Background: Rickettsiae infect the host cell by co-opting the actin cytoskeleton.
Results: The Rickettsiae invasin sca4 harbors two α-helices that bind and activate vinculin; the binding mode of one of these α-helices is unique.
Conclusion: Rickettsiae use mimicry of talin to subvert vinculin functions.
Significance: The unique nature of the sca4-vinculin interaction suggests it can be targeted by small molecules.
Keywords: Bacterial Toxins, Cell Adhesion, Crystal Structure, Cytoskeleton, Protein-Protein Interactions, Epidemic Typhus, sca4, Spotted Fever
Abstract
Pathogenic Rickettsia species cause high morbidity and mortality, especially R. prowazekii, the causative agent of typhus. Like many intracellular pathogens, Rickettsia exploit the cytoskeleton to enter and spread within the host cell. Here we report that the cell surface antigen sca4 of Rickettsia co-localizes with vinculin in cells at sites of focal adhesions in sca4-transfected cells and that sca4 binds to and activates vinculin through two vinculin binding sites (VBSs) that are conserved across all Rickettsia. Remarkably, this occurs through molecular mimicry of the vinculin-talin interaction that is also seen with the IpaA invasin of the intracellular pathogen Shigella, where binding of these VBSs to the vinculin seven-helix bundle head domain (Vh1) displaces intramolecular interactions with the vinculin tail domain that normally clamp vinculin in an inactive state. Finally, the vinculin·sca4-VBS crystal structures reveal that vinculin adopts a new conformation when bound to the C-terminal VBS of sca4. Collectively, our data define the mechanism by which sca4 activates vinculin and interacts with the actin cytoskeleton, and they suggest important roles for vinculin in Rickettsia pathogenesis.
Introduction
Pathogenic Rickettsia are non-motile, Gram-negative, obligate intracellular parasites that are the causative agents of epidemic typhus and spotted fevers (1, 2). Rickettsia rickettsii causes rocky mountain spotted fever with more than 20% mortality and no effective antibiotics (3), and Rickettsia prowazekii causes epidemic typhus, one of the most severe infectious diseases, with more than 3 million deaths during the last century and a mortality rate of 10–60%. Although several rickettsial genomes have been sequenced, including that of R. rickettsii, these pathogens are, unfortunately, intractable to classical genetics. Accordingly, our understanding of Rickettsia is in its infancy. For example, there are only six Rickettsia entries in the Protein Data Bank, and all are from R. prowazekii, the cause of typhus.
R. rickettsii and other spotted fever Rickettsia are disseminated to humans by tick bites in the skin (2) and then grow in epithelial and endothelial cells, triggering localized dermal and epidermal necrosis (4) and a dermal rash (4, 5). Rickettsia uses lipid raft-associated Ku70, an ATP-dependent DNA helicase, to facilitate its internalization (6), and their entry and spread within the host cell requires the cytoskeletal regulators Arp2/3 (actin-related protein 2/3), Cdc42 (cell division control protein 42), and cofilin (7–10). Rickettsia escape from a phagosomal compartment into the host cell cytosol and propel themselves using the actin network. Furthermore, the RickA protein, a Rickettsia conorii protein with similarity to the Wiskott-Aldrich syndrome protein family, directs migration of the pathogen within the cytoplasm by polymerizing actin filaments (8, 11).
Phylogenetic analysis of nine Rickettsia spp. showed strong positive selection on surface cell antigen (sca) genes, indicating the requirement of sca genes in Rickettsia life cycle (12). A few recent studies reported the involvement of sca proteins in the bacterial infection; sca0 (rOmpA (rickettsial outer-membrane protein A)) and sca1 are involved in the attachment to mammalian cells, sca5 (rOmpB) is involved in both attachment and the entry process of the bacteria into nonphagocytic mammalian cells (13, 14), and sca2 functions as a formin mimic that is responsible for actin-based motility of Rickettsia in the host cell cytosol (15).
Precisely how Rickettsia binds to the actin cytoskeleton is not known. Building on our findings in Shigella (16–18), we reasoned that Rickettsia encodes an invasin that would function either as an activator or mimic of vinculin. Vinculin is a globular helix bundle protein that is normally clamped in its inactive state via hydrophobic interactions of its N-terminal seven-helix bundle head (Vh1)3 domain with its five-helix bundle tail (Vt) domain (19–22). Vinculin activation requires severing the head-tail interaction, and studies initially from our laboratory (23–25) and then by others (26–30) have established that talin is a physiologic activator of vinculin, where it binds to the vinculin Vh1 domain via amphipathic α-helical vinculin binding sites (VBSs) present in its central talin rod domain. Notably, the VBSs of talin are normally buried within helix bundle domains (26, 29, 31), and talin needs to be force-activated via integrin receptors to release these VBSs so they can bind to and activate vinculin (26, 30, 32). In contrast, no “pre-activation” of the IpaA invasin of Shigella that binds to vinculin is necessary, as full-length IpaA binds to and activates vinculin (33, 34).
A search for VBSs encoded by Rickettsia led to the discovery that the cell surface antigen sca4, a 112-kDa protein (1024 residues) of Rickettsia, harbors two such VBSs in the C-terminal half of this protein that are conserved across Rickettsia and which are separated by 400 residues. Our studies show that full-length sca4 binds to and activates vinculin and that it co-localizes with vinculin in cells. Finally, the crystal structures of vinculin·sca4-VBS complexes reveal unique features of this interaction.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Chromosomal DNA of R. rickettsii, Iowa strain, was generated by polymerase chain reaction (PCR) and cloned into the pEGFP-N2-sca4 expression vector. The fragment was amplified with primers RCsca4-forward (CCGGAATTCATGAGTAAAGACGGTAACCTAG) and RCsca4-reverse (GCGGTACCGGCGTTGTGGAGGGGAAGAC), cloned in pCRBlunt, verified by sequencing using M13 and REV primers, and subcloned into the EcoRI-KpnI-digested pEGFP-N2 (Clontech) to generate pEGFP-N2-sca4. This construct was then used as a PCR template for all the subsequent sca4 constructs. The primers (supplemental table) were designed, and target genes were amplified to generate sca4 residues 412–434, 774–1008, 411–585, 21–360, and 21–1008. The amplified PCR products were digested with NdeI and XhoI and ligated into the pET-28a vector (Novagen) encoding a polypeptide with an N-terminal hexahistidine tag with a thrombin cleavage site for purification.
The human vinculin head domain (Vh1; residues 1–258) and full-length human vinculin were purified as described (19, 24). Recombinant sca4 proteins (residues 774–1008, 411–585, 21–360, or 21–1008) were expressed in Escherichia coli strain BL21(DE3) (Invitrogen) and grown at 37 °C in Luria-Bertani medium containing kanamycin (20 mg/l). Bacterial cultures were induced at A600 of 0.8 by adding isopropyl 1-thio-β-d-galactopyranoside to 0.5 mm and incubating at 30 °C for 24 h. Cells were lysed by sonication, and the lysate was loaded onto a HisTrap chelating nickel affinity chromatography column (GE Healthcare) and washed with 25 column volumes of 50 mm NaH2PO4, 300 mm NaCl, and 5 mm imidazole. The protein was eluted over a gradient of 1 m imidazole (pH 8). The eluted fractions were pooled and concentrated, and the buffer was exchanged to thrombin cleavage buffer (150 mm NaCl, 20 mm Tris-HCl, pH 8, 1 mm DTT). The hexahistidine tag was cleaved at 4 °C overnight with 2.5 units of bovine thrombin (Sigma) per mg of each protein. The proteins were further purified on a Superdex 75 or Superdex 200 gel filtration sizing chromatography column (Amersham Biosciences) equilibrated in 20 mm Tris-HCl (pH 8) supplemented with 150 mm NaCl.
For the actin co-sedimentation assays, sca4-VBS-N (residues 412–434) and sca4-VBS-C (residues 812–835) were synthesized and HPLC-purified (GenScript Corp.). The same regions were also PCR-amplified (supplemental table) and cloned into NheI/EcoRI of pMAL-c2e (New England Biolabs) to produce maltose-binding protein (MBP) fusions. The two constructs, pMAL-sca4-VBS-N and pMAL-sca4-VBS-C, were transformed into E. coli BL21(DE3) and expressed similarly as described above and purified by amylose chromatography column (New England Biolabs) following the manufacturer's instructions.
The pET-28a plasmid carrying the sca4-VBS-N construct (having a His tag and kanamycin resistance) was co-expressed with the pET-22b plasmid carrying the Vh1 construct (with no tag and ampicillin resistance) in E. coli BL21(DE3). The binary complex of Vh1·sca4-VBS-N was purified similarly as described previously (24).
Sca4 residues 20–1024, 20–360, 406–585, and 772–1008 were PCR amplified (supplemental table) and cloned into the mammalian expression vector pEGFP-N1 (Clontech) using the EcoRI restriction site. The PCR product for sca4 residues 772–1008 was cloned into pEGFP-C1 (Clontech) using the EcoRI and BamHI sites. For efficient translation of sca4 genes, with the exception of the 772–1008 construct, a Kozak sequence of GCCACC was added via the forward primers before the start codon.
Crystallization of the Vh1·Sca4-VBS-N and Vh1·Sca4-VBS-C Complexes
The purified Vh1·sca4-VBS-N complex was concentrated to 36 mg/ml and crystallized by hanging drop vapor diffusion. The Vh1·sca4-VBS-N complex was crystallized at room temperature by hanging-drop vapor diffusion. Crystals were grown on a siliconized coverslip by equilibrating a 1 μl of Vh1·sca4-VBS-N solution (36.4 mg ml−1 Vh1·sca4-VBS-N in 20 mm Tris-HCl (pH 8) and 150 mm NaCl) with 1 μl of reservoir solution (0.1 m MES monohydrate (pH 6.5), 14% [w/v] PEG 20,000) against 0.5 ml of reservoir solution. Single crystals formed after 2 days and grew to a largest dimension of 0.4 mm. The most suitable cryoprotectant was determined to be reservoir solution plus 20% (v/v) glycerol. The crystals were flash-cooled by plunging in liquid nitrogen for 1 min.
Sca4-VBS-C was dissolved in 20 mm Tris-HCl (pH 9) and 150 mm NaCl to a final concentration of 15 mm. For crystallizations, an excess of various sca4-VBS-C concentrations was added to Vh1 (purified as described) (24). Initial crystallization hits were obtained in a high salt condition using 100 mm sodium citrate tribasic dihydrate (pH 5.6) and 2.5 m ammonium phosphate, but these crystals diffracted poorly (to about 8 Å Bragg spacings). We therefore varied the Vh1·sca4-VBS-C ratio ranging from 1:5 to 1:3 and obtained a new cubic crystal form under different crystallization conditions. The final Vh1·sca4-VBS-C crystals (obtained at 1:3 molar ratio) were grown at room temperature by hanging drop vapor diffusion (0.5 mm Vh1 at 14.4 mg/ml and 1.5 mm sca4-VBS-C) against a reservoir solution of 2% (v/v) tacsimate (pH 5), 100 mm sodium citrate tribasic dihydrate (pH 5.6), and 16% (w/v) polyethylene glycol (3350). Successful flash-freezing was achieved when the crystals were transferred directly to the reservoir solution containing 20% glycerol and were allowed to equilibrate for 1 min.
X-ray Data Collection and Reduction
Diffraction data were collected for Vh1·sca4-VBS-N crystals at beamline 11-1 (wavelength of 1 Å) of the Stanford Synchrotron Radiation Laboratory. To reduce x-ray absorption and increase the spot quality, we focused the x-ray beam around the edge of the 0.6 × 0.6 × 0.4 mm3 crystal. The data were integrated and scaled using autoProc (35), which uses either XDS (36) or MOSFLM (37) as the data reduction engine. The crystals belonged to space group P212121 with the unit cell dimensions a = 54.4 Å, b = 76.3 Å, and c = 114.6 Å with one heterodimer in the asymmetric unit, resulting in a solvent content of 68% (Table 1).
TABLE 1.
X-ray data collection and reduction statistics
| Parameter | Vh1·sca4-VBS-N | Vh1·sca4-VBS-C |
|---|---|---|
| Space group | P212121 | I23 |
| Unit cell dimensions | ||
| a | 54.4 Å | 141.4 Å |
| b | 76.4 Å | 141.4 Å |
| c | 114.6 Å | 141.4 Å |
| α = β = γ | 90° | 90° |
| Asymmetric unit | 1 heterodimer | 1 heterodimer |
| Matthews coefficient | 3.82 | 3.85 |
| Solvent content | 0.678 | 0.68 |
| Resolution | 114 Å-1.99 Å | 100 Å-2.76 Å |
| Last shell | 2.1 Å-1.99 Å | 2.91 Å-2.76 Å |
| Total measurements | 196,687 | 182,498 |
| Number of unique reflections | 33,356 | 12,290 |
| Last shell | 4,774 | 1,759 |
| Wilson B | 37 Å2 | 71 Å2 |
| Wavelength | 1 Å | 0.97867 Å |
| R-mergea | 0.034 | 0.059 |
| Last shell | 0.499 | 0.49 |
| I/σ(I) | 26.3 | 36.4 |
| Last shell | 3.3 | 6.1 |
| Completeness | 0.999 | 1.0 |
| Last shell | 1.0 | 0.998 |
| Redundancy | 5.9 | 14.8 |
| Last shell | 5.3 | 15 |
a R-merge = ΣhklΣi|Ii(hkl) − ¯Īi¯(h̄kl̄)|/ΣhklΣiIi(hkl).
Vh1·sca4-VBS-C x-ray diffraction data were collected at beamline 11-1 (wavelength of 0.97867 Å) of the Stanford Synchrotron Radiation Laboratory to 2.8 Å Bragg spacings and processed using autoProc (35). The crystals belonged to the I213 space group with unit cell dimensions a = b = c = 141.4 Å and with one heterodimer in the asymmetric unit, resulting in a solvent content of 68% (Table 1).
Crystal Structure Determination and Crystallographic Refinement
Phases for Vh1·sca4-VBS-N were obtained by molecular replacement using MOLREP running in the CCP4 program interface with Vh1 from PDB entry 1rkc (24) as a search model. Molecular replacement using 30 to 2.4 Å data resulted in one top solution with an initial R-factor, correlation coefficient, and translation contrast of 0.48, 0.73, and 7.2, respectively, whereas the values of the second unrelated peak solution were 0.68, 0.39, and 2.2, respectively.
Crystallographic refinement of Vh1·sca4-VBS-N was performed using autoBUSTER with the TLS (translation, libration, and screw-motion) and water update options (38). After six cycles of refinement, the free and crystallographic R-factors dropped to 0.25 and 0.23, respectively. Further rounds of refinement were performed after manual adjustment and visual inspection of the model using the program COOT (39). The final crystallographic and free R-factors were 0.2 and 0.22, respectively. A Ramachandran plot analysis shows that 98.9% of all residues fall in the favored regions and that the remaining 1% fall in allowed regions. MolProbity (40) analysis resulted in a score of 1.45 (97th percentile) with a clash-score of 8.38 (89th percentile) (40). Crystallographic refinement statistics are summarized in Table 2. The final Vh1·sca4-VBS-N structure comprises Vh1 residues 2 to 255 and sca4 residues 411 to 434 and residues GSHM resulting from a cloning artifact at the N terminus of sca4 as well as six glycerol, one tetraethylene glycol, and one 2-(N-mopholino)-ethanesulfonic acid molecules and 316 ordered water molecules.
TABLE 2.
Crystallographic refinement statistics
| Parameter | Vh1·sca4-VBS-N | Vh1·sca4-VBS-C |
|---|---|---|
| Resolution | 23.39-1.99 Å | 44.71-2.76 Å |
| Last shell | 2.05-1.99 Å | 3.02-2.76 Å |
| Number of reflections (working set) | 33,271 | 12,251 |
| Number of reflections (test set) | 1,681 | 989 |
| R-factora | 0.1951 | 0.1996 |
| Last shell | 0.2361 | 0.2523 |
| R-freeb | 0.2180 | 0.2423 |
| Last shell | 0.2601 | 0.2786 |
| Root mean square deviation from ideal geometry | ||
| Bond length | 0.01 Å | 0.01 Å |
| Bond angle | 0.99° | 1.1° |
| No. of protein atoms | 2,176 | 2,150 |
| No. of solvent molecules | 316 | 141 |
| Average temperature factor | ||
| Vh1 | 51 Å2 | 78 Å2 |
| Sca4-VBS | 48 Å2 | 89 Å2 |
| Solvent | 61 Å2 | 67 Å2 |
a R-factor = Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖/Σhkl|Fobs(hkl)|, where 〈|Fcalc|〉 is the expectation of |Fcalc| under the error model used in maximum likelihood refinement.
b The free R-factor is a cross-validation residual calculated using 5% (sca4-VBS-N) and 8.5% (sca4-VBS-C) reflections, which were randomly chosen and excluded from the refinement.
Phases for Vh1·sca4-VBS-C were determined by molecular replacement using MOLREP (41) with the unliganded Vh1 structure (PDB entry 1rkc) as a search model. Molecular replacement using 44 to 3.2 Å data led to one correct solution with the initial R-factor, correlation coefficient, and translation contrast of 0.39, 0.79, and 10.1, respectively. The second unrelated peak produced the aforementioned values of 0.60, 0.46, and 3.1, respectively. This resulted in an unambiguous solution for the position of C-terminal four-helix bundle subdomain of Vh1. However, because of conformational change, the initial electron density map using phases of the starting model showed large discrepancies for the N-terminal four-helix bundle subdomain, which contains the binding site for VBSs. Even at this stage, strong continuous electron density for the omitted sca4-VBS-C was observed in the omit map. The bulky electron density of Tyr-815 provided clear direction of sca4-VBS-C for initial model building.
Multiple rounds of iterative manual fitting of sca4-VBS-C were carried out according to the 2Fobs − Fcalc and Fobs − Fcalc electron density maps by using COOT (39). Crystallographic refinement was performed using autoBUSTER (38) and resulted in crystallographic and free R-factors of 0.2 and 0.24, respectively (Table 2). The last five residues of Vh1 and the last residue of sca4-VBS-C were not included in the model due to disorder. In the Ramachandran plot, 96.3 and 3.3% of all residues fall in favored and allowed regions, respectively. Protein geometry analysis by Molprobity (40) resulted in a score of 2.16 (98th percentile) with a clash score of 8.98 (99th percentile) and with 3.7% violation in rotamers and bonds and 0% violation in angles. The final Vh1·sca4-VBS-C structure comprises Vh1 residues 1 to 30, 34 to 257, and sca4 residues 812 to 834 as well as 141 ordered water molecules.
Native-PAGE Shift Mobility Assays
To examine the interaction between sca4 proteins and Vh1, native-PAGE was performed using a Phastgel system (GE Healthcare) according to the manufacturer's instructions. 10 μm Vh1 was incubated with various sca4 proteins in ∼1:2 molar ratios at room temperature for 10 min before being run on an 8% to 25% gradient native gel. Vt displacement assays were performed as described (16).
F-actin Co-sedimentation Assay
Full-length human vinculin was prepared as described (19) and filtered with a 0.02-μm syringe filter and diluted with phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.47 mm KH2PO4 (pH 7.4)) buffer to a final concentration of 15 μm. Each of the VBSs of sca4 was dissolved in PBS and then added to vinculin at 1:2 (vinculin:sca4) molar ratio and incubated for 20 min at room temperature. F-actin co-sedimentation was performed as described (42).
Transient Transfection and Confocal Imaging
The sca4 expression vectors and pEGFP alone as a control were transfected into NIH 3T3 cells grown overnight on glass coverslips with Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After 16 h, the transfected NIH 3T3 cells were removed from the media and washed with PBS buffer. The cells were then fixed with PBS supplemented with 4% formaldehyde. The fixed cells were permeabilized and blocked in 3% BSA, 0.2% Triton X-100, 1 mm NaN3 in PBS. Immunostaining of endogenous vinculin was performed with an anti-vinculin mouse monoclonal antibody (V9131, 1:1000, Sigma) and an anti-mouse IgG conjugated with Alexa Fluor 568 (A10037, 1:1000, Molecular Probes) as a secondary antibody. The endogenous F-actin was stained with phalloidin-Alexa Fluor 633 (Invitrogen). Images were acquired on an Olympus FluoView 1000 Confocal Microscope, and the acquired images were processed using FluoView software (Olympus).
RESULTS
Identification of VBSs in the Cell Surface Antigen (Sca4) of Rickettsia
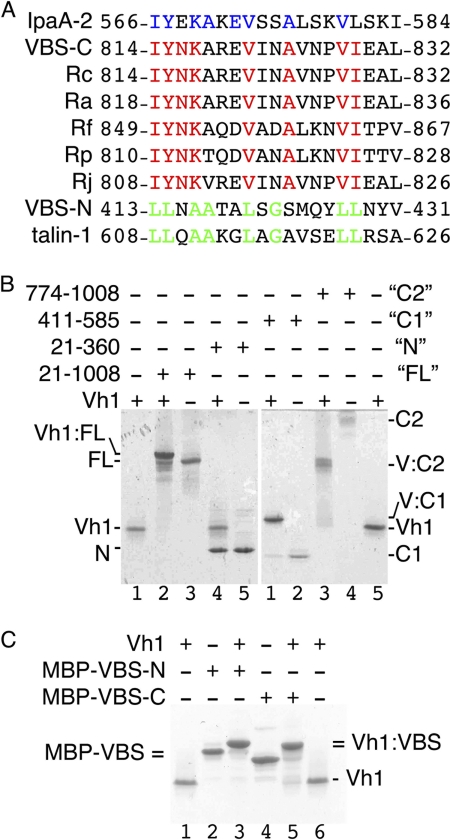
We identified a 19-residue consensus motif for VBS binding to the vinculin Vh1 domain by aligning six VBSs of talin, the two VBSs of IpaA, and the VBS of α-actinin, (ILM)(Y/L/V/T/A)XXA(K/R)X(V/L/I)XX(A/S/K/V)(L/T/V/E/I)(S/A/N/K)X(V/M/L/I)(L/V/I)XX(I/A/L/M), where X denotes any residue. Searches with this motif identified a potential VBS near the C terminus of the cell surface antigen sca4 (sca4-VBS-C) of R. rickettsii, where 8 of 19 residues of this putative VBS are identical to those of IpaA-VBS2 (32% sequence identity). Notably, this putative VBS was highly conserved in Rickettsia, with eight of 19 residues identical across six Rickettsia species (Fig. 1A).
FIGURE 1.
The cell surface antigen sca4 of R. rickettsii harbors two VBSs. A, sequence alignment of all 13 known VBSs (not shown) showed that sca4-VBS-C shares eight identical residues with IpaA-VBS2, shown in blue in the IpaA-VBS2 sequence. Sca4-VBS-C is highly conserved among Rickettsia, with eight identical residues (shown in red) among six species, including the spotted fever and the epidemic typhus group. Sca4-VBS-N is most similar to talin-VBS1, sharing eight residues, shown in green. IpaA-2, IpaA-VBS2; VBS-C, sca4-VBS-C; VBS-N, sca4-VBS-N; Rc, R. conorii; Ra, Rickettsia africae; Rf, Rickettsia felis; Rp, R. prowazekii; Rj, R. japonica. B, the sca4-VBS domains bind to the vinculin head domain Vh1 in solution. Native gel shift mobility assays (8–25% gradient native gel) showing that nearly full-length sca4 protein, residues 21–1008 (FL, lanes 2 and 3, left gel), sca4 residues 411–585 (C1, lanes 1 and 2, right gel), and sca4 residues 774–1008 (C2, lanes 3 and 4, right gel), bind to Vh1 as seen by the retarded (FL, C1, and C2) new complex bands appearing compared with unbound Vh1 (lanes 1 and 5, left and right gels, respectively). However, the N-terminal domain of sca4, residues 21–360 (N, lanes 4 and 5, left gel), does not bind to Vh1. C2 migrates poorly into the native gel probably due to its pI of 8.6. C, the VBSs of sca4 bind to Vh1 in solution. Native gel shift mobility assays show that binding of MBP fusions of sca4-VBS-N (lane 3) or sca4-VBS-C (lane 5) retards migration of Vh1 (lanes 1 and 6) or the MBP-sca4-VBSs (lanes 2 and 4). MBP-sca4-VBS-C (MBP-VBS-C) runs slightly farther (lane 2) than MBP-sca4-VBS-N (MBP-VBS-N, lane 2) and is indicated by MBP-VBS= due to their similar position. Vh1 in complex with MBP-sca4-VBS-C runs slightly farther (lane 5) versus MBP-sca4-VBS-N (lane 3), and both complexes are indicated by =Vh1:VBS due to their similar position.
The VBSs of the vinculin binding partners bind to the N-terminal seven-helix bundle domain of vinculin (Vh1) (16, 23, 24). Based on preliminary binding studies,4 we suspected that the C-terminal sca4 domain (residues 411–1008) had two binding sites for Vh1. Based on secondary structure predictions and domain predictions, we therefore constructed several sca4 deletions and found that residues 21–1008, 411–585, and 775–1008 all bound to Vh1, whereas residues 21–360 did not (Fig. 1B). Several other constructs did not result in soluble protein and were not pursued further. Secondary structure predictions4 suggested that the 411–585 sca4 domain that binds Vh1 is composed of five α-helices, and alignment with the VBSs of talin showed that sca4-VBS-N shares 32% sequence identity with talin-VBS1 (Fig. 1A) and 16% and 18% identity to talin-VBS2 and talin-VBS3, respectively, with 13 of 23 residues identical across six Rickettsia species.
Both of the VBSs of sca4 bound to Vh1, as did nearly full-length sca4 protein (residues 21–1008, Fig. 1C). Furthermore, recombinant sca4 protein having both VBSs (residues 411–1008) formed a 2:1 complex with Vh1 in solution (supplemental Figs. S1 and S2). Thus, sca4 harbors two vinculin binding sites.
Sca4 Activates Vinculin
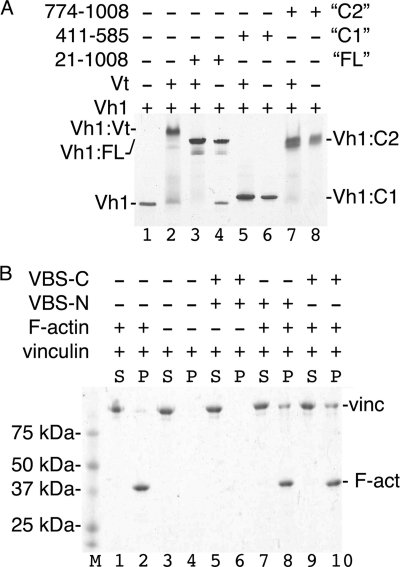
Activation of vinculin requires severing of the head-tail interaction, which exposes the cryptic actin binding sites within the Vt domain (30, 43–47). The sequence similarity between sca4-VBS-N and sca4-VBS-C and between talin-VBS1 and IpaA-VBS2, respectively, suggested that the VBSs of sca4 have similar functions. To test if the domains harboring the sca4-VBSs and full-length sca4 were sufficient to activate vinculin, we titrated the sca4-VBSs into preformed Vh1·Vt complexes. Indeed, native gel analyses showed that the nearly full-length sca4 (residues 21–1008) as well as the domains harboring the VBSs (residues 411–585 and 774–1008) readily displaced the Vt tail domain from pre-existing Vh1·Vt complexes by forming a new Vh1·sca4-VBS complex (Fig. 2A).
FIGURE 2.
Sca4 of R. rickettsii activates vinculin. A, native gel shift mobility assays are shown (8–25% gradient native gel) of Vh1 alone (lane 1), the preformed Vh1·Vt complex alone (lane 2), the Vh1·Vt complex with the addition of nearly full-length sca4 (residues 21–1008, lane 3), or of the sca4-VBS domains (lanes 5 and 7 for sca4-VBS-N and sca4-VBS-C domains, respectively). All three sca4 proteins displace Vt from the Vh1·Vt complex readily at equimolar ratios and form a Vh1·sca4 complex (lanes 3–8) that is retarded compared with Vh1 alone (lane 1). Unbound Vt does not show on a native gel because of its basic pI (24). B, actin co-sedimentation analyses (8–25% gradient native gel) of full-length vinculin in the presence (lanes 5–10) and absence (lanes 1–4) of the minimal sca4-VBSs are shown. Vinculin alone does not aggregate (lanes 1–4) and does not sediment with F-actin (lanes 1–2). The addition of a 2-fold molar excess of either sca4-VBSs to full-length vinculin does not lead to aggregates (lanes 5 and 6) but triggers co-sedimentation of vinculin with F-actin (lanes 7–10). F-act, F-actin; M, molecular weight markers; S, supernatant; P, pellet; VBS-C, sca4-VBS-C, residues 814–832; VBS-N, sca4-VBS-N, residues 413–431; vinc, vinculin.
A hallmark of vinculin activation is the exposure of cryptic F-actin binding sites (48, 49) that allow activated vinculin to bind to the cytoskeleton. We, therefore, tested whether the VBSs of sca4 could activate the latent F-actin binding functions of vinculin by co-sedimentation assays. Although the sca4-VBSs did not pellet full-length vinculin, the vinculin·sca4-VBS complexes co-sedimented with F-actin (Fig. 2B). Thus, both VBSs of sca4 are sufficient to activate vinculin, establishing their functions as bona fide vinculin binding sites.
Sca4 Co-localizes with Vinculin at Sites of Focal Adhesions
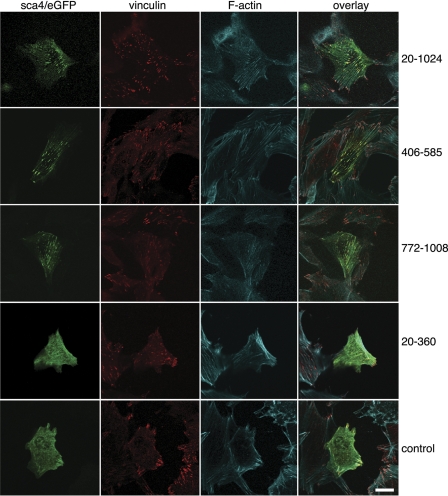
To assess the sca4-vinculin interaction in cells, we transfected NIH 3T3 fibroblasts with eGFP alone or with in-frame eGFP fusions of sca4 residues 21–1008 harboring both VBSs, sca4 residues 406–585 harboring sca4-VBS-N, sca4 residues 774–1008 containing sca4-VBS-C, or sca4 residues 21–360 that do not bind to vinculin in vitro (Fig. 3). Endogenous vinculin and F-actin were also visualized by fluorescence labeling. Expression of the various sca4 constructs did not induce morphological changes of the transfected cells. As shown previously by others (50, 51), vinculin was observed primarily at sites of focal adhesions. Confocal microscopy established that eGFP-sca4, residues 406–585, 776–1008, or 21–1008 were present in fibrous-like structures and at punctate that overlapped with focal adhesions. Notably, both nearly full-length sca4s and the domains harboring the sca4-VBSs co-localized with endogenous vinculin. In addition, the punctate patterns were predominantly at the inner basal surface of the cells. Only small amounts of sca4 co-localized with vinculin at the leading edge of the cell. The fibrous expression patterns of sca4 overlap with F-actin staining. By contrast, eGFP alone and e-GFP-sca4 (residues 21–360) did not co-localize with vinculin. Therefore, sca4 and vinculin can interact in cells.
FIGURE 3.
Sca4 and vinculin co-localize at focal adhesions in NIH 3T3 cells. Transient transfection of sca4 residues 20–1024 harboring both VBSs (top row), 406–585 harboring sca4-VBS-N (second row), 772–1008 harboring sca4-VBS-C (third row), the N-terminal non-binding 20–360 domain (fourth row), and pEGFP blank vector (bottom row) were analyzed on a fluorescence microscope. Endogenous vinculin (second column) and F-actin (third column) were stained with a fluorescence-conjugated vinculin antibody and phalloidin-Alexa Fluor 633, respectively. All images are at the same magnification. The scale bar represents 20 μm.
Vh1 Adopts a Novel Conformation When Bound to Sca4-VBS-C
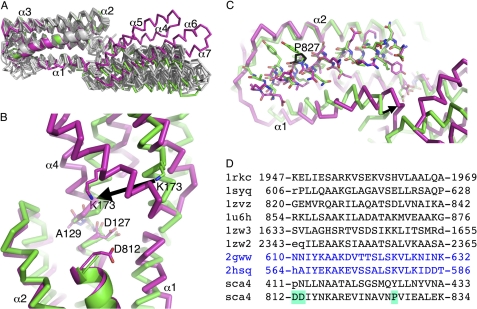
To define the molecular details of the sca4-vinculin interaction, we determined the crystal structures of the vinculin Vh1 domain in complex with either sca4-VBS to 2 and 2.8 Å, respectively (Tables 1 and 2, Fig. 4, A–C; supplemental Fig. S3–S6). Although we obtained several crystallization hits for the Vh1·sca4-VBS-N complex, these crystals diffracted only to about 8 Å Bragg spacings. However, by decreasing the molar ratio of Vh1 to sca4, we obtained cubic crystals that allowed complete data collection to 2 Å Bragg spacing on beamline 11-1 at the Stanford Linear Acceleration Center at Stanford Synchrotron Radiation Laboratory.
FIGURE 4.
Sca4 induces a novel conformation in vinculin. A, superposition of all known talin-VBSs and IpaA-VBSs in their Vh1-bound states (white) onto our 2 Å Vh1·sca4-VBS-N (green) and 2.8 Å Vh1·sca4-VBS-C (magenta) crystal structures. Vh1 is shown as a Cα trace and the VBSs as a schematic. Vh1 α-helices α4-α7 are labeled. All VBSs (talin-VBS1, residues 608–627; talin-VBS11, residues 822–841; talin-VBS2, residues 855–874; talin-VBS33, residues 1525–1544; talin-VBS36, residues 1633–1652; talin-VBS58, residues 2345–2364; IpaA-VBS1; IpaA-VBS2) were aligned to talin-VBS3 (residues 1949–1968) with root mean square deviations ranging from 0.5 Å to 1.1 Å. Sca4-VBS-C induces a significant movement of the C-terminal Vh1 four-helix bundle subdomain, which is unique versus all other Vh1·VBS structures. B, a zoomed view of the superposition of sca4-VBS-N (green) representative of all Vh1·VBS structures onto the sca4-VBS-C (magenta) in their Vh1-bound states. Unlike other VBSs, sca4-VBS-C has two acidic residues on its N terminus (Asp-812 is shown in sticks and is labeled), which might stabilize the sca4-VBS-C α-helix by neutralizing its dipole, and the large movement of the C-terminal Vh1 subdomain is indicated by the movement of Lys-173 and the arrow. C, a zoomed-in view showing the kink at sca4 Pro-827 (shown in black sticks, labeled) in sca4-VBS-C (magenta) compared with the sca4-VBS-N (green); the latter is representative of all other Vh1·VBS structures. Residues N-terminal to Pro-827 moved, whereas the C-terminal end superimpose well. This movement causes Vh1 α-helix α1 to shift down (in this view) and toward the C-terminal Vh1 subdomain as indicated by the arrow. D, structure-based sequence alignment of the talin-VBSs (first six black PDB entries 1rkc, 1syq, 1zvz, 1u6h, 1zw3, 1zw2), IpaA-VBSs (next two blue PDB entries 2gww, 2hsq), and the sca4-VBSs (last two black sequences). Residue numbers are indicated. The two acidic N-terminal residues found only in sca4-VBS-C and the Pro-827 found only in sca4-VBS-C are highlighted in cyan. Residues in lowercase are not present in the structure.
The Vh1·sca4-VBS-N crystal structures revealed that this VBS activates vinculin in a conventional fashion, akin to that of the VBSs of talin (24, 25), IpaA (16, 18) (Fig. 4A-C; supplemental Fig. S3–S6), or α-actinin even though the VBS of α-actinin has opposite directionality (23). Vh1 is composed of two four-helix bundle subdomains that are connected by a large shared central α-helix (α4). Binding of sca4-VBS to the N-terminal Vh1 four-helix bundle subdomain (residues 1–130) creates a five-helix bundle. As seen first in the Vh1·talin-VBS3 crystal structure (24), the Vh1·sca4-VBS interfaces are predominantly hydrophobic (supplemental Fig. S7) with buried solvent-accessible surface areas of 1152 and 1110 Å2 for sca4-VBS-N and sca4-VBS-C, respectively, upon complex formation. Furthermore, there are three hydrogen bond formations involving Vh1 residues Ile-12, Gln-19, and His-22 and sca4-VBS-N residues Ser-421, Gln-425, and Asn-432. In the Vh1·sca4-VBS-C structure only two hydrogen bond interactions are formed that are mediated by Vh1 residues Asp-127 and His-22 and sca4-VBS-C residues Tyr-815 and Glu-833, respectively.
Each of the two subdomains can be superimposed individually onto other known Vh1·VBS structures with root mean square deviations less than 1 Å, indicating that the subdomain structure is conserved. Indeed, a structural similarity search using the DALI server (52) for the N-terminal four-helix bundle Vh1 subdomain in complex with either sca4-VBS found the Vh1·talin-VBS1 structure (PDB entry 1syq; Z-scores of 1.2 and 2.2, respectively) (25) as the best match followed by the Vh1·IpaA-VBS2 structure (PDB entry 2ibf; Z-scores of 1.5 and 1.8, respectively) (42). Thus, like Shigella flexneri, R. rickettsii applies molecular mimicry of talin to bind to and activate vinculin.
Notably, the Vh1·sca4-VBS-C structure shows a novel subdomain movement of more than 35° relative to the sca4-VBS-N bound structure (as judged using the program Dyndom) where the hinge involves residues 8–13 and 126–129 and where the two 4-helix bundle Vh1 subdomains move into a unique and indeed more “closed” Vh1 conformation (Fig. 4A; supplemental Figs. S3, S8, and S9). A structure-based sequence alignment (Fig. 4D) reveals that the N terminus of sca4-VBS-C is acidic (Asp-811 and Asp-812) compared with all other VBSs, which might neutralize the helix dipole and stabilize the sca4-VBS-C α-helix. More importantly, these two aspartates could be responsible for the dramatic movement of Vh1 Lys-173 that may trigger subdomain movement in Vh1.
A second, somewhat subtler difference in the Vh1·sca4-VBS-C crystal structure compared with all other known Vh1·VBS structures is a movement of α-helix α1, which is probably caused by Pro-827 (Fig. 4C; supplemental Fig. S4). Indeed, mutating the talin-VBS1, which has a glutamate in the equivalent position (Glu-621; Fig. 4D) to a proline, abolishes its binding to Vh1 (28). The sca4-VBS-C superimposes well with the other VBSs up to residue Pro-827, which causes a kink in the sca4-VBS-C that probably pushes the Vh1 α-helix α1 somewhat further away from Vh1 α-helix α2 and almost half a turn closer to the C-terminal four-helix Vh1 subdomain, thus affecting its orientation relative to the N-terminal four-helix bundle subdomain (Fig. 4C, supplemental Figs. S4, S8, and S9). The distinct α-helix α1 conformation results in different interactions of residues residing on its N terminus with the C-terminal four-helix subdomain, in particular the C-terminal region of Vh1 α-helix α5. Superposition with full-length vinculin shows that α-helix α5 moves deeper into the core of vinculin (supplemental Figs. S8 and S9). For example, the side chain of His-5 that points toward the side chain of Leu-182 and is rotated almost 180° in Vh1·sca4-VBS-C to point toward the N-terminal four-helix bundle. This space is filled by Phe-4 (interacting with Ala-17 on α1 and Leu-116 on α4) in the other structures. In Vh1·sca4-VBS-C, Phe-4, rather, resides close to His-184, residing on Vh1 α-helix α6.
DISCUSSION
Obligate intracellular pathogens enter and spread within the host cell by co-opting the F-actin cytoskeleton, yet how these events are orchestrated is generally poorly understood. Our findings with sca4 suggest that this protein functions as an essential invasin for rickettsial species and that, in a remarkable case of convergent evolution, sca4 functions as an invasin akin to that of the IpaA protein of Shigella, where both invasins function as talin mimics that bind to and activate vinculin to provide connections of the respective pathogens with the F-actin network. Furthermore, both IpaA and sca4 invasins harbor at least two VBSs, suggesting they both employ a mechanism that amplifies vinculin activation and actin organization at sites of pathogen entry. Finally, both of the VBSs of sca4 are conserved across all rickettsial species, suggesting their essential roles in pathogenesis.
The mechanism by which sca4 activates vinculin occurs in a fashion described for IpaA (16), where the VBSs of both invasins bind to and activate vinculin. This scenario bypasses pre-activation steps that are required for the binding of talin or α-actinin to vinculin, where force-induced unraveling of helix bundles is needed to expose their VBSs to allow them to bind to and activate vinculin (24, 30, 42). This strategy employed by these pathogens ensures that they effectively compete with talin and α-actinin for vinculin and that they effectively sequester this essential F-actin binding protein for the uses of the pathogen in entry, motility, and spread.
Our Vh1·sca4-VBS crystal structures are the first reported structures from R. rickettsii, with only a handful structures determined from another Rickettsia species. Although both sca4-VBSs and IpaA-VBSs are talin-VBS mimics, it is notable that the sca4-VBS-C induces novel structural alterations in vinculin, in particular to the C-terminal Vh1 four-helix bundle subdomain that does not bind to sca4-VBS, a scenario unique among all vinculin·VBS structures. Two features of sca4-VBS-C appear to trigger this change. First, this is the only known VBS that has two negatively charged side chains on its N terminus, which we posit might stabilize the sca4-VBS-C α-helix. Second, sca4-VBS-C harbors a proline residue that likely contributes to the shift of α-helix α1 and to the observed contortions in the C-terminal four-helix bundle subdomain.
The fact that Rickettsia uses sca4 to bind to and activate vinculin suggests that this vinculin·sca4 interaction is involved in invasion and might be targeted as a therapeutic strategy for combating this deadly pathogen. In this regard, the unique interactions of sca4 and vinculin revealed in the vinculin·sca4 crystal structures may provide inroads for developing small molecule inhibitors that selectively disable the functions of sca4.
Supplementary Material
Acknowledgments
We are indebted to our colleagues at Scripps Florida, John L. Cleveland for discussions and critical review of the manuscript, Erumbi S. Rangarajan for fruitful discussions, and Philippe R. Bois and Zhen Wu for sequencing. We are grateful to the staff at Stanford Synchrotron Radiation Laboratory for synchrotron support.
This work was supported, in whole or in part, by the National Institutes of Health. This is publication 20902 from The Scripps Research Institute.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental table and Figs. S1–S9.
The atomic coordinates and structure factors (codes 3tj5 and 3tj6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. H. Lee, personal communication.
- Vh1
- vinculin seven-helix head domain
- VBS
- vinculin binding site
- eGFP
- enhanced green fluorescent protein
- F-actin
- filamentous actin
- IpaA
- invasion plasmid antigen
- MBP
- maltose-binding protein
- PDB
- Protein Data Bank
- sca
- cell surface antigen
- Vt
- vinculin five-helix bundle tail domain.
REFERENCES
- 1. Walker D. H., Ismail N. (2008) Nat. Rev. Microbiol. 6, 375–386 [DOI] [PubMed] [Google Scholar]
- 2. Hackstadt T. (1996) Infect Agents Dis. 5, 127–143 [PubMed] [Google Scholar]
- 3. Dantas-Torres F. (2007) Lancet Infect. Dis. 7, 724–732 [DOI] [PubMed] [Google Scholar]
- 4. Walker D. H., Occhino C., Tringali G. R., Di Rosa S., Mansueto S. (1988) Hum. Pathol. 19, 1449–1454 [DOI] [PubMed] [Google Scholar]
- 5. Hand W. L., Miller J. B., Reinarz J. A., Sanford J. P. (1970) Arch. Intern Med. 125, 879–882 [PubMed] [Google Scholar]
- 6. Martinez J. J., Seveau S., Veiga E., Matsuyama S., Cossart P. (2005) Cell 123, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 7. Martinez J. J., Cossart P. (2004) J. Cell Science 117, 5097–5106 [DOI] [PubMed] [Google Scholar]
- 8. Jeng R. L., Goley E. D., D'Alessio J. A., Chaga O. Y., Svitkina T. M., Borisy G. G., Heinzen R. A., Welch M. D. (2004) Cell. Microbiol. 6, 761–769 [DOI] [PubMed] [Google Scholar]
- 9. Tran Van Nhieu G., Bourdet-Sicard R., Duménil G., Blocker A., Sansonetti P. J. (2000) Cell. Microbiol. 2, 187–193 [DOI] [PubMed] [Google Scholar]
- 10. Cossart P. (2000) Cell. Microbiol. 2, 195–205 [DOI] [PubMed] [Google Scholar]
- 11. Gouin E., Egile C., Dehoux P., Villiers V., Adams J., Gertler F., Li R., Cossart P. (2004) Nature 427, 457–461 [DOI] [PubMed] [Google Scholar]
- 12. Blanc G., Ngwamidiba M., Ogata H., Fournier P. E., Claverie J. M., Raoult D. (2005) Mol. Biol. Evol. 22, 2073–2083 [DOI] [PubMed] [Google Scholar]
- 13. Chan Y. G., Cardwell M. M., Hermanas T. M., Uchiyama T., Martinez J. J. (2009) Cell. Microbiol. 11, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riley S. P., Goh K. C., Hermanas T. M., Cardwell M. M., Chan Y. G., Martinez J. J. (2010) Infect. Immun. 78, 1895–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haglund C. M., Choe J. E., Skau C. T., Kovar D. R., Welch M. D. (2010) Nat. Cell Biol. 12, 1057–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izard T., Tran Van Nhieu G., Bois P. R. (2006) J. Cell Biol. 175, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramarao N., Le Clainche C., Izard T., Bourdet-Sicard R., Ageron E., Sansonetti P. J., Carlier M. F., Tran Van Nhieu G. (2007) FEBS Lett. 581, 853–857 [DOI] [PubMed] [Google Scholar]
- 18. Nhieu G. T., Izard T. (2007) EMBO J. 26, 4588–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., Izard T. (2004) Structure 12, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 20. Rangarajan E. S., Lee J. H., Yogesha S. D., Izard T. (2010) PLoS ONE 5, e10679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 22. Bakolitsa C., de Pereda J. M., Bagshaw C. R., Critchley D. R., Liddington R. C. (1999) Cell 99, 603–613 [DOI] [PubMed] [Google Scholar]
- 23. Bois P. R., Borgon R. A., Vonrhein C., Izard T. (2005) Mol. Cell. Biol. 25, 6112–6122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 24. Izard T., Evans G., Borgon R. A., Rush C. L., Bricogne G., Bois P. R. (2004) Nature 427, 171–175 [DOI] [PubMed] [Google Scholar]
- 25. Izard T., Vonrhein C. (2004) J. Biol. Chem. 279, 27667–27678 [DOI] [PubMed] [Google Scholar]
- 26. Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 27. Gingras A. R., Vogel K. P., Steinhoff H. J., Ziegler W. H., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2006) Biochemistry 45, 1805–1817 [DOI] [PubMed] [Google Scholar]
- 28. Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 29. Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gingras A. R., Ziegler W. H., Bobkov A. A., Joyce M. G., Fasci D., Himmel M., Rothemund S., Ritter A., Grossmann J. G., Patel B., Bate N., Goult B. T., Emsley J., Barsukov I. L., Roberts G. C., Liddington R. C., Ginsberg M. H., Critchley D. R. (2009) J. Biol. Chem. 284, 8866–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grashoff C., Hoffman B. D., Brenner M. D., Zhou R., Parsons M., Yang M. T., McLean M. A., Sligar S. G., Chen C. S., Ha T., Schwartz M. A. (2010) Nature 466, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tran Van Nhieu G., Ben-Ze'ev A., Sansonetti P. J. (1997) EMBO J. 16, 2717–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park H., Valencia-Gallardo C., Sharff A., Tran Van Nhieu G., Izard T. (2011) J. Biol. Chem. 286, 23214–23221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vonrhein C., Bricogne G. (2008) Acta Crystallogr. A 64, C78 [Google Scholar]
- 36. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 37. Leslie A. G. (2006) Acta Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 38. Smart O. S., Brandl C., Flensburg P., Keller W., Paciorek C., Vonrhein C., Womack T., Bricogne G. (2008) Abstract of the Annual Meeting of the American Crystallogr Association, Knoxville, TN Abstr. TP139 [Google Scholar]
- 39. Emsley P., Cowtan K. (2004) Acta Crystallogr. D60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vagin A., Teplyakov A. (1997) J. Appl Crystallogr. 30, 1022–1025 [Google Scholar]
- 42. Bois P. R., O'Hara B. P., Nietlispach D., Kirkpatrick J., Izard T. (2006) J. Biol. Chem. 281, 7228–7236 [DOI] [PubMed] [Google Scholar]
- 43. Cohen D. M., Kutscher B., Chen H., Murphy D. B., Craig S. W. (2006) J. Biol. Chem. 281, 16006–16015 [DOI] [PubMed] [Google Scholar]
- 44. Gilmore A. P., Burridge K. (1996) Nature 381, 531–535 [DOI] [PubMed] [Google Scholar]
- 45. Johnson R. P., Craig S. W. (2000) J. Biol. Chem. 275, 95–105 [DOI] [PubMed] [Google Scholar]
- 46. Johnson R. P., Niggli V., Durrer P., Craig S. W. (1998) Biochemistry 37, 10211–10222 [DOI] [PubMed] [Google Scholar]
- 47. Winkler J., Lünsdorf H., Jockusch B. M. (1996) J. Struct. Biol. 116, 270–277 [DOI] [PubMed] [Google Scholar]
- 48. Critchley D. R. (2004) Biochem. Soc. Trans. 32, 831–836 [DOI] [PubMed] [Google Scholar]
- 49. Critchley D. R., Holt M. R., Barry S. T., Priddle H., Hemmings L., Norman J. (1999) Biochem. Soc. Symp. 65, 79–99 [PubMed] [Google Scholar]
- 50. Geiger B. (1979) Cell 18, 193–205 [DOI] [PubMed] [Google Scholar]
- 51. Geiger B., Tokuyasu K. T., Dutton A. H., Singer S. J. (1980) Proc. Natl. Acad. Sci. U. S. A. 77, 4127–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.