F., Sala S., Ballesta J. J., Criado M. (2002) Biochemistry 41, 7931–7938 [DOI] [PubMed] [Google Scholar]
- 25. Eiselé J. L., Bertrand S., Galzi J. L., Devillers-Thiéry A., Changeux J. P., Bertrand D. (1993) Nature 366, 479–483 [DOI] [PubMed] [Google Scholar]
- 26. Mihic S. J., Ye Q., Wick M. J., Koltchine V. V., Krasowski M. D., Finn S. E., Mascia M. P., Valenzuela C. F., Hanson K. K., Greenblatt E. P., Harris R. A., Harrison N. L. (1997) Nature 389, 385–389 [DOI] [PubMed] [Google Scholar]
- 27. Bouzat C., Gumilar F., Spitzmaul G., Wang H. L., Rayes D., Hansen S. B., Taylor P., Sine S. M. (2004) Nature 430, 896–900 [DOI] [PubMed] [Google Scholar]
- 28. Grutter T., Prado de Carvalho L., Virginie D., Taly A., Fischer M., Changeux J. P. (2005) C. R. Biol. 328, 223–234 [DOI] [PubMed] [Google Scholar]
- 29. Duret G., Van Renterghem C., Weng Y., Prevost M., Moraga-Cid G., Huon C., Sonner J. M., Corringer P. J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 12143–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jespersen T., Grunnet M., Angelo K., Klaerke D. A., Olesen S. P. (2002) BioTechniques 32, 536–540 [DOI] [PubMed] [Google Scholar]
- 31. Kunkel T. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jansen M., Akabas M. H. (2006) J. Neurosci. 26, 4492–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Halevi S., Yassin L., Eshel M., Sala F., Sala S., Criado M., Treinin M. (2003) J. Biol. Chem. 278, 34411–34417 [DOI] [PubMed] [Google Scholar]
- 34. Williams M. E., Burton B., Urrutia A., Shcherbatko A., Chavez-Noriega L. E., Cohen C. J., Aiyar J. (2005) J. Biol. Chem. 280, 1257–1263 [DOI] [PubMed] [Google Scholar]
- 35. Halevi S., McKay J., Palfreyman M., Yassin L., Eshel M., Jorgensen E., Treinin M. (2002) EMBO J. 21, 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nury H., Van Renterghem C., Weng Y., Tran A., Baaden M., Dufresne V., Changeux J. P., Sonner J. M., Delarue M., Corringer P. J. (2011) Nature 469, 428–431 [DOI] [PubMed] [Google Scholar]
- 37. Hilf R. J., Bertozzi C., Zimmermann I., Reiter A., Trauner D., Dutzler R. (2010) Nat. Struct. Mol. Biol. 17, 1330–1336 [DOI] [PubMed] [Google Scholar]
- 38. Bali M., Jansen M., Akabas M. H. (2009) J. Neurosci. 29, 3083–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li G. D., Chiara D. C., Cohen J. B., Olsen R. W. (2010) J. Biol. Chem. 285, 8615–8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li G. D., Chiara D. C., Sawyer G. W., Husain S. S., Olsen R. W., Cohen J. B. (2006) J. Neurosci. 26, 11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bali M., Akabas M. H. (2007) J. Gen. Physiol. 129, 145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schofield P. R., Darlison M. G., Fujita N., Burt D. R., Stephenson F. A., Rodriguez H., Rhee L. M., Ramachandran J., Reale V., Glencorse T. A., Seeburg P. H., Barnard E. A. (1987) Nature 328, 221–227 [DOI] [PubMed] [Google Scholar]
- 43. Grenningloh G., Rienitz A., Schmitt B., Methfessel C., Zensen M., Beyreuther K., Gundelfinger E. D., Betz H. (1987) Nature 328, 215–220 [DOI] [PubMed] [Google Scholar]
- 44. Grenningloh G., Gundelfinger E., Schmitt B., Betz H., Darlison M. G., Barnard E. A., Schofield P. R., Seeburg P. H. (1987) Nature 330, 25–26 [DOI] [PubMed] [Google Scholar]
- 45. Hales T. G., Dunlop J. I., Deeb T. Z., Carland J. E., Kelley S. P., Lambert J. J., Peters J. A. (2006) J. Biol. Chem. 281, 8062–8071 [DOI] [PubMed] [Google Scholar]
- 46. Carland J. E., Cooper M. A., Sugiharto S., Jeong H. J., Lewis T. M., Barry P. H., Peters J. A., Lambert J. J., Moorhouse A. J. (2009) J. Biol. Chem. 284, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 47. Bouzat C., Bren N., Sine S. M. (1994) Neuron 13, 1395–1402 [DOI] [PubMed] [Google Scholar]
- 48. Milone M., Wang H. L., Ohno K., Prince R., Fukudome T., Shen X. M., Brengman J. M., Griggs R. C., Sine S. M., Engel A. G. (1998) Neuron 20, 575–588 [DOI] [PubMed] [Google Scholar]
- 49. Akk G., Steinbach J. H. (2000) J. Physiol. 527, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu X. M., Hall Z. W. (1994) Neuron 13, 247–255 [DOI] [PubMed] [Google Scholar]
- 51. Kelley S. P., Dunlop J. I., Kirkness E. F., Lambert J. J., Peters J. A. (2003) Nature 424, 321–324 [DOI] [PubMed] [Google Scholar]
- 52. Peters J. A., Kelley S. P., Dunlop J. I., Kirkness E. F., Hales T. G., Lambert J. J. (2004) Biochem. Soc. Trans. 32, 547–552 [DOI] [PubMed] [Google Scholar]
- 53. Sadtler S., Laube B., Lashub A., Nicke A., Betz H., Schmalzing G. (2003) J. Biol. Chem. 278, 16782–16790 [DOI] [PubMed] [Google Scholar]
- 54. Belelli D., Balcarek J. M., Hope A. G., Peters J. A., Lambert J. J., Blackburn T. P. (1995) Mol. Pharmacol. 48, 1054–1062 [PubMed] [Google Scholar]
- 55. Miyake A., Mochizuki S., Takemoto Y., Akuzawa S. (1995) Mol. Pharmacol. 48, 407–416 [PubMed] [Google Scholar]
- 56. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 58. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chien E. Y., Liu W., Zhao Q., Katritch V., Han G. W., Hanson M. A., Shi L., Newman A. H., Javitch J. A., Cherezov V., Stevens R. C. (2010) Science 330, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kobe B., Center R. J., Kemp B. E., Poumbourios P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4319–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 63. Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Janovjak H., Szobota S., Wyart C., Trauner D., Isacoff E. Y. (2010) Nat. Neurosci. 13, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kottwitz D., Kukhtina V., Dergousova N., Alexeev T., Utkin Y., Tsetlin V., Hucho F. (2004) Protein Expr. Purif. 38, 237–247 [DOI] [PubMed] [Google Scholar]
- 66. Kukhtina V., Kottwitz D., Strauss H., Heise B., Chebotareva N., Tsetlin V., Hucho F. (2006) J. Neurochem. 97, Suppl. 1, 63–67 [DOI] [PubMed] [Google Scholar]
- 67. Castillo M., Mulet J., Gutiérrez L. M., Ortiz J. A., Castelán F., Gerber S., Sala S., Sala F., Criado M. (2005) J. Biol. Chem. 280, 27062–27068 [DOI] [PubMed] [Google Scholar]
- 68. Castelán F., Mulet J., Aldea M., Sala S., Sala F., Criado M. (2007) J. Neurochem. 100, 406–415 [DOI] [PubMed] [Google Scholar]