Abstract
The mechanism by which agonist binding to an ionotropic glutamate receptor leads to channel opening is a central issue in molecular neurobiology. Partial agonists are useful tools for studying the activation mechanism because they produce full channel activation with lower probability than full agonists. Structural transitions that determine the efficacy of partial agonists can provide information on the trigger that begins the channel-opening process. The ligand-binding domain of AMPA receptors is a bilobed structure, and the closure of the lobes is associated with channel activation. One possibility is that partial agonists sterically block full lobe closure but that partial degrees of closure trigger the channel with a lower probability. Alternatively, full lobe closure may be required for activation, and the stability of the fully closed state could determine efficacy with the fully closed state having a lower stability when bound to partial relative to full agonists. Disulfide-trapping experiments demonstrated that even extremely low efficacy ligands such as 6-cyano-7-nitroquinoxaline-2,3-dione can produce a full lobe closure, presumably with low probability. The results are consistent the hypothesis that the efficacy is determined at least in part by the stability of the state in which the lobes are fully closed.
Keywords: Crystal Structure; Glutamate Receptors; Glutamate Receptors Ionotropic (AMPA, NMDA); Neurotransmitter Receptors; NMR
Introduction
Ionotropic glutamate receptors are the major mediators of excitatory synaptic transmission in the vertebrate nervous system (1, 2). Glutamate receptor subunits are categorized by pharmacological properties, biological role, and sequence into those that are sensitive to the following: 1) α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA; GluA1–4)2; 2) the neurotoxin kainate (GluK1–5); and 3) N-methyl-d-aspartic acid (NMDA; GluN1, GluN2A-D, GluN3A-B). The important structural details of glutamate receptors were determined first by the solution of the structures of the ligand-binding domain (3–7) and the N-terminal domain (8, 9) followed by the structure of the full-length tetrameric GluA2 subtype of AMPA receptor (10). The structures in the presence of various agonists, partial agonists, and antagonists in combination with spectroscopic measurements, electrophysiological measurements, and site-directed mutagenesis have provided a wealth of information on the link between structure and function (11–13). The binding domain is a bilobed structure to which agonist binds in the cleft between the two lobes. Two linker peptide strands connect the lobes, and the lobes can close to envelope the agonist. One lobe (lobe 1) forms a dimer interface with a second copy of the ligand-binding domain within the tetrameric structure, and the second lobe (lobe 2) is linked to the ion channel domain. When the dimer interface is intact, the force generated by the closing of the lobes can affect the ion channel and presumably open a gate allowing ions to traverse the channel. Complexities arise due to the tetrameric structure and subtle differences between glutamate receptor subtypes, but the general outline of channel activation is likely to be similar across this class of receptor.
The mechanism of channel activation by partial agonists remains unclear. Single channel recording measurements of AMPA receptors have demonstrated that three or four conductance levels can be observed from a single channel, and these conductance levels are the same for both full and partial agonists (12, 14). Population of the higher conductance levels is favored at higher agonist concentrations, but at any given concentration, higher conductance levels are more prevalent for full than partial agonists. The concentration dependence is consistent with a model in which each subunit has a “gate” that contributes to ion conductance, and the more gates that are open, the higher the conductance. However, given the fact that the conductance levels are the same for all agonists, this would suggest that opening of a gate is an all-or-nothing process. That is, the signal from the ligand-binding domain results in a concerted change in the structure of the channel region. The question then is whether that change is produced by a specific conformation of the ligand-binding domain (such as a full lobe closure) or whether multiple conformations can produce the same change, perhaps with different probabilities, or possibly some combination of the two models can explain channel activation.
The initial crystal structures of the GluA2 ligand-binding domain in the presence of partial agonists suggested a correlation between the degree of lobe closure and the efficacy of the ligand, suggesting that multiple conformations can trigger channel gating. A full closure would result in a high probability of gating, and with a partial closure, the probability of gating would be much lower. However, subsequent studies indicated that at least some partial agonists could assume a range of lobe closures (15), and even the correlation in crystal structures between lobe opening and efficacy does not always hold (16–18). Most strikingly, crystal structures of partial agonists of the NMDA receptor show a fully closed lobe (19), leading to the suggestion that partial agonism is fundamentally different for NMDA versus AMPA receptors. An alternative view of partial agonism is that the stability of full lobe closure dictates efficacy (13, 15). That is, partial agonists could potentially exist in a dynamic equilibrium among two or more conformations. Some of the conformations may have relative lobe orientations that are open (similar to that seen in some crystal structures, 3, 12) and other conformations that may be closed to the same extent as that seen for full agonists (3). According to this hypothesis, channel activation would require the fully closed form, and the stability of that form would dictate efficacy. The binding of some weak partial agonists, such as iodowillardiine (IW), are consistent with this idea in that a wide range of lobe closures have been observed in crystal (12, 20) and NMR structures (15), and evidence for large scale dynamics is present in the NMR spectra (15, 21). Also, mutations that decrease the efficacy of AMPA show a range of lobe orientations as measured using single molecule FRET (22). In contrast, the weak partial agonist kainate presents a structural impediment to lobe closure. The isoprenyl group of kainate seems to block the closure of the lobe because of an apparent steric clash with the side chain of Leu-650 in GluA2 (3). Mutation of Leu-650 to threonine increases kainate efficacy presumably by reducing the steric interaction between this position and the isoprenyl of kainate (16, 18). Furthermore, little evidence for microseconds-to-milliseconds timescale dynamics in the presence of kainate have been observed in NMR studies (23). However, structures of GluA3 (24) and GluA4 (25) in the presence of kainate are more closed than GluA2 and the D651A mutation of GluA3 results in even further closure of the lobes due to a rotation of the side chain of Leu-650 (GluA2 numbering).3
Cysteine-trapping studies (i.e. the introduction of two cysteines to determine whether a disulfide can form) have been used to determine proximity of different portions of one protein or the proximity of two proteins or subunits. One criticism of this method is that proteins are dynamic structures, and very rare conformations can potentially be trapped. In this case, the shortcoming of the method can be an advantage. If, in fact, partial agonists can activate the channel by a relatively rare transition to a conformation with fully closed lobes, then cysteine trapping should be capable of stabilizing this form for further analysis by x-ray crystallography, NMR spectroscopy, and radioligand binding. We show here that with glutamate, iodowillardiine, kainate, and CNQX bound, the A452C/S652C GluA2 ligand-binding domain can be trapped in the closed form.
EXPERIMENTAL PROCEDURES
Materials
UBP282 and IW were purchased from Tocris (Ellisville, MO), and kainate, CNQX, and DNQX were purchased from Ascent Scientific (Princeton, NJ). Glutamate was purchased from Sigma-Aldrich. The GluA2 S1S2J construct was obtained from Eric Gouaux (Vollum Institute) (3). Stable isotopes were purchased from Cambridge Isotopes (Cambridge, MA). [3H]AMPA (40 Ci/mmol) was purchased from PerkinElmer Life Sciences.
Mutagenesis
Mutant receptors for both full-length GluA2 and the LBD of GluA2 were generated by QuikChange site-directed mutagenesis using Phusion or Pfu polymerase. The mRNAs for oocyte injection experiment were synthesized using a T7 high yield transcription kit following the instructions of the manufacturer (Aligent Technologies, Santa Clara, CA).
Cell Culture and Heterologous Expression
HEK293 cells were cultured with DMEM/F12 supplemented with 10% newborn calf serum and antibiotics. Cells were transfected with 500 ng of wild type or mutant receptors using Turbofect reagents (Fermentas, Inc.). Cells were plated on poly-d-lysine (0.1 mg/ml) and collagen (1:100 dilution of 10 mg/ml solution) double-coated chambered cover slides or 96-well plates for immunofluorescence or ratiometric fura-2 imaging experiments, respectively. Stage VI oocytes were prepared from isolated ovaries of Xenopus tropicalis (26) and injected with 7 ng of cRNA for each receptor (both wild type and mutants). Oocytes were kept in physiological OR-2 solution with antibiotics for 3–7 days before recording.
Electrophysiology
All electrophysiology experiments were conducted at room temperature (21 °C) using the two-electrode voltage clamp method. The standard extracellular solution was composed of 5 mm HEPES, 96 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, pH 7.4. 10 mm sodium glutamate was added for agonist-dependent activation. The membrane holding potential was −60 mV. A voltage ramp from −100 to +80 mV of 250 ms duration given at 1 Hz was used to continuously monitor the currents.
Ratiometric Ca2+ Imaging
Cells expressing wild type or mutant GluA2 were loaded with fura-2 AM (5 μm) for 2 h. Imaging experiments were carried out at room temperature (22 °C), with a sampling rate of one frame every 2 s with 100 ms of exposure time to either wavelength (340 and 380 nm). Images were acquired using a CCD camera driven by Slidebook software (version 4.2).
Immunofluorescence Imaging
HEK293 cells were transiently transfected with 500 ng of cDNA (WT, double mutant) using TurboFect (Fermentas) according to the manufacturer's instructions. Forty-eight h after transfection, cells were fixed in 2% paraformaldehyde in PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.47 mm KH2PO4, pH 7.4) for 15 m. Cells were blocked in 1× casein in TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) alone for surface staining or additional 0.1% Triton X-100 (for intracellular staining) at 37 °C for 1.5 h. After washing with TBS, cells were incubated with primary antibody (1:800 dilution; anti-glutamate receptor 2, extracellular, clone 6C4 from Chemicon, Millipore (MAB397)) in TBS containing 10% horse serum at 4 °C overnight. After washing with TBS, the cells were labeled with fluorescence-conjugated secondary antibody (Alexa Fluor 594 goat anti-mouse IgG from Invitrogen (A11005)) at 1:200 dilution in PBS containing 1× casein at room temperature for 30 m. The cells were washed extensively with TBS and then mounted with VECTASHIELD mounting medium with DAPI (Vector Laboratories, Inc.) and placed under a coverslip. Cells were examined under a fluorescence microscope (Olympus BX41, Olympus America, Inc.), and images were taken with Olympus Microfire digital camera.
Protein Preparation and Purification of LBD
The GluA2 LBD consists of residues Asn-392–Lys-506 and Pro-632–Ser-775 of the full rat GluA2-flop subunit (27) with and without the A452C/S652C mutations, a GA segment at the N terminus, and a GT linker connecting Lys-506 and Pro-632 (3). pET-22b(+) plasmids were transformed in Escherichia coli strain Origami B (DE3) cells and were grown at 37 °C to A600 of 0.9 to 1.0 in LB medium supplemented with antibiotics (ampicillin and kanamycin). The cultures were cooled to 20 °C for 20 min and isopropyl-β-d-thiogalactoside was added to a final concentration of 0.5 mm. Cultures were allowed to grow at 20 °C for 20 h. The cells were then pelleted. and the LBD was purified using a nickel-nitrilotriacetic acid column, followed by thrombin cleavage of the His6 tag, a sizing column (Superose 12, XK 26/100), and finally an HT SP ion exchange-Sepharose column (Amersham Biosciences Pharmacia). Glutamate (1 mm) was maintained in all buffers throughout purification. After the last column, the protein was concentrated and stored in 20 mm sodium acetate, 1 mm sodium azide, and 10 mm glutamate at pH 5.5. Final concentrations of CNQX, DNQX, kainate, IW, and UBP282 were 3 mm. Ligands were exchanged using successive dilution and concentration using an Amicon Ultra-4 (10 K) filter. For NMR spectroscopy, the bacteria were grown in M9 media with [15N]labeled ammonium chloride as the sole nitrogen source and [13C]glucose as the sole carbon source. The NMR buffer was an aqueous solution of 25 mm sodium acetate, 25 mm sodium chloride, 1 mm sodium azide, 12% D2O, pH 5.0.
NMR Spectroscopy
All measurements were made on a Varian Inova 500 spectrometer equipped with a triple resonance, z-gradient cryogenic probe. The standard double and triple resonance experiments in the BioPack software suite (Varian, Palo Alto) were used. Spectra were collected at 25 °C for glutamate, kainate, and IW, and at 13 °C for CNQX, DNQX, and UBP282. The data were apodized with a mixed exponential Gauss window function and zero-filled to double the original number of data points before Fourier transform (NMRPipe, version 1.6 (28)). Spectra were displayed, and the peaks were chosen and measured using Sparky (29).
Crystallography
For crystallization trials, the protein was concentrated to 0.3 to 0.4 mm using a Centricon 10 centrifugal filter (Millipore, Bedford, MA). The final protein concentration was ∼0.3 mm. Crystals were grown at 4 °C using the hanging drop technique, and the drops contained a 1:1 (v/v) ratio of protein solution to reservoir solution. The reservoir solution contained 16–18% PEG 8000, 0.1 m sodium cacodylate, 0.1–0.15 m zinc acetate, pH 6.5.
Data were collected at the Cornell High Energy Synchrotron Source beam line A1 using a Quantum-210 Area Detector Systems charge-coupled device detector. Data sets were indexed and scaled with HKL-3000 (30). Structures were solved with molecular replacement using Phenix (31). Refinement was performed with Phenix (31) and the CCP4 Program Suite 6.1.13 (32), and Coot 0.6.1 (33) was used for model building.
Radioligand Binding
The binding of [3H]AMPA (40 Ci/mmol; Amersham Biosciences) to GluA2 S1S2 was determined as described by Chen et al. (34). The binding buffer was 30 mm Tris-HCl, 100 mm potassium thiocyanate, 2.5 mm CaCl2, 10% glycerol, pH 7.2, maintained at all times at 4 °C. Glutamate was removed from the protein by successive concentration and dilution in the presence of β-mecaptoethanol and diluted to a final concentration of 70 nm. After the addition of the tested ligand, [3H]AMPA (10 nm) was added, the reaction (200 μl) proceeded for 1 h followed by filtration through Millipore GSWP filters and two 2-ml washes with binding buffer. All analysis was done using Kaleidagraph (Synergy Software).
RESULTS
Lobe-locking Mutations
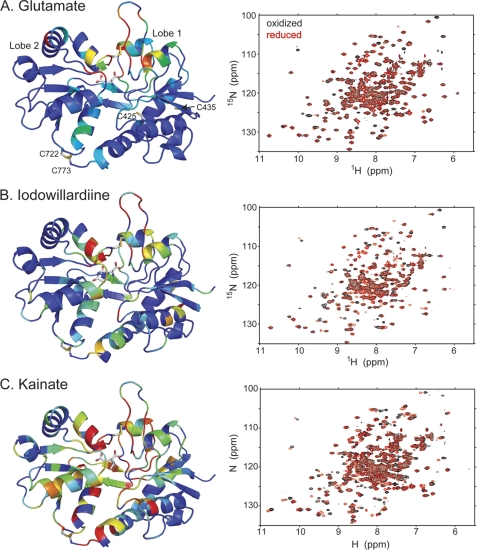
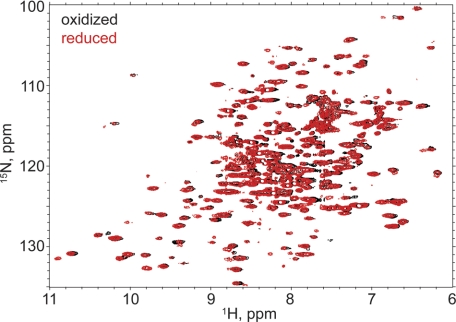
The ligand-binding domain of GluA2 contains four cysteines. Two (Cys-722 and Cys-773) are involved in a disulfide bond that ties lobe 2 to the C-terminal region of the domain in lobe 1 (Fig. 1A). The other two (Cys-425 and Cys-435) are not near the lobe interface and were predicted not to interfere with introduction of cysteines designed to maintain lobe closure. Two sets of double cysteine mutations were made on a wild type background of the GluA2 LBD: A452C/S652C and V484C/E657C. Both expressed well in E. coli and produced high quality NMR spectra when uniformly labeled with 15N. The 1H,15N-HSQC spectrum of V484C/E657C bound to glutamate was unchanged when exposed to either Cu-phenanthroline or DTT (supplemental Fig. S1A). On the other hand, A452C/S652C bound to glutamate was unchanged relative to untreated protein when exposed to Cu-phenanthroline, but the spectrum was markedly changed in the presence of DTT (Fig. 1A). The spectrum reverted to that seen in the presence of Cu-phenathroline when DTT was removed. The chemical shift changes were largely restricted to the lobe interface, suggesting that the disulfide bond was formed and broken by the reducing agent. This suggests that the disulfide bond is easily formed even in the absence of oxidizing agents when the protein is bound to glutamate but can be reduced. This was confirmed in the crystal structures of A452C/S652C, which showed a clear disulfide bond (Fig. 2A). In the case of V484C/E657C, the sulfur atoms of the two cysteines were separated by 5.41 ± 0.16 Å even after exposure to Cu-phenanthroline (supplemental Fig. S1B). All subsequent experiments employed the A452C/S652C mutation.
FIGURE 1.
Comparison of the amide backbone chemical shifts in the oxidized and reduced (DTT) form of A452C/S652C mutant of GluA2 LBD bound to glutamate (A; Protein Data Bank code 3T93), iodowillardiine (B; Protein Data Bank code 3T96), and kainate (C; Protein Data Bank code 3T9H). On the right is shown the 1H,15N-HSQC spectrum, and on the left, a representation of the resonances that have shifted upon reduction of the A452C/S652C disulfide bond is shown. The oxidized form is identical with no additions and in the presence of Cu-phenanthroline, suggesting that the disulfide bond forms spontaneously. The difference in chemical shift between the oxidized and reduced from was quantitated using the formula, chemical shift difference = . Residues were assigned colors with a chemical shift difference between 0 and 5 assigned to a rainbow of colors from blue to red.
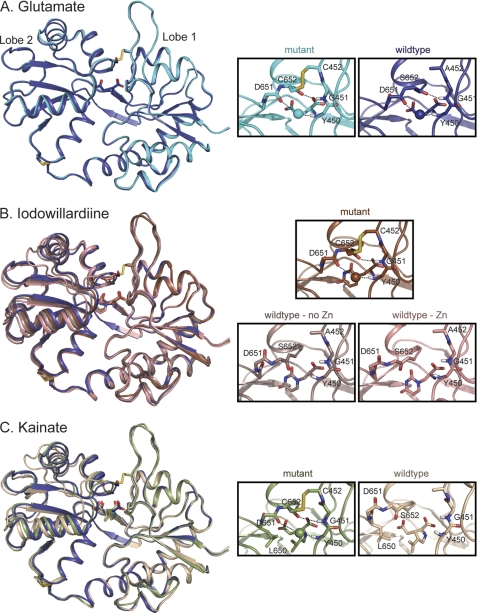
FIGURE 2.
Crystal structures of wild type and A452C/S652C mutant GluA2 LBD bound to glutamate (A), iodowillardiine (B), and kainate (C). The right panels show the agonist binding site. A, glutamate. The structure of wild type GluA2 bound to glutamate (Protein Data Bank code 3DP6; B protomer, 24) is shown in dark blue and the A452C/S652C mutant (Protein Data Bank code 3T93) is shown in cyan. The water molecule involved in the Asp-651–Tyr-450 H-bond is shown in the same color as the backbone in all parts of this figure. The Cys-452–Cys-652 disulfide bond is clearly present in the mutant. B, iodowillardiine. The structure bound to glutamate is shown in blue to indicate the orientation of a fully closed lobe. The iodowillardiine-bound structures are shown in shades of brown/pink. Two wild type iodowillardiine-bound structures are shown, one crystallized in the presence of zinc (Protein Data Bank code 1MY4, A protomer; 20) and one in the absence of zinc (Protein Data Bank code 1MQG, A protomer; 12). Both are shown to illustrate the variability seen in different crystal structures. The mutant structure is more closed and the Asp-651-H2O-Tyr-450 and Cys-652–Gly-451 H-bonds are clearly present as is the Cys-452–Cys-652 disulfide bond (Protein Data Bank code 3T96). C, kainate. The glutamate-bound structure is shown in dark blue, and the wild type kainate-bound structure is shown in tan (Protein Data Bank code 1FW0; 3). As seen for iodowillardiine, the A452C/S652C mutant structure (shown in green) is more closed and the Asp-651–H2O–Tyr-450 and Cys-652–Gly-451 H-bonds are clearly present as is the Cys-452–Cys-652 disulfide bond (Protein Data Bank code 3T9H). Note the change in the rotameric state of the side chain of Leu-650.
Reduction of Disulfide Bond Revealed by Radioligand Binding
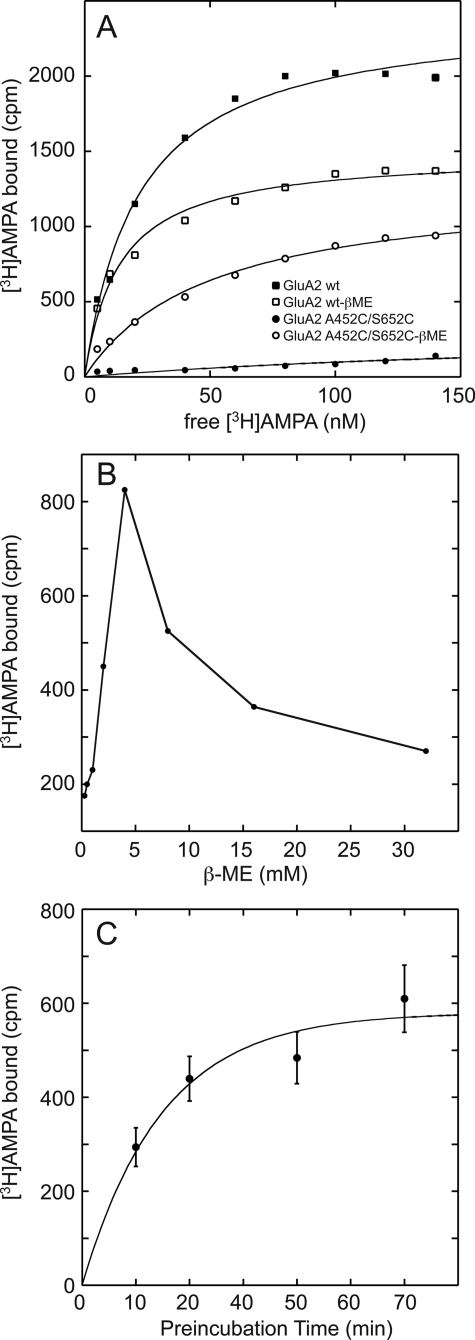
Radioligand binding was used to demonstrate that the lobes could be locked in a conformation that prevents further binding. Both wild type and A452C/S652C GluA2 LBD were diluted to 40 nm, reduced with either DTT or β-metcaptoethanol, and washed by successive dilution and concentration to remove residual glutamate. The sample was divided into two. One-half was washed further with DTT or β-metcaptoethanol, and the other was washed simply with buffer. Binding of [3H]AMPA was then measured as a function of AMPA concentration. In the case of wild type LBD, the binding was decreased somewhat by the reducing agent (Fig. 3A), possibly due to a partial reduction of the native Cys-718/Cys-773 disulfide bond in the apo state. For the A452C/S652C mutation, binding was only observed in the reduced state, with no binding detected in the absence of reducing agent. Reduction of the disulfide bond by β-metcaptoethanol peaked at 5 mm with a decrease in binding at higher concentrations of the reducing agent (Fig. 3B). Also, the reduction of the disulfide bond was relatively slow, with a maximum reduction occurring after ∼1 h, as judged by the increase in binding sites for [3H]AMPA (Fig. 3C).
FIGURE 3.
A, binding of [3H]AMPA to both wild type and the A452C/S652C mutant GluA2 LBD. For the wild type, a decrease in binding was observed in the presence of 5 mm β-metcaptoethanol (apparent decrease in Bmax). In the case of the mutant, binding is only observed when the disulfide bond is reduced by β-metcaptoethanol (β-ME). The KD for binding of [3H]AMPA to the reduced form of the mutant is 2-fold higher than that observed for wild type. B, treatment with β-metcaptoethanol increases binding of the A452C/S652C mutant up to 5 mm, and above that concentration, binding decreases. This may be due to the initial selective reduction of the A452C/S652C followed by the reduction of the Cys-722/Cys-773 disulfide bond. C, the time course for reduction of the disulfide bond shows that it is complete within 60 min in the presence of 5 mm β-metcaptoethanol.
Lobe Locking with Partial Agonists
Although full agonists seem to act by binding and closing the lobes of the LBD fully, the action of partial agonists is less well understood. One possibility is that the bulkier partial agonists induce a stable partial lobe closing that, by some unknown mechanism, results in a lower probability of channel gating. Another alternative is that despite the apparent steric conflict implied by crystal structures, lobes can close fully (or almost fully) around a partial agonist, although less often than seen with full agonists. Nevertheless, a full closure may be required to activate the channel gate. Because the gate would be less likely to be activated, the efficacy of the agonist would be decreased relative to full agonists. The question asked here is whether partial agonists can close the channel fully, even if rarely.
Iodowillardiine and Kainate (NMR Spectroscopy)
Two weak partial agonists were tested: IW and kainate. In both cases, NMR spectroscopy was used to determine whether effects were uniform or whether both oxidized and reduced protein could be detected. Exchange of ligand was done in the presence of 50 mm DTT to remove glutamate and allow binding of the partial agonist. The 1H,15N-HSQC (TROSY) NMR spectra (after the removal of DTT) are shown in Fig. 1, B (IW) and C (kainate). Addition of Cu-phenanthroline had no effect on the spectra, suggesting that either the disulfide bond formed spontaneously or not at all. However, addition of DTT, resulted in a clear change in the spectrum. Because of the mutations, the chemical shifts of the mutant were changed sufficiently relative to the wild type (35) to make assignment by proximity problematic in some regions of the spectrum. For this reason, assignments were verified with a combination of triple resonance experiments (HNCO, HN(CO)CA, and HNCA). Mapping of the chemical shift changes to the structure of the protein suggests that most changes occur near the introduced disulfide and much less so at the native disulfide, suggesting that the interlobe disulfide is formed spontaneously but can be reduced. Furthermore, the spectra do not show multiple peaks for a given correlation, suggesting that the disulfide was largely formed. Given the sensitivity of the measurements, less than ∼15% was in the reduced form in the absence of reducing agent or in the oxidized form in the presence of reducing agent.
Iodowillardiine and Kainate (X-ray Crystallography)
The crystal structures of the IW- and kainate-bound proteins were then determined. In both cases, the disulfide bond between A452C and S652C was clearly formed in all three copies in the asymmetric unit (Fig. 2, B, IW, and C, kainate). Interestingly, even in the presence of reducing agents, the disulfide bond was formed, suggesting that this form of the protein, although not the predominant form in solution, was preferentially crystallized.
In the case of kainate, the A452C/S652C protein crystallizes in the P22121 space group with three copies in the asymmetric unit (similar to the wild type bound to glutamate) in contrast to the wild type kainate-bound protein, which crystallizes as P2, with one or two copies in the asymmetric unit (supplemental Table S1). The lobes are open by 3.0 ± 1.0° relative to the most closed wild type protein bound to glutamate (Protein Data Bank code 3DP6, B protomer), in contrast to 10.1° found in the kainate-bound wild type protein. Although the lobes are slightly open relative to glutamate, two interlobe H-bonds (Gly-451–Gly-653 and Tyr-450-H2O-Ser-652) produced by a 180° rotation of the peptide bond are present in all three copies. These H-bonds are absent in most crystal structures of partial agonists and have been absent in all previously determined structures of kainate-bound AMPA receptors (3, 18, 24, 25). Previous crystal structures have suggested that kainate prevents further lobe closure due to the steric clash between the side chain of Leu-650 with the isoprenyl group of kainate (foot-in-the-door mechanism) (18). The more fully closed structure is produced largely by the rotation of the side chain of Leu-650 and a small movement of kainate toward lobe 1. With these rather subtle movements, the distribution of structures of the kainate-bound protein approaches the degree of closure of the glutamate-bound protein, although the average structure is still slightly more open in the presence of kainate. Although the crystal packing differs in the wild type versus the double cysteine mutant, the lattice packing does not seem to be the distorting the structure, resulting in the differences seen here. Average lobe orientation of the wild type protein bound to both kainate (P2 symmetry) and glutamate (P222 symmetry) in solution is similar to that seen in the crystals (15).
In the case of iodowillardiine bound to wild type GluA2 LBD, the distribution of lobe orientations is greater, with the zinc-free structure having a lobe opening of 6.9 ± 1.8° (12) and a zinc-containing structure having a lobe opening of 2.2 ± 0.5° (20), with the NMR structure falling between these two values (5.3 ± 1.5°; Ref. 15). In the mutant structure bound to IW, the lobes are open by 0.8 ± 0.9°, essentially identical to the glutamate-bound structure (Fig. 2B). Likewise, the H-bonds described above for the kainate-bound structure have not been observed in IW-bound wild type structures but are present in all three structures in the mutant. The willardiine ring and the iodo group are accommodated in the binding site by a rotation of the side chain of Met-708.
CNQX and DNQX
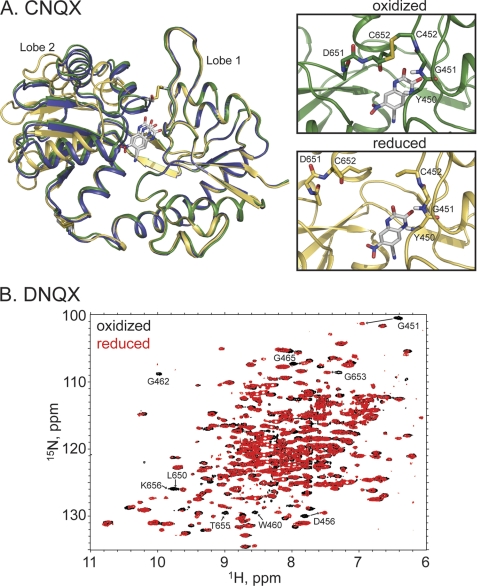
CNQX and DNQX have classically been considered antagonists of AMPA and kainate receptors, but both compounds can activate AMPA receptors under some circumstances (36–38). In this regard, they might be considered very weak partial agonists. Thus, one might expect that if a full lobe closure is required for activation, the lobes might close, albeit rarely, with CNQX or DNQX bound. The introduced cysteines may then trap the protein in the closed conformation. Indeed, the oxidized, CNQX-bound structure was crystallized with P21212 symmetry, and the lobes were open only by 1.27 ± 1.36° relative to glutamate, with two of the three structures in the asymmetric unit having the lobe orientation within the range observed for the wild type LBD bound to glutamate (Fig. 4A). The A452C/S652C disulfide was formed in all three copies in the asymmetric unit. Unlike the IW- and kainate-bound oxidized structures, the Asp-651/Ser-652 peptide bond was not flipped, and the interlobe H-bonds were not formed. In the presence of reducing agent (4 mm β-mercaptoethanol), the protein crystallized in the P21 space group with two copies in the asymmetric unit. The reduced protein was significantly more open (19.0 ± 0.4°) than the oxidized form and somewhat more open than the wild type CNQX (15.0 ± 0.4°; Ref. 37) structures. In contrast, the structures obtained with DNQX were all in the open form, and the disulfide bond was not formed. To determine whether the lack of disulfide-linked structures was simply due to selection by crystallization or if the disulfide could form in solution, the protein was uniformly labeled with 15N, and 1H,15N-HSQC spectra were collected in the presence and absence of DTT (Fig. 4B). Because of stability issues of the protein bound to DNQX, a full assignment with triple resonance experiments was problematic, but the spectrum clearly changed in the presence of DTT, suggesting that the disulfide can be formed. Tentative assignments based on the glutamate-bound spectra, would place most of the changed resonances near the presumed disulfide bond. These results suggest that, for both DNQX and CNQX, the lobes can transiently assume a fully closed structure, consistent with their very weak agonist activity.
FIGURE 4.
A, crystal structures of the reduced (yellow, Protein Data Bank code 3T9V) and oxidized (green, Protein Data Bank code 3T9U) forms of the A452C/S652C GluA2 LBD mutant bound to CNQX. Also shown is the wild type structure bound to glutamate in blue. The disulfide bond is clearly formed in the oxidized case, with a dramatic change in the relative lobe orientations. B, the 1H,15N-HSQC spectra of the reduced (red) and oxidized (black) forms of the A452C/S652C GluA2 LBD mutant bound to DNQX. Specific chemical shift changes, consistent with the formation of the disulfide bond, are seen.
No Lobe Locking with Antagonist
If activation of the channel requires full lobe closure, then antagonists should prevent the formation of the interlobe disulfide. This was tested by reducing and exchanging the ligand for UBP282. UBP282 is a bulky willardiine derivative that produces lobe extension (i.e. lobe opening greater than the apo form; 39) and functions as a relatively low affinity antagonist of GluA2 (40). The 1H-15N HSQC NMR spectra of Cu-phenantroline-oxidized and DTT-reduced GluA2 LBD in the presence of UBP282 are shown in Fig. 5. Unlike a full agonist (glutamate), partial agonists (iodowillardiine and kainate) and very weak partial agonists (CNQX and DNQX), no difference was observed between the oxidized and reduced spectra, suggesting that the disulfide bond was not formed.
FIGURE 5.
The 1H,15N-HSQC spectra of the reduced (red) and oxidized (black; Cu-phenanthroline) forms of the A452C/S652C GluA2 LBD mutant bound to UBP282. No chemical shift changes are observed, consistent with the absence of the disulfide bond in the presence of Cu-phenanthroline.
Effect of A452C/S652C Mutation on Cell Surface Expression
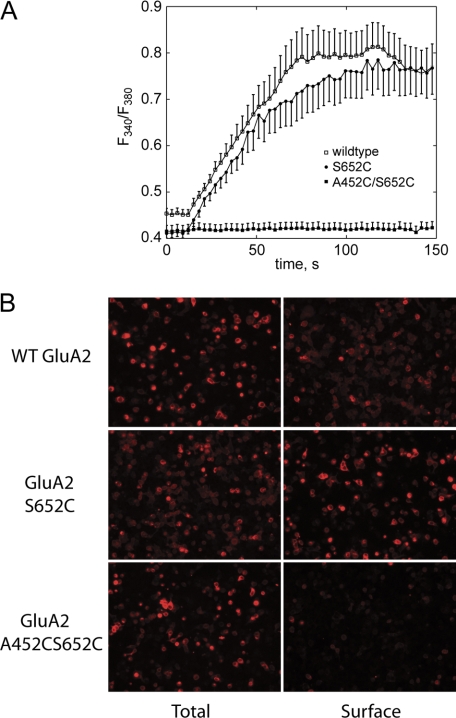
Individual mutations (A452C and S652C) were made in full-length GluA2i, and the proteins were expressed in Xenopus oocytes. We recorded glutamate-induced cationic currents in ooctyes expressing A452C mutant channels, which were of smaller amplitude than wild type GluA2. In contrast, oocytes injected with S652C RNA had no recordable currents. We, therefore, tested whether the lack of functional current of S652C is dependent on the expression system and discovered that S652C did express in HEK293 cells as a functional channel, shown using glutamate-evoked Ca2+ entry in ratiometric fura-2 imaging experiments (Fig. 6A). In the case of the double mutation (A452C/S652C) expressed in either expression system, no currents or Ca2+ transients were observed in response to glutamate even when cyclothiazide (an inhibitor of desensitization) and DTT or β-mercaptoethanol were included in the solution. This could be due to either a lack of expression, a dysfunctional receptor at the cell surface, or the failure of the protein to reach the cell surface. Using an antibody that recognizes an extracellular determinant, we compared the cell surface and total expression of A452C/S652C, S652C, and wild type GluA2i, which are shown in Fig. 6B. Clearly, the wild type and single mutation traffic to the cell surface, but the double mutation (A452C/S652C), while being robustly expressed, is not found at the cell surface. Similar results were observed when the A452C/S652C mutation was expressed with stargazin, a transmembrane AMPAR regulatory protein that can increase the surface expression of AMPA receptors (41).
FIGURE 6.
A, full-length wild type, S652C, or A452C/S652C GluA2 was transfected into HEK293 cells, and the response to glutamate was measured using a ratiometric fura-2 imaging. Clear channel activation was observed for wild type and S652C but not for the double mutant. B, immunofluorescent labeling of HEK293 cells transfected with full-length wild type, S652C, or A452C/S652C GluA2. In both cases, the cells were fixed in 2% paraformaldehyde prior to labeling, but the total staining was done in the presence of 0.1% Triton X-100 to allow access of the antibody to the cytoplasm. Surface staining was done in the absence of Triton X-100. The primary antibody recognized an extracellular determinant on GluA2.
DISCUSSION
The activation of AMPA receptors begins with binding of agonist and a global motion of the LBD, which in turn, produces a change in the ion channel gate, allowing cations to flow through the channel pore. The closing of the bilobed structure of the LBD is at least in part responsible for channel opening. Single channel recording experiments (12) have demonstrated that full and partial agonists can activate AMPA receptor channels to the same three or four distinct conductance levels. At saturating agonist concentrations in the absence of desensitization, partial agonists exhibit lower currents than full agonists because the lower conductance levels are preferentially populated relative to that seen with full agonists (12, 14, 42). The different conductance levels have been assumed to arise from the activation of individual gates on each of four subunits (43). That is, the highest conductance level is achieved by opening the gates for all four subunits, the next highest conductance has three open gates and so on. At saturating concentrations of agonist, all four subunits are occupied, so in the absence of desensitization, the population of lower conductance states by partial agonists would suggest that the channel gate is not automatically triggered by the binding of agonist but rather the opening of the channel gate for an individual subunit is associated with a conformational equilibrium whose energy levels are changed upon binding agonist. Jin et al. (12) provided an elegant explanation of partial agonism based on a series of crystal structures that correlated lobe orientation in the GluA2 LBD with efficacy. The greater the closure of the lobes observed in a series of crystal structures, the greater the efficacy. The conductance states for both full and partial agonists were shown to be identical and the population of the conductance levels followed a bionomial distribution. The probability of success for the bionomial can be taken as a measure of efficacy (“efficacy factor”). The efficacy factor is in turn correlated with the relative lobe orientation of the LBD. This hypothesis has been termed a “static” explanation in that a relatively fixed degree of lobe closure sets the probability of the activation of a gate (44). An alternative model (“dynamic”), but not mutually exclusive, is that each subunit exhibits a conformational ensemble that is modified by the binding of full and partial agonists. For full agonists, the conformational ensemble would favor mainly the closed lobe state and activation of the gate for the subunit, whereas partial agonists would include a fully closed state as well as distribution of more open states, with a less frequent activation of the gate for that subunit. In the simplest version of the model, the fully closed state of the LBD would be the trigger for the activation of the channel gate and the probability of attaining a fully closed LBD would determine the efficacy.
Landes et al. (22) have shown, using single molecule FRET, that a distribution of lobe orientations can be observed in the GluA2 LBD construct, and that, compared with the wild type protein bound to glutamate, both the apo form and the T686S mutant bound to glutamate have wider distributions, with the more open states populated to a greater extent. Also, for partial agonists such as IW, a wide range of lobe orientations has been observed using crystallography (12, 20) and NMR spectroscopy (15). In addition, dynamics near the lobe interface suggest that large-scale lobe motions may be present when the LBD is bound to IW (21, 23). In contrast, when the wild type LBD is bound to kainate, the lobes are in a relatively open orientation in all crystal structures to date (4, 18, 24, 25), and this is supported by the average structure determined using NMR spectroscopy (15). The idea has been that the lobes remain separated by a steric clash between the isoprenyl group of kainate and the side chain of Leu-650 (3, 18). In addition, no evidence for lobe motions on the microsecond-to-millisecond time scale has been observed in the GluA2 LBD when bound to kainate (23). Thus, the binding of kainate to the GluA2 LBD has been a strong argument against the dynamic hypothesis. We show here, however, that the LBD can be trapped in an almost fully closed form when bound to kainate, and the binding pocket accommodates by a small shift in the position of kainate and a change in the rotameric state of Leu-650 (Fig. 2C). This would suggest that, when bound to kainate, the LBD does transition to a fully closed state, albeit rarely, consistent with the low efficacy of kainate at GluA2 receptors. The caveat in all of these experiments is that the cross-linking is done using the isolated LBD, which is not constrained by the remainder the protein. Taken together with single molecule FRET experiments (22), which clearly show a large distribution of lobe orientations in some cases, these results strongly suggest that one conformation among the ensemble is the closed lobe form. Although this does not prove that it is the closed lobe form that results in channel activation, it does show that the LBD bound to even very weak partial agonists can attain this conformation and is consistent with the notion that the stability of the fully closed lobe may contribute to efficacy.
If the hypothesis described above is true, then antagonists should not be trapped in the fully closed state. The definition of an antagonist is somewhat arbitrary in the sense that CNQX and DNQX, which are classical antagonists of AMPA receptors, can activate the channel under some conditions (36–38). This would suggest that, in the isolated LBD, it should be possible to detect the formation of the disulfide in the A452C/S652C mutant in the presence of CNQX and DNQX. This is clearly the case for CNQX, where a crystal structure of the closed, disulfide-bonded form was obtained. For DNQX, no disulfide-bonded crystal structure was observed, but the 1H,15N-HSQC spectrum of the protein differed in the presence and absence of reducing agent, suggesting that the disulfide could be formed. On the other hand, the binding of UBP282 results in hyperextension of the lobes due to rather bulky groups that interact specifically with sites on both lobes (39). Although this compound has a relatively low affinity, it is clearly an antagonist (40). The 1H,15N-HSQC spectra were unchanged in the presence and absence of reducing agents, suggesting that the A452C/S652C disulfide did not form. These results further support the notion that only when bound to agonists can the protein assume a fully closed state, and, in fact, even very weak partial agonists, such as CNQX and DNQX, are capable of assuming a fully closed state.
Although NMDA receptors are expressed well on the cell surface and are functional when double cysteine mutations are introduced in either NR1 or NR2 to lock the lobes closed (45, 46), the A452C/S652C mutation of GluA2 is robustly expressed but does not reach the cell surface. However, when expressed in bacteria, the GluA2 LBD with these mutations folds correctly and the agonist binding site is intact. Assuming the protein is properly folded but does not traffic to the cell surface, the defect in trafficking might be due either to impairment of the formation of dimers or tetramers or to the conformational state (e.g. desensitized). The L483Y mutation seems to promote tetramerization and stabilizes the interface between LBD dimers. Although the tetramers are formed, the lack of desensitization of the L483Y mutants limits trafficking to the cell surface (47, 48). Formation of the A452C/S652C disulfide would destabilize the interface between LBD dimers and perhaps have the opposite effect on tetramerization (or even dimerization). Although desensitization may promote trafficking to the cell surface, if a tetrameric receptor were not formed, then the protein would not exit the ER.
The use of disulfide trapping has demonstrated that an essentially fully closed form of the LBD of GluA2 can be obtained in the presence of a range of partial agonists. This suggests that the lobes can show transitions to multiple conformations, as suggested previously by NMR dynamics measurements (21, 23) and single molecule FRET experiments (22). Although these experiments do not address directly the conformation required to activate the channel, previous studies showing that partial agonists can assume a range of conformations would suggest that it is this ensemble that may determine efficacy. The finding that the fully closed form is part of that ensemble is consistent with the idea that it is the stability of the fully closed form that determines efficacy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM068935. This work was also supported in part by research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by National Science Foundation Award DMR 0225180, and by using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by National Institutes of Health Award RR-01646, through its National Center for Research Resources.
The atomic coordinates and structure factors (codes 3T93, 3T96, 3T9H, 3T9U, 3T9V, and 3T9X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
Sandra M. Holley, Ahmed H. Ahmed, Jayasri Srinivasan, Swetha E. Murthy, Gregory A. Weiland, Robert E. Oswald, and Linda M. Nowak, submitted for publication.
- GluA1–4
- four subtypes of AMPA receptor
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid
- CNQX
- 6-cyano-7-nitroquinoxaline-2,3-dione
- DNQX
- 6,7-dinitroquinoxaline-2,3-dione
- flip and flop
- alternatively spliced versions of AMPA receptors that vary in rates of desensitization and sensitivity to allosteric modulators
- HSQC
- heteronuclear correlation spectroscopy
- IW
- (S)-5-iodowillardiine
- LBD
- extracellular ligand-binding domain of GluA2
- TROSY
- transverse relaxation optimized spectroscopy
- UBP282
- (S)-3-(4-carboxybenzyl)willardiine.
REFERENCES
- 1. Dingledine R., Borges K., Bowie D., Traynelis S. F. (1999) Pharmacol. Rev. 51, 7–61 [PubMed] [Google Scholar]
- 2. Traynelis S. F., Wollmuth L. P., McBain C. J., Menniti F. S., Vance K. M., Ogden K. K., Hansen K. B., Yuan H., Myers S. J., Dingledine R., Sibley D. (2010) Pharmacol. Rev. 62, 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armstrong N., Gouaux E. (2000) Neuron 28, 165–181 [DOI] [PubMed] [Google Scholar]
- 4. Armstrong N., Sun Y., Chen G. Q., Gouaux E. (1998) Nature 395, 913–917 [DOI] [PubMed] [Google Scholar]
- 5. Mayer M. L. (2005) Neuron 45, 539–552 [DOI] [PubMed] [Google Scholar]
- 6. Furukawa H., Gouaux E. (2003) EMBO J. 22, 2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nanao M. H., Green T., Stern-Bach Y., Heinemann S. F., Choe S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin R., Singh S. K., Gu S., Furukawa H., Sobolevsky A. I., Zhou J., Jin Y., Gouaux E. (2009) EMBO J. 28, 1812–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar J., Schuck P., Jin R., Mayer M. L. (2009) Nat. Struct. Mol. Biol. 16, 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobolevsky A. I., Rosconi M. P., Gouaux E. (2009) Nature 462, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun Y., Olson R., Horning M., Armstrong N., Mayer M., Gouaux E. (2002) Nature 417, 245–253 [DOI] [PubMed] [Google Scholar]
- 12. Jin R., Banke T. G., Mayer M. L., Traynelis S. F., Gouaux E. (2003) Nat. Neurosci. 6, 803–810 [DOI] [PubMed] [Google Scholar]
- 13. Zhang W., Cho Y., Lolis E., Howe J. R. (2008) J. Neurosci. 28, 932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poon K., Nowak L. M., Oswald R. E. (2010) Biophys. J. 99, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maltsev A. S., Ahmed A. H., Fenwick M. K., Jane D. E., Oswald R. E. (2008) Biochemistry 47, 10600–10610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mankiewicz K. A., Rambhadran A., Du M., Ramanoudjame G., Jayaraman V. (2007) Biochemistry 46, 1343–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robert A., Armstrong N., Gouaux J. E., Howe J. R. (2005) J. Neurosci. 25, 3752–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Armstrong N., Mayer M., Gouaux E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5736–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inanobe A., Furukawa H., Gouaux E. (2005) Neuron 47, 71–84 [DOI] [PubMed] [Google Scholar]
- 20. Jin R., Gouaux E. (2003) Biochemistry 42, 5201–5213 [DOI] [PubMed] [Google Scholar]
- 21. Fenwick M. K., Oswald R. E. (2008) J. Mol. Biol. 378, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Landes C. F., Rambhadran A., Taylor J. N., Salatan F., Jayaraman V. (2011) Nat. Chem. Biol. 7, 168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahmed A. H., Loh A. P., Jane D. E., Oswald R. E. (2007) J. Biol. Chem. 282, 12773–12784 [DOI] [PubMed] [Google Scholar]
- 24. Ahmed A. H., Wang Q., Sondermann H., Oswald R. E. (2009) Proteins 75, 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gill A., Birdsey-Benson A., Jones B. L., Henderson L. P., Madden D. R. (2008) Biochemistry 47, 13831–13841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldin A. L. (1992) Methods Enzymol. 207, 266–279 [DOI] [PubMed] [Google Scholar]
- 27. Hollmann M., Heinemann S. (1994) Annu. Rev. Neurosci. 17, 31–108 [DOI] [PubMed] [Google Scholar]
- 28. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29. Goddard T. D., Kneller D. G. SPARKY 3. University of California, San Francisco [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) in Methods in Enzymology, Macromolecular Crystallography, part A (Carter C. W., Sweet R. M. eds.) Vol. 276, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 31. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32. Collaborative Computational Project (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Chen G. Q., Gouaux E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fenwick M. K., Oswald R. E. (2007) Biomolecular NMR Assignments 1, 241–243 [DOI] [PubMed] [Google Scholar]
- 36. Coki B., Stein V. (2008) Neuropharmacology 54, 1062–1070 [DOI] [PubMed] [Google Scholar]
- 37. Menuz K., Stroud R. M., Nicoll R. A., Hays F. A. (2007) Science 318, 815–817 [DOI] [PubMed] [Google Scholar]
- 38. Taverna F., Xiong Z. G., Brandes L., Roder J. C., Salter M. W., MacDonald J. F. (2000) J. Biol. Chem. 275, 8475–8479 [DOI] [PubMed] [Google Scholar]
- 39. Ahmed A. H., Thompson M. D., Fenwick M. K., Romero B., Loh A. P., Jane D. E., Sondermann H., Oswald R. E. (2009) Biochemistry 48, 3894–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. More J. C., Troop H. M., Dolman N. P., Jane D. E. (2003) Br. J. Pharmacol. 138, 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomita S., Adesnik H., Sekiguchi M., Zhang W., Wada K., Howe J. R., Nicoll R. A., Bredt D. S. (2005) Nature 435, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 42. Poon K., Ahmed A. H., Nowak L. M., Oswald R. E. (2011) Mol. Pharmacol. 80, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenmund C., Stern-Bach Y., Stevens C. F. (1998) Science 280, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 44. Lau A. Y., Roux B. (2011) Nat. Struct. Mol. Biol. 18, 283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanke M. L., VanDongen A. M. (2008) J. Biol. Chem. 283, 21519–21529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kussius C. L., Popescu G. K. (2010) J. Neurosci. 30, 12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greger I. H., Akamine P., Khatri L., Ziff E. B. (2006) Neuron 51, 85–97 [DOI] [PubMed] [Google Scholar]
- 48. Greger I. H., Esteban J. A. (2007) Curr. Opin. Neurobiol. 17, 289–297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.