Abstract
The progesterone receptor is able to bind to a large number and variety of ligands that elicit a broad range of transcriptional responses ranging from full agonism to full antagonism and numerous mixed profiles inbetween. We describe here two new progesterone receptor ligand binding domain x-ray structures bound to compounds from a structurally related but functionally divergent series, which show different binding modes corresponding to their agonistic or antagonistic nature. In addition, we present a third progesterone receptor ligand binding domain dimer bound to an agonist in monomer A and an antagonist in monomer B, which display binding modes in agreement with the earlier observation that agonists and antagonists from this series adopt different binding modes.
Keywords: Drug Design, Endocrinology, Nuclear Receptors, Progesterone, Protein Structure
Introduction
The progesterone receptor (PR)2 is a nuclear receptor, related most closely to the glucocorticoid receptor, androgen receptor, and mineralocorticoid receptor (1). PR has a primary role in pregnancy, both the initiation and maintenance of gestation (2), as well as other functions related to menstruation and the endometrium (3). PR drug discovery has historically focused on three areas as follows: the discovery of pure agonists for contraception, discovery of anti-progestins blocking the function of endogenous progesterone for pregnancy termination, and more recently, the development of so-called selective progesterone receptor modulators (SPRMs) for a number of indications, including uterine fibroids and endometriosis (2). PR modulators continue to provide important medical interventions in a number of areas of reproductive medicine (4, 5) with an outlook for application in other fields, including oncology (6). Clinically relevant PR modulators span a spectrum of function from full agonists through to full antagonists (7), with interest growing particularly in the development of compounds with mixed profiles (8, 9). The most comprehensively studied SPRMs are all steroidal, including compounds such as asoprisnil (10), but early accounts of nonsteroidal SPRMs have been reported as well (11, 12).
Identifying the molecular determinants for the ligand-induced switch between agonism and antagonism, and the infinite number of mixed profiles between, is an important step in enabling rational design of novel SPRMs and is facilitated by structural biology studies of the progesterone receptor (13). Nuclear hormone receptor structural biology has proven to be important in helping to understand the biology and function of this protein family, including ligand-driven effects (14–19).
X-ray crystallography studies of PR bound to the 11β-substituted steroids, asoprisnil (20) and RU486 (21), along with the full PR agonist levonorgestrel (22) clearly indicate a crucial function for Met909 in the agonism/antagonism balance. This has also been shown to be true for nonsteroidal SPRMs (11, 12). Met909 sits within helix-12 at the C-terminal end of the ligand binding domain (LBD), and in the classic agonist conformation of the receptor, is oriented toward the ligand binding pocket. Met909 is typically the only helix-12 residue directly in contact with ligands, and the nature of these interactions appears to be a key determinant of the function of the receptors. Clashes between Met909 and ligands are likely to destabilize helix-12 (21), which results in a reduced agonistic response. It has even been suggested that the degree of clash with Met909 might correspond directly to the reduction in agonism (2), but this has yet to be shown categorically.
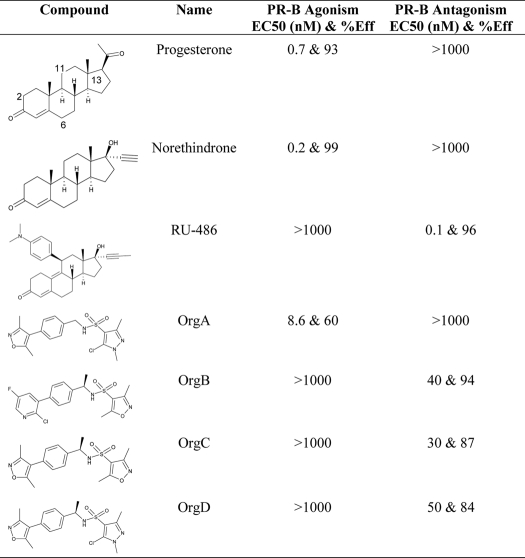
Our earlier work on the discovery and optimization of a selective nonsteroidal glucocorticoid receptor antagonist (23) resulted in the synthesis of a number of moderately active PR modulators, including OrgA, OrgB, and OrgC (see Table 1). Interestingly, OrgA is a partial PR agonist despite high chemical similarity to OrgB and OrgC, which are both full PR antagonists. Furthermore, earlier descriptions of nonsteroidal PR antagonists tended to include mimics of the dimethylaniline moiety of RU486 (24), but neither OrgB nor OrgC has a likely replacement for this group.
TABLE 1.
Cell-based agonism and antagonism, shown for OrgA, OrgB, and OrgC with data and structures for progesterone, norethindrone, and RU486 shown as references
Agonism and antagonism are shown as EC50 values (in nanomolar) with percentage efficacies associated. Efficacy is measured by curve height at maximal effect and expressed as percentage of reference compound effect. >1000 nm indicates no response at the highest tested concentration.
To investigate the molecular basis for antagonism in OrgB and OrgC and the lack of antagonism in OrgA, we have determined their x-ray crystal structures in complex with the agonist conformation of PR following a previously described soaking method (21). Comparison of these structures, along with earlier structures of PR bound to norethindrone and RU486, indicate a basis for the antagonism of OrgB and OrgC, with a clear role for Met909. We also show that OrgA adopts a remarkably different binding mode to the other compounds from the series explaining its lack of antagonism.
MATERIALS AND METHODS
Expression and Purification of PR LBD
The PR LBD, including residues 678–933, was cloned in pET15b (Novagen). Expression was performed in Escherichia coli BL21 Star (DE3) (Invitrogen) in 2× YT medium by overnight induction at 20 °C in the presence of 10 μm OrgA. Bacteria were lysed in buffer A (50 mm Tris, pH 7.8, 250 mm NaCl, 10% glycerol, 10 mm β-mercaptoethanol) with 0.4 mm Pefabloc (Roche Applied Science) and 50 μm OrgA and purified on nickel-nitrilotriacetic acid. Fractions were eluted with buffer A with 100 mm imidazole. Elution fractions were collected and treated with 2.5% w/w thrombin (Kordia) overnight at 4 °C to cleave the N-terminal His tag. Thrombin was removed by adding benzamidine-Sepharose (GE Healthcare), centrifuging for 10 min at 5000 × g, and harvesting the supernatant. To make the final crystallization sample, the protein was dialyzed to buffer A to which 1 mm EDTA, 1 mm dithiothreitol, and 10 μm OrgA were added and subsequently concentrated in a stirring cell to about 4 mg/ml as measured by its absorption at 280 nm. The sample was stored at −70 °C in aliquots of 50 μl.
Crystallization
Crystals of the PR LBD in complex with OrgA were grown at room temperature from 3.5-μl drops hanging over a mother liquor of 20–30% polyethylene glycol 3350, 0.1 m Hepes, pH 6.5, 100 mm Mg2SO4, and 10% glycerol. Drops consisted of 2 μl of protein sample and 1 μl of mother liquor and 0.5 μl of 40% 1,3-propanediol. Crystals usually appeared after about 3 days and were kite-shaped. One crystal was transferred to a cryoprotectant solution of 80% mother liquor and 20% glycerol, dipped in liquid nitrogen, and shipped for data collection to 1.65 Å at 100 K.
Ligand Replacement
Additional crystals of the PR LBD in complex with OrgA were collected and transferred to mother liquor to which 0.25 mm of either compound OrgB or OrgC were added. 0.25 mm is a significant excess of either compound. The crystals were stored in a sitting drop at room temperature. The solutions surrounding the crystals were replaced by fresh solution 10 times over a period of 2 weeks. After this period, the crystals were frozen as described above and shipped for data collection. The dataset for OrgB complex was collected to 2.0 Å at 100 K, and the data for OrgC complex was collected to 1.8 Å at 100 K.
Data Collection
All data were collected by the MXpress service of the European Synchrotron Radiation Facility, Grenoble, France. Structures were solved and refined using the CCP4i interface of the CCP4 software suite (25). All figures have been generated using PyMOL (PyMOL molecular graphics system, Schrodinger, LLC). Data are summarized in Table 2.
TABLE 2.
Final crystallographic data and refinement statistics
Data in parentheses indicate the last resolution shell. r.m.s.d. means root mean square deviation.
| Structure | PR-OrgA | PR-OrgB | PR-OrgC |
|---|---|---|---|
| Compound(s) | OrgA | OrgB | OrgC & OrgA |
| Space group | P21 | P21 | P21 |
| Unit cell | 57.79 × 64.24 × 69.98 Å3 | 56.305 × 64.114 × 69.932 Å3 | 56.25 × 64.06 × 69.69 Å3 |
| β-Angle | 95.67° | 96.53° | 96.62° |
| Resolution | 47.193 to 1.65 Å (1.74 to 1.65 Å) | 28.12 to 1.9 Å (1.97 to 1.9 Å) | 46.17 to 1.80 Å (1.9 to 1.8 Å) |
| Completeness | 95.2% (63.1%) | 97.4% (95.5%) | 96.5% (95.8) |
| Rpima | 0.038 (0.291) | 0.067 (0.388) | 0.042 (0.206) |
| Mn(I/σ) | 14.4 (2.8) | 9.3 (2.6) | 12.9 (3.5) |
| Rfactor/Rfreeb | 0.177/0.206 | 0.213/0.248 | 0.184/0.221 |
| Atoms | 4571 | 4206 | 4384 |
| Protein atoms | 4135 | 4062 | 4077 |
| Ligand atoms | 52 | 54 | 52 |
| Water molecules | 350 | 85 | 226 |
| Other molecules (no. of atoms) | |||
| Glycerol | 64 | 5 | 29 |
| r.m.s.d. bonds Å | 0.01 Å | 0.01 Å | 0.007 Å |
| r.m.s.d. angles | 1.2° | 1.128° | 1.17° |
| B-factors (average Å2) | |||
| Main chain | 5.4 | 20.7 | 12.2 |
| Side chain | 7.8 | 22.2 | 14.3 |
| Water molecules | 17.4 | 24.3 | 21 |
| Ligand A-chain | 12.3 | 37.5 | 18.5 |
| Ligand B-chain | 7.2 | 45.6 | 25 |
| Wilson B-factor (Å2) | 18.3 | 30 | 21.8 |
| Protein Data Bank code | 3ZR7 | 3ZRA | 3ZRB |
a Rpim = Σh((1/(N − 1))1/2Σi|Ii(h) − 〈I(h)〉|)/ΣhΣiIi(h), where I is the observed intensity, and 〈I〉 is the average intensity of multiple observations from symmetry-related reflections. It is an indicator of the precision of the final merged and averaged dataset.
b Rfree was determined using 5% of the data.
Cell-based Activity Measurements
PR agonist and antagonist data for OrgA, OrgB, OrgC, and the reference compounds progesterone and RU486 have been measured following procedures outlined in earlier publications (26–28). Agonism was measured via PR-induced activation of a stably transfected murine mammary tumor virus-luciferase reporter in Chinese hamster ovary cells (27). Org 2058, a PR agonist, (29) was used as a reference. Antagonism was determined by a decrease in Org 2058-induced activation, by competition with the compound of interest. Assays are run in a standardized fashion, and results presented are the average of a duplicate test in two different experiments.
RESULTS AND DISCUSSION
Agonism and Antagonism Are Not Consistent across Series
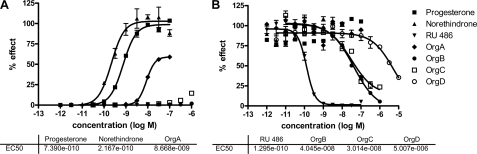
In vitro agonism and antagonism of OrgA, OrgB, and OrgC has been measured in cell-based assays alongside progesterone, norethindrone, and RU486 as references with a summary of results shown in Table 1. Fig. 1A shows the agonistic dose-response curves for all six compounds with progesterone and norethindrone displaying a full agonist effect compared with no effect from RU486, OrgB, and OrgC. The reverse is seen in the antagonistic assay with RU486, OrgB, and OrgC displaying fully antagonistic responses and norethindrone and progesterone showing no effect. OrgA has the unique (among this set) profile of showing a partial agonistic response in the agonistic setting and no response in the antagonistic setting. The molecular basis of the PR partial agonism of OrgA compared with the full PR antagonism of OrgB and OrgC was not apparent, and their x-ray structures were determined to explain the basis for the differing functionality.
FIGURE 1.
Compound activities on PR-driven murine mammary tumor virus-luciferase reporter activity. Assays were run in an agonistic mode (A) or an antagonistic mode (B). In the antagonistic mode, 0.1 nm Org 2058 was added to activate the reporter. Reporter activities are expressed relative to the activity of a saturating concentration of Org 2058, which was set at 100%. The figure shows mean values and standard deviations from four replicate experiments.
Structure of PR LBD Dimer Complexed to OrgA
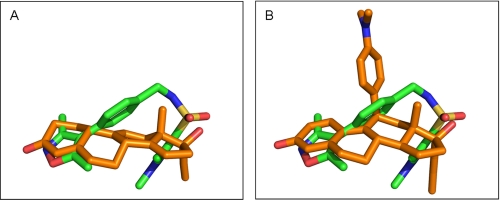
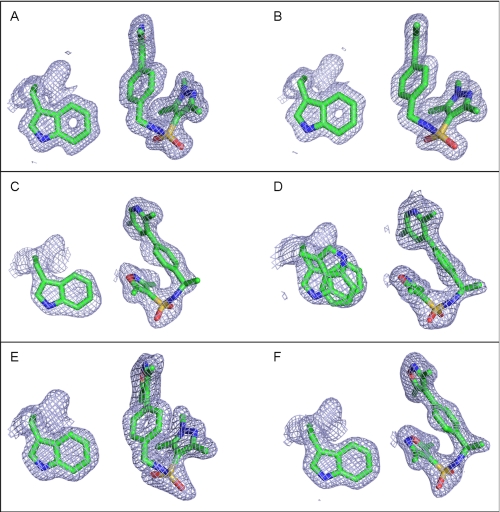
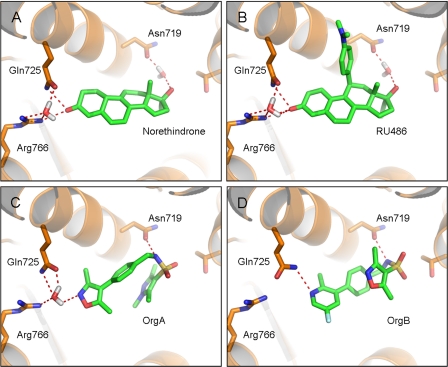
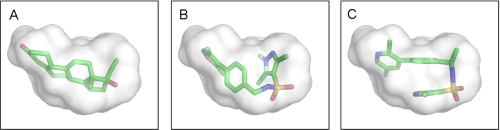
The overall fold of the PR LBD homodimer bound to OrgA is identical to the structure bound to norethindrone with an average root mean square deviation of 0.21 Å for all equivalent Cα atoms. As for the previously described PR x-ray structures bound to nonsteroidal ligands (12, 30, 31), OrgA binds within the traditional ligand-binding pocket in a similar manner to reference steroids as shown in Fig. 2. Electron density maps around the ligand in monomers A and B of the classic dimer are shown in Fig. 3, A and B. The maps account for the entire ligand and enable precise positioning of the ligand with high confidence. Published x-ray structures of PR bound to norethindrone (32) and RU486 (21) show direct interactions between the 3-keto groups from each ligand to Gln725 and Arg766 as well as a structurally conserved water molecule completing complex H-bonding networks as shown in Fig. 4, A and B. OrgA, as shown in Fig. 4C, makes no direct H-bond to either Gln725 or Arg766 but rather relies on water-mediated interactions to these two residues via the previously described conserved water. This appears to be a result of the nitrogen overlaying more closely with the carbon at position 3 in the steroid than the carbonyl oxygen attached at this position as seen in Fig. 2. Although this surely has an effect on the strength of interaction with Gln725 and Arg766, it does allow the methyl groups on the isoxazole to overlay well with the steroid scaffold, specifically positions 2 and 6 (labeled in Table 1). This may have benefits in terms of potency by maximizing hydrophobic van der Waals contacts between the ligand and receptor. The implications of the modified interaction of OrgA with Gln725 and Arg766 are discussed below.
FIGURE 2.
Overlay of OrgA (carbons colored orange) compared with norethindrone (carbons colored green) in A and RU486 in B (carbons colored green). Protein-ligand complexes aligned using the Motif function in Yasara (33) and images were generated using PyMOL.
FIGURE 3.
Shown are the 2Fo − DFc OMIT electron density maps around the ligand and Trp755 in monomers A and B for each of the three complexes shown at 1. 0 σ. Trp755 is included as three-dimensional reference to demonstrate that each of the ligands is shown in a comparable orientation. A is monomer A from the PR-OrgA complex, and B is monomer B from the same complex. C is monomer A from the PR-OrgB complex, and D is monomer B from the same complex. Trp755 is shown in a dual conformation for this monomer as indicated by the electron density map. E is monomer A from the PR-OrgC complex, and F is monomer B from the same complex. Images were generated using PyMOL. The ligand in monomer B is OrgC indicating a successful exchange, but the electron density indicates that monomer A still contains OrgA, indicating that exchange did not occur in this monomer.
FIGURE 4.
Overview of the binding modes of norethindrone (A), RU486 (B), OrgA (C), and OrgB (D). The three key H-bonding residues, Asn719, Gln725, and Arg766 are shown, along with water molecules directly H-bonding to ligands. The remainder of the protein is hidden for clarity. The norethindrone-PR complex is from Protein Data Bank code 1SQN (20), and the RU486-PR complex is also available in the Protein Data Bank, code 1W8Y (21). Protein residues are shown with carbons in orange, and ligands are shown with carbons in green. All structures have been energy-minimized using Yasara (see Ref. 33), and images were generated using PyMOL.
OrgA makes a direct H-bond to Asn719 via its sulfonamide nitrogen in contrast to norethindrone and RU486, which both require the presence of a water molecule to mediate interactions to this residue. Interaction with the equivalent residue in the glucocorticoid receptor, Asn564, is a prerequisite for potent glucocorticoid activity. H-bond networks between ligands (normally 11-OH groups in steroids) and Asn564 and from Asn564 to Glu748 in the loop between helix-11 and helix-12 also appear crucial for mediating a full agonistic response in glucocorticoid receptor (34). This interaction appears less important for progestagenic activity as demonstrated by the lack of any interaction between the endogenous ligand progesterone and Asn719 in PR (13).
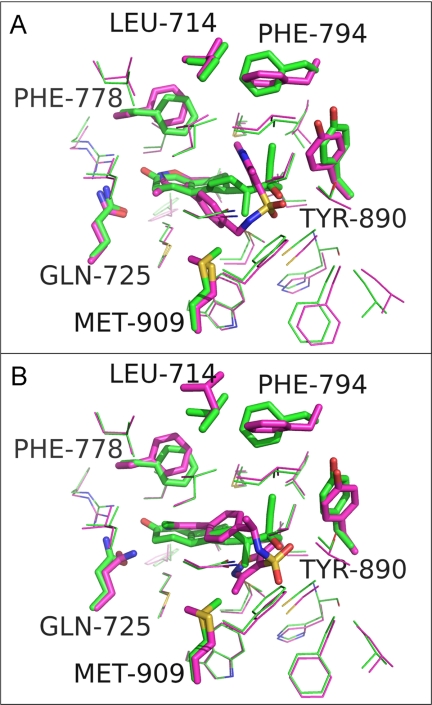
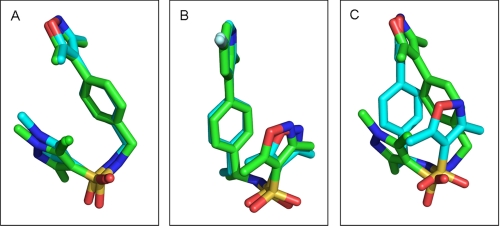
Despite the obvious differences between OrgA and norethindrone, OrgA is generally well tolerated within the same binding pocket as the steroid (Fig. 5, A and B) with just the need for a very small number of side chains to move a short distance as shown in Fig. 6A. Fig. 6A also shows the pyrazole ring of OrgA reaching into the pocket occupied by 17α groups from steroids such as the ethinyl group of norethindrone (13). OrgA also makes use of the space normally filled by 13-methyl groups in classic progestagenic steroids with one of the sulfonamide carbonyl oxygens oriented in close proximity and direction to the hydroxyl oxygen on the 17β position of norethindrone. The sulfonamide carbonyl actually overlays closely with the carbonyl oxygen in the 17β-acetyl group of progesterone with one the methyl group in the acetyl overlaying with one of the methyl substituents on the pyrazole of OrgA (data not shown). Fig. 7A shows an overlay OrgA from monomer A and monomer B demonstrating that no significant difference appears to exist between the two monomers in terms of ligand binding.
FIGURE 5.
A, norethindrone is shown as sticks with the binding pocket of the PR-norethindrone complex shown as a surface. B, PR-OrgA complex overlaid with PR-norethindrone complex using the Motif function in Yasara (33). OrgA is shown in stick format with the binding pocket of the PR-norethindrone complex shown as a surface for comparison. C, PR-OrgB complex overlaid with PR-norethindrone complex using the Motif function in Yasara (33). OrgB is shown in stick format with the binding pocket of the PR-norethindrone complex shown as a surface for comparison. Surface and images were generated in PyMOL.
FIGURE 6.
A, overlaid binding pockets of PR-norethindrone complex (carbons colored green) with PR-OrgA complex (carbons colored magenta). Residues with the largest deviation in position between these two complexes are shown as sticks and labeled, and the remaining residues around the binding pocket are shown as lines. B, as per image A, but PR-OrgA complex replaced by PR-OrgB complex (carbons colored blue). Images were generated in PyMOL.
FIGURE 7.
A, overlay of OrgA from monomer A (shown with carbons colored green) and OrgA from monomer B (shown with carbons colored cyan) from x-ray structure 1. B, overlay of OrgB from monomer A (shown with carbons colored green) and OrgA from monomer B (shown with carbons colored cyan) from x-ray structure 2. C, overlay of OrgA from monomer A (shown with carbons colored green) and OrgC from monomer B (shown with carbons colored cyan) from x-ray structure 3. Images were generated in PyMOL.
Clash with Met909 as a Contributing Factor to Partial Agonism of OrgA
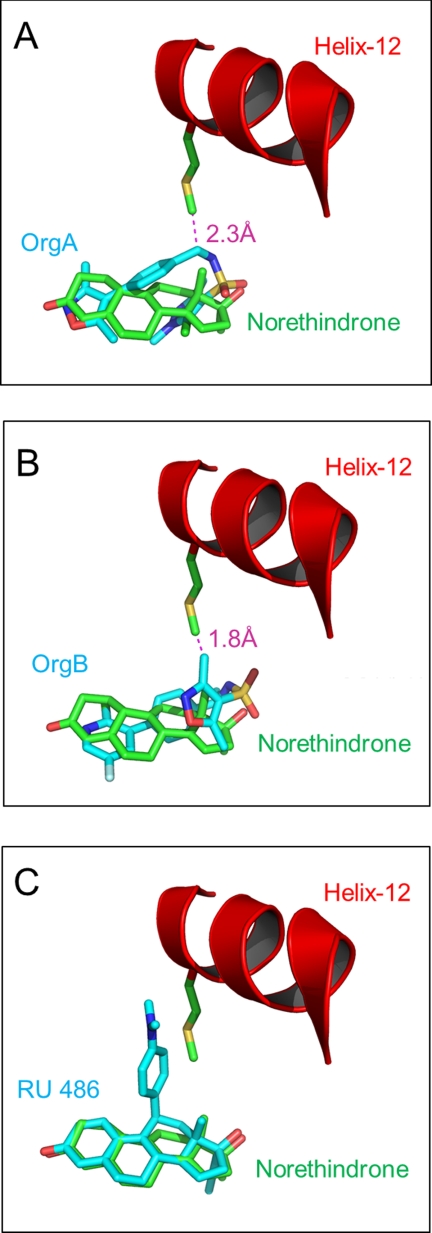
OrgA is a PR agonist but shows reduced efficacy (60%) compared with norethindrone and progesterone (Fig. 1). It has previously been shown that clashes between ligands and Met909 in helix-12 of PR are a key determinant for reduced agonistic activity in steroidal PR antagonists and partial agonists (21). OrgA lacks an obvious structural mimic of the dimethylaniline of RU486, or any of the other bulky 11β groups common to PR antagonists of SPRMs (2). We did consider the possibility that either the isoxazole end of the ligand or more likely the pyrazole end might be oriented to overlay with the 11β-substituent of RU486, but comparison of PR LBDs bound to both ligands demonstrates that this is not case as can be seen in Fig. 2B. However, despite the lack of the classic bulky side chain, OrgA does still impede the agonist position of Met909 as determined by its position bound to norethindrone, resulting in the residue having to adopt a modified orientation. As can be seen in Fig. 8A, the methylene linker between the phenyl ring and the sulfonamide nitrogen in OrgA is only 2.3 Å from the terminal carbon in the side chain of Met909 from the previously described full agonist conformation. This may be sufficient to partially disrupt the ideal agonistic packing of helix-12 and result in the reduced agonistic efficacy of OrgA (60%) compared with the reference steroids progesterone and norethindrone. The fact that the clash with Met909 is made by part of the central scaffold of the ligand might explain why of the nearly 1000 compounds tested in this series, none have been identified as full PR agonists. The clash is not as significant as the clash between Met909 and RU486, shown in Fig. 8C, in line with the observation that OrgA is a partial agonist and not a full antagonist.
FIGURE 8.
A, overlay of PR-norethindrone complex (carbons colored green) with PR-OrgA complex (carbons colored cyan). Met909 from the PR-norethindrone complex is shown along with the ligands. Helix-12 is shown as a red schematic. Distance between the ϵ-carbon in Met909 and the nearest heavy atom in OrgA is shown. B, as per image A, but PR-OrgA complex replaced by PR-OrgB. C, as per image A, but PR-OrgA complex replaced by RU486. Measurements were made and images generated in PyMOL.
Soaking Experiments Resulted in Complete Exchange of OrgA by OrgB
To generate co-crystals of OrgB bound to PR, we have taken PR LBD crystals bound to OrgA and soaked in OrgB as described in earlier experiments (21). To be sure complete exchange has occurred, it was important to carefully analyze the electron density defining the position of the ligand to ensure that it is only compatible with the soaked ligand. OrgB has a disubstituted pyridine replacing the dimethylisoxazole of OrgA, with the larger ring system of the pyridine resulting in a different orientation of the attachments than we would expect to see if the isoxazole was still in place. Next, the pyrazole of OrgA has three substituents (two methyl groups and a chlorine), but the electron density (as shown in Fig. 3, C and D) indicates a ring with two substituents at this end of the structure compatible with the dimethylisoxazole in OrgB. Finally, OrgB has an additional methyl group between the biaryl and sulfonamide ends of its structure, which can be seen in the electron density of the ligand in monomer B most clearly but also in monomer A at lower σ levels. These observations together indicate that OrgB has completely displaced OrgA in the crystals.
Structure of PR LBD Dimer Complexed to OrgB
The successful soaking of OrgB into the OrgA co-expressed PR LBD x-ray structures did not result in any significant change in the structure of the receptor, despite the fact that OrgB is a full antagonist and OrgA is a partial agonist. As discussed in previously (21), it is likely that the receptor exists in equilibrium between agonistic and antagonistic states with ligands at least partly driving that balance. Therefore, although the agonistic conformation of the receptor will not be the lowest energy conformation when bound to OrgB, it does remain a thermodynamically accessible conformation, even if only present at low concentrations. A number of clashes between OrgB and the receptor are present in this structure. These include a clash between the fluorine from the ligand with Val760, a clash with the backbone oxygen of Leu715, and a final clash with Met756. This is in contrast to the OrgA structure, which includes only a moderate clash with Cys891, indicating that OrgA is better tolerated in the agonistic form of PR than OrgB. Physiologically it is likely that these clashes are relieved as helix-12 is displaced on binding of OrgB with the receptor adopting its antagonist conformation.
Comparison of Binding Mode of OrgA Compared with OrgB
As for OrgA, OrgB makes a direct H-bond to Asn719 via the nitrogen of its sulfonamide group shown in Fig. 3D. Unlike for OrgA, the conserved water molecule between Gln725 and Arg766 is not present. Instead, the pyridine nitrogen of OrgB makes a direct H-bond to Gln725, an interaction only possible due to the movement of the residue in this structure compared with PR-OrgA, PR-RU486, and PR-norethindrone structures described earlier. Despite this interesting difference, the most striking change between the binding of OrgA and OrgB was observed around their sulfonamides and the hetero-aryl groups attached to them.
As outlined earlier, the pyrazole group of OrgA is directed toward the so-called 17α pocket, but for OrgB, the isoxazole equivalent to this group is directed in a distinctly different direction toward Trp755. Despite this significant difference, the two observed binding modes for the series share a number of key characteristics. Both are able to mimic the 3-keto group of steroids, and they both make use of the hydrophobic region around the steroidal position 13, make a direct H-bond to Asn719 via the nitrogen of the sulfonamide group, and position a carbonyl from the sulfonamide in the same region as the oxygen in either of the two different 17β groups of progesterone and norethindrone.
Fig. 7B shows the overlay of OrgB from monomer-A and monomer-B, demonstrating that they are practically identical. Despite this, their binding mode, directed toward Trp755, has a different effect depending on the monomer. Specifically, in monomer-B, Trp755 shows a double conformation, presumably as a result of the close proximity of the ligand. This “flipping” of Trp755 has been observed previously (30) but does not appear to correlate with any change in agonism or antagonism.
Clash with Met909 as a Contributing Factor to Full Antagonism of OrgB
Despite adopting a different binding mode to OrgA, OrgB still clashes with Met909 in the classic agonistic position, but now via one of the methyl groups on the isoxazole ring as shown in Fig. 8B. In fact, compared with a partial agonist like OrgA, the clash between OrgB and Met909 appears slightly more significant and likely to be a contributing factor to the full antagonism seen in this series. The clash is still not as significant as the clash between Met909 and RU486, and it is therefore likely that there are other additional factors that render OrgB as a full antagonist compared with the partial agonist OrgA.
Disrupted Interaction between Helix-3 and Helix-5 as a Contributing Factor in the Reduced Agonism of OrgA and OrgB
Agonism in steroid receptors is dependent, at least in part, on the stabilization by interactions between helices 3 and 5. This has most clearly been shown for the mineralocorticoid receptor (35–39), a very close relation of PR. Ligand-mediated (and often water-bridged) interactions between Gln725 in helix-3 and Arg766 in helix-5 are key components in the interaction between these helices. As discussed previously, and shown in Fig. 4A, agonistic steroids such as norethindrone participate in directly mediating an H-bond network between these two residues via their 3-keto groups. This appears to result in the bending of helix-3, and the correct orientation of the charge-clamp residue Lys734. Lys734 in helix-3 and Glu911 in helix-12 are in part responsible for the binding of co-modulating proteins by capping either end of classic LXXLL-binding helices present in co-modulators. Neither OrgA nor OrgB faithfully reproduced the important H-bond network between Gln725 and Arg766, which may certainly contribute to their reduced agonistic efficacy compared with full agonists. For the full antagonist OrgB, no interaction between Gln725 and Arg766 is retained, but the partial agonist OrgA does retain the often conserved water molecule, allowing for a partial retention of the network. It may be possible that this difference contributes to the difference we see in the function of OrgA compared with OrgB.
Molecular Basis for Differences in Binding Modes
Understanding the basis for the switch in binding mode, which appears to result in a switch from partial agonism to full antagonism, was our next goal. An obvious difference between OrgA and the two full antagonists is the presence of an extra stereo center in the latter, due to the presence of an additional methyl group between the biaryl and sulfonamide substructures. The binding mode of OrgB orients this methyl group toward Leu715 in an area of the pocket well able to tolerate its presence. However, examination of the binding mode of OrgA shows that the methyl group of OrgB would not be tolerated in this binding mode, most likely clashing with Cys891, Trp755, and Met756. Although Trp755 could adopt a second position that would relieve this clash, the Cys891 clash would remain unresolved. We therefore conclude that the presence of this methyl group results in the compound series adopting a second binding mode that is only compatible with a displacement of helix-12 and therefore induce full antagonism.
Antagonism of OrgD Confirms Importance of Methyl Group in Determining Function
OrgD is identical to the agonist OrgA except for the addition of a methyl group branched from the carbon atom next to the nitrogen atom in the sulfonamide substructure. PR agonism and antagonism were measured for OrgD (Fig. 1) following the earlier described procedure that clearly identifies this compound as a PR antagonist. This experiment confirms the importance of the methyl group in differentiating between PR agonism and antagonism and therefore supports the structural biology conclusion that this methyl induces a different binding mode for the series that results in a switch from agonism to antagonism.
Structure of PR LBD Dimer Complexed to OrgA and OrgC
Following the same procedure described for OrgB, we attempted to soak OrgC into PR LBD co-crystallized with OrgA. Analysis of the resulting electron density indicated clearly that OrgC had fully replaced OrgA in monomer-B as hoped but that OrgA was still present in monomer-A. In our previous study, RU486 was able to exchange fully with norethindrone in monomer-B but did not replace the co-expressed ligand in monomer-A (21). It has previously been postulated that this difference in success is a result of monomer-B being more open than monomer-A (21).
OrgA in monomer-A of this third x-ray structure remains in the same orientation that we have seen earlier for the binding of OrgA in PR. OrgC, however, adopts the binding orientation of OrgB as shown in Fig. 3, E and F. The overlay of the ligands from the two monomers, demonstrating the divergent binding modes of compounds from the same series bound to the same dimer, is shown in Fig. 7C. As the monomer with the successful soaking of the full antagonist OrgC adopts the same binding mode as the full antagonist OrgB, we presume that they also share the mechanism for their antagonism. Interestingly, although we see a flip of Trp755 in monomer-B bound to OrgB, we do not see the same flipping of Trp755 in the same monomer when bound to OrgC.
Conclusion
We describe here three new PR LBD x-ray structures bound to three structurally related but pharmacologically different compounds, including the first reported structures of PR bound to nonsteroidal antagonists. The agonist and antagonist compounds display distinct binding modes, with the antagonists displaying a more profound clash with Met909 from helix-12 than a partial agonist from the same series. The clash with Met909 reinforces the key role of this residue in the switch between agonism and antagonism of PR ligands as previously shown for 11β-steroids (21) and other nonsteroidal antagonists. The ability to tolerate two distinct binding modes is further evidence of the plasticity of the steroid receptor binding pocket.
Acknowledgments
We thank Joost Uitdehaag and Judith Versteegh for their earlier contribution to the soaking strategy for the progesterone receptor as well as Michael Speake and Angela Cowley for the measurement of in vitro pharmacology data.
The atomic coordinates and structure factors (codes 3ZR7, 3ZRA, and 3ZRB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PR
- progesterone receptor
- LBD
- ligand binding domain
- SPRM
- selective progesterone receptor modulator.
REFERENCES
- 1. Lu N. Z., Wardell S. E., Burnstein K. L., Defranco D., Fuller P. J., Giguere V., Hochberg R. B., McKay L., Renoir J. M., Weigel N. L., Wilson E. M., McDonnell D. P., Cidlowski J. A. (2006) Pharmacol. Rev. 58, 782–797 [DOI] [PubMed] [Google Scholar]
- 2. Madauss K. P., Stewart E. L., Williams S. P. (2007) Med. Res. Rev. 27, 374–400 [DOI] [PubMed] [Google Scholar]
- 3. Spitz I. M., Chwalisz K. (2000) Steroids 65, 807–815 [DOI] [PubMed] [Google Scholar]
- 4. Benagiano G., Bastianelli C., Farris M. (2008) Expert Opin. Pharmacother. 9, 2459–2472 [DOI] [PubMed] [Google Scholar]
- 5. Benagiano G., Bastianelli C., Farris M. (2008) Expert Opin. Pharmacother. 9, 2473–2485 [DOI] [PubMed] [Google Scholar]
- 6. Benagiano G., Bastianelli C., Farris M. (2008) Expert Opin. Pharmacother. 9, 2487–2496 [DOI] [PubMed] [Google Scholar]
- 7. Spitz I. M. (2003) Steroids 68, 981–993 [DOI] [PubMed] [Google Scholar]
- 8. Winneker R. C., Fensome A., Zhang P., Yudt M. R., McComas C. C., Unwalla R. J. (2008) Steroids 73, 689–701 [DOI] [PubMed] [Google Scholar]
- 9. Chabbert-Buffet N., Meduri G., Bouchard P., Spitz I. M. (2005) Hum. Reprod. Update 11, 293–307 [DOI] [PubMed] [Google Scholar]
- 10. DeManno D., Elger W., Garg R., Lee R., Schneider B., Hess-Stumpp H., Schubert G., Chwalisz K. (2003) Steroids 68, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 11. Thompson S. K., Washburn D. G., Frazee J. S., Madauss K. P., Hoang T. H., Lapinski L., Grygielko E. T., Glace L. E., Trizna W., Williams S. P., Duraiswami C., Bray J. D., Laping N. J. (2009) Bioorg. Med. Chem. Lett. 19, 4777–4780 [DOI] [PubMed] [Google Scholar]
- 12. Kallander L. S., Washburn D. G., Hoang T. H., Frazee J. S., Stoy P., Johnson L., Lu Q., Hammond M., Barton L. S., Patterson J. R., Azzarano L. M., Nagilla R., Madauss K. P., Williams S. P., Stewart E. L., Duraiswami C., Grygielko E. T., Xu X., Laping N. J., Bray J. D., Thompson S. K. (2010) Bioorg. Med. Chem. Lett. 20, 371–374 [DOI] [PubMed] [Google Scholar]
- 13. Williams S. P., Sigler P. B. (1998) Nature 393, 392–396 [DOI] [PubMed] [Google Scholar]
- 14. Bain D. L., Heneghan A. F., Connaghan-Jones K. D., Miura M. T. (2007) Annu. Rev. Physiol. 69, 201–220 [DOI] [PubMed] [Google Scholar]
- 15. Kumar R., Thompson E. B. (2003) Mol. Endocrinol. 17, 1–10 [DOI] [PubMed] [Google Scholar]
- 16. Egea P. F., Klaholz B. P., Moras D. (2000) FEBS Lett. 476, 62–67 [DOI] [PubMed] [Google Scholar]
- 17. Moras D., Gronemeyer H. (1998) Curr. Opin. Cell Biol. 10, 384–391 [DOI] [PubMed] [Google Scholar]
- 18. Bourguet W., Germain P., Gronemeyer H. (2000) Trends Pharmacol. Sci. 21, 381–388 [DOI] [PubMed] [Google Scholar]
- 19. Weatherman R. V., Fletterick R. J., Scanlan T. S. (1999) Annu. Rev. Biochem. 68, 559–581 [DOI] [PubMed] [Google Scholar]
- 20. Madauss K. P., Grygielko E. T., Deng S. J., Sulpizio A. C., Stanley T. B., Wu C., Short S. A., Thompson S. K., Stewart E. L., Laping N. J., Williams S. P., Bray J. D. (2007) Mol. Endocrinol. 21, 1066–1081 [DOI] [PubMed] [Google Scholar]
- 21. Raaijmakers H. C., Versteegh J. E., Uitdehaag J. C. (2009) J. Biol. Chem. 284, 19572–19579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petit-Topin I., Turque N., Fagart J., Fay M., Ulmann A., Gainer E., Rafestin-Oblin M. E. (2009) Mol. Pharmacol. 75, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 23. Brown A. R., Bosies M., Cameron H., Clark J., Cowley A., Craighead M., Elmore M. A., Firth A., Goodwin R., Goutcher S., Grant E., Grassie M., Grove S. J., Hamilton N. M., Hampson H., Hillier A., Ho K. K., Kiczun M., Kingsbury C., Kultgen S. G., Littlewood P. T., Lusher S. J., Macdonald S., McIntosh L., McIntyre T., Mistry A., Morphy J. R., Nimz O., Ohlmeyer M., Pick J., Rankovic Z., Sherborne B., Smith A., Speake M., Spinks G., Thomson F., Watson L., Weston M. (2011) Bioorg. Med. Chem. Lett. 21, 137–140 [DOI] [PubMed] [Google Scholar]
- 24. Jones D. G., Liang X., Stewart E. L., Noe R. A., Kallander L. S., Madauss K. P., Williams S. P., Thompson S. K., Gray D. W., Hoekstra W. J. (2005) Bioorg. Med. Chem. Lett. 15, 3203–3206 [DOI] [PubMed] [Google Scholar]
- 25. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 26. Dijkema R., Schoonen W. G., Teuwen R., van der Struik E., de Ries R. J., van der Kar B. A., Olijve W. (1998) J. Steroid Biochem. Mol. Biol. 64, 147–156 [DOI] [PubMed] [Google Scholar]
- 27. Schoonen W. G., Dijkema R., de Ries R. J., Wagenaars J. L., Joosten J. W., de Gooyer M. E., Deckers G. H., Kloosterboer H. J. (1998) J. Steroid Biochem. Mol. Biol. 64, 157–170 [DOI] [PubMed] [Google Scholar]
- 28. Schoonen W. G., Deckers G., de Gooijer M. E., de Ries R., Mathijssen-Mommers G., Hamersma H., Kloosterboer H. J. (2000) J. Steroid Biochem. Mol. Biol. 74, 109–123 [DOI] [PubMed] [Google Scholar]
- 29. Keightley D. D. (1979) Eur. J. Cancer 15, 785–790 [DOI] [PubMed] [Google Scholar]
- 30. Washburn D. G., Hoang T. H., Frazee J. S., Johnson L., Hammond M., Manns S., Madauss K. P., Williams S. P., Duraiswami C., Tran T. B., Stewart E. L., Grygielko E. T., Glace L. E., Trizna W., Nagilla R., Bray J. D., Thompson S. K. (2009) Bioorg. Med. Chem. Lett. 19, 4664–4668 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Z., Olland A. M., Zhu Y., Cohen J., Berrodin T., Chippari S., Appavu C., Li S., Wilhem J., Chopra R., Fensome A., Zhang P., Wrobel J., Unwalla R. J., Lyttle C. R., Winneker R. C. (2005) J. Biol. Chem. 280, 28468–28475 [DOI] [PubMed] [Google Scholar]
- 32. Madauss K. P., Deng S. J., Austin R. J., Lambert M. H., McLay I., Pritchard J., Short S. A., Stewart E. L., Uings I. J., Williams S. P. (2004) J. Med. Chem. 47, 3381–3387 [DOI] [PubMed] [Google Scholar]
- 33. Krieger E., Darden T., Nabuurs S. B., Finkelstein A., Vriend G. (2004) Proteins 57, 678–683 [DOI] [PubMed] [Google Scholar]
- 34. Bledsoe R. K., Stewart E. L., Pearce K. H. (2004) Vitam. Horm. 68, 49–91 [DOI] [PubMed] [Google Scholar]
- 35. Bledsoe R. K., Madauss K. P., Holt J. A., Apolito C. J., Lambert M. H., Pearce K. H., Stanley T. B., Stewart E. L., Trump R. P., Willson T. M., Williams S. P. (2005) J. Biol. Chem. 280, 31283–31293 [DOI] [PubMed] [Google Scholar]
- 36. Fagart J., Huyet J., Pinon G. M., Rochel M., Mayer C., Rafestin-Oblin M. E. (2005) Nat. Struct. Mol. Biol. 12, 554–555 [DOI] [PubMed] [Google Scholar]
- 37. Geller D. S., Farhi A., Pinkerton N., Fradley M., Moritz M., Spitzer A., Meinke G., Tsai F. T., Sigler P. B., Lifton R. P. (2000) Science 289, 119–123 [DOI] [PubMed] [Google Scholar]
- 38. Huyet J., Pinon G. M., Fay M. R., Fagart J., Rafestin-Oblin M. E. (2007) Mol. Pharmacol. 72, 563–571 [DOI] [PubMed] [Google Scholar]
- 39. Li Y., Suino K., Daugherty J., Xu H. E. (2005) Mol. Cell 19, 367–380 [DOI] [PubMed] [Google Scholar]