Abstract
Mucolipidosis II (ML-II) is a fatal inherited metabolic disease caused by deficiency of GlcNAc-phosphotransferase, which plays a role in generating the mannose 6-phosphate recognition marker on lysosomal enzymes. In ML-II, many lysosomal acid hydrolases are mistargeted out of cells, and lysosomes become filled with undigested substrates, which explains inclusion cell disease as an alternative name for this disease. In this study, we revealed various cellular phenotypes in ML-II skin fibroblasts. We quantitated phospholipid and cholesterol within cells and showed ∼2-fold accumulation in ML-II as compared with normal cells. Lysosomal pH of ML-II cells was higher than that of normal cells (5.29 ± 0.08 versus 4.79 ± 0.10, p < 0.001). The proliferated lysosomes in ML-II cells were accumulated ∼3-fold in amount as compared with normal cells. Intracellular logistics including endocytosis and mannose 6-phosphate receptor recycling were impaired in ML-II cells. To confirm whether these ML-II cellular phenotypes derive from deficient lysosomal acid hydrolases within lysosomes, we performed supplementation of lysosomal enzymes using a partially purified total enzyme mixture, which was derived from the conditioned culture medium of normal skin fibroblasts after NH4Cl treatment. This supplementation corrected all of the previously described ML-II phenotypes. In addition, the autophagic and mitochondrial impairment that we have previously reported improved, and inclusion bodies disappeared on electron micrography following total lysosomal enzyme supplementation. Our results indicate that various cellular phenotypes in ML-II are caused by the deficiency of many lysosomal enzymes and massive accumulation of undigested substrates.
Keywords: Fibroblast, Inborn Errors of Metabolism, Lysosomal Acidification, Lysosomal Storage Disease, Membrane Trafficking, Mucolipidosis II
Introduction
The first step of mannose 6-phosphorylation of lysosomal enzymes, which is required for their localization in lysosomes, is mediated by GlcNAc-phosphotransferase (UDP-N-acetylglucosamine:lysosomal-enzyme N-acetylglucosaminephosphotransferase; EC 2.7.8.17) (1, 2). This enzyme is located in the endoplasmic reticulum or cis-Golgi network and transfers GlcNAc-phosphate to the sugar chain of lysosomal acid hydrolases followed by removal of GlcNAc by uncovering enzyme (3). GlcNAc-phosphotransferase is composed of six subunits, α2β2γ2. The α and β subunits are encoded by a single gene GNPTAB (4), which has two transmembrane domains, whereas the γ subunit is encoded by GNPTG. Mucolipidosis II (ML-II)2 alpha/beta (Mendelian Inheritance in Man (MIM) number 252500) and its milder forms, mucolipidosis III alpha/beta (MIM number 252600) and mucolipidosis III gamma (MIM number 252605), are caused by mutations in GNPTAB (5) or GNPTG (6) and are inherited as an autosomal recessive trait. In these diseases, mannose 6-phosphorylation of many lysosomal enzymes is impaired, which finally causes massive accumulation of undigested substrates in lysosomes. These lysosomes are observed in skin fibroblasts as phase-dense bodies, which is the origin of the term “inclusion cell disease.”
ML-II patients show developmental delay and severe bone deformities (dysostosis multiplex), which partially overlap with mucopolysaccharidoses, and life expectancy is limited to ∼10 years. Enzyme replacement therapy (ERT) using recombinant enzyme, which is designed to be incorporated by the mannose 6-phosphate (M6P) receptor and targeted to lysosomes, is available in some lysosomal storage disorders such as Fabry disease, Gaucher disease, Pompe disease, and various mucopolysaccharidoses, whereas many other therapies that enhance enzyme activities or reduce substrates are undergoing clinical trials for many different lysosomal storage disorders (7–13). In the case of ML-II, however, there is no ERT to date as lysosomes lack dozens of enzymes targeted by the M6P receptor-dependent pathway. Extrinsic replacement of GlcNAc-phosphotransferase also seems to be difficult because of its hexameric composition and localization.
In this study, we analyzed ML-II skin fibroblasts and clarified various cellular phenotypes including the pH increase of lysosomes, accumulation of cholesterol, and impaired intracellular trafficking. These impaired cellular functions were corrected and inclusion bodies disappeared following extrinsic total lysosomal enzyme supplementation.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
We obtained skin fibroblasts from an unaffected individual and three individuals affected with ML-II. Samples were collected after obtaining written informed consent according to the institute's regulations for using human cells. We also purchased two normal skin fibroblast cell lines (HDFn from Invitrogen and NHDF from Lonza). The ML-II patients had typical ML-II phenotype and were diagnosed both enzymatically and genetically (14). Mutations in GNPTAB gene were c.3565C>T(p.R1189X)/c.3565C>T(p.R1189X), c.310C>T(p.Q104X)/c.3428_3429insA(p.N1143fs), and c.310C>T(p.Q104X)/c.2544delA(p.K848fs)in respective ML-II patients. Standard culture medium was Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) with heat-inactivated 10% fetal bovine serum (Sigma) and antibiotic-antimycotic (Invitrogen) according to the manufacturers' instructions. Heat inactivation of fetal bovine serum was performed at 56 °C for 30 min. 250× cholesterol lipid concentrate (Invitrogen) was added at a ratio of 1/1000 and 1/100 to standard culture medium for cholesterol loading.
Antibodies
Purified mouse anti-LBPA (Z-PLBPA; Echelon Biosciences), polyclonal anti-LC3 (PM036; MBL International), polyclonal anti-β-actin HRP DirecT (PM053-7; MBL International), monoclonal anti-LAMP-2 (H4B4; Santa Cruz Biotechnology), polyclonal anti-cathepsin B (S-12; Santa Cruz Biotechnology), polyclonal anti-cathepsin D (H-75; Santa Cruz Biotechnology), polyclonal anti-β-glucosidase (H-300; Santa Cruz Biotechnology), monoclonal anti-mannose-6-phosphate receptor (2G11; Abcam), polyclonal anti-ATP6V1B2 (ab73404; Abcam), rabbit polyclonal anti-ATP6V0D1 (18274-1-AP; Proteintech Group), and rabbit anti-mucolipin-1 (N-terminal) (M1696; Sigma) antibodies were purchased.
Measurement of Lysosomal pH
The pH of lysosomes was measured with LysoSensor Yellow/Blue DND-160 (Molecular Probes) according to the manufacturer's protocol. In short, skin fibroblasts were collected by trypsin and stained with LysoSensor at 1:200 concentration in standard culture medium for 10 min followed by washing with PBS. Fluorescence of cell suspension in 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 7.0) was measured by microplate reader (MTP810Lab; Corona) at excitation 365 nm/emission 450 nm (for blue) and 385 nm/emission 550 nm (for yellow). Standard curves were obtained in each sample using pH-fixed MES buffer (pH 4.0–6.5) with 10 μm nigericin (Sigma) and 10 μm monensin (Sigma).
Fluorescence Staining and Micrography
Cells were cultured and fixed with 3.7% formaldehyde for 1 h followed by permeabilization with 0.1% Triton X-100 for 15 min and blocking with 1% bovine serum albumin for 1 h at room temperature. Primary antibody was used at a concentration of 1:100 for all antibodies at room temperature for 1 h. Secondary antibodies (Alexa Fluor 488 or 555; Molecular Probes) were used at a 1:1000 dilution at room temperature for 1 h. MitoTracker Red CMXRos (Molecular Probes) and LysoTracker Red DND-99 (Molecular Probes) were purchased and used at a concentration of 0.2 μm at 37 °C for 1 h before fixation. Filipin III (Sigma) was diluted in PBS to 100 μg/ml for staining at room temperature for 1 h. All fluorescence images were acquired using a fluorescence microscope (BX51; Olympus) or confocal laser scan microscopy system (TCS SP-2; Leica Microsystems). Electron micrographs were taken by Tokai Electron Micrograph Analysis Co. We tested all skin fibroblast cell lines (i.e. three normal and three ML-II) and confirmed similarities within each group. In Figs. 1–6, we show an unaffected individual as an example of a normal case and the first patient (see above) as an ML-II case.
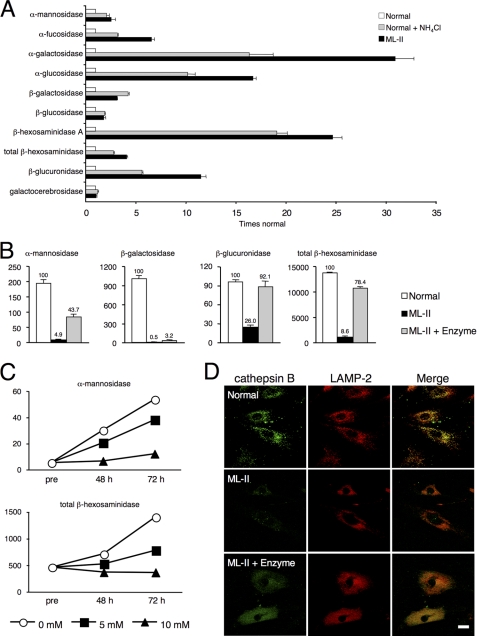
FIGURE 1.
Total lysosomal enzymes are supplied to ML-II skin fibroblasts. A, multiple lysosomal enzymes were excreted into culture supernatant in NH4Cl-treated normal cells and untreated ML-II cells. Activities of 10 major lysosomal enzymes were measured in culture supernatants of normal cells, ML-II cells, and normal cells treated with 20 mm NH4Cl for 7 days. Relative activities as compared with those of normal cells (means ± S.D.) are shown. B, activities of major lysosomal enzymes were measured in skin fibroblasts of normal, ML-II, and enzyme-treated ML-II cells. Measurements were carried out in triplicate for each sample, and means ± S.D. are indicated. Units are nmol/h/mg of protein. Relative activities (%) as compared with normal cells are shown above each bar. C, to elucidate whether this enzyme uptake was mediated by the M6P receptor, we performed enzyme treatment with various concentrations of M6P (0, 5, and 10 mm). ML-II skin fibroblasts were collected before treatment (pre), and at 48 and 72 h after treatment, and major enzyme activities were measured. Means are indicated, and units are nmol/h/mg of protein. Additional data are available for B and C (supplemental Fig. 1). D, micrographs showing localization of enzymes within normal, ML-II, and enzyme-treated ML-II skin fibroblasts evaluated by cathepsin B and LAMP-2 immunocytochemistry co-staining. Merging of cathepsin B and LAMP-2 signals indicates that incorporated enzymes were targeted to lysosomes. Scale bars, 20 μm. We also confirmed these results by immunocytochemical co-staining using cathepsin D and LAMP-2 (data not shown).
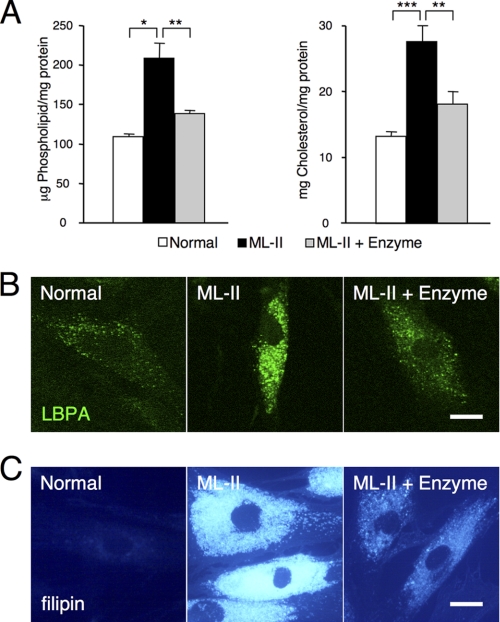
FIGURE 2.
Phospholipid and cholesterol are accumulated in ML-II cells. A, storage materials were measured as phospholipid and cholesterol in normal, ML-II, and enzyme-treated ML-II skin fibroblast total lysates (n = 3). Enzyme treatment of ML-II cells decreased phospholipid and cholesterol contents. *, p < 0.01, **, p < 0.005, ***, p < 0.001. B and C, LBPA (B) and filipin (C) staining micrographs of normal, ML-II, and enzyme-treated ML-II cells are shown. Accumulation of LBPA and cholesterol in endosomes and lysosomes of ML-II cells was corrected by enzyme treatment. Scale bars, 20 μm.
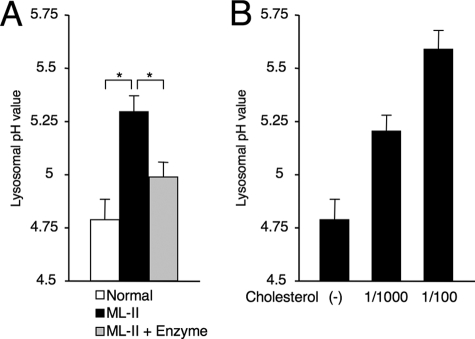
FIGURE 3.
Lysosomal acidification is impaired in ML-II. A, lysosomal pH was measured in skin fibroblasts of normal, ML-II, and enzyme-treated ML-II cells (n = 3). ML-II cells showed a significant increase in lysosomal pH value, and the pH value was normalized by enzyme treatment. *, p < 0.001. B, to elucidate whether storage materials affect the acidifying function, we loaded the cholesterol concentration in culture medium of normal cells. As a result, cholesterol increased lysosomal pH dose dependently in normal cells.
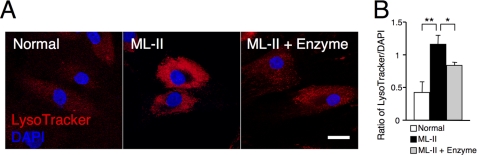
FIGURE 4.
Lysosomes are proliferated in ML-II skin fibroblasts. A, LysoTracker and DAPI fluorescent micrographs of normal, ML-II, and enzyme-treated ML-II cells, indicating a reduction in total lysosomal amount. Scale bar, 20 μm. B, to quantify the amount of lysosomes in one cell, the ratio of LysoTracker/DAPI fluorescence was measured with a plate reader after dual staining of a living suspension of normal, ML-II, and enzyme-treated ML-II cells (n = 3). The lysosomal amount was estimated to be accumulated ∼3-fold in ML-II cells. *, p < 0.05, **, p < 0.005.
FIGURE 5.
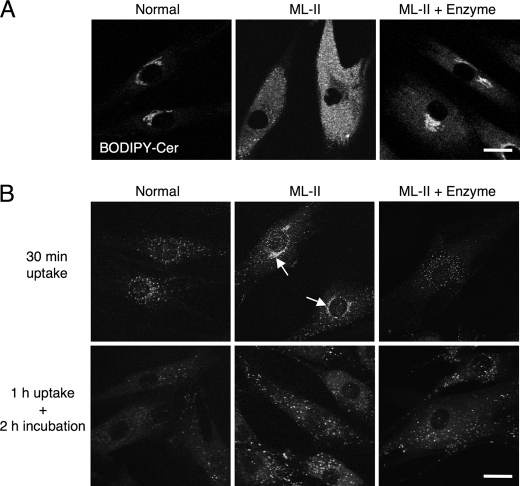
Intracellular logistics are impaired in ML-II. A, endocytic targeting of ceramide in normal, ML-II, and enzyme-treated ML-II cells was evaluated by BODIPY-ceramide targeting. Micrographs show fluorescence of living cells using confocal microscopy after treatment with BODIPY-ceramide for 30 min. Scale bars, 20 μm. B, micrographs showing results of MP6 receptor antibody uptake testing of normal, ML-II, and enzyme-treated ML-II cells. In normal and enzyme-treated ML-II cells, M6P receptors appear to be cycling and distributing broadly. Arrows indicate the Golgi pattern of M6P receptor localization in ML-II cells. Scale bars, 20 μm.
FIGURE 6.
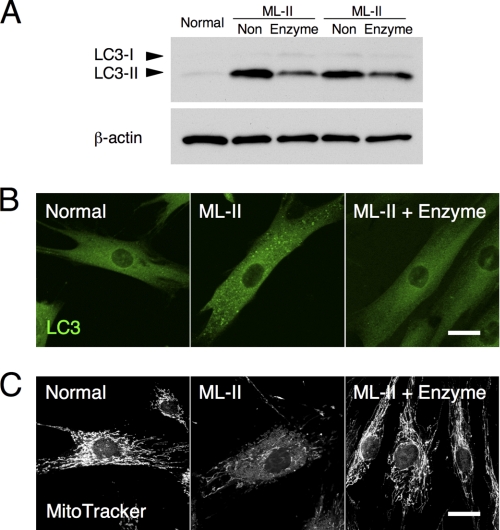
Enzyme treatment improves autophagic and mitochondrial status. A, LC3 immunoblotting of total lysates of normal, ML-II (non), and enzyme-treated ML-II cells indicates a decrease in the specific autophagosome marker, LC3-II, after enzyme treatment of ML-II cells. B and C, LC3 (B) and MitoTracker (C) staining micrographs of normal, ML-II, and enzyme-treated ML-II cells. Scale bars, 20 μm. Enzyme treatment of ML-II cells decreased the number of LC3-positive large vesicles and showed a remarkable recovery of mitochondrial structure.
Measurement of Lysosome Amount in Each Cell
We harvested skin fibroblasts using the standard trypsin method with collection into small tubes. We resuspended the cellular pellet with standard culture medium containing both 1 μm LysoTracker Red DND-99 and 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) followed by incubation at 37 °C for 1 h. After incubation, cells were collected by centrifugation, washed once with PBS, and finally resuspended in PBS. We measured the fluorescence intensity of the cell suspension with a fluorescence microplate reader at excitation 530 nm/emission 590 nm (for LysoTracker) and excitation 365 nm/emission 450 nm (for DAPI). We calculated the LysoTracker/DAPI intensity ratio, which estimates the lysosomal amount (i.e. number and size) in each cell.
Endocytic Targeting of BODIPY-Cer
We prepared skin fibroblasts in glass bottom dishes. BODIPY FL C5-ceramide (BODIPY-Cer) complexed to BSA (Invitrogen) was purchased and diluted in standard culture medium at a concentration of 2.5 nm. Cells were cultured in this medium for 30 min, washed once with PBS, and directly observed by confocal microscopy. We also tested the attachment of BODIPY-Cer to plasma membrane in ice-cold medium, which showed no difference between normal, ML-II, and enzyme-treated ML-II cells.
M6P Receptor Antibody Uptake Test
Cells were incubated in standard culture medium containing M6P receptor antibody at 1:75 concentration. For the 30-min uptake test, cells were incubated with antibody-containing medium for 30 min, washed twice with ice-cold PBS, and quickly fixed with 3.7% formaldehyde. Cells were treated with secondary antibody following permeabilization with 0.1% Triton X-100 for 15 min and blocking with 1% BSA for 1 h at room temperature. For examination of uptake and subcellular distribution, we incubated cells with antibody-containing medium for 1 h, washed twice with PBS, and then followed by incubation in standard culture medium without antibody for 2 h. Subsequent processing was the same as for the 30-min assay described above. We used Alexa Fluor 488 (Molecular Probes) as the secondary antibody against the M6P receptor antibody.
Western Blotting
Skin fibroblasts were collected and subjected to sonication in pure water with Complete protease inhibitor mixture (Roche Applied Science). Protein concentration was measured (protein assay rapid kit; Wako) and mixed with sample buffer containing SDS. For each lane, 10 μg was applied to a 16% polyacrylamide gel. After SDS-PAGE, protein was transferred to a PVDF membrane followed by blocking in 5% skim milk. For the primary antibodies, anti-LC3 was used at a 1:2000 dilution, and anti-ATP6V1B2, anti-ATP6V0D1, and anti-mucolipin-1 were used at 1:5000 dilution followed by the secondary antibody conjugated with HRP. Detection was performed with chemical luminescence (SuperSignal West Dura; Thermo Scientific).
Preparation of Total Enzyme Mixture and Enzyme Supplementation
We cultured normal skin fibroblasts with standard culture medium containing 20 mm NH4Cl. After 7 days of culture, supernatant was retrieved and passed through a 0.2-μm sterilization filter to remove large debris and living cells, and lysosomal enzyme activities were measured. Filtered supernatant was centrifuged in a 5000-Da molecular mass filter column (Vivaspin 15R; Sartorius Stedim Biotech) at 0 °C. As a result, this enzyme mixture was condensed into ∼0.025 volume. Finally, we diluted the total lysosomal enzyme mixture to a concentration of 600 nmol/h/mg of protein of α-mannosidase activity with serum-free DMEM. Total lysosomal enzyme supplementation was performed by culturing cells with this enzyme mixture for 7 days. After 7 days, medium was changed to standard culture medium, and samples were subjected to each assay within 24 h. Subsequently within this study, we describe this total lysosomal enzyme supplementation method simply as “enzyme treatment.” We used normal and ML-II cells cultured with standard culture medium for untreated groups, which were divided and seeded at the same time as treated cells.
Other Methods
Phospholipid (phospholipids C-test; Wako) and cholesterol (Amplex Red cholesterol assay kit; Molecular Probes) measurements were performed according to the manufacturers' instructions. Activities of lysosomal enzymes were measured by the standard method using artificial 4-methylumbelliferyl substrates. In short, samples were incubated with artificial substrates in acidic phosphate or citrate buffer at 37 °C for 1 h, and fluorescence of excitation at 365 nm/emission at 450 nm was measured with a microplate reader. In the case of galactocerebrosidase, we used 6-hexadecanoylamino-4-methylumbelliferyl β-d-galactopyranoside (Slater & Frith Ltd.) and measured fluorescence of excitation at 385 nm/emission at 450 nm. Enzyme activities were calculated as nmol/h/mg of protein or nmol/h/μl.
Statistical Analysis
We first confirmed the normality of values by Shapiro-Wilk W test for each assessment. Next, the distribution of values was tested by F test. For testing significant differences between the samples, we applied two-tailed Student's t test or two-tailed Welch's t test especially in samples that were not equally distributed. All data were obtained from three independent measurements for each sample and reported as means ± S.D. Significant differences are indicated in Figs. 2–4.
RESULTS
Total Lysosomal Enzyme Supplementation Mixture Is Targeted to Lysosomes via the M6P Receptor-dependent Pathway
We first examined the validity of the total lysosomal enzyme supplementation method (enzyme treatment). NH4Cl is known to cause mistargeting of lysosomal enzymes, resulting in their extracellular secretion (15). We generated total lysosomal enzyme mixture from the supernatant of normal skin fibroblasts treated with NH4Cl. The supernatant contained many kinds of lysosomal enzymes, and their secretion profile was similar to that of natural ML-II culture supernatant not treated with NH4Cl (Fig. 1A). After enzyme treatment, the activities of major lysosomal acid hydrolases increased in ML-II cells, although the degree of the increase for each enzyme differed between the enzymes (from 3.2% of normal for β-galactosidase to 92.1% of normal for β-glucuronidase) (Fig. 1B and supplemental Fig. 1A). As enzyme uptake was inhibited by M6P in a concentration-dependent manner, it is considered that the lysosomal enzymes were taken up by the M6P receptor (Fig. 1C and supplemental Fig. 1B). Incorporated lysosomal enzymes were co-localized with the lysosomal marker, lysosomal-associated membrane protein 2 (LAMP-2) (Fig. 1D). These results show that lysosomal enzymes were supplied to the defective lysosomes of ML-II skin fibroblasts. β-Glucosidase activity is not defective by nature in ML-II skin fibroblasts as it is targeted to lysosomes by lysosomal integral membrane protein 2 (LIMP-2) (16). We confirmed that β-glucosidase was not defective in ML-II and not influenced by the addition of M6P (supplemental Fig. 2). This result indicates that these experiments were appropriately performed.
Phospholipid and Cholesterol Are Accumulated in ML-II Cells
To quantitate lysosomal storage materials, we measured phospholipid and cholesterol in total cellular lysate. Phospholipid and total cholesterol were accumulated about 2-fold in ML-II skin fibroblasts as compared with normal cells, and enzyme treatment decreased these amounts to near normal levels (Fig. 2A). Next, we investigated the amount and localization of these storage materials using fluorescent micrography. Lysobisphosphatidic acid (LBPA), a phospholipid and marker of late endosome and lysosome (17, 18), accumulated in ML-II cells and showed a decrease after enzyme treatment on immunocytochemistry (Fig. 2B). Storage of free cholesterol in natural ML-II skin fibroblasts and its clearance by enzyme treatment were also confirmed by filipin staining (Fig. 2C).
Lysosomal Acidification Is Impaired in ML-II
Lysosomal acidification is essential for the proper function of lysosomes and is one of the definitions of lysosomes. Lysosomal pH of ML-II cells was significantly higher than that of normal cells (5.29 ± 0.08 versus 4.79 ± 0.10, p < 0.001). Enzyme treatment recovered lysosomal acidification in ML-II cells toward a near normal level (4.99 ± 0.07, p < 0.001) (Fig. 3A). To elucidate the effect of storage materials on lysosomal pH, we loaded cholesterol in culture medium of normal cells. Cholesterol loading increased the lysosomal pH dose dependently: from 4.79 ± 0.10 without loading to 5.20 ± 0.08 and 5.59 ± 0.09 with 1/1000 and 1/100 cholesterol loading, respectively (Fig. 3B). Lysosomal acidic pH is produced and modulated by vacuolar-type H+-ATPase (V-ATPase) and mucolipin-1. However, analysis of V-ATPase V0d1 subunit, V1B2 subunit, and mucolipin-1 by Western blotting of total cellular lysate from normal and ML-II skin fibroblasts cultured under standard culture conditions revealed no clear difference in V1/V0 protein ratio and the amount of mucolipin-1 protein (controlled by β-actin) between normal and ML-II cells (supplemental Fig. 4A). On immunocytochemistry, V-ATPase V1B2 subunit was co-localized with LysoTracker in both normal and ML-II cells and showed a similar pattern (supplemental Fig. 4B).
Lysosomal Amount Is Increased in ML-II Cells
One specific feature of ML-II skin fibroblasts is the inclusion bodies that represent proliferated lysosomes filled with undigested substrates. These proliferated lysosomes can be observed in ML-II cells using LysoTracker staining (Fig. 4A). To evaluate the lysosomal amount in each cell numerically, we determined the intensity ratio of LysoTracker and DAPI fluorescence of living cells; the lysosomal amount was about 3-fold higher in ML-II cells as compared with normal cells. LysoTracker fluorescence on micrography and the estimated lysosomal amount were decreased after enzyme treatment in ML-II cells (Fig. 4B).
Endocytic Pathway and M6P Receptor Recycling Are Inhibited in ML-II
Intracellular logistics including the endocytic pathway and M6P receptor movement were analyzed by BODIPY-Cer targeting (Fig. 5A) and M6P receptor antibody uptake tests (Fig. 5B), respectively. Similar to a previous study using BODIPY-lactosylceramide (19), fluorescence was observed in the Golgi apparatus of normal cells and in the endosome and lysosome of ML-II cells 30 min after BODIPY-Cer treatment. This endosomal and lysosomal pattern was not changed by the duration of BODIPY-Cer uptake from 30 min up to 6 h and was congruent with LysoTracker staining (supplemental Fig. 3). Enzyme treatment of ML-II cells changed the pattern of ceramide toward Golgi distribution, although slight endosomal and lysosomal entrapment remained. The M6P receptor antibody uptake test showed a vesicular and Golgi pattern in ML-II cells but only a vesicular pattern in normal cells. This pattern was not changed by the duration of antibody uptake from 30 min up to 6 h. After enzyme treatment of ML-II cells, the M6P receptor distribution changed from the Golgi pattern to a vesicular pattern, similar to that in normal cells. When we incubated ML-II cells for 1 h with antibody followed by 2 h without antibody, the antibody was only detected as a vesicular pattern. In normal cells, antibody was hardly detected as it is likely to be degraded within 2 h following incubation without antibody.
Impaired Autophagy and Mitochondria Are Corrected by Total Lysosomal Enzyme Supplementation
We have previously reported the impairment of autophagy and mitochondria in ML-II skin fibroblasts (20). After enzyme treatment of ML-II skin fibroblasts, LC3-II, a specific autophagosome marker (21), was decreased in immunoblotting of total cell lysates (Fig. 6A). Immunocytochemistry also showed a decreased number of LC3-positive large vesicles after enzyme treatment, which were highly accumulated without treatment (Fig. 6B). Mitochondria were usually tubular and showed a constructive network in normal cells but were thinned and fragmented in ML-II cells. Enzyme treatment of ML-II cells resulted in a remarkable recovery of mitochondrial structure on MitoTracker staining, showing a normal tubular form and network (Fig. 6C).
Total Lysosomal Enzyme Supplementation Clears Inclusion Bodies in ML-II Skin Fibroblasts
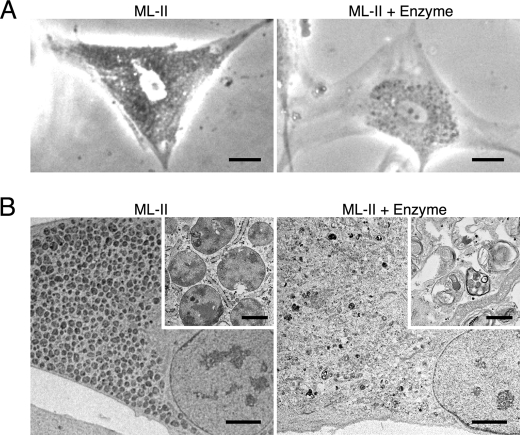
Many inclusion bodies were seen in ML-II skin fibroblasts using phase-contrast micrography, and they were decreased following enzyme treatment (Fig. 7A). Electron micrography also showed many vacuoles containing dense, uniform materials in untreated ML-II skin fibroblasts. These vacuoles, thought to be inclusion bodies, disappeared after enzyme treatment together with the recovery of many intracellular structures including mitochondria and some void vacuoles (Fig. 7B).
FIGURE 7.
Inclusion bodies disappear after enzyme treatment in ML-II. A, phase-contrast micrographs of ML-II and enzyme-treated ML-II cells. Scale bars, 20 μm. B, electron micrographs of ML-II and enzyme-treated ML-II cells. Scale bars, 5 μm and 500 nm (inset). Micrographs show accumulation of inclusion bodies in ML-II cells and their reduction following enzyme treatment.
DISCUSSION
In this study, we describe two important results for understanding the pathogenesis of ML-II. First are the various cellular phenotypes of ML-II skin fibroblasts including quantitative analyses and imaging of impaired cellular functions; second is their correction by total lysosomal enzyme supplementation.
Phase-contrast micrography of ML-II skin fibroblasts shows accumulation of dense vacuoles, inclusion bodies, that are considered to be swollen and proliferated lysosomes filled with undigested substrates resulting from the lack of lysosomal acid hydrolases. Storage materials in lysosomes are thought to be a mixture of a range of various substrates as ML-II lysosomes lack dozens of lysosomal enzymes. Using lectin staining, Kawashima et al. (22) reported that GM2 ganglioside and some glycoconjugates were accumulated in ML-II cells because of the lack of β-hexosaminidase A, sialidase, β-galactosidase, and α-mannosidase. On the other hand, ML-II skin fibroblasts are well known to be stained by filipin, which indicates free cholesterol accumulation within endosomes and lysosomes (23). In this study, we quantitated the amounts of phospholipid and total cholesterol in ML-II total cellular lysate for the first time. In addition, to see the localization of these lipids, LBPA and filipin staining was performed with fluorescence microscopy. LBPA, which is a particular kind of phospholipid that is a marker of late endosomes and lysosomes, has never been investigated in ML-II cells. Phospholipid and LBPA were accumulated in ML-II skin fibroblasts. Together with the result of measurement of lysosomal amount, the inclusion bodies accumulated in ML-II skin fibroblasts originated from late endosomes and lysosomes, although we still believe that these two organelles are a hybrid and cannot be clearly distinguished from each other. Free cholesterol accumulation was obvious by filipin fluorescence staining in ML-II fibroblasts, which is similar to Niemann-Pick type C disease. In Niemann-Pick type C disease, the total amount of cholesterol is not clearly increased; only its distribution is changed to accumulation in late endosomes (17). In our study, we measured the total amount of cholesterol in ML-II cells and showed a significant increase. Both the amount and the distribution of cholesterol are changed in ML-II as compared with normal cells, which is different from the results seen in Niemann-Pick type C disease.
In this study, we found that intracellular logistics, such as endocytosis and M6P receptor recycling, were impaired in ML-II cells. There is no primary defect of M6P receptor movement in ML-II cells (24). However, there seems to be a secondary impairment of M6P receptor trafficking, probably due to the pre-existing massive storage materials such as cholesterol in endosomes and lysosomes as cholesterol has been reported to modulate membrane traffic along the endocytic pathway in sphingolipid storage diseases (19). M6P receptors cycle between membranes such as plasma membrane, Golgi, and endosomes bidirectionally. In our M6P receptor antibody uptake study, the amount of M6P receptor in the perinuclear region in ML-II is higher than normal after 30 min of uptake. This indicates that M6P receptor movement through the perinuclear compartment appears delayed. This delay would seem to be cleared after 2 h of incubation. We suggest that M6P receptors moved from the perinuclear region to endosomes. However, we cannot exclude other potential explanations, such as the return of perinuclear antibody-labeled M6P receptor to the plasma membrane or elsewhere possibly causing detachment between M6P receptor and antibody or the bivalent antibody possibly cross-linking with the receptor and affecting the intracellular movement of M6P receptor. Intracellular M6P receptor recycling is important for the efficient uptake of lysosomal enzymes from the cell surface, and a smooth endocytic pathway is also important for the targeting of lysosomal enzymes from the cell surface to lysosomes. These two essential pathways are indirectly impaired in ML-II cells, and enzyme treatment was able to correct both pathways gradually. This improvement promotes efficient enzyme uptake and eventually makes total enzyme supplementation more effective.
Lysosomal acid pH is mainly produced by the lysosomal vacuolar-type proton pump, V-ATPase, and mucolipin-1 has recently been reported to modulate lysosomal pH (25, 26). In several lysosomal storage disorders, abnormal lysosomal pH has been reported (27, 28). This is the first study showing impaired lysosomal acidification in ML-II. Lysosomal pH was significantly increased in ML-II skin fibroblasts, and it was normalized by total lysosomal enzyme supplementation. Cholesterol loading effectively increased lysosomal pH in normal cells. These results indicate that storage materials, such as cholesterol within lysosomes, increase lysosomal pH by an unknown mechanism. Accumulated substrates may be at a higher pH, or storage materials may inhibit the function of the proton pump. It is also possible that cholesterol accumulation changes the composition of the membrane lipid raft, which influences proton pump function. V-ATPase is made up of various subunits, and one study has revealed the importance of the lipid raft and V1/V0 subunit ratio in modulating its acidifying function (29). As mentioned above, protein analysis of V-ATPase and mucolipin-1 showed no clear differences in Western blotting and localization between normal and ML-II cells. From these results, lysosomal maturation is impaired by defective lysosomal acid hydrolases and acidification, although the mechanism of increased lysosomal pH remains unclear.
Lysosomal storage possibly causes autophagic impairment and leads to accumulation of aberrant mitochondria as secondary storage (30). In the case of ML-II, autophagic and mitochondrial impairment has been reported in skin fibroblasts, mice, and an autopsy case (20, 31, 32). Autophagic status and mitochondrial structure recovered after enzyme treatment of ML-II cells in our study. It is therefore suggested that mitochondrial turnover is also returned to a normal state combined with the functional recovery of autophagy and lysosomes following clearance of storage materials within lysosomes by enzyme treatment. Lysosomal accumulation drives lysosome-related gene expression and increases the amount of lysosomes (33). On the contrary, clearance of accumulation by enzyme treatment significantly decreased the amount of lysosomes in our study, which is clearly demonstrated on electron micrography. For the first time, we succeeded in the reversal of inclusion bodies by total lysosomal enzyme supplementation.
Total lysosomal enzyme supplementation is based on the idea that the enzyme mixture is taken up by M6P receptors on the cell surface and targeted to lysosomes because there is no primary defect of the M6P receptor-dependent targeting pathway itself in ML-II. This concept is similar to that of other commercially available ERTs. Our procedure clearly reduced lysosomal storage and improved cellular phenotypes in ML-II cells and is applicable for the functional correction of ML-II cells. This may imply a potential method for “total lysosomal ERT” in ML-II patients. Animal studies will be indispensable to confirm the in vivo applicability of this enzyme treatment as a therapeutic concept. Furthermore, the classical and convenient method for obtaining the total enzyme mixture using NH4Cl may provide ERT for not only ML-II but also for other lysosomal storage disorders for which ERT has not been developed.
In conclusion, storage materials and various cellular phenotypes of ML-II are demonstrated quantitatively in our study. It is presumed that the excessive lysosomal storage materials are the main cause of the diseased state in ML-II and induce various cellular phenotypes that are intricately intertwined with each other. Finally, these phenotypes were reversed by supplementation with an enriched total lysosomal enzyme mixture derived from normal skin fibroblasts, which suggests a potential new, alternative therapeutic approach for ML-II.
Supplementary Material
This work was supported by grants from the Ministry of Education, Culture, Science, Sports and Technology of Japan (Grants 20790728, 18390299, 21659257, and 21591322) and the Ministry of Health, Labour and Welfare of Japan (Grant H20-Kokoro-022 and a grant for Research for Intractable Diseases).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- ML-II
- mucolipidosis II
- BODIPY-Cer
- BODIPY FL C5-ceramide
- Cer
- ceramide
- ERT
- enzyme replacement therapy
- LAMP-2
- lysosome-associated membrane protein 2
- LBPA
- lysobisphosphatidic acid
- M6P
- mannose 6-phosphate
- V-ATPase
- vacuolar-type H+-ATPase.
REFERENCES
- 1. Kornfeld S., Sly W. S. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds) pp. 3421–3452, 8th Ed., McGraw-Hill, New York [Google Scholar]
- 2. Storch S., Braulke T. (2005) in Lysosomes (Saftig P. ed) pp. 17–26, 1st Ed., Eurekah, Landes Bioscience, Springer and Business Media, New York [Google Scholar]
- 3. Kornfeld R., Bao M., Brewer K., Noll C., Canfield W. (1999) J. Biol. Chem. 274, 32778–32785 [DOI] [PubMed] [Google Scholar]
- 4. Kudo M., Bao M., D'Souza A., Ying F., Pan H., Roe B. A., Canfield W. M. (2005) J. Biol. Chem. 280, 36141–36149 [DOI] [PubMed] [Google Scholar]
- 5. Tiede S., Storch S., Lübke T., Henrissat B., Bargal R., Raas-Rothschild A., Braulke T. (2005) Nat. Med. 11, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 6. Raas-Rothschild A., Cormier-Daire V., Bao M., Genin E., Salomon R., Brewer K., Zeigler M., Mandel H., Toth S., Roe B., Munnich A., Canfield W. M. (2000) J. Clin. Invest. 105, 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schiffmann R., Murray G. J., Treco D., Daniel P., Sellos-Moura M., Myers M., Quirk J. M., Zirzow G. C., Borowski M., Loveday K., Anderson T., Gillespie F., Oliver K. L., Jeffries N. O., Doo E., Liang T. J., Kreps C., Gunter K., Frei K., Crutchfield K., Selden R. F., Brady R. O. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barton N. W., Furbish F. S., Murray G. J., Garfield M., Brady R. O. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1913–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kakkis E. D., Muenzer J., Tiller G. E., Waber L., Belmont J., Passage M., Izykowski B., Phillips J., Doroshow R., Walot I., Hoft R., Neufeld E. F. (2001) N. Engl. J. Med. 344, 182–188 [DOI] [PubMed] [Google Scholar]
- 10. Amalfitano A., Bengur A. R., Morse R. P., Majure J. M., Case L. E., Veerling D. L., Mackey J., Kishnani P., Smith W., McVie-Wylie A., Sullivan J. A., Hoganson G. E., Phillips J. A., 3rd, Schaefer G. B., Charrow J., Ware R. E., Bossen E. H., Chen Y. T. (2001) Genet. Med. 3, 132–138 [DOI] [PubMed] [Google Scholar]
- 11. Desnick R. J. (2004) J. Inherit. Metab. Dis. 27, 385–410 [DOI] [PubMed] [Google Scholar]
- 12. Schiffmann R. (2010) J. Inherit. Metab. Dis. 33, 373–379 [DOI] [PubMed] [Google Scholar]
- 13. Beck M. (2007) Hum. Genet. 121, 1–22 [DOI] [PubMed] [Google Scholar]
- 14. Otomo T., Muramatsu T., Yorifuji T., Okuyama T., Nakabayashi H., Fukao T., Ohura T., Yoshino M., Tanaka A., Okamoto N., Inui K., Ozono K., Sakai N. (2009) J. Hum. Genet. 54, 145–151 [DOI] [PubMed] [Google Scholar]
- 15. Brown J. A., Novak E. K., Swank R. T. (1985) J. Cell Biol. 100, 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reczek D., Schwake M., Schröder J., Hughes H., Blanz J., Jin X., Brondyk W., Van Patten S., Edmunds T., Saftig P. (2007) Cell 131, 770–783 [DOI] [PubMed] [Google Scholar]
- 17. Kobayashi T., Beuchat M. H., Lindsay M., Frias S., Palmiter R. D., Sakuraba H., Parton R. G., Gruenberg J. (1999) Nat. Cell Biol. 1, 113–118 [DOI] [PubMed] [Google Scholar]
- 18. Kolter T., Sandhoff K. (2010) FEBS Lett. 584, 1700–1712 [DOI] [PubMed] [Google Scholar]
- 19. Puri V., Watanabe R., Dominguez M., Sun X., Wheatley C. L., Marks D. L., Pagano R. E. (1999) Nat. Cell Biol. 1, 386–388 [DOI] [PubMed] [Google Scholar]
- 20. Otomo T., Higaki K., Nanba E., Ozono K., Sakai N. (2009) Mol. Genet. Metab. 98, 393–399 [DOI] [PubMed] [Google Scholar]
- 21. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawashima I., Ohsawa M., Fukushige T., Nagayama Y., Niida Y., Kotani M., Tajima Y., Kanekura T., Kanzaki T., Sakuraba H. (2007) Clin. Chim. Acta 378, 142–146 [DOI] [PubMed] [Google Scholar]
- 23. Vanier M. T., Rodriguez-Lafrasse C., Rousson R., Gazzah N., Juge M. C., Pentchev P. G., Revol A., Louisot P. (1991) Biochim. Biophys. Acta 1096, 328–337 [DOI] [PubMed] [Google Scholar]
- 24. Braulke T., Bonifacino J. S. (2009) Biochim. Biophys. Acta 1793, 605–614 [DOI] [PubMed] [Google Scholar]
- 25. Casey J. R., Grinstein S., Orlowski J. (2010) Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 26. Soyombo A. A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. (2006) J. Biol. Chem. 281, 7294–7301 [DOI] [PubMed] [Google Scholar]
- 27. Bach G., Chen C. S., Pagano R. E. (1999) Clin. Chim. Acta 280, 173–179 [DOI] [PubMed] [Google Scholar]
- 28. Kogot-Levin A., Zeigler M., Ornoy A., Bach G. (2009) Pediatr. Res. 65, 686–690 [DOI] [PubMed] [Google Scholar]
- 29. Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., van der Goot F. G. (2008) PLoS One 3, e2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D., de Pablo R., Tacchetti C., Rubinsztein D. C., Ballabio A. (2008) Hum. Mol. Genet. 17, 119–129 [DOI] [PubMed] [Google Scholar]
- 31. Boonen M., van Meel E., Oorschot V., Klumperman J., Kornfeld S. (2011) Mol. Biol. Cell 22, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi H., Takahashi-Fujigasaki J., Fukuda T., Sakurai K., Shimada Y., Nomura K., Ariga M., Ohashi T., Eto Y., Otomo T., Sakai N., Ida H. (2011) Mol. Genet. Metab. 102, 170–175 [DOI] [PubMed] [Google Scholar]
- 33. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.